Abstract
Mutualistic interactions are often subject to exploitation by species that are not directly involved in the mutualism. Understanding which organisms act as such ‘third-party’ species and how they do so is a major challenge in the current study of mutualistic interactions. Here, we show that even species that appear ecologically similar can have contrasting effects as third-party species. We experimentally compared the effects of nectar-inhabiting bacteria and yeasts on the strength of a mutualism between a hummingbird-pollinated shrub, Mimulus aurantiacus, and its pollinators. We found that the common bacterium Gluconobacter sp., but not the common yeast Metschnikowia reukaufii, reduced pollination success, seed set and nectar consumption by pollinators, thereby weakening the plant–pollinator mutualism. We also found that the bacteria reduced nectar pH and total sugar concentration more greatly than the yeasts did and that the bacteria decreased glucose concentration and increased fructose concentration whereas the yeasts affected neither. These distinct changes to nectar chemistry may underlie the microbes' contrasting effects on the mutualism. Our results suggest that it is necessary to understand the determinants of microbial species composition in nectar and their differential modification of floral rewards to explain the mutual benefits that plants and pollinators gain from each other.
Keywords: microbial ecology, hummingbird pollination, chemical ecology, mutualism breakdown
1. Introduction
Mutualisms are increasingly recognized as essential inter-specific interactions that affect populations, communities and ecosystems [1]. However, the consequences of mutualisms are often difficult to predict because they operate within complex webs of ecological interactions [2]. For example, many pairwise mutualisms are subject to exploitation by ‘third-party’ species that are not directly involved in the mutualistic relationship [3,4]. When exploitation is substantial enough, these species can potentially cause the breakdown of mutualism [5–7]. Understanding which organisms act as such third-party species and by what mechanisms they exert their effects is a major challenge in the current study of mutualisms [2,8]. Much remains unknown about these questions because even species that appear ecologically similar may vary in their effects as third-party species [4].
In plant–pollinator mutualisms, the strength of the interactions may be affected by the various species of micro-organisms that colonize floral nectar [9–11]. Floral microbes are thought to potentially weaken plant–pollinator mutualisms by decreasing floral attractiveness through consumption of nectar resources [12] and interfering with pollen germination and damaging pollen tubes [13]. Alternatively, it has also been suggested that micro-organisms may enhance pollination by producing volatiles or fermentation by-products that attract pollinators [14–16]. However, direct evidence for these potential effects is still scanty. Further, most studies have focused on yeasts, even though another group of microbes, bacteria, are also frequently found in floral nectar [17,18]. Nectar-inhabiting yeasts and bacteria are likely to be similar in resource use (e.g. sugar consumption) and other ecological traits (e.g. tolerance to high osmotic pressure, hydrogen peroxide and oxygen limitation) that allow them to thrive in the rather unique habitat [11,19,20]. Nonetheless, given the physiological and metabolic diversity of microbial species, their effects on plant–pollinator mutualisms may still differ. To our knowledge, however, no study has experimentally compared the effects of yeasts and bacteria on the strength of plant–pollinator mutualisms.
In this paper, we test the hypothesis that nectar-dwelling yeasts and bacteria differentially affect the strength of a plant–pollinator mutualism. Specifically, we conducted three experiments using a combination of artificial and real flowers of a hummingbird-pollinated shrub in California, Mimulus aurantiacus. First, we investigated the effect of microbes on pollination and seed set across the flowering season, using real M. aurantiacus flowers inoculated with the common yeast, Metschnikowia reukaufii, or the common bacterium, Gluconobacter sp. Second, we examined the effect of the two microbes on nectar consumption by pollinators, using artificial flowers mimicking real M. aurantiacus flowers. Lastly, in the laboratory, we inoculated M. aurantiacus nectar with the two microbes and quantified modification of the chemical properties of nectar in order to identify potential mechanisms underlying the microbial effects that we detected on pollination, seed set and nectar consumption by pollinators.
2. Methods
(a). Study organisms
In M. aurantiacus, seed set is limited by pollen availability and requires pollination for outcrossing [21]. Stigmas of M. aurantiacus flowers close upon contact and stay closed if much pollen is received, but reopen if little pollen is received [21]. For this reason, stigma closure can be used as an indicator of pollination in this species [22]. At the Jasper Ridge Biological Preserve (JRBP), located in the Santa Cruz Mountains of California (37°24′ N, 122°13′30″ W), we frequently observe floral visitation by Anna's hummingbird (Calypte anna), although Allen's hummingbird (Selasphorus sasin), Rufous hummingbird (Selasphorus rufus) and occasionally bees (e.g. Bombus vosnesenskii and Xylocopa micans) may also visit M. aurantiacus flowers. Flowers can persist for approximately 6–10 days [22] and contain up to 10 μl of nectar [21], in which both yeasts [23] and bacteria frequently attain densities of 104 CFUs (colony forming units) per μl, similar to densities reported in other systems [24]. At JRBP, bacterial densities ranged from 0 to 104 CFUs per μl, with an average of 350 CFUs per μl among flowers exposed to pollinators (n = 82 flowers). Previously, we found lower microbial densities when nectar was sampled from flowers in an experimental cage that excluded large pollinators like hummingbirds (but not bees and other smaller flower visitors) compared with flowers outside the cage, indicating that the micro-organisms mainly colonize flowers via large pollinators [23].
Metschnikowia reukaufii is the most common yeast species in M. aurantiacus flowers at JRBP [23]. It is also commonly found in the nectar of other plant species [24] and on bees, ants, hummingbirds and other pollinators [23,25,26]. We isolated M. reukaufii from M. aurantiacus flowers at JRBP in 2010 by plating diluted nectar samples on yeast–malt agar (YMA; Difco, Sparks, MD, USA). Resulting colonies were identified by sequencing the D1/D2 domains of the large subunit nuclear ribosomal RNA gene [27], as described previously [22,23]. Strains were stored at −80°C in 20 per cent glycerol and freshly streaked on YMA plates 2–4 days prior to each experiment described below.
The bacterial taxon used in this study, hereafter referred to as Gluconobacter, is also common in M. aurantiacus flowers at JRBP (see the electronic supplementary material, S1). To sample culturable bacteria from M. aurantiacus nectar, in 2011 and 2012, we plated diluted nectar samples on either YMA plates or R2A plates supplemented with 20 per cent sucrose [18] and 100 mg l−1 of the anti-fungal cycloheximide. The plating yielded bacterial colonies from approximately 30–70% of the flowers sampled, depending on flower age (see the electronic supplementary material, S2). The DNA of two to four colonies per plate, covering approximately 50 flowers, was extracted using Extract-N-Amp (Sigma-Aldrich, Saint Louis, MO, USA) and a portion of the 16S rRNA gene amplified using primers E343F (5′-TACGGRAGGCAGCAG-3′) and E1099FRC (5′-GGGTTGCGCTCGTTRC-3′). Resulting amplicons were sequenced by ElimBio (Hayward, CA, USA). Sequences were clustered into OTUs based on 97 per cent similarity and were identified by search against GenBank with the Basic Local Alignment Search Tool (BLAST) and placement in a phylogenetic tree with reference sequences gathered from the literature. This analysis was conducted using the program Geneious Pro v. 5.1.7 (Biomatters, Auckland, New Zealand). Results revealed that sequences recovered from one of the most common OTUs clustered with those of Gluconobacter spp. (see the electronic supplementary material, S1). Isolated colonies belonging to this OTU were used in the experiments described below.
(b). Experiment 1: microbial effects on pollination and seed set
To test whether M. reukaufii and Gluconobacter affect pollination and seed set, we collected cuttings at JRBP and propagated approximately 80 M. aurantiacus plants in pots, in a common garden at the Stock Farm plant growth facility on the Stanford University campus, located about 5 km from JRBP. In summer 2011, approximately 100 flower buds were chosen haphazardly among mature plants in the common garden and monitored daily for anthesis. Upon anthesis, flowers with open stigmas were tagged and inoculated with 4 μl of a suspension of either M. reukaufii, Gluconobacter, or a control solution. Inoculation solutions were prepared by suspending single colonies of M. reukaufii or Gluconobacter in sterile water with 20 per cent sucrose, diluted to 104 cells μl, and incubated for 2 days at 25°C. Stigma status (open or closed) was recorded for each flower 2, 4 and 6 days after inoculation. We used data from day 4 for analysis; not many flowers had been visited by day 2, and many flowers had wilted by day 6. This experiment was performed seven times between June 2011 and September 2011, and involved a total of 449 flowers.
Flowers from the first two trials were collected to measure microbial abundance at the end of the experiment (see the electronic supplementary material, S3). Flowers from the final five trials were left on the plants to assess seed set: mature seed capsules were collected 3–4 months after the inoculations, but before the capsules split. Some capsules could not be recovered due to early abscission or loss, but no bias was detected in the number of capsules recovered per treatment. In the laboratory, seeds were removed from each capsule and counted under a dissecting microscope. During the final two trials, half of the experimental plants were covered with a net (mesh size: 3 cm), which allowed access by small animals, such as bees, ants and other insects, but not by large animals, including Anna's hummingbird (C. anna), several individuals of which were regularly present around the common garden.
Stigma closure data were analysed using logistic regression, with microbial treatment and net presence as fixed effects and trial number as a random effect, followed by a likelihood ratio (LR) test to assess significance of the fixed effects. Because of a zero-inflated distribution [28], a two-part approach [29,30] was taken to analysing seed set data. First, we used a logistic regression to model the probability of setting seeds (yes or no), with the same predictors as for stigma closure, followed by a LR test. Second, using only flowers that set seeds, we assessed microbial effects on log-transformed seed number by a linear mixed model with the same predictors as above, using packages lme4 [31] and nlme [32] in R v. 2.15.0 [33].
(c). Experiment 2: microbial effects on nectar removal
To test whether M. reukaufii and Gluconobacter affect nectar consumption by pollinators, we constructed synthetic flowers using 200-μl pipette tips (VWR, Radnor, PA, USA) wrapped in orange and green tapes to appear similar in colour, shape and size to real M. aurantiacus flowers. Nine of these flowers were wired to a green stake approximately 1 m tall, typical of real M. aurantiacus plants (see the electronic supplementary material, S4). Six stakes each with nine synthetic flowers were placed in the ground at the Stock Farm plant growth facility, within 5 m from the M. aurantiacus plants in the common garden, where Anna's hummingbirds (C. anna) frequently visited the plants. A rubber ring (diameter: 3 cm) was attached to each stake at about 10 cm from the ground, and glycerol applied to the ring, which minimized access to the flowers by ants. Nine 200 μl PCR tubes containing conditioned nectar (prepared as described below) were placed in the flowers, with three replicates of each treatment (M. reukaufii, Gluconobacter, or water as control) randomly arranged on each stand.
To assess microbial effects on pollinators, we prepared conditioned nectar using single colonies of each microbial taxon selected from single-species cultures on YMA plates. The selected colonies were each diluted to 104 cells μl−1 of deionized water, and 1 μl of this suspension (or water as a control) added to 60 μl of 0.2 μm-filter-sterilized nectar in 200 μl GeneMate PCR tubes (BioExpress, UT, USA) and incubated for 2 days at 25°C. Bagged tubes containing nectar (bag mesh size: 1 mm) were also placed on the stakes during the experiment as a control to account for evaporation while preventing nectar removal by birds, insects and other animals. All tubes were left open for 1 day, and remaining nectar weighed. Anna's hummingbirds were frequently observed to visit the artificial flowers to consume nectar in them (see the electronic supplementary material, S4). Nectar removal from each tube was calculated by subtracting the mass of nectar remaining in experimental tubes from the average mass of nectar in bagged controls for each trial. This experiment was repeated five times during June and July of 2011. Because floral nectar is a complex solution [14,34–36], we chose to use real nectar rather than a synthetic analogue for removal experiments. However, since it was difficult to obtain the high volume of nectar needed for the experiment, we used nectar collected from Musa spp. (banana) flowers in Costa Rica, which produce copious amounts of nectar. Musa nectar was used for the first three trials, but locally collected nectar from M. aurantiacus flowers was used for the remaining two trials. We detected no significant difference in results between Musa spp. and M. aurantiacus nectar (nectar type F1,209 = 0.08, p = 0.79, nectar type × treatment F2,209 = 1.27, p = 0.28).
We analysed nectar removal data using a linear mixed effects model, with microbial treatment as a main effect and trial number as a random effect using package nlme [32] in R v. 2.15.0 [33].
(d). Experiment 3: microbial effects on nectar chemistry
To assess the effects of the microbes on nectar chemistry, single colonies of M. reukaufii and two isolates of Gluconobacter were introduced to filter-sterilized M. aurantiacus nectar, incubated and chemical characteristics of nectar quantified after incubation. Nectar used in this experiment was collected from M. aurantiacus plants grown in a greenhouse, from seeds collected at JRBP. Nectar was collected using glass pipette tips, immediately filtered through a 0.2 μm filter and stored at −80°C until use. We examined the pre-filtered nectar for the presence of culturable microbes by plating on YMA; no yeasts or bacteria were detected. Nine μl of the sterilized M. aurantiacus nectar was added to 200-μl PCR tubes and separately inoculated with 1 μl of Gluconobacter, M. reukaufii, or the control solution, with five replicates per treatment. The inoculation suspensions were prepared by diluting individual colonies of each strain to 104 cells μl−1 in filter-sterilized 15 per cent w/v sucrose solution supplemented with 0.32 mM amino acids from digested casein to mimic real M. aurantiacus nectar. Inoculated nectar in the PCR tubes was incubated at 25°C for 4 days, after which hydrogen peroxide (H2O2) concentration, pH and sucrose, glucose and fructose concentrations were quantified as detailed below. These chemical properties of nectar have been demonstrated or hypothesized to influence pollinator attraction to flowers [37,38] and plant–microbe interactions in nectar [19,20].
To quantify H2O2 concentration, we used a Peroxide Assay Kit for aqueous samples (Thermo Scientific, Rockford, IL, USA). Briefly, 2 μl of nectar or H2O2 standard solution was added to 100 μl of reaction solution and absorbance measured at 560 nm using a plate reader (TECAN, San Jose, CA, USA). To measure pH, we applied 0.5 μl of nectar to each of three sections of a pH strip (EMD Millipore, Darmstadt, Germany). To quantify sucrose, glucose and fructose concentrations, we diluted 1 μl of nectar in 200 μl of 50 : 50 acetonitrile : water containing 0.5 mg ml−1 maltose (Sigma-Aldrich, St Louis, MO, USA) as an internal standard. Sugars were then separated by UPLC (Waters, Milford, MA, USA) on a Luna amide column (50 × 2 mm, 3 μm, Phenomenex, Torrance, CA, USA). An acetonitrile : water mobile phase with a 4.5-min linear gradient beginning at 80 : 20 MeCN : H2O and ending at 30 : 70 MeCN:H2O was used, with a 10 min equilibration at initial conditions between samples. Mono- and disaccharides were quantified using an ELS detector (Waters), and the concentration of sucrose, glucose and fructose in each sample was calculated using the internal standard and a series of external standards. Microbial abundance was also quantified in order to verify growth in nectar. For this purpose, 1 μl of nectar was diluted in sterile 20 per cent sucrose and plated on YMA plates, and colonies counted after 5 days of incubation at 25°C. We confirmed that CFUs corresponded well to the number of cells in solution for the focal yeast [22] and bacterium (see the electronic supplementary material, S5).
Nectar chemistry data were analysed using a series of one-way ANOVAs, with a Bonferonni correction to control for multiple tests, in R v. 2.15.0 [33]. Because the effects of the two Gluconobacter strains were indistinguishable, we combined the two strains in all analyses.
3. Results
(a). Experiment 1: microbial effects on pollination and seed set
Inoculation with Gluconobacter decreased the proportion of closed stigmas by an average of 23 per cent, compared with flowers inoculated with yeasts or the sucrose control solution (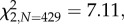 p = 0.02; figure 1a). This effect was consistent across trials despite high among-trial variation in the total proportion of closed stigmas, ranging from 27 to 84 per cent (see the electronic supplementary material, S6). Neither microbial treatment significantly affected the likelihood of a flower setting seed (
p = 0.02; figure 1a). This effect was consistent across trials despite high among-trial variation in the total proportion of closed stigmas, ranging from 27 to 84 per cent (see the electronic supplementary material, S6). Neither microbial treatment significantly affected the likelihood of a flower setting seed (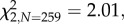 p = 0.36), but inoculation with Gluconobacter decreased the number of seeds produced, by an average of 18 per cent, compared with the control treatment (F2,243 = 4.78, p = 0.009; figure 1). In contrast, M. reukaufii inoculation did not significantly affect stigma closure or seed set compared with the control treatment (figure 1). Netting decreased the probability of stigma closure (
p = 0.36), but inoculation with Gluconobacter decreased the number of seeds produced, by an average of 18 per cent, compared with the control treatment (F2,243 = 4.78, p = 0.009; figure 1). In contrast, M. reukaufii inoculation did not significantly affect stigma closure or seed set compared with the control treatment (figure 1). Netting decreased the probability of stigma closure (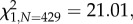 p < 0.001), from 60 to 28 per cent on average, as well as the probability of a capsule containing any seeds (
p < 0.001), from 60 to 28 per cent on average, as well as the probability of a capsule containing any seeds (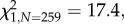 p < 0.001) and the number of seeds produced (F1,243 = 14.04, p < 0.001), confirming that large animals, most probably Anna's hummingbirds, were major pollinators (see the electronic supplementary material, S7).
p < 0.001) and the number of seeds produced (F1,243 = 14.04, p < 0.001), confirming that large animals, most probably Anna's hummingbirds, were major pollinators (see the electronic supplementary material, S7).
Figure 1.

Effects of inoculation with control solution (C), the yeast M. reukaufii (M), or the bacterium Gluconobacter (G) on (a) the proportion of floral stigmas closed four days after anthesis and inoculation and (b) the number of seeds per capsule produced. Bars indicate mean±1 s.e. Letters above bars indicate treatments that differ when compared using Wald contrasts. Numbers at the bottom of bars indicate the number of replicates (flowers) for each treatment.
(b). Experiment 2: microbial effects on nectar removal
After accounting for evaporation using the bagged controls, an average of 27 per cent less nectar was removed from Gluconobacter-inoculated flowers than either control or yeast-inoculated flowers when considered across all trials (F2,199 = 34.2, p < 0.001; figure 2), even though the magnitude of this effect varied among trials (F12,199 = 56.73, p < 0.001, electronic supplementary material, S8). Nectar removal did not depend on nectar identity (i.e. Mimulus versus Musa nectar), and in the two trials that used Mimulus nectar, Gluconobacter inoculation decreased nectar removal compared to control or yeast-inoculated flowers (F2,85 = 10.42, p < 0.001), consistent with the experiment-wide results.
Figure 2.
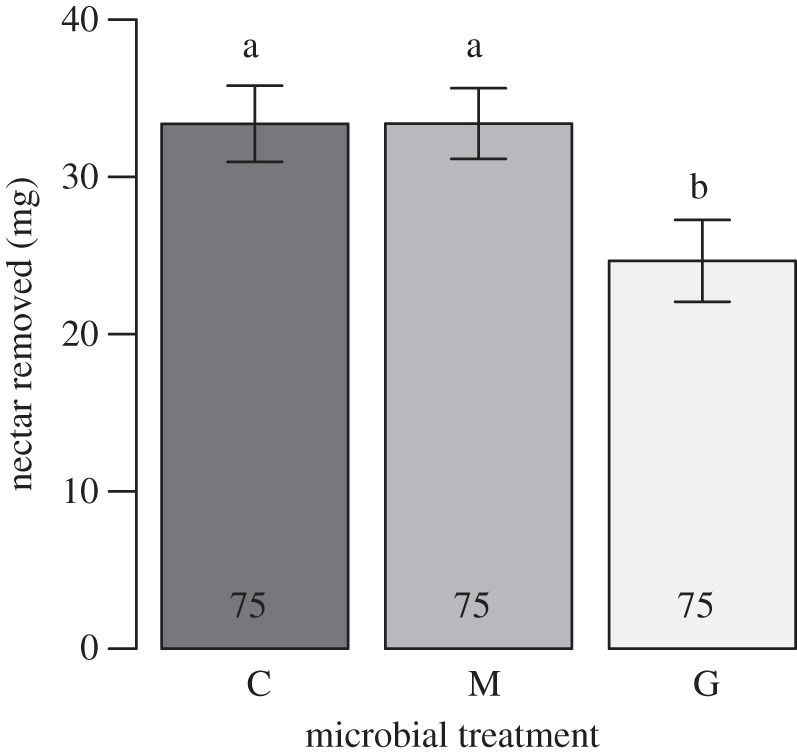
Effects of microbial inoculation on nectar mass removed by pollinators in synthetic flowers. Reduction in nectar mass because of evaporation, not by pollinators, was accounted for (see methods for detail). Symbols are as in figure 1.
(c). Experiment 3: microbial effects on nectar chemistry
In M. aurantiacus nectar, both Gluconobacter and M. reukaufii grew in number (see the electronic supplementary material, S9) and decreased H2O2 concentration by nearly 80 per cent (figure 3a). Their effects differed on all other chemical properties of nectar measured, however. Gluconobacter decreased pH by 5 units (a 105 increase in H+ concentration) and sucrose concentration by 35 per cent, whereas M. reukaufii decreased pH by 2 units and sucrose by 17 per cent (figure 3b,c). Gluconobacter reduced glucose concentration by 64 per cent and increased fructose concentration by 42 per cent, whereas M. reukaufii had little effect on glucose concentration and tended to decrease fructose concentration (figure 3d,e). Both microbes reduced total sugar concentration, but Gluconobacter-inoculated flowers had 27 per cent lower concentration of all sugars than the control, compared with a 16 per cent reduction by M. reukaufii (figure 3f).
Figure 3.
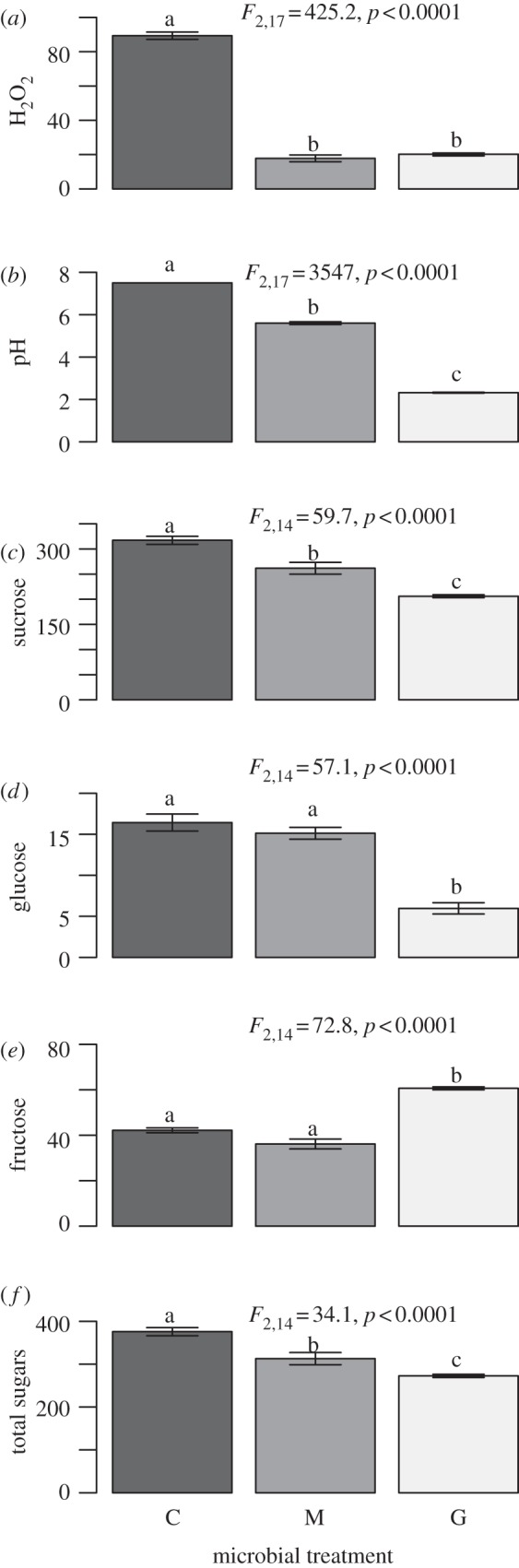
Effects of microbial inoculation on nectar chemistry, including (a) hydrogen peroxide (µM), (b) pH, (c) sucrose (mg/ml), (d) glucose (mg/ml), (e) fructose (mg/ml) and (f) total sugars (sum of sucrose, glucose and fructose). Bars indicate mean±1 s.e. Letters above bars indicate Tukey HSD test results. Symbols are as in figure 1.
4. Discussion
Taken together, our results demonstrate that a common bacterial species, but not a common yeast species, reduces pollination success, seed set and nectar consumption by pollinators, thereby weakening the mutual benefits that the plants and the pollinators gain from each other. Furthermore, our results also show that the two microbial species cause distinct changes in nectar chemistry, indicating possible mechanisms for their contrasting effects on the plant–pollinator mutualism.
(a). Possible mechanisms
Hummingbirds are known for strong preferences with respect to sugars, favouring concentrated solutions [39], particularly sucrose-dominant nectar [39,40], while avoiding high fructose concentrations [37]. Therefore, given the contrasting effects of bacteria and yeasts on sucrose (figure 3c), glucose (figure 3d) and fructose (figure 3e), changes in sugar composition may underlie the microbial effects we observed on pollination. In addition, some birds are known to avoid highly acidic solutions [41,42], although little work has been done on hummingbirds specifically. Thus, the precipitous pH reduction by Gluconobacter—a species that has been used in vinegar production since prehistoric times [43]—may also have been responsible for the bacterial effects on pollination. Although striking, the level of Gluconobacter-induced reduction in pH (figure 3b) is within the range recorded in field-collected M. aurantiacus nectar (between 2.5 and 8.0).
Hummingbirds respond more strongly to compounds within nectar than those emitted as volatiles [44] and may be unable to detect the presence of microbes in flowers without tasting nectar [45]. If so, this may explain why some nectar was removed from bacteria-inoculated flowers (figure 2) and why only the number of seeds, but not the probability of a flower setting seed, was affected by bacterial inoculation. If hummingbirds forage less when nectar tastes less favourably, they may still transfer some pollen, but not a sufficient amount for maximum seed set (female fitness of plants), a possibility consistent with our results (figure 1). To investigate this possibility further, we are currently using camera trapping to directly observe hummingbird behaviour. Although not measured in this study, the amount of pollen transport (male fitness of plants) may also be affected by microbial colonization of nectar.
Yeast and bacteria were present in the non-inoculated, control flowers by the end of our experiments (see the electronic supplementary material, S3). The measured effects of the focal microbes on pollination and seed set are, therefore, in comparison to the nectar in which microbial species colonized naturally, rather than to the nectar that was kept sterile for the duration of the experiment. For this reason, our experiment may have underestimated the effect of microbial inoculation.
(b). Generality of results
Because nectar yeasts and bacteria are widespread in many species of flowering plants worldwide, both insect- and vertebrate-pollinated [11,17,23,24], it is possible that our findings apply broadly to many cases of plant–pollinator mutualisms. Indeed, preliminary results of our ongoing research indicate that the honeybee, Apis mellifera, also prefers yeast-colonized nectar to bacteria-colonized nectar. Moreover, although our study involved only one species each of yeasts and bacteria, M. reukaufii is the dominant yeast species at our field site [23] and commonly found in the nectar of many other plants [24]. Further, Gluconobacter is a member of the acetic acid bacteria, many of which frequently occur in nectar (e.g. Acinetobacter, Asaia) [17,18, electronic supplementary material, table S1] and cause similar chemical changes in solutions analogous to nectar [43,46]. In addition, microbial species richness is generally low within individual flowers, which often contain only one or a few species [23,26,47], perhaps due to strong competition and priority effects [22]. For these reasons, our single-species inoculation treatments are relevant to understanding naturally assembling microbial communities in nectar, and our findings may well represent typical outcomes of nectar colonization by bacteria and yeasts.
Nevertheless, other species of yeasts and bacteria inhabit floral nectar [10,17,18], and some of these species are likely to differ in their effects on nectar chemistry [22]. It remains unknown to what extent these species-specific effects translate into variation in their effects on the strength of plant–pollinator mutualisms. Likewise, species of plants and pollinators may vary in their response to microbe-induced changes to nectar. Future research should investigate how different species of plants, pollinators and nectar microbes interact with one another in order to assess the generality of our findings.
(c). Plant–pollinator–microbe interactions
It has been pointed out that the community ecology of nectar microbes cannot be understood without considering plant–pollinator interactions [11,19]. This is because many nectar microbes depend on pollinators to disperse between flowers [11,26]. Our results indicate that the reverse is also true: plant–pollinator interactions may not be fully understood without considering the determinants of microbial species composition in nectar. This is because microbial species vary in their effects on pollination, as we have shown here. As a consequence, factors affecting microbial community assembly in nectar, including habitat filtering [19], niche breadth [48], dispersal [23,25] and competition and priority effects [22] should no longer be of interest only to microbial ecologists, but also to pollination biologists.
Our findings suggest several directions of future research on plant–pollinator–microbe interactions. One direction concerns microbial dispersal among flowers. The degree to which nectar microbes depend on pollinators for dispersal may have major implications for understanding pollination mutualisms. For example, if a microbial species depends heavily on pollinators, that species should fare well if it minimized any negative effect on pollinator visitation. On the other hand, if a species was capable of dispersing to flowers at a reasonable rate without pollinators’ help, it could exploit nectar to its own benefit as much as possible even if its growth degraded nectar and discouraged pollinator visits. Given our results, one might hypothesize that bacteria are less reliant on pollinators for dispersal than yeasts, but this hypothesis awaits further investigation.
Another hypothesis may be that, with all else equal, the role of pollinators as dispersal vectors is more important to the persistence of specialist microbes (e.g. those species to which nectar is the primary habitat) than to that of generalist counterparts (e.g. those to which nectar is only a minor part of the range of their habitats). If so, evolutionary selective pressure for minimizing detrimental effects on nectar should be stronger in specialists than in generalists and may lead to divergent effects of different microbial species on plant–pollinator mutualisms depending on their habitat breadth. Testing this and other hypotheses linking the dispersal and habitat breadth of nectar microbes to their effects on pollination mutualisms should contribute to placing plant–pollinator–microbe interactions in a coherent eco-evolutionary context.
Microbial growth in nectar may in turn elicit evolutionary and ecological responses by plants, pollinators and other floral visitors. Some plants are thought to exhibit specific traits that contribute to protecting nectar resources [34]. For example, the presence of nectarin proteins [49], H2O2 [20] and other metabolites in nectar [35] may reduce microbial growth, protecting the integrity of floral rewards. As for responses by pollinators, we have noticed that hummingbirds sometimes visit unopened flowers (see the electronic supplementary material, S10). Given our results, it seems plausible that this behaviour is adaptive because unopened flowers are less likely to contain microbe-degraded nectar than open flowers. At the same time, however, preferential visits to unopened flowers may reduce the pollinators' efficacy as the plants' partner because pollen transfer may be less effective. Furthermore, mites, thrips, nitidulid beetles and other arthropods that visit flowers of some plants phoretically on hummingbirds or via their own power [50–53] may also serve as microbial vectors and modify plant–pollinator–microbe interactions. It remains poorly known how these insects may affect plant–pollinator mutualisms indirectly via nectar microbes. We believe that consideration of these possibilities regarding plant–pollinator–microbe interactions will lead to a better understanding of the conditions under which strong pollination mutualisms are maintained.
5. Conclusion
We have provided experimental demonstration of how even seemingly similar species can have contrasting effects as third-party species influencing the strength of a mutualistic relationship. The differences that we found between the effects of bacteria and yeasts suggest that understanding the determinants of microbial species composition in nectar and their differential modification of floral rewards is necessary to fully explain the mutual benefits exchanged between plants and pollinators. Species-specific third-party effects of nectar microbes make their community assembly not only interesting from basic ecological perspectives, but also potentially important from applied agricultural standpoints, given the large number of crops that rely on pollination [54] and the diverse group of pollinators that depend on floral nectar.
Acknowledgements
We thank Simone Barley-Greenfield, Melinda Belisle, Nona Chiariello, Ashley Good, Trevor Hebert, Chase Mendenhall, Kabir Peay, Rachel Powell, Ray Von Itter, Dave Wilson, and the students and teaching staff of the 2012 Biology 44Y class for assistance with laboratory and fieldwork; Joel Sachs, three anonymous reviewers, and the members of the community ecology group at Stanford University for comments; and the Department of Biology and the Terman Fellowship of Stanford University and the National Science Foundation (award number: DEB1149600) for financial support. R.L.V. is a Gordon and Betty Moore Foundation Fellow of the Life Sciences Research Foundation. Data accession: Dryad DOI: 10.5061/dryad.3db81.
References
- 1.Stachowicz JJ. 2001. Mutualism, facilitation, and the structure of ecological communities. Bioscience 51, 235–246 10.1641/0006-3568(2001)051[0235:mfatso]2.0.co;2 (doi:10.1641/0006-3568(2001)051[0235:mfatso]2.0.co;2) [DOI] [Google Scholar]
- 2.Strauss SY, Irwin RE. 2004. Ecological and evolutionary consequences of multispecies plant–animal interactions. Annu. Rev. Ecol. Evol. Syst. 35, 435–466 10.1146/annurev.ecolsys.35.112202.130215 (doi:10.1146/annurev.ecolsys.35.112202.130215) [DOI] [Google Scholar]
- 3.Biere A, Honders SC. 2006. Coping with third parties in a nursery pollination mutualism: Hadena bicruris avoids oviposition on pathogen-infected, less rewarding Silene latifolia. New Phytol. 169, 719–727 10.1111/j.1469-8137.2005.01511.x (doi:10.1111/j.1469-8137.2005.01511.x) [DOI] [PubMed] [Google Scholar]
- 4.Bronstein JL. 2001. The exploitation of mutualisms. Ecol. Lett. 4, 277–287 10.1046/j.1461-0248.2001.00218.x (doi:10.1046/j.1461-0248.2001.00218.x) [DOI] [Google Scholar]
- 5.Sachs JL, Simms EL. 2006. Pathways to mutualism breakdown. Trends Ecol. Evol. 21, 585–592 10.1016/j.tree.2006.06.018 (doi:10.1016/j.tree.2006.06.018) [DOI] [PubMed] [Google Scholar]
- 6.Yu DW, Pierce NE. 1998. A castration parasite of an ant–plant mutualism. Proc. R. Soc. Lond. B 265, 375–382 10.1098/rspb.1998.0305 (doi:10.1098/rspb.1998.0305) [DOI] [Google Scholar]
- 7.Bronstein JL. 1994. Conditional outcomes in mutualistic interactions. Trends Ecol. Evol. 9, 214–217 10.1016/0169-5347(94)90246-1 (doi:10.1016/0169-5347(94)90246-1) [DOI] [PubMed] [Google Scholar]
- 8.Heath KD, Lau JA. 2011. Herbivores alter the fitness benefits of a plant–rhizobium mutualism. Acta Oecol-Int. J. Ecol. 37, 87–92 10.1016/j.actao.2010.12.002 (doi:10.1016/j.actao.2010.12.002) [DOI] [Google Scholar]
- 9.Jimbo T. 1926. Yeasts isolated from flower nectar. Sci. Rept. Tohoku Imp. Univ. 2, 161–187 [Google Scholar]
- 10.Lachance MA, Starmer WT, Rosa CA, Bowles JM, Barker JSF, Janzen DH. 2001. Biogeography of the yeasts of ephemeral flowers and their insects. FEMS Yeast Res. 1, 1–8 10.1111/j.1567-1364.2001.tb00007.x (doi:10.1111/j.1567-1364.2001.tb00007.x) [DOI] [PubMed] [Google Scholar]
- 11.Brysch-Herzberg M. 2004. Ecology of yeasts in plant–bumblebee mutualism in Central Europe. FEMS Microbiol. Ecol. 50, 87–100 10.1016/j.femsec.2004.06.003 (doi:10.1016/j.femsec.2004.06.003) [DOI] [PubMed] [Google Scholar]
- 12.Herrera CM, Garcia IM, Perez R. 2008. Invisible floral larcenies: microbial communities degrade floral nectar of bumble bee-pollinated plants. Ecology 89, 2369–2376 10.1890/08-0241.1 (doi:10.1890/08-0241.1) [DOI] [PubMed] [Google Scholar]
- 13.Eisikowitch D, Lachance MA, Kevan PG, Willis S, Collinsthompson DL. 1990. The effect of the natural assemblage of microorganisms and selected strains of the yeast Metschnikowia reukaufii in controlling the germination of pollen of the common milkweed Asclepias syriaca. Can. J. Bot-Rev. Can. Bot. 68, 1163–1165 10.1139/b90-147 (doi:10.1139/b90-147) [DOI] [Google Scholar]
- 14.Raguso RA. 2004. Why are some floral nectars scented? Ecology 85, 1486–1494 10.1890/03-0410 (doi:10.1890/03-0410) [DOI] [Google Scholar]
- 15.Pozo MI, de Vega C, Canto A, Herrera CM. 2009. Presence of yeasts in floral nectar is consistent with the hypothesis of microbial-mediated signaling in plant–pollinator interactions. Plant Signal. Behav. 4, 1102–1104 10.4161/psb.4.11.9874 (doi:10.4161/psb.4.11.9874) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrera CM, Pozo MaI. 2010. Nectar yeasts warm the flowers of a winter-blooming plant. Proc. R. Soc. B 277, 1827–1834 10.1098/rspb.2009.2252 (doi:10.1098/rspb.2009.2252) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Álvarez-Pérez S, Herrera CM, de Vega C. 2012. Zooming-in on floral nectar: a first exploration of nectar-associated bacteria in wild plant communities. FEMS Microbiol. Ecol. 80, 591–602 10.1111/j.1574-6941.2012.01329.x (doi:10.1111/j.1574-6941.2012.01329.x) [DOI] [PubMed] [Google Scholar]
- 18.Fridman S, Izhaki I, Gerchman Y, Halpern M. 2012. Bacterial communities in floral nectar. Environ. Microbiol. Rep. 4, 97–104 10.1111/j.1758-2229.2011.00309.x (doi:10.1111/j.1758-2229.2011.00309.x) [DOI] [PubMed] [Google Scholar]
- 19.Herrera CM, Canto A, Pozo MaI, Bazaga P. 2010. Inhospitable sweetness: nectar filtering of pollinator-borne inocula leads to impoverished, phylogenetically clustered yeast communities. Proc. R. Soc. B 277, 747–754 10.1098/rspb.2009.1485 (doi:10.1098/rspb.2009.1485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter C, Thornburg RW. 2004. Is the nectar redox cycle a floral defense against microbial attack? Trends Plant Sci. 9, 320–324 10.1016/j.tplants.2004.05.08 (doi:10.1016/j.tplants.2004.05.08) [DOI] [PubMed] [Google Scholar]
- 21.Fetscher AE, Kohn JR. 1999. Stigma behavior in Mimulus aurantiacus (Scrophulariaceae). Am. J. Bot. 86, 1130–1135 10.2307/2656976 (doi:10.2307/2656976) [DOI] [PubMed] [Google Scholar]
- 22.Peay KG, Belisle M, Fukami T. 2012. Phylogenetic relatedness predicts priority effects in nectar yeast communities. Proc. R. Soc. B 279, 749–758 10.1098/rspb.2011.1230 (doi:10.1098/rspb.2011.1230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belisle M, Peay KG, Fukami T. 2012. Flowers as islands: spatial distribution of nectar-inhabiting microfungi among plants of Mimulus aurantiacus, a hummingbird-pollinated shrub. Microb Ecol. 63, 711–718 10.1007/s00248-011-9975-8 (doi:10.1007/s00248-011-9975-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrera CM, de Vega C, Canto A, Pozo MI. 2009. Yeasts in floral nectar: a quantitative survey. Ann. Bot. 103, 1415–1423 10.1093/aob/mcp026 (doi:10.1093/aob/mcp026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pozo MI, Lachance M-A, Herrera CM. 2012. Nectar yeasts of two southern Spanish plants: the roles of immigration and physiological traits in community assembly. FEMS Microbiol. Ecol. 80, 281–293 10.1111/j.1574-6941.2011.01286.x (doi:10.1111/j.1574-6941.2011.01286.x) [DOI] [PubMed] [Google Scholar]
- 26.de Vega C, Herrera CM. 2012. Relationships among nectar-dwelling yeasts, flowers and ants: patterns and incidence on nectar traits. Oikos 121, 1878–1888 10.1111/j.1600-0706.2012.20295.x (doi:10.1111/j.1600-0706.2012.20295.x) [DOI] [Google Scholar]
- 27.O'Donnell K. 1993. Fusarium and its near relatives. In The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics (eds Reynolds DR, Taylor JW.), pp. 225–233 Wallingford, WA: CAB International [Google Scholar]
- 28.Heilbron DC. 1994. Zero-altered and other regression models for count data with added zeros. Biomet. J. 36, 531–547 10.1002/bimj.4710360505 (doi:10.1002/bimj.4710360505) [DOI] [Google Scholar]
- 29.Martin TG, Wintle BA, Rhodes JR, Kuhnert PM, Field SA, Low-Choy SJ, Tyre AJ, Possingham HP. 2005. Zero tolerance ecology: improving ecological inference by modelling the source of zero observations. Ecol. Lett. 8, 1235–1246 10.1111/j.1461-0248.2005.00826.x (doi:10.1111/j.1461-0248.2005.00826.x) [DOI] [PubMed] [Google Scholar]
- 30.Fletcher D, MacKenzie D, Villouta E. 2005. Modelling skewed data with many zeros: a simple approach combining ordinary and logistic regression. Environ. Ecol. Stat. 12, 45–54 10.1007/s10651-005-6817-1 (doi:10.1007/s10651-005-6817-1) [DOI] [Google Scholar]
- 31.Bates D, Maechler M, Bolker B. 2011. lme4: linear mixed-effects models using S4 classes. R package v. 0.999375-42 [Google Scholar]
- 32.Pinheiro J, Bates D, DebRoy S, Sarkar D. 2012. nlme: linear and nonlinear mixed effects Models. R package v. 3.1-105 [Google Scholar]
- 33.R Development Core Team 2012. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 34.Heil M. 2011. Nectar: generation, regulation, and ecological functions. Trends Plant Sci. 16, 191–200 10.1016/j.tplants.2011.01.003 (doi:10.1016/j.tplants.2011.01.003) [DOI] [PubMed] [Google Scholar]
- 35.Adler LS. 2000. The ecological significance of toxic nectar. Oikos 91, 409–420 10.1034/j.1600-0706.2000.910301.x (doi:10.1034/j.1600-0706.2000.910301.x) [DOI] [Google Scholar]
- 36.Kessler D, Baldwin IT. 2007. Making sense of nectar scents: the effects of nectar secondary metabolites on floral visitors of Nicotiana attenuata. Plant J. 49, 840–854 10.1111/j.1365-313X.2006.02995.x (doi:10.1111/j.1365-313X.2006.02995.x) [DOI] [PubMed] [Google Scholar]
- 37.Stiles FG. 1976. Taste preferences, color preferences, and flower choice in hummingbirds. Condor 78, 10–26 10.2307/1366912 (doi:10.2307/1366912) [DOI] [Google Scholar]
- 38.Olesen JM, Ronsted N, Tolderlund U, Cornett C, Molgaard P, Madsen J, Jones CG, Olsen CE. 1998. Mauritian red nectar remains a mystery. Nature 393, 529–529 10.1038/31128 (doi:10.1038/31128) [DOI] [Google Scholar]
- 39.Hainsworth FR, Wolf LL. 1976. Nectar characteristics and food selection by hummingbirds. Oecologia 25, 101–113 10.1007/BF00368847 (doi:10.1007/BF00368847) [DOI] [PubMed] [Google Scholar]
- 40.Martínez del Rio CM, Baker HG, Baker I. 1992. Ecological and evolutionary implications of digestive processes: bird preferences and the sugar constituents of floral nectar and fruit pulp. Experientia 48, 544–550 10.1007/BF01920237 (doi:10.1007/BF01920237) [DOI] [Google Scholar]
- 41.Prŷs-Jones OE, Willmer PG. 1992. The biology of alkaline nectar in the purple toothwort (Lathraea clandestina): ground level defences. Biol. J. Linnean Soc. 45, 373–388 10.1111/j.1095-8312.1992.tb00650.x (doi:10.1111/j.1095-8312.1992.tb00650.x) [DOI] [Google Scholar]
- 42.Harriman AE. 1968. Rejection thresholds for citric acid solutions in cowbirds, starlings, and red-wing blackbirds. Amer. Midland Nat. 79, 240–242 10.2307/2423169 (doi:10.2307/2423169) [DOI] [Google Scholar]
- 43.Deppenmeier U, Hoffmeister M, Prust C. 2002. Biochemistry and biotechnological applications of Gluconobacter strains. Appl. Microbiol. Biotechnol. 60, 233–242 10.1007/s00253-002-1114-5 (doi:10.1007/s00253-002-1114-5) [DOI] [PubMed] [Google Scholar]
- 44.Kessler A, Halitschke R. 2007. Specificity and complexity: the impact of herbivore-induced plant responses on arthropod community structure. Curr. Opin. Plant Biol. 10, 409–414 10.1016/j.pbi.2007.06.001 (doi:10.1016/j.pbi.2007.06.001) [DOI] [PubMed] [Google Scholar]
- 45.Irwin RE. 2000. Hummingbird avoidance of nectar-robbed plants: spatial location or visual cues. Oikos 91, 499–506 10.1034/j.1600-0706.2000.910311.x (doi:10.1034/j.1600-0706.2000.910311.x) [DOI] [Google Scholar]
- 46.Crotti E, et al. 2010. Acetic acid bacteria, newly emerging symbionts of insects. Appl. Environ. Microbiol. 76, 6963–6970 10.1128/aem.01336-10 (doi:10.1128/aem.01336-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pozo MI, Herrera CM, Bazaga P. 2011. Species richness of yeast communities in floral nectar of southern spanish plants. Microb. Ecol. 61, 82–91 10.1007/s00248-010-9682-x (doi:10.1007/s00248-010-9682-x) [DOI] [PubMed] [Google Scholar]
- 48.Herrera CM, Pozo MI, Bazaga P. 2012. Jack of all nectars, master of most: DNA methylation and the epigenetic basis of niche width in a flower-living yeast. Mol. Ecol. 21, 2602–2616 10.1111/j.1365-294X.2011.05402.x (doi:10.1111/j.1365-294X.2011.05402.x) [DOI] [PubMed] [Google Scholar]
- 49.Harper AD, Stalnaker SH, Wells L, Darvill A, Thornburg R, York WS. 2010. Interaction of Nectarin 4 with a fungal protein triggers a microbial surveillance and defense mechanism in nectar. Phytochemistry 71, 1963–1969 10.1016/j.phytochem.2010.09.009 (doi:10.1016/j.phytochem.2010.09.009) [DOI] [PubMed] [Google Scholar]
- 50.Grant V, Connell WA. 1979. The association between Carpophilus beetles and cactus flowers. Plant Syst. Evol. 133, 99–102 10.1007/bf00985884 (doi:10.1007/bf00985884) [DOI] [Google Scholar]
- 51.Lachance MA, Starmer WT. 2008. Kurtzmaniella gen. nov and description of the heterothallic, haplontic yeast species Kurtzmaniella cleridarum sp nov., the teleornorph of Candida cleridarum. Int. J. Syst. Evol. Microbiol. 58, 520–524 10.1099/ijs.0.65460-0 (doi:10.1099/ijs.0.65460-0) [DOI] [PubMed] [Google Scholar]
- 52.Colwell RK. 1995. Effects of nectar consumption by the hummingbird flower mite Proctolaelaps kirmsei on nectar availability in Hamelia patens. Biotropica 27, 206–217 10.2307/2388996 (doi:10.2307/2388996) [DOI] [Google Scholar]
- 53.Lara C, Ornelas JF. 2002. Flower mites and nectar production in six hummingbird-pollinated plants with contrasting flower longevities. Can. J. Bot-Rev. Can. Bot. 80, 1216–1229 10.1139/b02-109 (doi:10.1139/b02-109) [DOI] [Google Scholar]
- 54.Klein AM, Vaissiere BE, Cane JH, Steffan-Dewenter I, Cunningham SA, Kremen C, Tscharntke T. 2007. Importance of pollinators in changing landscapes for world crops. Proc. R. Soc. B 274, 303–313 10.1098/rspb.2006.3721 (doi:10.1098/rspb.2006.3721) [DOI] [PMC free article] [PubMed] [Google Scholar]


