Abstract
The concentration of CO2 in the atmosphere is expected to double by the end of the century. Experiments have shown that this will have important effects on the physiology and ecology of photosynthetic organisms, but it is still unclear if elevated CO2 will elicit an evolutionary response in primary producers that causes changes in physiological and ecological attributes. In this study, we cultured lines of seven species of freshwater phytoplankton from three major groups at current (approx. 380 ppm CO2) and predicted future conditions (1000 ppm CO2) for over 750 generations. We grew the phytoplankton under three culture regimes: nutrient-replete liquid medium, nutrient-poor liquid medium and solid agar medium. We then performed reciprocal transplant assays to test for specific adaptation to elevated CO2 in these lines. We found no evidence for evolutionary change. We conclude that the physiology of carbon utilization may be conserved in natural freshwater phytoplankton communities experiencing rising atmospheric CO2 levels, without substantial evolutionary change.
Keywords: functional group, global change, nutrient enrichment, carbon, drawdown, experimental evolution
1. Introduction
(a). The response of primary producers to rising CO2
The concentration of CO2 in the atmosphere is expected to reach levels between 700 and 1000 ppm by the end of the century [1]. The physiological response, from altered stoichiometry to increased whole-plant growth, is well documented in all major groups of plants [2,3]. Differences between plant taxa in their physiological response to CO2 are likely to lead to altered community composition [4]. Experiments with plants have even detected differences among genotypes within a species with respect to their response to elevated CO2 [4,5]. The presence of genotypic variance in the response to elevated CO2 suggests that populations are likely to adapt to elevated CO2 through the sorting of standing genetic variation within populations, which could be expanded by new beneficial mutations. This evolutionary response may alter the physiological response of species to future CO2 levels. Moreover, if species evolve to rising CO2 at different rates, evolutionary changes may alter the course of ecological shifts in species composition. Thus, the evolutionary response of populations may affect species and communities, and thereby the properties of whole ecosystems.
Despite the potential importance of evolutionary change in response to elevated CO2, it has rarely been investigated [4]. Experiments using reciprocal transplantation after selection at elevated CO2 are the clearest way of demonstrating an evolutionary response [6], but few have been reported for elevated CO2 [7–11]. Some reciprocal transplant experiments in plants have shown adaptation to CO2 concentration below current levels, but they have consistently failed to detect specific adaptation to elevated CO2 [8,9]. The only study of land plants that has shown an effect of high CO2 across generations was a single-generation reciprocal transplant experiment that was not capable of distinguishing between maternal effects and genetic change [7].
The few selection experiments on land plants that have been reported were limited to very short time scales (only a dozen generations of selection at most) and involved very small populations (fewer than 100 individuals per population [4]). These conditions greatly restrict the potential for an adaptive response [12]. In these conditions, an evolutionary response would be detected only if a genotype with a large selective advantage at elevated CO2 were already segregating at high frequency in the population.
Selection experiments extending over hundreds of generations with effective population sizes of more than a million individuals are feasible in microbial systems. There has been comparatively little work on phytoplankton, even though photosynthetic microbes make roughly the same contribution as land plants to the global carbon cycle [13]. This neglect may be rooted in the general belief that most phytoplankton systems are not limited by carbon supply. This assumption is challenged by the presence of inducible carbon concentration mechanisms (CCMs), which increase the local concentration of CO2 around RuBisCo in the chloroplast when the external concentration is low [14]. Moreover, there is a growing body of evidence that enrichment with CO2 increases productivity in a diversity of aquatic systems [15–17]. We are gaining a growing understanding of the growth response of phytoplankton to CO2, including the effect of nutrient availability and the physiological mechanism underlying it [18,19]. Experiments have shown that species and groups differ in their response to elevated CO2, leading to changes in community dynamics when CO2 concentration is increased [20–22].
Despite the importance of phytoplankton in carbon cycles and food chains, and the growing evidence of phytoplankton sensitivity to atmospheric CO2 concentrations, only two selection experiments have been reported. Long-term experiments with the unicellular chlorophyte Chlamydomonas reinhardtii failed to show any specific adaptation to elevated CO2 [10]. Some lines evolved higher rates of photosynthesis, but these were offset by photorespiration and carbon leakage from the cell [23]. Some high-CO2 selection lines grew slowly at ambient CO2 concentration, as do natural populations of soil algae collected from CO2 springs [23]. Further experiments suggest that the evolution of strains differing in their response to elevated CO2 is constrained by competition with other strains [24]. The coccolithophore Emiliania huxleyi is detrimentally affected by elevated CO2, which impedes formation of the calcium carbonate scales that cover the cell surface. Long-term selection of this species under elevated CO2 yielded cells with enhanced calcification at the low pH caused by the treatment [11,25].
A general prediction of how elevated CO2 will affect primary productivity requires understanding the evolutionary response of phytoplankton groups with different histories and physiological attributes. We have measured the evolutionary response of seven species of phytoplankton from three major groups, all of which benefit to different extents from the rise in atmospheric CO2 [22]. They were cultured in growth regimes differing in how the atmospheric concentration affected the CO2 concentration experienced by the organisms (directly or through diffusion into liquid) and in the expected constraints on growth (low and elevated nutrients). We cultured these species for between 750 and 1050 generations at ambient and elevated CO2 concentrations in all culture regimes before measuring their growth in reciprocal transplant assays so as to identify any specific genetic adaptation to elevated CO2.
(b). Mechanisms of evolutionary response to elevated CO2
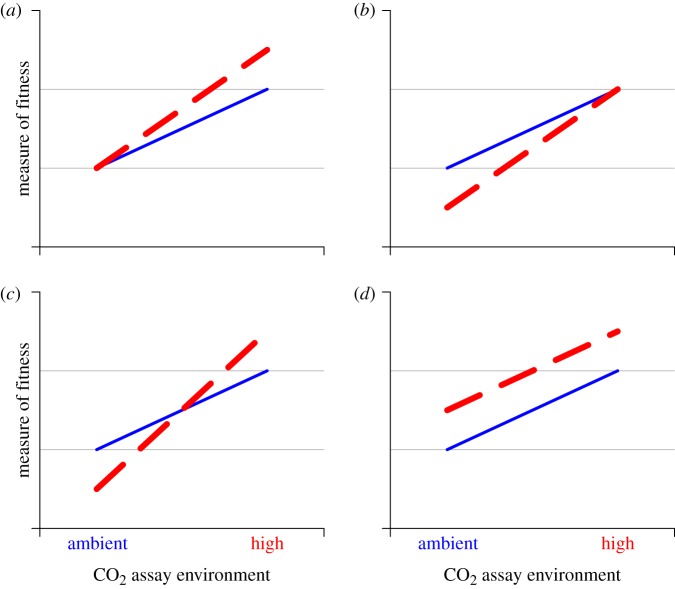
The direct response to elevated CO2 is specific adaptation to the new environment. The hypothesis that photosynthetic organisms may adapt specifically to elevated CO2 would be supported by reports of differences between genotypes in CO2 response. Under elevated CO2, carbon is no longer limiting for growth. The increased growth made possible by greater carbon availability will increase the demand for other nutrients and light. Genotypes with mutations conferring increased uptake of resources other than CO2, or reduced needs for these resources, would be at a selective advantage. Such phenotypes contribute to the physiological response to elevated CO2, including increased quantum yield, and thereby enhanced light use efficiency [18]. The mean fitness of the population selected at elevated CO2 would increase as these types spread (figure 1a).
Figure 1.
Possible outcomes of a reciprocal transplant experiment showing the expected effect of different mechanisms of evolutionary change. Solid grey line is ambient CO2 selection line; dashed black line is elevated CO2 selection line. (a) Beneficial mutations in elevated CO2 lines could lead to specific adaptation to elevated CO2. (b) Conditionally deleterious mutations in genes regulating carbon uptake and efficient use could accumulate at elevated CO2, lowering fitness of high lines in ambient CO2. (c) A trade-off in adapting to elevated CO2 or a combination of specific response and conditionally deleterious mutation could lead to lines from each environment becoming adapted only to their environment. (d) Our selection lines might diverge due to factors not directly related to CO2.
The indirect antagonistic response to elevated CO2 is the impairment of growth at ambient CO2. This might be caused by the accumulation of mutations that are neutral at elevated CO2 but harmful at ambient CO2. Growth is partly limited by carbon availability at ambient CO2, as shown by the increased growth of many photosynthetic organisms at elevated CO2. There is thus selective pressure to increase the carbon supply rate through CCMs and to use fixed carbon efficiently. Elevated CO2 leads to downregulation of genes governing the CCM [18], and the increased availability of CO2 might relax or eliminate the selective pressure on the maintenance of CCMs. Conditionally deleterious mutations in genes regulating carbon uptake and efficient use would accumulate at elevated CO2, an evolutionary response we can readily detect in our reciprocal transplant assay: lines selected at elevated CO2 will have reduced growth, relative to controls maintained at ambient CO2, when grown in ambient CO2 (figure 1b). Collins & Bell [10] postulated that this mechanism explains the results of their long-term selection experiment in which two of ten elevated CO2 lines could not grow at ambient CO2 concentrations. The accumulation of conditionally deleterious mutations would be harder to detect in diploid species as a functional allele could mask the effect of a deleterious allele in the same organism. Similar results to those expected from the accumulation of conditional deleterious mutations could also be obtained if ambient selection lines became better adapted to ambient CO2 conditions, but we assume that the ancestral populations are optimally adapted to current conditions.
The antagonistic indirect response, or specific adaptation with trade-offs, might instead arise because alleles have different effects at ambient and elevated CO2. Carbon uptake, and the uptake and use of other nutrients, may be functionally antagonistic in such a way that relaxed selection for carbon uptake at elevated CO2 would lead to selection for the alteration of other physiological traits. If these traits had been optimized in the ancestral population, adaptation to elevated CO2 would be accompanied by reduced fitness at ambient CO2 (figure 1c). A combination of conditionally deleterious mutations and specific adaptation would lead to a similar outcome.
Finally, selection lines might adapt to some general feature of the culture environment independent of the CO2 treatment, such as temperature or light level. The treatment and control lines would then both respond in the same way to elevated CO2 (figure 1d; note that this figure could equally be drawn with the ambient lines uppermost).
2. Material and methods
(a). Selection experiments
We set up a selection experiment with three major groups of freshwater phytoplankton: cyanobacteria, diatoms and chlorophytes. We chose two species of different growth form or size, where possible, as representatives of each group (Canadian Phycological Culture Centre number is given in brackets). The species were the cyanobacteria (haploid) Synechococcus leopoliensis (102) and Anabaena variabilis (105), the diatoms (diploid) Navicula pelliculosa (552) and Nitzschia palea (160), and the chlorophytes (haploid) Pseudokirchneriella subcapitata (37) and Scenedesmus acutus (10). To allow for comparison with previous studies of adaptation to elevated CO2 in phytoplankton, Chlamydomonas reinhardtii (haploid; Chlamydomonas Centre no. 2936 [26]) was included in the experiment.
We propagated a treatment set of three axenic (i.e. free from living organisms other than the organism of interest) selection lines of each species in atmospheres with 1000 ppm of CO2. We maintained a control set of three replicate axenic lines per species at ambient CO2 levels (375–400 ppm) throughout the experiment.
Cultures were grown under 100 μE m–2 s−1 of continuous light at 25°C in growth chambers at the McGill University Phytotron, using the WMA-4 CO2 analyser (PP Systems Inc.) to control CO2. We used modified Bold's Basal Medium (BBM) with pH adjusted to 7 using HCl , supplemented with silicate (0.58 g l−1 Na2SiO3) and vitamins (2 ml l−1 of the vitamin mix from F/2 Medium; for further information on the medium and references see [22]). We allowed the medium to equilibrate with chamber air for 5 days before inoculation. BBM is devoid of carbon, making CO2 the sole available carbon source. For liquid culture, the medium was buffered against changes in pH using HEPES buffer (4.766 g l−1) in 125 ml glass flasks. Cultures were continuously shaken at 250 r.p.m. with a 3 mm rotation diameter. An inoculum of 500 μl was transferred to 50 ml fresh medium after at each transfer. The cultures were not bubbled.
Three culture regimes were devised to modulate the availability of CO2 and limitation by other mineral nutrients.
(i). Regime 1
Nutrient-replete medium (undiluted modified BBM) with long transfer cycles of 5 days. In this regime, the CO2 concentration of the elevated treatment increased gradually, rising over 20 transfers and about 100 generations from ambient levels to 1000 ppm (less than 7 ppm per generation). This regime was maintained for over 1050 generations (160 transfers over 800 days).
(ii). Regime 2
Nutrient-poor medium (10% BBM) with short transfer cycles of 3 days. In this regime, elevated lines were directly exposed to 1000 ppm. This regime was maintained for over 750 generations (118 transfers over 413 days), with the exception of Navicula which was maintained for 340 generations (53 transfers over 184 days) owing to initial poor growth in diluted medium.
(iii). Regime 3
Nutrient-replete solid medium (undiluted BBM with 2.5 g l−1 agar and no HEPES) with short transfer cycles of 3 days. The added agar is not a source of available carbon. As in regime 2, elevated lines were directly exposed to 1000 ppm. Less than 1 per cent of the grown culture was transferred to a new agar plate flooded with 0.5 ml of undiluted BBM at each transfer. A single species of each group followed this regime (the cyanobacterium Synechococcus leopoliensis, the diatom Navicula pelliculosa and the chlorophyte Pseudokirchneriella subcapitata). This regime was maintained for over 750 generations (118 transfers over 413 days).
(b). Dissolved CO2 measurement and other physical parameters
Aqueous CO2 was measured in the final transfer in each regime. A 30 ml sample of culture was vigorously shaken with 30 ml of air lacking CO2 (using Sofnolime indicator by Molecular Products Inc.). The equilibrated air sample was injected through a hydrophobic 0.2 µm filter into an infrared gas analyser (IRGA, EGM-4 by PP-Systems Inc.) that was set up for static sampling. Measurements of salinity and temperature were made with a YSI-63 probe (YSI Inc.). Aqueous CO2 concentrations were calculated from the measure of CO2 in the equilibrated CO2 samples, adjusted for temperature and salinity, using methods provided by Yves Prairie, Biology Department, Université du Québec à Montréal [27]. These values were comparable with values calculated from the method provided by E. Lewis, Atmospheric Sciences Division, Brookhaven National Laboratory [28].
(c). Assay of growth by reciprocal transplant experiments
We conducted reciprocal transplants at intervals during the selection experiments; only the final assays are described here, but all gave similar results. The growth of lines with histories of ambient and elevated CO2 was measured at both ambient and elevated CO2 assay concentrations simultaneously. We used samples from each selection line to inoculate two cultures, one for each assay environment. All lines were tested in liquid cultures, as growth could not be measured accurately on solid medium. One culture was grown at elevated CO2, while the other culture was grown at ambient CO2. These cultures were allowed to acclimate to the CO2 environment for one transfer cycle to remove any potential non-evolutionary effect carried over from the selection line (e.g. the nutrient status of cells in the inoculum). The assay cultures whose growth was measured were inoculated from each of these acclimated cultures.
Reciprocal transplants were conducted both in flasks and in 48-well microplates (Costar plates by Corning Inc.) sealed with highly permeable membrane (Aeraseal by Excel Scientific Inc.). Growth in microplates allowed for an increased measurement rate. For microplates, 10 μl was used to inoculate 1 ml culture volume (1%, same as for flasks). Only the results of the microplate transplants are presented here; results from flask culture were similar. Absorbance through time was measured throughout the duration of growth (10–12 days) using an optical plate reader (Synergy-HT, BioTek, Winooski, VT).
(d). Analysis
All analyses and curve-fitting were conducted using the R statistical package [29]. All data and analysis scripts are available in the electronic supplementary material. Logistic growth Nt = KN0/[N0+(K – N0) exp(−rt)] was fitted using bounded maximum-likelihood estimation assuming normality of error with Simplex/Nelder-Mead optimization (optim) to estimate the limiting growth rate (r) and limiting density (K).
Specific adaptation to an environment, the accumulation of conditionally deleterious mutations owing to the relaxation of a stress, or both together, would lead to an interaction between the selection environment and the assay environment (figure 1a–c) [6]. Thus, to test for the presence of an evolutionary response, we focused on the interaction term containing selection and assay environment in ANOVAs conducted on growth parameters (r and K). Selection lines (A–F) are nested within selection history. We used a repeated-measures analysis, because a given line is tested in both CO2 environments simultaneously, with the variance among lines treated as the appropriate error term for testing hypotheses.
We shall report on estimates of the limiting growth rate, r (the estimates of K are reported in the electronic supplementary material and lead to the same conclusions). This parameter is the most important determinant of fitness, as cultures were transferred during the exponential growth phase in all regimes. The estimates we shall use are those for the last reciprocal transplant for each regime. This is the transplant for which lines had the most time to adapt to the treatment conditions. The same conclusions were obtained from all previous transplants (results available in the electronic supplementary material). Thus, our measure of fitness is based on growth in pure culture, not growth in competition.
3. Results
(a). Dissolved CO2 measurement and other physical parameters
At inoculation, all media were in equilibrium with treatment levels of CO2 (ambient approx. 380 ppm, elevated 1000 ppm).
In liquid cultures, however, the biological uptake of CO2 exceeded the maximum flux of CO2 through the stopper and through the air–liquid interface, despite thorough mixing. This caused CO2 concentration in the headspace and the aqueous CO2 concentrations to decrease during the transfer cycle. However, CO2 treatments remained different throughout the transfer cycle (for regime 1, ANOVA, F1,24 = 6.70, p = 0.016, average 51 ppm in the control culture and 72 ppm in the elevated CO2 cultures, Chlamydomonas was not measured; for regime 2, ANOVA, F1,28 = 771.31, p < 0.0001, average 131 ppm in the control culture and 626 ppm in the elevated CO2 cultures; only Anabaena decreased CO2 to equal levels across treatments in regime 2; figures provided in the electronic supplementary material).
Drawdown—the amount of CO2 being taken up by phytoplankton from the medium and air—is related to the amount of growth and photosynthesis in each culture. Species differed in growth and their growth was also greatly affected by culture regime (figure 2). Culture regimes thus differed in their drawdown. In culture regime 1, with strong growth permitted by high nutrient availability, CO2 uptake by the phytoplankton was much higher than the influx into the media resulting in a steeper decline in aqueous CO2 concentration across all treatments and species (average final concentration was 13.4% of initial concentration in ambient and 7.1% in elevated). In regime 2, where growth was more strongly limited by nutrient availability, the decline was lower (average final concentration was 34.4% of initial concentration in ambient and 62.9% in elevated). The regimes were thus effectively distinct environments to test for the evolutionary effect of elevated atmospheric CO2.
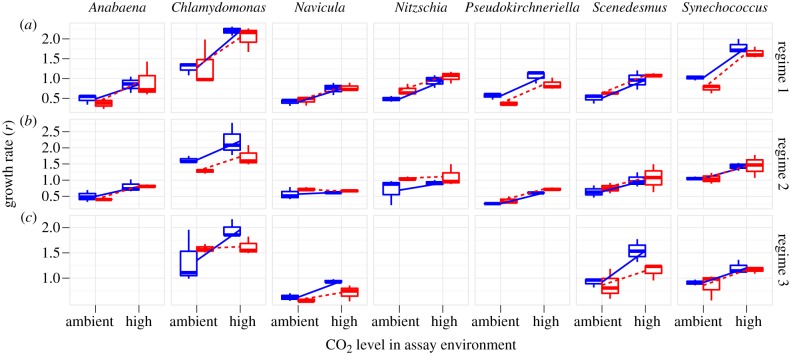
Figure 2.
Boxplot (hinges are first and third quartile, midline is the median) of limiting growth rate (r) in ambient and elevated CO2 assay environment. Grey and solid lines indicate ambient CO2 selection history, black and dashed lines indicate elevated CO2 selection history. Interaction due to specific adaptation would be shown by black above grey at right (figure 1a,c). Results are shown for (a) regime 1, (b) regime 2 and (c) regime 3.
Species differed in their drawdown of CO2 in both liquid regimes, however; Anabaena stood out with the highest drawdown in both regimes (see the electronic supplementary material, figures S1–S4). Anabaena may be less nutrient-limited owing to its ability to fix nitrogen.
The buffering was effective: pH rose marginally during transfer cycles and differed significantly with treatment (ambient compared with elevated) only in regime 2, which had a lower buffer concentration (ANOVA, F1,61 = 5.20, p < 0.0001; pH 7.11 in control cultures and pH 7.03 in elevated treatment cultures). The lower pH in the elevated-CO2 cultures would lead to a greater CO2 availability in the elevated CO2 conditions, increasing the effect of this treatment.
There was no liquid phase in the solid culture, and we presume that chamber CO2 concentrations are representative of CO2 available to cultures and individual cells.
(b). Overall effect of elevated CO2 on growth
In all regimes, elevated CO2 increased growth rate (figure 2). This effect was strongest in regime 1, in which elevated CO2 almost doubled growth rates (average increase by a factor of 1.90; ANOVA, F1,6 > 13, p < 0.02). Effects were smaller but generally remained significant in regimes 2 and 3. In regime 2, the average increase was 1.56; the estimate was not significantly different from 1.00 for diatoms, Chlamydomonas and Scenedesmus (ANOVA, F1,6 < 6.44, p > 0.064), but was significant for all other species in both regimes (ANOVA, F1,6 > 13.74, p < 0.021). In regime 3, the average factor of increase was 1.403, which was significant for all species (ANOVA, F1,6 > 7.84, p < 0.049).
(c). Evolutionary response to elevated CO2
No evolutionary response was detected in any of our regimes in any of our assays. Any general evolutionary response is reflected by the main effect of selection environment (figure 1d). This was not consistent across species and was generally not significant, showing that there was no predictable divergence between ambient and high-CO2 lines (figure 2). For regime 1, the ANOVA main effect had F1,4 < 6.31, p > 0.07. For regime 2, the main effect was significant only for Chlamydomonas, (ANOVA, F1,4 = 8.06, p = 0.0469; all others, F1,4 < 4.82, p > 0.0932). For regime 3, the main effect had F1,4 < 4.90, p > 0.0912.
Specific adaptation to CO2 concentration is reflected by the interaction of assay and selection (figure 1a–c). This was also non-significant for all regimes (figure 2; ANOVA interaction term, regime 1, F1,6 < 1.82, p > 0.24; regime 2, F1,6 < 2.25, p > 0.21; regime 3, F1,6 < 6.01, p > 0.070). No consistent trend could be found in any of our transplant assays for any of the three culture regimes in any of the seven species.
4. Discussion
Despite long-term selection with a relatively large population, we failed to detect any form of evolutionary response to CO2 by any of our species in any of the test conditions. We discuss this result in the light of the effectiveness of the CO2 treatment imposed, the lack of specific adaptation and the lack of accumulation of conditionally deleterious mutations. We shall then compare our system of experimental evolution with other experimental systems in which evolution was detected and with what we expect to happen in natural phytoplankton assemblages.
(a). Effectiveness of CO2 treatment
The atmospheric CO2 concentrations that we imposed as treatments in our growth chambers were found in the media surrounding the phytoplankton cells at inoculation; however, these treatment levels in the media were not maintained throughout the growth cycle (in regimes 1 and 2). This was due to rapid uptake by phytoplankton of CO2 from the medium. CO2 concentrations in the medium, however, were at treatment levels during the initial phase of growth and were consistently different, representative of the high versus low CO2 conditions we sought to establish in our cultures throughout the growth cycle (figure 2; Anabaena in regime 2 is an exception). Comparable falls in CO2 concentrations have been reported in large-scale marine mesocosm studies, with CO2 concentration in elevated CO2 mesocosms falling to half the treatment concentration of 1000 ppm [30]. In natural marine systems under current CO2 concentrations, algal blooms can reduce CO2 concentration by over 90 ppm [31]. In freshwater systems, phytoplankton can completely deplete dissolved CO2 concentrations [32,33]. The contrast between high and low CO2 treatments was more pronounced under regime 2 (low nutrients) than regime 1, reflecting the lesser capability of nutrient limited cultures to reduce aqueous CO2 concentrations. We conclude that our ambient versus elevated treatments provided environments that differ both in flux of CO2 through the cultures and in aqueous CO2 concentrations around the cells, so that our phytoplankton populations could be expected, in principle, to evolve specific adaptations to elevated CO2. The lack of an evolutionary response in this experiment does not preclude the possibility of specific adaptation arising in conditions where CO2 concentrations are much higher or are artificially maintained at the treatment level, as in experiments using bubbling.
The difference in CO2 availability between treatments affected the growth of all species and major groups, showing that carbon is a limiting resource in our system (figure 2), and consequently that the capacity to take up CO2 and use it more efficiently is potentially exposed to selection at ambient CO2 concentrations. Our findings are consistent with previous reports that CO2 availability can limit phytoplankton productivity in a wide range of conditions, including nutrient-replete freshwater cultures [16] and nutrient-poor oceans [15].
(b). Lack of specific adaptation
We cultured seven phytoplankton species at elevated CO2; the species differed substantially in their ancestry, physiology and ability to assimilate CO2, and were cultured in three different regimes that affected the availability of CO2. We were unable to detect evolutionary adaptation in any combination of species and culture regime: in no case did elevated CO2 lines have consistently higher growth than ambient lines at elevated CO2. We were unable to perform competitive assays as we do not have genetically marked strains in these non-model organisms, but we have no reason to suppose that competition between genotypes would depend on anything other than differential growth. Our results are consistent with other investigations into adaptation to elevated CO2 in plants (reviewed in [4]).
In lines maintained in liquid culture (regimes 1 and 2), where CO2 concentration fell during the transfer cycle, selection would have favoured types capable of higher peak CO2 uptake rates at peak CO2 concentrations. The change in selection pressure through the transfer cycle associated with changes in CO2 availability might retard the process of adaptation to directional change in CO2 concentrations [34]. The potential for specific adaptation was greatest in regime 3, where CO2 concentrations were maintained at treatment level throughout the transfer cycle. Growth as a biofilm may cause local CO2 depletion, however, and decrease flux [35]. In these lines, specific adaptation to CO2 could have arisen through peak CO2 uptake or through mechanisms to minimize the barrier to CO2 in biofilms. Phytoplankton in these cultures was selected in environments similar to that of photosynthetically active cells in land plants. The lack of adaptation in these cultures may suggest that direct adaptation to CO2 conditions may not arise in phytoplankton or land plants, and that evolutionary changes under rising CO2 may occur through indirect consequences, such as acidification for phytoplankton and decreased water loss in land plants.
It is unusual for selection experiments to fail as comprehensively as ours. Mutations that confer specific adaptation to elevated CO2 must be correspondingly scarce. The number of divisions occurring in each cycle in each line is approximately equal to the final number of cells, so that a conservative figure for cell density of 106 ml−1 leads to a rule-of-thumb estimate of 7 (species) × 3 (replicate lines per species) × 3 (culture regimes) × 50 ml (culture volume) × 106 (cells ml−1) × 100 (transfer cycles) ≈ 3 × 1011 divisions during which a beneficial mutation might have occurred. Many new beneficial mutations will be lost soon after appearing, through demographic stochasticity or dilution at transfer. The probability of fixation is proportional to the selection coefficient and the dilution rate [36]; for a modest selection coefficient of 0.05 and a dilution rate of 0.01 the probability of fixation is about 0.02 (it is higher for mutations of larger effect). The frequency of beneficial mutations is then less than about 2 × 10−10 per genome per generation. The distribution of rates and effects of mutations have not been studied in any of our focal species, but even the highest rate leading to our observations is much lower than estimates from other microbial populations in permissive conditions (e.g. 2 × 10−5 in E. coli [37] and 4.8 × 10−4 in Streptococcus [38]). As a broad general rule, the overall rate of mutation is about 3 × 10−3 per genome per DNA replication, independent of genome size [39], implying that the fraction of all mutations that are beneficial under elevated CO2 conditions in phytoplankton is less than about 10−7. This is much less than estimates of 0.13 for yeast [40] and 0.027 for Pseudomonas [41] in permissive conditions. Comparable estimates for viruses are 0.07 for a chimaeric RNA vesicular stomatitis virus (VSV) in permissive conditions [42], 0.0009 for VSV adapting to a new host cell type [43] and 0.0034 for phage φX174 adapting to high temperature [44]. Further experimental evolution studies focusing on a diversity of phytoplankton, which may have been neglected by experimental evolutionary biologists [45], would be required to understand patterns of adaptive potential across species for a diversity of conditions. However, it seems likely that contemporary phytoplankton species are almost perfectly adapted for photosynthesis over a broad range of conditions close to contemporary values with little if any room for improvement. In natural phytoplankton assemblages, the standing genetic variance may be far larger than in our axenic laboratory cultures, which were started from a single clone. It is true that, 50 million years ago, phytoplankton encountered atmospheric CO2 concentrations similar to the predicted conditions (1000 ppm), but they have not encountered such atmospheric conditions for at least 500 000 years [12,13]. It is thus conceivable (but unlikely) that genotypes adapted to these ancestral conditions are still present in contemporary populations. It is also possible that mutations conferring higher fitness in these conditions are found in contemporary organisms. Thus, adaptation to elevated CO2 concentrations cannot be ruled out in nature, but if it occurs at all it will involve very rare genotypes or very rare beneficial mutations that are unlikely to be observed in laboratory studies.
(c). Lack of accumulation of conditionally deleterious mutations
The accumulation of conditionally deleterious mutations is the only evolutionary response specific to CO2 concentrations (as opposed to adaptation to pH changes) for which there is experimental evidence at present [10]. Our experiment failed to elicit such a response either in diploid (diatoms) or in haploid species (chlorophytes and cyanobacteria), in which we would be more likely to detect this phenomenon. This may be attributable to the culture conditions we used. Most phytoplankton CO2 perturbation experiments so far reported, and both long-term selection experiments [10,11], have been conducted by bubbling the media with air having a controlled concentration of CO2. Bubbling creates an aqueous environment in constant equilibrium with the atmosphere. Furthermore, bubbling provides a far greater CO2 flux than would be available even in a well-aerated natural system. Thus, these experiments maximize the potential for the accumulation of conditionally deleterious mutations for carbon uptake and use mechanisms by maximizing the availability of CO2. In our liquid cultures, on the other hand, CO2 concentration was drawn down by algal growth so that all liquid lines were exposed to relatively low levels of aqueous CO2, reducing the potential to detect the accumulation of conditionally deleterious mutations in genes affecting carbon uptake.
There are few situations where natural water is in equilibrium with the atmosphere; that is, where it is neither a source nor a sink. Relative saturation with CO2 is strongly affected by biological activity, which can deplete CO2 by photosynthesis or produce CO2 by respiration. Freshwater systems tend to be supersaturated with CO2 produced by bacterial respiration [46], with a large portion of primary productivity being attributable to this supersaturation [47]. Many populations of freshwater phytoplankton may thus already experience chronically elevated CO2. In the oceans, relative saturation varies over time and space [48], with strong local decreases caused by blooming [49,50]. The availability of CO2 for phytoplankton is further modified by factors including pH, ionic strength and temperature [51]. It is thus expected that even in a future atmosphere with elevated CO2, marine phytoplankton would periodically face low-CO2 conditions comparable with the endpoint in our experiment. Hence, the accumulation of conditionally deleterious mutations depends on the local availability of carbon and will not necessarily occur as a general response to global increases in CO2 concentration [35].
(d). Adaptation to elevated nutrients, including CO2
For most phytoplankton, with the notable exception of calcifying phytoplankton [11], increasing CO2 to high but not extreme values (less than 1%) is not expected to have an adverse effect on growth. In the absence of acidification, even calcifying organisms would be expected to benefit from an increase in the availability of CO2. Adaptation to acidification would represent an evolutionary response to stress, whereas adaptation to elevated CO2 would represent an evolutionary response to fertilization. In this case, we were unable to identify an evolutionary response to fertilization.
More broadly, adaptation has usually been investigated as a response to stress rather than to fertilization. Bacteria cultured for thousands of generations with sugars such as lactose or glucose as a source of carbon and energy readily evolve higher fitness [52,53], often through loss-of-function mutations in genes regulating transport [54]. It is not clear, however, whether there is a specific adaptation to high-sugar conditions as opposed to low-sugar conditions. There has been great interest in the physiological response of plants and phytoplankton to non-toxic nutrient enrichment [55–57], including elevated CO2 [2], but there have been few, if any, convincing studies of specific adaptation to fertilization. In the long-term Park Grass Experiment at Rothamsted, the vegetation has been treated with different levels of fertilization for over 150 years. The physiological differences between populations of Anthoxanthum odoratum L. exposed to different treatments have been investigated through a modified reciprocal transplant experiment [58–60]. Populations occupying sites enriched for a given nutrient are more responsive to high levels of that nutrient in some cases, although not in all. However, these studies investigate the physiological response of plants directly extracted from the treatment population—a single-generation transplant that may carry maternal effects—and thus do not provide an unambiguous test of adaptation. New cultivars of crop plants may be selected for their responsiveness to higher inputs of fertilizers, and this provides evidence for specific adaptation through artificial selection. Comparisons of the yield of obsolete and modern cultivars of maize have shown that modern cultivars have greater yield under modern high-input management regimes, but they retain most of this advantage under the low-input regimes of the past [61]. In brief, experimental evolution, long-term field experiments and artificial selection experiments have all so far failed to provide unequivocal examples of specific adaptation to improved conditions of growth. Our failure to detect specific adaptation by phytoplankton to elevated CO2 may be only one example of a general tendency.
It has been argued that rapid evolutionary change may alter the ecological attributes of species, so that predicting changes in populations or communities requires understanding their combined eco-evolutionary dynamics. In many cases, this point of view offers a novel perspective on ecological processes. For the case of elevated CO2 in freshwater, however, there is good evidence that little or no evolutionary change will occur in the short term, so that ecological predictions can be made assuming that current ecological attributes are likely to remain stable.
(e). Data and analysis scripts
Data and analysis scripts are available at datadryad.org.
Acknowledgements
This work was supported by the Natural Science and Engineering Research Council of Canada (NSERC) through grants to GFF and GB and through scholarships to E.L.-D. and by the Fonds de recherche du Québec—Nature et technologies (FRQNT) through a scholarship to E.L.-D. We thank Yves Prairie for help measuring dissolved CO2 concentrations. We thank Janane Maheswaran, Katherine Huebner, Gregory (Adam) Meyer, Tyler Moulton and Kathy Tallon for help with culture maintenance. We thank two anonymous reviewers for comments on the manuscript.
References
- 1.IPCC 2007. Climate change 2007—the physical science basis. Contribution of working group I to the fourth assessment report of the Intergovernmental Panel on Climate Change, p. 996 Cambridge, UK: Cambridge University Press [Google Scholar]
- 2.Ainsworth EA, Long SP. 2005. What have we learned from 15 years of free-air CO2 enrichment (FACE)? A meta-analytic review of the responses of photosynthesis, canopy properties and plant production to rising CO2. New Phytol. 165, 351–372 10.1111/j.1469-8137.2004.01224.x (doi:10.1111/j.1469-8137.2004.01224.x) [DOI] [PubMed] [Google Scholar]
- 3.Urban O. 2003. Physiological impacts of elevated CO2 concentration ranging from molecular to whole plant responses. Photosynthetica 41, 9–20 10.1023/A:1025891825050 (doi:10.1023/A:1025891825050) [DOI] [Google Scholar]
- 4.Potvin C, Chapin F, Gonzalez A, Leadley P, Reich P, Roy J. 2006. Plant biodiversity and responses to elevated carbon dioxide. In Terrestrial ecosystems in a changing world (eds Canadell JG, Pataki DE, Pitelka LF.), pp. 103–112 Berlin, Germany: Springer [Google Scholar]
- 5.Rae AM, Tricker PJ, Bunn SM, Taylor G. 2007. Adaptation of tree growth to elevated CO. New Phytol. 175, 59–69 10.1111/j.1469-8137.2007.02091.x (doi:10.1111/j.1469-8137.2007.02091.x) [DOI] [PubMed] [Google Scholar]
- 6.Kawecki TJ, Ebert D. 2004. Conceptual issues in local adaptation. Ecol. Lett. 7, 1225–1241 10.1111/j.1461-0248.2004.00684.x (doi:10.1111/j.1461-0248.2004.00684.x) [DOI] [Google Scholar]
- 7.Lau J, Peiffer J, Reich P, Tiffin P. 2008. Transgenerational effects of global environmental change: long-term CO2 and nitrogen treatments influence offspring growth response to elevated CO2. Oecologia 158, 141–150 10.1007/s00442-008-1127-6 (doi:10.1007/s00442-008-1127-6) [DOI] [PubMed] [Google Scholar]
- 8.Potvin C, Tousignant D. 1996. Evolutionary consequences of simulated global change: genetic adaptation or adaptive phenotypic plasticity. Oecologia 108, 683–693 10.1007/BF00329043 (doi:10.1007/BF00329043) [DOI] [PubMed] [Google Scholar]
- 9.Ward JK, Antonovics J, Thomas RB, Strain BR. 2000. Is atmospheric CO2 a selective agent on model C3 annuals? Oecologia 123, 330–341 10.1007/s004420051019 (doi:10.1007/s004420051019) [DOI] [PubMed] [Google Scholar]
- 10.Collins SS, Bell G. 2004. Phenotypic consequences of 1,000 generations of selection at elevated CO2 in a green alga. Nature 431, 566–569 10.1038/nature02945 (doi:10.1038/nature02945) [DOI] [PubMed] [Google Scholar]
- 11.Lohbeck KT, Riebesell U, Reusch TBH. 2012. Adaptive evolution of a key phytoplankton species to ocean acidification. Nat. Geosci. 5, 1–6 10.1038/ngeo1441 (doi:10.1038/ngeo1441) [DOI] [Google Scholar]
- 12.Samani P, Bell G. 2010. Adaptation of experimental yeast populations to stressful conditions in relation to population size. J. Evol. Biol. 23, 791–796 10.1016/j.ecolecon.2008.08.022 (doi:10.1016/j.ecolecon.2008.08.022) [DOI] [PubMed] [Google Scholar]
- 13.Field CB, Behrenfield MJ, Randerson JT, Falkowski P. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240 10.1126/science.281.5374.237 (doi:10.1126/science.281.5374.237) [DOI] [PubMed] [Google Scholar]
- 14.Kaplan A, Reinhold L. 1999. CO2 concentrating mechanisms in photosynthetic microorganisms. Annu. Rev. Plant Biol. 50, 539–570 10.1162/jiec.1998.2.4.61 (doi:10.1162/jiec.1998.2.4.61) [DOI] [PubMed] [Google Scholar]
- 15.Hein M, Sand-Jensen K. 1997. CO2 increases oceanic primary production. Nature 388, 526. 10.1111/j.1530-9290.2011.00347.x (doi:10.1111/j.1530-9290.2011.00347.x) [DOI] [Google Scholar]
- 16.Schippers P, Lurling M, Scheffer M. 2004. Increase of atmospheric CO2 promotes phytoplankton productivity. Ecol. Lett. 7, 446–451 10.1006/jema.2002.0578 (doi:10.1006/jema.2002.0578) [DOI] [Google Scholar]
- 17.Ramos JBE, Biswas H, Schulz KG, LaRoche J, Riebesell U. 2007. Effect of rising atmospheric carbon dioxide on the marine nitrogen fixer Trichodesmium. Global Biogeochem. Cycles 21 10.1029/2006gb002898 (doi:10.1029/2006gb002898) [DOI] [Google Scholar]
- 18.Spijkerman E. 2008. What physiological acclimation supports increased growth at high CO2 conditions? Physiol. Plantarum 133, 41–48 10.1111/j.1399-3054.2008.01062.x (doi:10.1111/j.1399-3054.2008.01062.x) [DOI] [PubMed] [Google Scholar]
- 19.Beardall J, Stojkovic S, Larsen S. 2009. Living in a high CO2 world: impacts of global climate change on marine phytoplankton. Plant Ecol. Divers. 2, 191–205 10.1080/17550870903271363 (doi:10.1080/17550870903271363) [DOI] [Google Scholar]
- 20.Tortell PD, et al. 2008. CO2 sensitivity of southern ocean phytoplankton. Geophys. Res. Lett. 35, L04605. 10.1111/j.1530-9290.2011.00366.x (doi:10.1111/j.1530-9290.2011.00366.x) [DOI] [Google Scholar]
- 21.Paulino AI, Egge JK, Larsen A. 2007. Effects of increased atmospheric CO2 on small and intermediate sized osmotrophs during a nutrient induced phytoplankton bloom. Biogeosci. Discuss. 4, 4173–4195 10.5194/bgd-4-4173-2007 (doi:10.5194/bgd-4-4173-2007) [DOI] [Google Scholar]
- 22.Low-Décarie E, Fussmann GF, Bell G. 2011. The effect of elevated CO2 on growth and competition in experimental phytoplankton communities. Global Change Biol. 17, 2525–2535 10.1111/j.1365-2486.2011.02402.x (doi:10.1111/j.1365-2486.2011.02402.x) [DOI] [Google Scholar]
- 23.Collins S, Bell G. 2006. Evolution of natural algal populations at elevated CO2. Ecol. Lett. 9, 129–135 [DOI] [PubMed] [Google Scholar]
- 24.Collins S. 2010. Competition limits adaptation and productivity in a photosynthetic alga at elevated CO2. Proc. R. Soc. B 278, 247–255 10.1098/rspb.2010.1173 (doi:10.1098/rspb.2010.1173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohbeck KT, Riebesell U, Collins S, Reusch TBH. 2012. Functional genetic divergence in high CO2 adapted Emiliania huxleyi populations. Evolution (doi:10.1111/j.1558-5646.2012.01812.x) [DOI] [PubMed] [Google Scholar]
- 26.Sack L, Zeyl C, Bell G, Sharbel T, Reboud X, Bernhardt T, Koelewyn H. 1994. Note. Isolation of four new strains of Chlamydomonas reinhardtii (Chlorophyta) from soil samples. J. Phycol. 30, 770–773 10.1111/j.0022-3646.1994.00770.x (doi:10.1111/j.0022-3646.1994.00770.x) [DOI] [Google Scholar]
- 27.Cole JJ, Prairie YT. 2009. Dissolved CO2. In Encyclopedia of inland waters (ed Gene EL.), pp. 30–34 Oxford, UK: Academic Press [Google Scholar]
- 28.Lewis E, Wallace D, Allison L. 1998. Program developed for CO2 system calculations. See http://cdiac.esd.ornl.gov/oceans/co2rprtnbk.html [Google Scholar]
- 29.R Development Core Team 2009. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. (doi:10.1021/es802345a) [Google Scholar]
- 30.Bellerby RGJ, Schulz KG, Riebesell U, Neill C, Nondal G, Heegaard E, Johannessen T, Brown KR. 2008. Marine ecosystem community carbon and nutrient uptake stoichiometry under varying ocean acidification during the PeECE III experiment. Biogeosciences 5, 1517–1527 10.5194/bg-5-1517-2008 (doi:10.5194/bg-5-1517-2008) [DOI] [Google Scholar]
- 31.Robertson JE, Robinson C, Turner DR, Holligan P, Watson AJ, Boyd P, Fernandez E, Finch M. 1994. The impact of a coccolithophore bloom on oceanic carbon uptake in the northeast Atlantic during summer 1991. Deep Sea Res. Part I: Oceanogr. Res. Pap. 41, 297–314 10.1016/0967-0637(94)90005-1 (doi:10.1016/0967-0637(94)90005-1) [DOI] [Google Scholar]
- 32.Gu B, Schelske CL, Coveney MF. 2010. Low carbon dioxide partial pressure in a productive subtropical lake. Aquat. Sci. 73, 317–330 10.1007/s00027-010-0179-y (doi:10.1007/s00027-010-0179-y) [DOI] [Google Scholar]
- 33.Balmer MB, Downing JA. 2011. Carbon dioxide concentrations in eutrophic lakes: undersaturation implies atmospheric uptake. Inland Waters 2, 125–132 10.5268/IW-1.2.366 (doi:10.5268/IW-1.2.366) [DOI] [Google Scholar]
- 34.Bell G. 2008. Selection: the mechanism of evolution. Oxford, UK: Oxford University Press [Google Scholar]
- 35.Raven JA, Giordano M, Beardall J, Maberly SC. 2012. Algal evolution in relation to atmospheric CO2: carboxylases, carbon-concentrating mechanisms and carbon oxidation cycles. Phil. Trans. R. Soc. B 367, 493–507 10.1098/rstb.2011.0212 (doi:10.1098/rstb.2011.0212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wahl LM, Gerrish PJ, Saika-Voivod I. 2002. Evaluating the impact of population bottlenecks in experimental evolution. Genetics 162, 961–971 10.1162/1088198054084635 (doi:10.1162/1088198054084635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perfeito L, Fernandes L, Mota C, Gordo I. 2007. Adaptive mutations in bacteria: high rate and small effects. Science 317, 813–815 10.1126/science.1142284 (doi:10.1126/science.1142284) [DOI] [PubMed] [Google Scholar]
- 38.Stevens KE, Sebert ME. 2011. Frequent beneficial mutations during single-colony serial transfer of Streptococcus pneumoniae. PLoS Genet. 7, e1002232. 10.1371/journal.pgen.1002232 (doi:10.1371/journal.pgen.1002232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drake JW. 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl Acad. Sci. USA 88, 7160–7164 10.1146/annurev-matsci-062910-095759 (doi:10.1146/annurev-matsci-062910-095759) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall DW, Mahmoudizad R, Hurd AW, Joseph SB. 2008. Spontaneous mutations in diploid Saccharomyces cerevisiae: another thousand cell generations. Genet. Res. 90, 229–241 10.1017/S0016672308009324 (doi:10.1017/S0016672308009324) [DOI] [PubMed] [Google Scholar]
- 41.Kassen R, Bataillon T. 2006. Distribution of fitness effects among beneficial mutations before selection in experimental populations of bacteria. Nat. Genet. 38, 484–488 10.1038/ng1751 (doi:10.1038/ng1751) [DOI] [PubMed] [Google Scholar]
- 42.Sanjuán R, Moya A, Elena SF. 2004. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc. Natl Acad. Sci. USA 101, 8396–8401 10.1073/pnas.0400146101 (doi:10.1073/pnas.0400146101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cuevas JM, Elena SF, Moya A. 2002. Molecular basis of adaptive convergence in experimental populations of RNA viruses. Genetics 162, 533–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wichman HA, Badgett MR, Scott LA, Boulianne CM, Bull JJ. 1999. Different trajectories of parallel evolution during viral adaptation. Science 285, 422–424 10.1111/j.1530-9290.2010.00283.x (doi:10.1111/j.1530-9290.2010.00283.x) [DOI] [PubMed] [Google Scholar]
- 45.Chepurnov VA, Mann DG, von Dassow P, Vanormelingen P, Gillard J, Inzé D, Sabbe K, Vyverman W. 2008. In search of new tractable diatoms for experimental biology. BioEssays 30, 692–702 10.1002/bies.20773 (doi:10.1002/bies.20773) [DOI] [PubMed] [Google Scholar]
- 46.Ritchie RJ. 2008. Universal chlorophyll equations for estimating chlorophylls a, b, c, and d and total chlorophylls in natural assemblages of photosynthetic organisms using acetone, methanol, or ethanol solvents. Photosynthetica 46, 115–126 10.1007/s11099-008-0019-7 (doi:10.1007/s11099-008-0019-7) [DOI] [Google Scholar]
- 47.Jansson M, Karlsson J, Jonsson A. 2012. Carbon dioxide supersaturation promotes primary production in lakes. Ecol. Lett. 15, 527–532 10.1111/j.1461-0248.2012.01762.x (doi:10.1111/j.1461-0248.2012.01762.x) [DOI] [PubMed] [Google Scholar]
- 48.Takahashi T, et al. 2002. Global sea–air CO2 flux based on climatological surface ocean pCO2, and seasonal biological and temperature effects. Deep-Sea Res. Part II 49, 1601–1622 10.1016/S0967-0645(02)00003-6 (doi:10.1016/S0967-0645(02)00003-6) [DOI] [Google Scholar]
- 49.Boyd PW, et al. 2000. A mesoscale phytoplankton bloom in the polar Southern Ocean stimulated by iron fertilization. Nature 407, 695–702 10.1038/35037500 (doi:10.1038/35037500) [DOI] [PubMed] [Google Scholar]
- 50.Buitenhuis ET, van der Wal P, de Baar HJW. 2001. Blooms of Emiliania huxleyi are sinks of atmospheric carbon dioxide: a field and mesocosm study derived simulation. Global Biogeochem. Cycles 15, 577. 10.1029/2000GB001292 (doi:10.1029/2000GB001292) [DOI] [Google Scholar]
- 51.Beardall J, Johnston A, Raven J. 1998. Environmental regulation of CO2-concentrating mechanisms in microalgae. Botany 76, 1010–1017 [Google Scholar]
- 52.Dykhuizen DE, Dean AM. 1990. Enzyme activity and fitness: evolution in solution. Trends Ecol. Evol. 5, 257–262 10.1016/0169-5347(90)90067-N (doi:10.1016/0169-5347(90)90067-N) [DOI] [PubMed] [Google Scholar]
- 53.Cooper VS, Lenski RE. 2000. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407, 736–739 [DOI] [PubMed] [Google Scholar]
- 54.Notley-McRobb L, Ferenci T. 1999. The generation of multiple co-existing mal-regulatory mutations through polygenic evolution in glucose-limited populations of Escherichia coli. Environ. Microbiol. 1, 45–52 [DOI] [PubMed] [Google Scholar]
- 55.DiTommaso A, Aarssen LW. 1989. Resource manipulations in natural vegetation: a review. Plant Ecol. 84, 9–29 10.1007/BF00054662 (doi:10.1007/BF00054662) [DOI] [Google Scholar]
- 56.Downing JA, Osenberg CW, Sarnelle O. 2008. Meta-analysis of marine nutrient-enrichment experiments: variation in the magnitude of nutrient limitation. Ecology 80, 1157–1167 10.1890/0012-9658(1999)080[1157:MAOMNE]2.0.CO;2 (doi:10.1890/0012-9658(1999)080[1157:MAOMNE]2.0.CO;2) [DOI] [Google Scholar]
- 57.Elser JJ, et al. 2007. Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol. Lett. 10, 1135–1142 10.1111/j.1461-0248.2007.01113.x (doi:10.1111/j.1461-0248.2007.01113.x) [DOI] [PubMed] [Google Scholar]
- 58.Davies MS. 1975. Physiological differences among populations of Anthoxanthum odoratum L. collected from the Park Grass Experiment, Rothamsted. IV. Response to potassium and magnesium. J. Appl. Ecol. 12, 953–964 10.2307/2402101 (doi:10.2307/2402101) [DOI] [Google Scholar]
- 59.Davies MS, Snaydon RW. 1973. Physiological differences among populations of Anthoxanthum odoratum L. collected from the Park Grass Experiment, Rothamsted. I. Response to calcium. J. Appl. Ecol. 10, 33–45 10.2307/2404713 (doi:10.2307/2404713) [DOI] [Google Scholar]
- 60.Davies MS, Snaydon RW. 1974. Physiological differences among populations of Anthoxanthum odoratum L. collected from the Park Grass Experiment, Rothamsted. III. Response to phosphate. J. Appl. Ecol. 11, 699–707 10.2307/2402220 (doi:10.2307/2402220) [DOI] [Google Scholar]
- 61.Castleberry R, Crum C, Krull CF. 1984. Genetic yield improvement of US maize cultivars under varying fertility and climatic environments. Crop Sci. 24, 33–36 10.2135/cropsci1984.0011183X002400010008x (doi:10.2135/cropsci1984.0011183X002400010008x) [DOI] [Google Scholar]