Abstract
We previously described the RiboPuromyclation method (RPM) to visualize and quantitate translating ribosomes in fixed and permeabilized cells by standard immunofluorescence. RPM is based on puromycylation of nascent chains bound to translating ribosomes followed by detection of puromycylated nascent chains with a puromycin-specific mAb. We now demonstrate that emetine optimally enhances nascent chain puromycylation, and describe a modified RPM protocol for identifying ribosome-bound nascent chains in metabolically inert permeabilized cells.
Keywords: ribopuromycylation, puromycin, translation, ribosome
Introduction
Puromycin (PMY) is an aminonucleoside Tyr-tRNA mimetic antibiotic that enters the A site of prokaryotic and eukaryotic ribosomes, covalently incorporating into nascent chains, terminating translation [1]. Eggers et al. pioneered PMY as a nascent protein tag, using polyclonal Abs to PMY to immunoblot and immunoprecipitate nascent proteins [2]. To circumvent non-specific reactivity of polyclonal anti-PMY antibodies (Abs), Schmidt et al. generated anti-PMY monoclonal Abs (mAbs) and applied them in flow cytometry to measure relative translation rates in living cells exposed to PMY based on expression of PMY-terminated cell surface proteins [3]. More recently, we used these mAbs and generated several new mAbs to localize puromycylated nascent chains on ribosomes immobilized by chain elongation inhibitors in living cells prior to processing for intracellular immunofluorescence [4,5]. We described this method, termed the RiboPuromycylation Method (RPM), as a simple and generally applicable method to localize and quantitate actively translating ribosomes by immunofluorescence.
Here, we show that emetine, an irreversible translation elongation inhibitor, enhances in situ nascent chains puromycylation, explaining our previous observation that emetine gives a brighter RPM-based signal than cycloheximide. We also describe a new RPM protocol to localize translation sites in permeabilized cells with a labeling efficiency similar to the regular RPM.
Material and Methods
Cells and treatments
We cultured HeLa cells in DMEM, 7.5% FBS, splitting cultures the day before each experiment to achieve log phase growth concomitant with high translation rate. We used drugs at the indicated concentrations: CHX (355 M, Sigma), emetine (208 M, Calbiochem), anisomycin (9.4 M, Calbiochem), pactamycin (0.5 M), sodium arsenite (500 M, Sigma), CCCP (10 M), deoxyglucose (50mM), sodium azide (10mM). We infected HeLa cells with VV at a multiplicity of 1 pfu/cell in saline supplemented with 0.1% BSA. After adsorption at 37°C for 1h, we overlaid infected monolayers with DMEM containing 7.5% FBS, and incubated for 6 more h.
RPM
RPM on ice
We transferred HeLa cells to coverslips the day before the experiment to be 60–80% confluent at the time of the experiment. Depending on the experiment, we pre-treated cells with different inhibitors. Optimal staining with this protocol is obtained by pre-treating cells with 208 M of emetine diluted in warm DMEM 7.5% FBS for 15 min at 37°C prior to PMY labeling. We transferred coverslips to 24 well plates and washed cells twice with cold PBS supplemented with CHX (355 M, Sigma). All extraction/labeling procedures are performed on ice, using reagents pre-chilled to ice temperature. We incubated cells for 2 min with 500 L/well of Permeabilization Buffer (50mM Tris-HCl pH 7.5, 5mM MgCl2, 25mM KCl, 355 M CHX, EDTA-free protease inhibitors (Roche), 10U/ML RNAse Out (Invitrogen) containing 0.015% digitonin (Wako)). Following this extraction step, we washed cells once with 500 L of polysome buffer, added 500 L of polysome buffer supplemented with 91 M of PMY, and incubated for 10 min. After washing with 500 L of cold polysome buffer, we fixed cells with 500 L of 3% paraformaldehyde (PFA) for 15 min at RT, aspirated the PFA, added PBS, and incubated cells in a 4 °C refrigerator prior to immunofluorescence staining.
RPM at 37°C
HeLa cells on coverslips were incubated with DMEM 7.5% FBS supplemented with 91 M of PMY and 355 M of CHX for 5 min at 37°C. We then performed the extraction procedure described for RPM on ice.
Abs
The mouse PMY-specific mAb (clone 2A4) has been described [4] Anti-ribosomal P antibody is a human polyclonal autoimmune antisera (Immunovision). As secondary Abs we used goat anti-mouse A488 (Molecular Probes), donkey anti-rabbit Texas Red and donkey anti-human Cy5 (Jackson ImmunoResearch). To label DNA we used Hoechst 3358 (Molecular Probes). For immunoblotting we used polyclonal rabbit anti-mouse Ig–HRP (Dako) and goat anti-human Ig-HRP (Jackson ImmunoResearch).
Immunofluorescence and Microscopy
All staining was performed using staining buffer (SB, 0.05% saponin, 10mM Glycine, 5% FBS, PBS), as previously described [4]. Coverslips were incubated in SB for 15 min, and then incubated with Abs diluted in SB for 1 h at RT. After washing 3 times with PBS coverslips were incubated with secondary Abs diluted in SB for 1 h at RT. Coverslips were washed twice and incubated with 1 g/mL of Hoechst 3358 diluted in SB for 5 min at RT. Coverslips were washed twice with distilled water and mounted on slides with Fluoromount-G (Southern Biotech). Images were acquired using a Leica TCS SP5 laser scanning confocal microscope with a HCX PL APO lambda blue 63.0×1.40 OIL UV objective, Type FF immersion liquid (Cargille), 152.4 m pinhole. Images were processed using LAS AF software (Leica), Imaris (Bitplane), Huygens Essentials Software (Version 3.6, Scientific Volume Imaging BV), and Photoshop CS4 (Adobe) without manipulating the gamma function. Each set of images for a given experiment were processed identically to maintain the image intensity ratio. ImageJ and Prism software were used for quantitation and statistical analysis. Groups were analyzed for statistical significance with a two-tailed unpaired t-test; error bars represent the s.e.m.
Results and Discussion
Emetine optimizes puromycylation at 4 °C
The puromycylation reaction occurs spontaneously in cell free extracts [1]. To correlate this finding with RPM staining, we depleted cellular energy stores by treating cells with deoxyglucose to block glycolysis in combination with either CCCP or azide to block oxidative phosphorylation. We then incubated cells with PMY for 5 min at 37 °C and performed anti-PMY immunoblots using the 12D10 anti-PMY mAb. Using either combination of energy depleting drugs only slightly reduces puromycylation (Figure 1A), consistent with the energy-independence of the cell free puromycylation reaction. While in vivo puromycylation is energy independent, it is undetectable when live cells are incubated with PMY at 4 °C (Fig. 1A). This low temperature sensitivity is not due to the temperature dependence of puromycylation per se, but to the accessibility of PMY to ribosome bound-nascent chains probably caused by a general decrease of cell membrane passive diffusion [6]. 4°C puromycylation is clearly seen when cells are permeabilized with 0.015% digitonin prior to PMY addition (Fig. 1B), and strikingly enhanced when cells were pretreated with emetine prior to permeabilization, which increases puromycylation to levels obtained with live cells at 37°C. While the precise mechanism is uncertain, the enhancing effect of emetine on low temperature puromycylation clearly demonstrates that first, ribosomal catalysis is essential for protein puromycylation, and second confirms the energy-independence of nascent chain puromycylation in cells.
Figure 1.
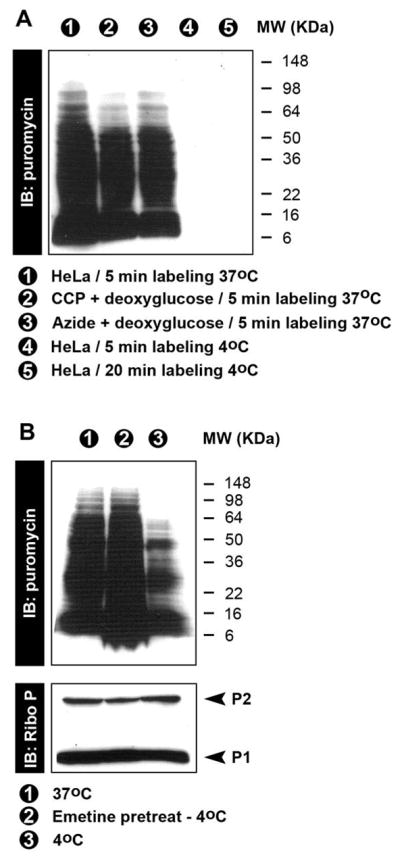
A. Immunoblotting of total cell lysates demonstrates energy independence of puromycylation and also the temperature dependence (37°C vs 4°C) when intact cells are labeled with PMY.
B. Immunoblotting of NP-40 extracts form HeLa labeled at 37 °C in culture or at 4 °C after digitonin treatment. Pre-incubation of cells with emetine prior to digitonin treatment greatly increases puromycylation.
RiboPuromycylation Method for Permeabilized Cells
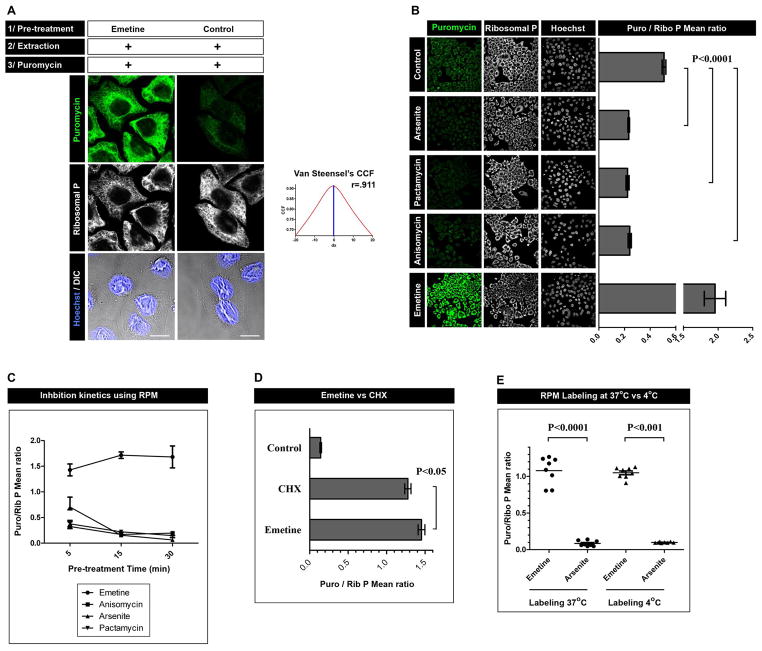
We adapted the permeabilized cells protocol to enable RPM staining cells cultured on coverslips. We permeabilized cells with digitonin, incubated with PMY on ice for 10 min, paraformaldehyde fixed, and stained with anti-PMY mAb. We obtained relatively weak PMY staining (Fig. 2A) under these conditions, which is consistent with the immunoblotting data (Fig. 1B). As predicted from immunoblotting, pre-treating living cells with emetine at 37°C greatly increases PMY staining, which extensively co-localizes with ribosomes, as expected (Fig. 2A).
Figure 2.
A. HeLa cells were incubated or not for 15 min with emetine, were washed with PBS supplemented with CHX, permeabilized with digitonin and labeled with PMY in the presence of CHX. Cells were then fixed and stained for PMY (green) and large ribosomal subunit (white). Co-localization was quantitated using Van Steensel’s cross-correlation coefficient (graph) and Pearson’s coefficient (r; 1=complete co-localization, −1 = complete non-colocalization). Bar scale: 10 m.
B. HeLa cells pre-treated for 15 min with the inhibitor indicated were labeled as in A. For each condition, multiple fields were acquired and the mean fluorescence ratio of PMY/ribosome staining for each field was quantitated using ImageJ. Values are plotted on the right. Statistical analyses, two-tailed unpaired t-test; ***, P < 0.0001.
C. As in B, but with the treatment times indicated
D. As in B with CHX and emetine pre-treatments. Statistical analyses, two-tailed unpaired t-test; *, P < 0.05.
E. Direct comparison of RPM labeling using the 4 °C or 37 °C protocol. Statistical analyses, two-tailed unpaired t-test; ***, P < 0.0001.
We next treated live cells for 15 min at 37°C with various translation inhibitors prior to permeabilization and 4 °C PMY staining (Fig. 2B). Arsenite induced translation shutdown [7] reduces anti-PMY staining to near background levels, while anti-ribosome staining is unaffected. Pactamycin, a translation initiation inhibitor [8], completely abrogated staining. By competing with PMY, anisomycin [9] had the same effect. By contrast, emetine pre- treatment increased staining 4-fold. Emetine, anisomycin and pactamycin achieved near maximal effects within 5 min of pre-treatment, while arsenite required 15 min treatment, paralleling the known kinetics of translation inhibition (Fig. 2C).
In direct comparison, emetine is slightly but clearly superior to CHX at enhancing RPM staining at 4 °C (Figure 2D). Importantly, using emetine, inert cell staining equals standard live cell RPM staining performed at 37 °C (Fig. 2E)
Emetine irreversibly binds the ribosomal 40S subunit [10], inhibiting aminoacyl-tRNA transfer [11] but unlike CHX, does not prevent transpeptidation or translocation of peptidyl-tRNA from acceptor to donor sites [1]. The superior properties of emetine vs. CHX may be related to its irreversibility or a differential effect on ribosome puromycylation activity. In any event, the ability to achieve a maximal RPM signal on inert cells following digitonin permeabilization is a twist on the RPM that reduces ambiguity regarding the potential release of nascent from ribosomes, which while unlikely based on our previous findings [4], cannot be unequivocally ruled out.
Acknowledgments
Glennys Reynoso provided outstanding technical support. This work was generously supported by the Division of Intramural Research, NIAID.
References
- 1.Pestka S. Inhibitors of ribosome functions. Annu Rev Microbiol. 1971;25:487–562. doi: 10.1146/annurev.mi.25.100171.002415. [DOI] [PubMed] [Google Scholar]
- 2.Eggers DK, Welch WJ, Hansen WJ. Complexes between nascent polypeptides and their molecular chaperones in the cytosol of mammalian cells. Mol Biol Cell. 1997;8:1559–1573. doi: 10.1091/mbc.8.8.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 4.David A, Dolan BP, Hickman HD, Knowlton JJ, Clavarino G, et al. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol. 2012;197:45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David A, Netzer N, Strader MB, Das SR, Chen CY, et al. RNA binding targets aminoacyl-tRNA synthetases to translating ribosomes. J Biol Chem. 2011;286:20688–20700. doi: 10.1074/jbc.M110.209452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fallon AM, Stollar V. Isolation and characterization of puromycin-resistant clones from cultured mosquito cells. Somatic Cell Genet. 1982;8:521–32. doi: 10.1007/BF01538712. [DOI] [PubMed] [Google Scholar]
- 7.Kedersha N, Chen S, Gilks N, Li W, Miller IJ, et al. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kappen LS, Suzuki H, Goldberg IH. Inhibition of reticulocyte peptide-chain initiation by pactamycin: accumulation of inactive ribosomal initiation complexes. Proc Natl Acad Sci U S A. 1973;70:22–26. doi: 10.1073/pnas.70.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen JL, Moore PB, Steitz TA. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J Mol Biol. 2003;330:1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- 10.Jimenez A, Carrasco L, Vazquez D. Enzymic and nonenzymic translocation by yeast polysomes. Site of action of a number of inhibitors. Biochemistry. 1977;16:4727–4730. doi: 10.1021/bi00640a030. [DOI] [PubMed] [Google Scholar]
- 11.Grollman AP. Inhibitors of protein biosynthesis. V. Effects of emetine on protein and nucleic acid biosynthesis in HeLa cells. J Biol Chem. 1968;243:4089–4094. [PubMed] [Google Scholar]