Abstract
The ecological success of giant celled, siphonous green algae in coastal habitats has repeatedly been linked to endophytic bacteria living within the cytoplasm of the hosts. Yet, very little is known about the relative importance of evolutionary and ecological factors controlling the intracellular bacterial flora of these seaweeds. Using the marine alga Bryopsis (Bryopsidales, Chlorophyta) as a model, we explore the diversity of the intracellular bacterial communities and investigate whether their composition is controlled by ecological and biogeographic factors rather than the evolutionary history of the host. Using a combination of 16S rDNA clone libraries and denaturing gradient gel electrophoresis analyses, we show that Bryopsis harbours a mixture of relatively few but phylogenetically diverse bacterial species. Variation partitioning analyses show a strong impact of local environmental factors on the presence of Rickettsia and Mycoplasma in their association with Bryopsis. The presence of Flavobacteriaceae and Bacteroidetes, on the other hand, reflects a predominant imprint of host evolutionary history, suggesting that these bacteria are more specialized in their association. The results highlight the importance of interpreting the presence of individual bacterial phylotypes in the light of ecological and evolutionary principles such as phylogenetic niche conservatism to understand complex endobiotic communities and the parameters shaping them.
Keywords: algae, bacteria, biogeography, endosymbiosis, seaweed, variation partitioning
1. Introduction
Variation in traits across species or populations is influenced by their ecology and evolutionary history [1]. Organisms are shaped by the environment in which they live, with species residing in similar environments having common adaptations [2]. They are also the product of their evolutionary history, and closely related species have the tendency to be more similar than distantly related species [3]. This tendency for related species to resemble each other more in a trait than expected by chance is referred to as phylogenetic signal or phylogenetic conservatism [4]. Applying these principles to host–bacterial relationships, one might presume that obligate bacteria are phylogenetically structured, while facultative endobiotic bacteria are expected to be more randomly dispersed among host species ([5]; figure 1). In this study, we assess for the first time, to our knowledge, the combined effect of host dependency, ecology and biogeography on the structure of a complex endobiotic community in an algal model.
Figure 1.
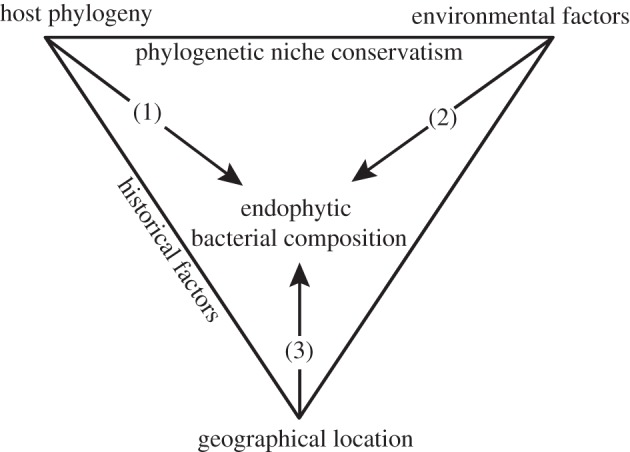
Relationships between host phylogeny, environment and geography on endophytic bacterial composition in Bryopsis seaweeds and relations between these three factors. (1) Phylogenetic structured variation, (2) ecological structured variation and (3) geographical structured variation. The shared influence of phylogeny and environment (1+2) is known as ‘phylogenetically structured environmental variation’.
Marine macroalgae (seaweeds) are commonly associated with bacteria that either live on the surface or in the cytoplasm and/or vacuolar systems of the cells [6–8]. These bacteria are able to influence the morphogenesis and life cycle of their algal host [9–11] and are linked with various metabolic functions such as the production of growth factors, fixed nitrogen and antimicrobial compounds [12–14]. Siphonous green seaweeds, belonging to the Bryopsidales and Dasycladales of the chlorophytan class Ulvophyceae, consist of a single giant tubular cell and form a benevolent biotic environment for endobiotic bacterial communities [15–17]. The siphonous cells, which range from centimetres to metres in length, typically exhibit vigorous cytoplasmic streaming to transport organelles, photosynthates and nutrients [18]. Chisholm et al. [19] demonstrated that siphonous algae take up nutrients from the sediment by a root-like system containing intracellular bacteria and translocate them throughout the thallus. These cellular innovations alongside unique mechanisms of wounding response [20,21] and the close interactions with bacteria may provide a physiological explanation for the successful spread of invasive siphonous green algae such as Caulerpa and Codium in marine coastal habitats [19,22,23].
Very little is known about the factors controlling the presence of bacteria inside siphonous seaweeds. Two host-related mechanisms may affect the intracellular bacterial composition. First, siphonous seaweeds readily regenerate from protoplasts, facilitating environmental uptake of bacteria into the cell [24,25]. Second, endogenous bacteria can persist by vertical inheritance through gametes [26]. Beside the question of whether the endobionts are acquired vertically or horizontally from the environment, ecological parameters and geographical aspects may also need to be considered to explain the bacterial composition, as some bacteria (or hosts) are likely to be geographically restricted or occur only in particular niches. Although a previous study suggested that seaweed-associated bacterial communities are biogeographically structured [23], it is not known whether ecological or historical factors cause this structure.
The goal of this study is to investigate the relative roles of host, environment and geography in determining the intracellular bacterial flora of siphonous seaweeds, focusing on the genus Bryopsis (Bryopsidales, Chlorophyta) as a case study. This genus is known to harbour several types of endogenous bacteria and protocols are in place to study them [17,27]. Bryopsis is known to possess mechanisms for environmental uptake as well as vertical inheritance of bacteria [25,26]. This combination of features, combined with the large collection of available cultures, makes the genus an ideal case study to address our goal. The experimental approach consisted of molecular characterization of host samples and their intracellular bacterial flora. The molecular identification of bacterial phylotypes, along with the host phylogeny and environmental data, was explored and analysed with statistical techniques designed to disentangle the effects of host phylogeny, geography and the external environment on the intracellular bacterial composition.
2. Material and methods
(a). Algal material
The 20 Bryopsis samples analysed in this study are listed in the electronic supplementary material, table S1, and their sampling sites are depicted in the electronic supplementary material, figure S1. All samples were transferred to and maintained as unialgal cultures under the conditions described by Hollants et al. [17].
(b). Molecular approach
Bryopsis samples were subjected to a surface sterilization step to eliminate epiphytic bacterial contamination [27] prior to total DNA extraction [28]. The host rbcL and bacterial 16S rRNA genes were PCR amplified as described by Hollants et al. [17]. The endophytic bacterial diversity was assessed by creating 16S rRNA gene clone libraries and performing nested PCR denaturing gradient gel electrophoresis (DGGE) analyses as described previously [17,25]. Sequences were submitted to EMBL under accession numbers HE648924–HE648948.
(c). Sequence data analyses
Bryopsis rbcL and bacterial 16S rRNA gene sequences were assembled, checked for chimaeras, compared with nucleotide databases and aligned as previously described [17]. Phylogenetic trees were inferred with maximum-likelihood implemented in PhyML v. 3.0 [29] and Bayesian inference using MrBayes [30], via the University of Oslo Bioportal website [31]. Both the analyses were performed under a HKY+G model as determined by the Akaike Information Criterion in JModeltest v. 0.1.1 [32]. Bacterial phylotypes or operational taxonomic units (OTUs) were identified based on 97 per cent sequence similarity.
(d). Statistical analysis
The influence of environmental, geographical and host phylogenetic factors on the endophytic bacterial diversity in Bryopsis was analysed using multivariate statistical and comparative phylogenetic approaches. The response table was represented by a presence/absence matrix of the seven bacterial phylotypes in the 20 host samples (figure 2). The three explanatory matrices (environment, geography and phylogeny) were prepared as follows. The environmental component was represented by seven macro-ecological variables (figure 2) extracted from Bio-ORACLE [33]. The geographical component was represented by a set of orthogonal spatial variables extracted from geographical coordinates by Moran's Eigenvector Maps (MEM) analysis [34] using ‘codep’ in R [35]. The geographical matrix was represented by the first two eigenvectors, which were the only ones having positive eigenvalues (6.54 and 1.52). The phylogenetic component was expressed as principal coordinates via a principal coordinate analysis (PCoA) [36] computed from a distance matrix [37]. A corrected distance matrix of the Bryopsis rbcL alignment was calculated in MEGA [38]; the PCoA analysis was performed in PCO [39]. The phylogenetic matrix was represented by the first four principal coordinates, representing 98 per cent of the total variation.
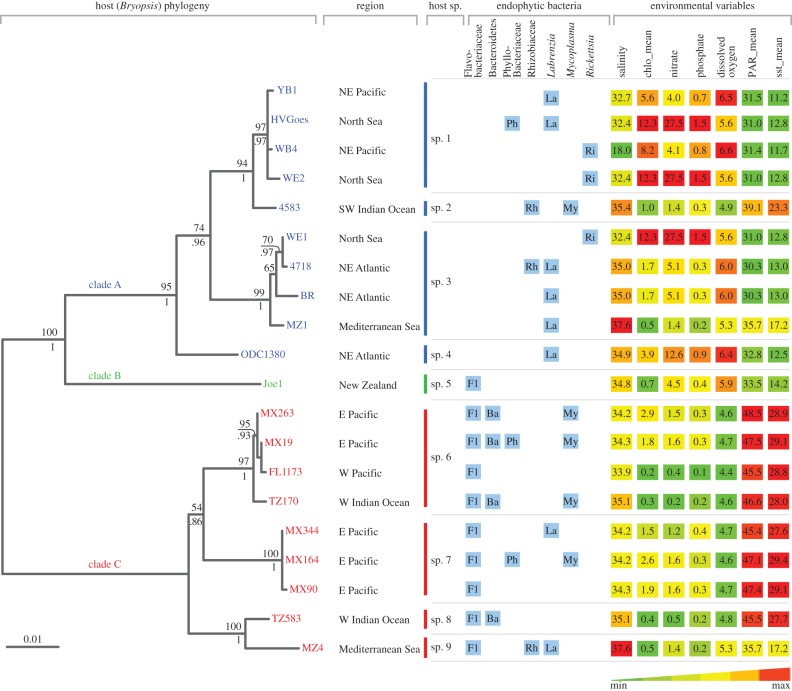
Figure 2.
Endophytic diversity results, geographical data and environmental variables plotted against the Bryopsis host phylogram. The endophytic bacterial diversity, displayed by blue boxes, summarizes the diversity results from the 16S rRNA gene clone libraries and DGGE analyses. Environmental variables were extracted from the host sampling sites using Bio-ORACLE, salinity (PSS); chlo_mean, annual mean chlorophyll (mg m−3), nitrate (μmol l−1), phosphate (μmol l−1), dissolved oxygen (ml l−1); PAR_mean, annual mean photosynthetically available radiation (µmol m−2 s−1); sst_mean, annual mean sea surface temperature (°C). The phylogram on the left classifies the 20 algal samples for which endophytic bacterial data are available in nine different Bryopsis species and three distinct clades (i.e. A, B and C). These clades seem more consistent with the ecology of the host samples (environmental variables depicted on the right) than with their geographical origin (sample region). Maximum-likelihood bootstrap values and Bayesian inference posterior probabilities, respectively, are indicated above and below the branch nodes. The scale bar indicates 0.01 nucleotide changes per nucleotide position. NE, northeast; SW, southwest; E, east; W, west.
To study the influence of environment, geography and host phylogeny on the endophytic bacterial diversity, we first performed data ordinations and calculated phylogenetic signals of the bacterial community composition. Ordination of Bryopsis samples based on endophytic bacterial community composition was performed using a principal component analysis (PCA) in Canoco for Windows v. 4.5 [40]. Environmental variables were plotted on the PCA graph as electronic supplementary material. Phylogenetic signal was assessed for: (i) the environmental variables, (ii) the geography, (iii) the total endophytic bacterial community (i.e. represented by principal components 1 and 2 calculated as described above), and (iv) the presence/absence data of each of the endophytic bacterial OTUs separately. p-values were calculated using randomizations of the K-statistic [41] in the R package Picante [42] (for (i)–(iii)) and the D statistic [43] in the R package ‘caper’ (http://cran.r-project.org/web/packages/caper/) (for (iv)). We quantified the common and unique influences of host phylogeny, geography and environment on the endophytic flora variation using variation partitioning analyses [2,44], using the varpart function in the R package ‘vegan’. The total bacterial diversity as well as presence/absence data of the seven individual phylotypes was considered separately as response tables. We performed variation partitioning analyses using three (phylogeny, environment and geography) and two (phylogeny and environment) explanatory tables, respectively.
3. Results
(a). Bryopsis host phylogeny
Based on the phylogenetic analysis of host rbcL sequences (figure 2), we assigned the seaweed samples to nine Bryopsis species, numbered sp. 1–9. The host phylogeny shows three main clades. Clades A and B include Bryopsis samples isolated from cold to temperate regions, whereas clade C is warm-temperate to tropical. The phylogenetic signal in annual mean sea surface temperature, as well as annual mean photosynthetically available radiation and dissolved oxygen levels, which are inversely proportional to each other, is statistically significant (p < 0.01, electronic supplementary material, table S2), suggesting that the structure of the Bryopsis phylogeny reflects temperature-related environmental variables. Conversely, geographical location (represented by MEM) did not show a significant phylogenetic structure (see the electronic supplementary material, table S2).
(b). Endophytic bacterial diversity
The results from the clone libraries and DGGE analyses showed the presence of seven unique endophytic bacterial phylotypes or OTUs within Bryopsis (see the electronic supplementary material, table S3). Five could be identified as Flavobacteriaceae (OTU-1), Mycoplasma (OTU-2), Bacteroidetes (OTU-3), Phyllobacteriaceae (OTU-4) and Labrenzia (OTU-7) species, which were previously shown to occur in Bryopsis [17] (see the electronic supplementary material, table S3 and figure S2). In addition, two new endophytic phylotypes were identified, OTU-5 and OTU-6 (see the electronic supplementary material, table S3 and figure S2). OTU-5 showed high sequence similarities with Rhizobiaceae strains isolated from root nodules of leguminous plants, and represents two distinct clusters that include Rhizobium leguminosarum and Ensifer meliloti-type strains, respectively. OTU-6 is allied to uncultured Rickettsiales bacteria associated with the coral Montastraea faveolata and the marine ciliate Diophrys appendiculata. All OTU-6 sequences formed a distinct and well-supported clade closely related to the genus Rickettsia, and most probably represent a new species based on their low sequence similarities (less than or equal to 93%) with Rickettsia-type strains.
(c). Endophytic bacterial composition
Figure 2 schematizes the endophytic bacterial diversity (blue boxes) in Bryopsis. Composition of the endophytic community varied between host species, and samples from the same host species harboured diverse combinations of one to four different endophytic phylotypes. Different host species with the same geographical origin commonly displayed differences in their intracellular bacterial community composition (e.g. samples MZ1 and MZ4). This apparent lack of correlation between total bacterial diversity and Bryopsis host species and geography is confirmed by the PCA plot, which illustrates that the ordination of the different Bryopsis species is not fully explained by their similarity in endophytic bacterial community composition (figure 3). This PCA plot, however, clearly indicates a correlation between the presence of individual endophytic phylotypes and certain environmental variables. Flavobacteriaceae, Bacteroidetes and Mycoplasma endophytes were only present in Bryopsis species isolated from tropical or warm-temperate seas, Labrenzia species were more often found in algal samples isolated from temperate regions, and Rickettsia endophytes were only present in Bryopsis species inhabiting seas with a low mean sea surface temperature (11.7°C–12.8°C) and high chlorophyll, nitrate and phosphate levels (figures 2 and 3). These correlations suggest that the distribution of individual bacterial OTUs may be more predictable than the total bacterial community composition. Individual bacterial endophyte groups also appear to be more strongly correlated with the host phylogeny than the overall bacterial composition. Flavobacteriaceae and Bacteroidetes species displayed a significant phylogenetic signal (p ≤ 0.01, see electronic supplementary material, table S2), while Rhizobiaceae, Phyllobacteriaceae, Mycoplasma, Rickettsia and Labrenzia species did not. Because the host phylogeny is correlated with ecological features as a consequence of niche conservatism (figure 1), it is not obvious whether the latter pattern is due to ecological preferences of the endophytic bacteria or their host.
Figure 3.
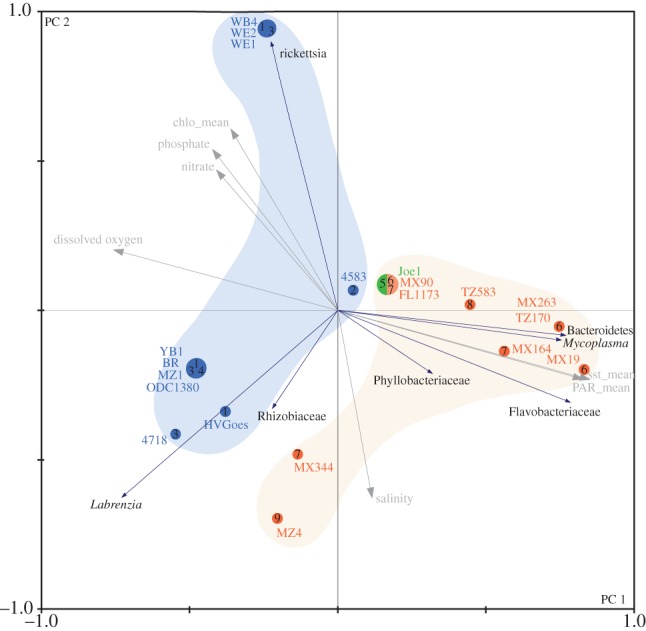
PCA of the 20 Bryopsis samples for which endophytic bacterial information is available. The PCA plot spreads the host samples in direction of maximum variance in endophytic bacterial community composition with principal component 1 explaining 41.7% and principal component 2 explaining 19.9% of the variance. Bryopsis species are indicated as numbers 1–9 and phylogenetic clades A, B and C are shown in blue, green and red, respectively. Environmental variables (in grey) were plotted on the PCA graph in the electronic supplementary material.
(d). Host versus environmental influences
In order to disentangle the influences of different factors shaping the endophytic bacterial diversity, we performed variation partitioning analyses. In the first set of analyses, we partitioned the variation of the bacterial diversity data with respect to the ecological, geographical and host phylogenetic factors into different portions: a part strictly influenced by environmental variables, a part strictly influenced by the Bryopsis host phylogeny, a part strictly explained by geography, four parts explained by the shared influence of these three factors and an unexplained part of the variation. When considering the total endophytic bacterial diversity, approximately equal parts of the variation (ca 30%) were explained by environmental and phylogenetic factors, while the strict influence of geography was low; most of the variance, however, remained unexplained (figure 4). Analyses of the seven bacterial phylotypes separately showed that the influence of environment, phylogeny and geography was very different between the seven phylotypes. The influence of geography was, in most cases, low and highly correlated with environment and/or host phylogeny (see figure 4 and electronic supplementary material, table S4). For this reason, we excluded geography in a second set of analyses (figure 5). The independent effects of host phylogeny and environment had little influence on the presence of Phyllobacteriaceae, Rhizobiaceae and Labrenzia phylotypes. The shared influence of host phylogeny and environment was larger than their individual effects for these bacterial types. The occurrence of Mycoplasma and Rickettsia species, on the other hand, was in part strictly determined by environmental factors, whereas the distribution of Bacteroidetes could to a large extent be explained by host phylogenetic factors only. Most of the variance in presence of these six endophytic phylotypes, however, remained unexplained, suggesting that factors other than host phylogeny and environment (at least the seven variables sampled) determine their occurrence within particular Bryopsis samples (figure 4). This is in contrast with the situation for Flavobacteriaceae endophytes, whose presence could be almost entirely explained by host phylogenetic factors, which partly overlapped with environmental factors.
Figure 4.
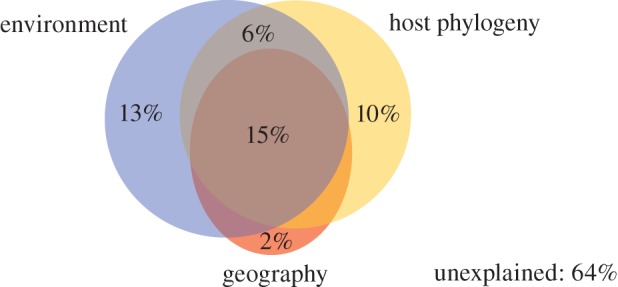
Partitioning the variation in total bacterial species composition explained by phylogenetic, environmental and geographical variables. Adjusted R2 values are given. A detailed overview of the variation partitioning results is provided in the electronic supplementary material, table S4.
Figure 5.
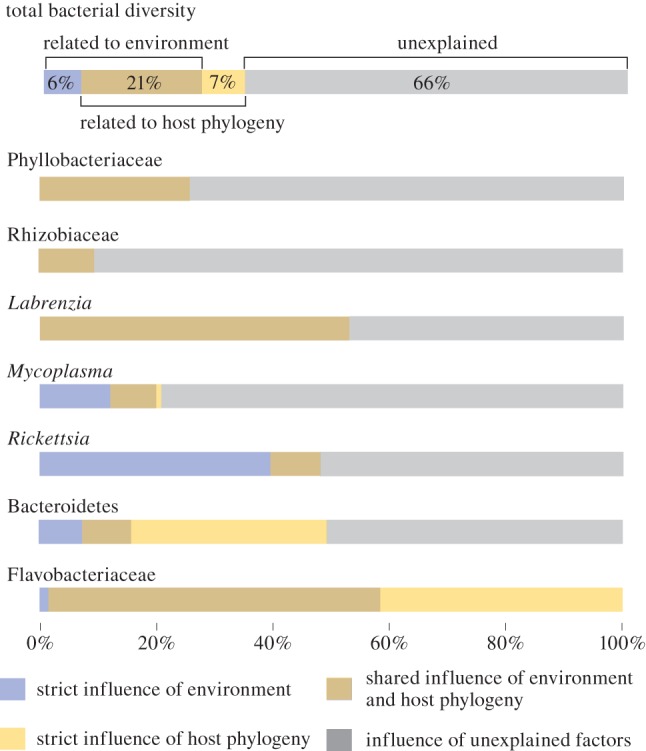
Partitioning the variation in bacterial species composition explained by phylogenetic and environmental variables. Diagrams show the unique and shared influences of both factors on the variation in total endophytic bacterial diversity and the individual endophytic phylotypes. Adjusted R2 values are given or illustrated. Negative fractions (which indicate that two explanatory variables have strong and opposite effects on the dependant variable) are treated as zeros in the graphs. A detailed overview of the variation partitioning results is given in the electronic supplementary material, table S4.
4. Discussion
Community structure and variation in traits across species are the outcome of environmental, geographical and historical factors which are clearly interwoven with each other. Bacterial communities associated with eukaryotic hosts are influenced by similar factors which need to be identified separately. Besides serving as baseline knowledge of the bacterial diversity occurring inside the siphonous cells of Bryopsis, our results provide insights into the various elements that contribute to the composition of the endogenous bacterial flora of siphonous green seaweeds.
(a). Diversity of endogenous bacteria
Characterization of the endobiotic microbial communities of 20 Bryopsis samples from widely divergent geographical localities indicates that Bryopsis harbours endophytic bacterial communities that are not very complex, but nevertheless taxonomically diverse. The constraint of living, unialgal samples as starting material necessitated extended cultivation of the algal samples prior to molecular processing [17]. These artificial culture conditions might have unknown effects on the endophytic bacterial community, potentially altering its complexity and this needs to be taken into account when interpreting these findings. Although they may not fully represent the total microbial variety present within the alga in its natural environment, the bacteria identified in this study are part of the natural Bryopsis endobiotic flora. Supplementary time stability experiments [25] demonstrated that Bryopsis endophytic bacterial communities are rather stable and clearly distinct from the epiphytic and surrounding cultivation water bacterial communities.
Besides the five bacterial phylotypes that were previously characterized in Bryopsis (Labrenzia, Mycoplasma, Phyllobacteriaceae, Bacteroidetes and Flavobacteriaceae) [17], we identified two additional phylotypes related to Rhizobiaceae and Rickettsia species. These bacteria have been especially well studied from terrestrial habitats [45,46], but have also been reported from marine habitats. Rhizobiales are common epiphytes of Ulva seaweeds [47–50] and have also been isolated from the surface of kelps where they display antimicrobial activity [13]. Additionally, a Rhodopseudomonas species with the potential to fix nitrogen was isolated from the rhizoidal cytoplasm of the siphonous green seaweed Caulerpa taxifolia [19]. We presume that Bryopsis also hosts Rhizobiaceae species with nitrogen fixing capacities, as we were able to amplify Ensifer-like nitrogenase reductase genes (EMBL accession numbers HE649370–HE649371) from Bryopsis samples 4718 and MZ4 [51]. Obligate intracellular Rickettsia species, on the other hand, have not previously been described from macroalgae but have been characterized through 16S rRNA gene analysis within freshwater green microalgae [52], marine ciliates [53] and coral tissue [54].
(b). Factors affecting bacterial composition
Even though each bacterial phylotype was encountered in at least three Bryopsis samples, the total endophytic bacterial diversity per host sample showed no clear pattern. All algal samples harboured diverse combinations of one to four endophytic phylotypes regardless of their phylogenetic affiliation, geographical origin or macro-ecological niche. On the other hand, when the presence of individual endophytic phylotypes rather than the total bacterial composition was analysed, host phylogenetic, geographical and environmental influences could be determined more clearly. These three factors, however, are inevitably interrelated as a result of phylogenetic niche conservatism, i.e. the tendency of closely related species to be ecologically similar [55], and historical factors such as dispersal limitation, resulting in geographical proximity of closely related species (figure 1). The Bryopsis host phylogeny was found to be mainly correlated with temperature-dependent variables and to a lesser extent with geography (see the electronic supplementary material, table S2). To disentangle the effects of host phylogeny, geography and environment on the endophytic bacterial community, we performed a variation partitioning analysis [2,56]. This technique has proved useful in quantifying independent influences of host phylogeny and other traits such as habitat and morphology in host–parasite [57–59] and arbuscular mycorrhizal symbiosis studies [60]. Our analyses shed light on the symbiotic nature and on potential modes of transmission of the individual endophytic phylotypes.
(c). Geography and environment on the endophytic bacterial community
The presence of endophytic Phyllobacteriaceae, Rhizobiaceae and Labrenzia phylotypes was not separately determined by host phylogenetic, geographical and ecological factors, suggesting that these endophytes are true generalists adapted to both free-living and host associated lifestyles along with a wide variety of environmental conditions. This is consistent with our previous observations that Labrenzia and Phyllobacteriaceae endophytes can survive outside their Bryopsis host and are reacquired from the local environment after repeated wounding events in culture [25]. Also the close phylogenetic relatedness of all three endophytic phylotypes with sequences from free-living bacteria (see the electronic supplementary material, figure S2) indicates a facultative association with the Bryopsis host. These generalist phylotypes may be selectively acquired by Bryopsis hosts to fulfil specific metabolic requirements, such as nitrogen-fixation (Rhizobiaceae; [45]), anoxygenic photosynthesis (Phyllobacteriaceae; [17]) or CO-oxidation (Labrenzia; [61]).
The occurrence of Mycoplasma and Rickettsia endophytes was to some extent strictly influenced by environmental factors. Mycoplasma endophytes were only present in Bryopsis samples from tropical regions, whereas Rickettsia bacteria were only found in algal samples isolated from temperate seas. This environmental influence suggests the acquisition of habitat-specific endophytes by Bryopsis hosts. In addition, the phylogenies of these more specialized endophytic phylotypes show a close relatedness with symbiotic Rickettsia and Mycoplasma species isolated from the cytoplasm of the marine ciliate D. appendiculata [53] and the intestinal bacterial flora of the Bryopsis-feeding abalone Haliotis diversicolor [62], respectively, suggesting the uptake of these endophytes could be vector dependant. This hypothesis is likely as both endophytes belong to the orders that are well known as obligate intracellular parasites of plants and animals [63,64]. Also within sponge hosts, horizontal symbiont transmission has been proposed to occur through vectors, including sponge-feeding animals [65].
The presence of Bacteroidetes endophytes within Bryopsis was to a large degree influenced by host phylogenetic factors, which may result from the fact that related algae have evolved similar traits that select for the uptake of these bacteria. Alternatively, the presence of Bacteroidetes endophytes in related hosts may be explained through vertical inheritance of bacteria either during sexual reproduction or asexual proliferation by fragmentation or extruded protoplasts that regenerate into new Bryopsis plants [66]. The absence of Bacteroidetes endophytes in the culture medium suggests that they may be obligate and hence vertically inherited endosymbionts [25], but further research is needed to exclude alternative hypotheses.
The presence of Flavobacteriaceae was found to be influenced by host phylogenetic factors in part combined with environmental influences, suggesting that these bacteria are specialized and obligate endosymbionts [67]. This is in line with results from culture experiments, which showed that these bacterial species are strictly dependant on the Bryopsis host for their growth and survival [25].
Overall, the variation partitioning analyses showed a high fraction of unexplained variation. This was true when considering the total endophytic bacterial diversity and the individual bacterial phylotypes, with the notable exception of the Flavobacteriaceae endophytes. This unexplained variation indicates that other factors may be important in explaining endophytic bacterial composition in Bryopsis. The variables included in this study are situated at the macro-ecological level and are considered suitable for explaining broad scale distribution patterns [33]. Ecological variables associated with microhabitat preferences, biotic interactions (e.g. bacterial transmission as a result of grazer-induced wounding) and physiological state of the host might also be important for interpreting fine scale bacterial community structure. In addition, endophytic community composition may be a result of stochastic processes. The few samples collected from one region and the same host species in this study indicate that a considerable variation in bacterial community composition may exist. Further studies, including many samples from a single host species at a single locality will be required to shed light on variation between co-occurring Bryopsis plants. However, to do so, technical difficulties inherent to this study (e.g. time consuming culturing and surface sterilization of host plants, and a large (greater than 95%) fraction of host chloroplast 16S rDNA in clone libraries) will need to be overcome, for example by applying species specific primers or high-throughput sequencing techniques. The unexplained variation in endophytic bacterial species composition is also relevant in the light of recent studies providing evidence that functional genes and transcriptomes, rather than species identified through rRNA taxonomy may be important in understanding bacterial community structure [68,69]. For example, bacterial communities associated with the green seaweed Ulva were found to be largely determined by function, rather than taxonomic identity [68]. It might thus be possible that functional genes rather than symbiont species or phylotypes are influenced by evolutionary (host phylogeny) and ecological factors [69].
In conclusion, characterization of Bryopsis algae sampled worldwide revealed the presence of variable endobiotic communities of relatively few but taxonomically diverse bacterial species. Variation partitioning analyses show a strong impact of local environmental factors on the presence of some bacteria, while the presence of others reflects a predominant imprint of host evolutionary history. These observations, however, were only evident when subdividing the total endophytic diversity into its individual bacterial phylotypes, suggesting that both the whole community and individual community members need to be considered in host–symbiont studies.
Acknowledgements
This research was funded by Research Foundation—Flanders project G.0045.08. We sincerely thank Gayle Hansen, Lennert Tyberghein, John West and Joe Zuccarello for providing Bryopsis cultures or for collecting specimens. We acknowledge Frederik Hendrickx, Liam Revell and Guillaume Guénard for useful comments on the statistical design. F.L. is a postdoctoral fellow of the Research Foundation—Flanders.
References
- 1.Freckleton RP, Jetz W. 2009. Space versus phylogeny: disentangling phylogenetic and spatial signals in comparative data. Proc. R. Soc. B 276, 21–30 10.1098/rspb.2008.0905 (doi:10.1098/rspb.2008.0905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borcard D, Legendre P, Drapeau P. 1992. Partialling out the spatial component of ecological variation. Ecology 73, 1045–1055 10.2307/1940179 (doi:10.2307/1940179) [DOI] [Google Scholar]
- 3.Harvey PH, Pagel MD. 1991. The comparative method in evolutionary biology. Oxford, UK: Oxford University Press [Google Scholar]
- 4.Blomberg SP, Garland T. 2002. Tempo and mode in evolution: phylogenetic inertia, adaptation and comparative methods. J. Evol. Biol. 15, 899–910 10.1046/j.1420-9101.2002.00472.x (doi:10.1046/j.1420-9101.2002.00472.x) [DOI] [Google Scholar]
- 5.Ronquist F. 1997. Phylogenetic approaches in coevolution and biogeography. Zool. Scr. 26, 313–322 10.1111/j.1463-6409.1997.tb00421.x (doi:10.1111/j.1463-6409.1997.tb00421.x) [DOI] [Google Scholar]
- 6.Goecke F, Labes A, Wiese J, Imhoff JF. 2010. Chemical interactions between marine macroalgae and bacteria. Mar. Ecol. Prog. Ser. 409, 267–299 10.3354/meps08607 (doi:10.3354/meps08607) [DOI] [Google Scholar]
- 7.Hollants J, Leliaert F, De Clerck O, Willems A. 2012. What we can learn from sushi: a review on seaweed–bacterial associations. FEMS Microbiol. Ecol. 83, 1–16 10.1111/j.1574-6941.2012.01446.x (doi:10.1111/j.1574-6941.2012.01446.x) [DOI] [PubMed] [Google Scholar]
- 8.Friedrich MW. 2012. Bacterial communities on macroalgae. In Seaweed biology (eds Wiencke C, Bischof K.), pp. 189–201 Berlin, Germany: Springer [Google Scholar]
- 9.Marshall K, Joint I, Callow ME, Callow JA. 2006. Effect of marine bacterial isolates on the growth and morphology of axenic plantlets of the green alga Ulva linza. Microb. Ecol. 52, 302–310 10.1007/s00248-006-9060-x (doi:10.1007/s00248-006-9060-x) [DOI] [PubMed] [Google Scholar]
- 10.Patel P, Callow ME, Joint I, Callow JA. 2003. Specificity in the settlement: modifying response of bacterial biofilms towards zoospores of the marine alga Enteromorpha. Environ. Microbiol. 5, 338–349 10.1046/j.1462-2920.2003.00407.x (doi:10.1046/j.1462-2920.2003.00407.x) [DOI] [PubMed] [Google Scholar]
- 11.Weinberger F, Beltran J, Correa JA, Lion U, Pohnert G, Kumar N, Steinberg P, Kloareg B, Potin P. 2007. Spore release in Acrochaetium sp. (Rhodophyta) is bacterially controlled. J. Phycol. 43, 235–241 10.1111/j.1529-8817.2007.00329.x (doi:10.1111/j.1529-8817.2007.00329.x) [DOI] [Google Scholar]
- 12.Dimitrieva GY, Crawford RL, Yüksel GÜ. 2006. The nature of plant growth-promoting effects of a pseudoalteromonad associated with the marine algae Laminaria japonica and linked to catalase excretion. J. Appl. Microbiol. 100, 1159–1169 10.1111/j.1365-2672.2006.02831.x (doi:10.1111/j.1365-2672.2006.02831.x) [DOI] [PubMed] [Google Scholar]
- 13.Wiese J, Thiel V, Nagel K, Staufenberger T, Imhoff JF. 2009. Diversity of antibiotic-active bacteria associated with the brown alga Laminaria saccharina from the Baltic Sea. Mar. Biotechnol. 11, 287–300 10.1007/s10126-008-9143-4 (doi:10.1007/s10126-008-9143-4) [DOI] [PubMed] [Google Scholar]
- 14.Rosenberg G, Paerl HW. 1981. Nitrogen fixation by blue-green algae associated with the siphonous green seaweed Codium decorticatum, effects on ammonium uptake. Mar. Biol. 61, 151–158 10.1007/bf00386654 (doi:10.1007/bf00386654) [DOI] [Google Scholar]
- 15.Colombo PM. 1978. Occurrence of endophytic bacteria in siphonous algae. Phycologia 17, 148–151 10.2216/i0031-8884-17-2-148.1 (doi:10.2216/i0031-8884-17-2-148.1) [DOI] [Google Scholar]
- 16.Delbridge L, Coulburn J, Fagerberg W, Tisa LS. 2004. Community profiles of bacterial endosymbionts in four species of Caulerpa. Symbiosis 37, 335–344 [Google Scholar]
- 17.Hollants J, Leroux O, Leliaert F, Decleyre H, De Clerck O, Willems A. 2011. Who is in there? Exploration of endophytic bacteria within the siphonous green seaweed Bryopsis (Bryopsidales, Chlorophyta). PLoS ONE 6, e26458. 10.1371/journal.pone.0026458 (doi:10.1371/journal.pone.0026458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cocquyt E, Verbruggen H, Leliaert F, De Clerck O. 2010. Evolution and cytological diversification of the green seaweeds (Ulvophyceae). Mol. Biol. Evol. 27, 2052–2061 10.1093/molbev/msq091 (doi:10.1093/molbev/msq091) [DOI] [PubMed] [Google Scholar]
- 19.Chisholm JRM, Dauga C, Ageron E, Grimont PAD, Jaubert JM. 1996. ‘Roots’ in mixotrophic algae. Nature 381, 382. 10.1038/381382a0 (doi:10.1038/381382a0) [DOI] [Google Scholar]
- 20.Menzel D. 1988. How do giant plant-cells cope with injury: the wound response in siphonous green algae. Protoplasma 144, 73–91 10.1007/BF01637240 (doi:10.1007/BF01637240) [DOI] [Google Scholar]
- 21.Welling M, Pohnert G, Kupper FC, Ross C. 2009. Rapid biopolymerisation during wound plug formation in green algae. J. Adhes. 85, 825–838 10.1080/00218460903291452 (doi:10.1080/00218460903291452) [DOI] [Google Scholar]
- 22.Williams SL, Smith JE. 2007. A global review of the distribution, taxonomy, and impacts of introduced seaweeds. Annu. Rev. Ecol. Evol. Syst. 38, 327–359 10.1146/annurev.ecolsys.38.091206.095543 (doi:10.1146/annurev.ecolsys.38.091206.095543) [DOI] [Google Scholar]
- 23.Meusnier I, Olsen JL, Stam WT, Destombe C, Valero M. 2001. Phylogenetic analyses of Caulerpa taxifolia (Chlorophyta) and of its associated bacterial microflora provide clues to the origin of the Mediterranean introduction. Mol. Ecol. 10, 931–946 10.1046/j.1365-294X.2001.01245.x (doi:10.1046/j.1365-294X.2001.01245.x) [DOI] [PubMed] [Google Scholar]
- 24.Kim GH, Klotchkova TA, West JA. 2002. From protoplasm to swarmer: regeneration of protoplasts from disintegrated cells of the multicellular marine green alga Microdictyon umbilicatum (Chlorophyta). J. Phycol. 38, 174–183 10.1046/j.1529-8817.2002.01053.x (doi:10.1046/j.1529-8817.2002.01053.x) [DOI] [Google Scholar]
- 25.Hollants J, Decleyre H, Leliaert F, De Clerck O, Willems A. 2011. Life without a cell membrane: challenging the specificity of bacterial endophytes within Bryopsis (Bryopsidales, Chlorophyta). BMC Microbiol. 11, e255. 10.1186/1471-2180-11-255 (doi:10.1186/1471-2180-11-255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burr FA, West JA. 1970. Light and electron microscope observations on the vegetative and reproductive structures of Bryopsis hypnoides. Phycologia 9, 17–37 10.2216/i0031-8884-9-1-17.1 (doi:10.2216/i0031-8884-9-1-17.1) [DOI] [Google Scholar]
- 27.Hollants J, Leliaert F, De Clerck O, Willems A. 2010. How endo- is endo-? Surface sterilization of delicate samples: a Bryopsis (Bryopsidales, Chlorophyta) case study. Symbiosis 51, 131–138 10.1007/s13199-010-0068-0 (doi:10.1007/s13199-010-0068-0) [DOI] [Google Scholar]
- 28.Doyle JL, Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15 [Google Scholar]
- 29.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 10.1080/10635150390235520 (doi:10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- 30.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 10.1093/bioinformatics/btg180 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 31.Kumar S, Skjaeveland A, Orr R, Enger P, Ruden T, Mevik B-H, Burki F, Botnen A, Shalchian-Tabrizi K. 2009. AIR: a batch-oriented web program package for construction of supermatrices ready for phylogenomic analyses. BMC Bioinformatics 10, 357. 10.1186/1471-2105-10-357 (doi:10.1186/1471-2105-10-357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 10.1093/molbev/msn083 (doi:10.1093/molbev/msn083) [DOI] [PubMed] [Google Scholar]
- 33.Tyberghein L, Verbruggen H, Pauly K, Troupin C, Mineur F, De Clerck O. 2012. Bio-ORACLE: a global environmental dataset for marine species distribution modelling. Glob. Ecol. Biogeogr. 21, 272–281 10.1111/j.1466-8238.2011.00656.x (doi:10.1111/j.1466-8238.2011.00656.x) [DOI] [Google Scholar]
- 34.Dray S, Legendre P, Peres-Neto PR. 2006. Spatial modelling: a comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecol. Model. 196, 483–493 10.1016/j.ecolmodel.2006.02.015 (doi:10.1016/j.ecolmodel.2006.02.015) [DOI] [Google Scholar]
- 35.Guénard G, Legendre P, Boisclair D, Bilodeau M. 2010. Multiscale codependence analysis: an integrated approach to analyze relationships across scales. Ecology 91, 2952–2964 10.1890/09-0460.1 (doi:10.1890/09-0460.1) [DOI] [PubMed] [Google Scholar]
- 36.Gower JC. 1966. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53, 325–338 [Google Scholar]
- 37.Diniz-Filho JAF, De Sant'Ana CER, Bini LM. 1998. An eigenvector method for estimating phylogenetic inertia. Evolution 52, 1247–1262 10.2307/2411294 (doi:10.2307/2411294) [DOI] [PubMed] [Google Scholar]
- 38.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 10.1093/molbev/msm092 (doi:10.1093/molbev/msm092) [DOI] [PubMed] [Google Scholar]
- 39.Anderson MJ. 2003. PCO: a FORTRAN computer program for principal coordinate analysis. Auckland, New Zealand: Department of Statistics, University of Auckland [Google Scholar]
- 40.Ter Braak CJF. 1986. Canonical correspondence analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology 67, 1167–1179 10.2307/1938672 (doi:10.2307/1938672) [DOI] [Google Scholar]
- 41.Blomberg SP, Garland T, Ives AR. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745 10.1111/j.0014-3820.2003.tb00285.x (doi:10.1111/j.0014-3820.2003.tb00285.x) [DOI] [PubMed] [Google Scholar]
- 42.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464 10.1093/bioinformatics/btq166 (doi:10.1093/bioinformatics/btq166) [DOI] [PubMed] [Google Scholar]
- 43.Fritz SA, Purvis A. 2010. Selectivity in mammalian extinction risk and threat types: a new measure of phylogenetic signal strength in binary traits. Conserv. Biol. 24, 1042–1051 10.1111/j.1523-1739.2010.01455.x (doi:10.1111/j.1523-1739.2010.01455.x) [DOI] [PubMed] [Google Scholar]
- 44.Peres-Neto PR, Legendre P, Dray S, Borcard D. 2006. Variation partitioning of species data matrices: estimation and comparison of fractions. Ecology 87, 2614–2625 10.1890/0012-9658(2006)87[2614:VPOSDM]2.0.CO;2 (doi:10.1890/0012-9658(2006)87[2614:VPOSDM]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 45.Kuykendall LD. 2005. Order VI. Rhizobiales ord. nov. In Bergey's manual of systematic bacteriology (the proteobacteria), part C (the alpha-, beta-, delta-, and epsilonproteobacteria) (eds Brenner DJ, Krieg NR, Staley JT, Garrity GM.), pp. 324 New York, NY: Springer [Google Scholar]
- 46.Dale C, Moran NA. 2006. Molecular interactions between bacterial symbionts and their hosts. Cell 126, 453–465 10.1016/j.cell.2006.07.014 (doi:10.1016/j.cell.2006.07.014) [DOI] [PubMed] [Google Scholar]
- 47.Tujula NA, Crocetti GR, Burke C, Thomas T, Holmstrom C, Kjelleberg S. 2010. Variability and abundance of the epiphytic bacterial community associated with a green marine Ulvacean alga. ISME J. 4, 301–311 10.1038/ismej.2009.107 (doi:10.1038/ismej.2009.107) [DOI] [PubMed] [Google Scholar]
- 48.Lachnit T, Meske D, Wahl M, Harder T, Schmitz R. 2011. Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol. 13, 655–665 10.1111/j.1462-2920.2010.02371.x (doi:10.1111/j.1462-2920.2010.02371.x) [DOI] [PubMed] [Google Scholar]
- 49.Burke C, Thomas T, Lewis M, Steinberg P, Kjelleberg S. 2011. Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 5, 590–600 10.1038/ismej.2010.164 (doi:10.1038/ismej.2010.164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Longford SR, Tujula NA, Crocetti GR, Holmes AJ, Holmström C, Kjelleberg S, Steinberg PD, Taylor MW. 2007. Comparisons of diversity of bacterial communities associated with three sessile marine eukaryotes. Aquat. Microb. Ecol. 48, 217–229 10.3354/ame048217 (doi:10.3354/ame048217) [DOI] [Google Scholar]
- 51.Hollants J. 2012. Endophytic bacteria within the green siphonous seaweed Bryopsis: exploration of a partnership. Dissertation, Ghent University, Ghent, Belgium [Google Scholar]
- 52.Kawafune K, Hongoh Y, Hamaji T, Nozaki H. 2012. Molecular identification of rickettsial endosymbionts in the non-phagotrophic volvocalean green algae. PLoS ONE 7, e31749. 10.1371/journal.pone.0031749 (doi:10.1371/journal.pone.0031749) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vannini C, Petroni G, Verni F, Rosati G. 2005. A bacterium belonging to the Rickettsiaceae family inhabits the cytoplasm of the marine ciliate Diophrys appendiculata (Ciliophora, Hypotrichia). Microb. Ecol. 49, 434–442 10.1007/s00248-004-0055-1 (doi:10.1007/s00248-004-0055-1) [DOI] [PubMed] [Google Scholar]
- 54.Sunagawa S, DeSantis TZ, Piceno YM, Brodie EL, DeSalvo MK, Voolstra CR, Weil E, Andersen GL, Medina M. 2009. Bacterial diversity and white plague disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 3, 512–521 10.1038/ismej.2008.131 (doi:10.1038/ismej.2008.131) [DOI] [PubMed] [Google Scholar]
- 55.Losos JB. 2008. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett. 11, 995–1003 10.1111/j.1461-0248.2008.01229.x (doi:10.1111/j.1461-0248.2008.01229.x) [DOI] [PubMed] [Google Scholar]
- 56.Desdevises Y, Legendre P, Azouzi L, Morand S. 2003. Quantifying phylogenetically structured environmental variation. Evolution 57, 2647–2652 10.1111/j.0014-3820.2003.tb01508.x (doi:10.1111/j.0014-3820.2003.tb01508.x) [DOI] [PubMed] [Google Scholar]
- 57.Timi JT, Lanfranchi AL. 2009. The importance of the compound community on the parasite infracommunity structure in a small benthic fish. Parasitol. Res. 104, 295–302 10.1007/s00436-008-1191-1 (doi:10.1007/s00436-008-1191-1) [DOI] [PubMed] [Google Scholar]
- 58.Krasnov BR, Poulin R, Mouillot D. 2011. Scale-dependence of phylogenetic signal in ecological traits of ectoparasites. Ecography 34, 114–122 10.1111/j.1600-0587.2010.06502.x (doi:10.1111/j.1600-0587.2010.06502.x) [DOI] [Google Scholar]
- 59.Desdevises Y, Morand S, Legendre P. 2002. Evolution and determinants of host specificity in the genus Lamellodiscus (Monogenea). Biol. J. Linnean Soc. 77, 431–443 10.1046/j.1095-8312.2002.00114.x (doi:10.1046/j.1095-8312.2002.00114.x) [DOI] [Google Scholar]
- 60.Stajerova K, Smilauerova M, Smilauer P. 2009. Arbuscular mycorrhizal symbiosis of herbaceous invasive neophytes in the Czech Republic. Preslia 81, 341–355 [Google Scholar]
- 61.Weber CF, King GM. 2007. Physiological, ecological, and phylogenetic characterization of Stappia, a marine CO-oxidizing bacterial genus. Appl. Environ. Microbiol. 73, 1266–1276 10.1128/aem.01724-06 (doi:10.1128/aem.01724-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang Z-B, Guo F, Zhao J, Li W-D, Ke C-H. 2010. Molecular analysis of the intestinal bacterial flora in cage-cultured adult small abalone, Haliotis diversicolor. Aquacult. Res. 41, e760–e769 10.1111/j.1365-2109.2010.02577.x (doi:10.1111/j.1365-2109.2010.02577.x) [DOI] [Google Scholar]
- 63.Weinert L, Werren J, Aebi A, Stone G, Jiggins F. 2009. Evolution and diversity of Rickettsia bacteria. BMC Biol. 7, 6. 10.1186/1741-7007-7-6 (doi:10.1186/1741-7007-7-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fraune S, Zimmer M. 2008. Host-specificity of environmentally transmitted Mycoplasma-like isopod symbionts. Environ. Microbiol. 10, 2497–2504 10.1111/j.1462-2920.2008.01672.x (doi:10.1111/j.1462-2920.2008.01672.x) [DOI] [PubMed] [Google Scholar]
- 65.Taylor MW, Radax R, Steger D, Wagner M. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71, 295–347 10.1128/mmbr.00040-06 (doi:10.1128/mmbr.00040-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morabito M, Gargiulo G, Genovese G. 2010. A review of life history pathways in Bryopsis. AAPP|Phys. Math. Nat. Sci. 88 10.1478/C1471A1002005 (doi:10.1478/C1471A1002005) [DOI] [Google Scholar]
- 67.Vignon M, Pariselle A, Vanhove MPM. 2011. Modularity in attachment organs of African Cichlidogyrus (Platyhelminthes: Monogenea: Ancyrocephalidae) reflects phylogeny rather than host specificity or geographic distribution. Biol. J. Linnean Soc. 102, 694–706 10.1111/j.1095-8312.2010.01607.x (doi:10.1111/j.1095-8312.2010.01607.x) [DOI] [Google Scholar]
- 68.Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T. 2011. Bacterial community assembly based on functional genes rather than species. Proc. Natl Acad. Sci. USA 108, 14 288–14 293 10.1073/pnas.1101591108 (doi:10.1073/pnas.1101591108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fan L, Reynolds D, Liu M, Stark M, Kjelleberg S, Webster NS, Thomas T. 2012. Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc. Natl Acad. Sci. USA 109, E1878–E1887 10.1073/pnas.1203287109 (doi:10.1073/pnas.1203287109) [DOI] [PMC free article] [PubMed] [Google Scholar]