Abstract
Interactions between species are important catalysts of the evolutionary processes that generate the remarkable diversity of life. Symbioses, conspicuous and inherently interesting forms of species interaction, are pervasive throughout the tree of life. However, nearly all studies of the impact of species interactions on diversification have concentrated on competition and predation leaving unclear the importance of symbiotic interaction. Here, I show that, as predicted by evolutionary theories of symbiosis and diversification, multiple origins of a key innovation, symbiosis between gall-inducing insects and fungi, catalysed both expansion in resource use (niche expansion) and diversification. Symbiotic lineages have undergone a more than sevenfold expansion in the range of host-plant taxa they use relative to lineages without such fungal symbionts, as defined by the genetic distance between host plants. Furthermore, symbiotic gall-inducing insects are more than 17 times as diverse as their non-symbiotic relatives. These results demonstrate that the evolution of symbiotic interaction leads to niche expansion, which in turn catalyses diversification.
Keywords: species interactions, plant–insect interactions, symbioses, diversification, cecidomyiidae, gall-inducing insects
1. Introduction
Species interactions are a fundamental driver of evolutionary change. Competition, one form of species interaction, can drive the evolution of both novel phenotypic and species diversity [1]. Studies of other forms of species interactions have also illuminated symbiotic interactions (intimate, often long-term interaction between different species) as motivators of evolutionary change, spurring novel phenotypes, life-histories and developmental pathways [2,3]. Symbioses have played a critical role in the ecological diversification of many organisms by facilitating expansion into novel ecological niches [2,4], and this expansion is theorized to catalyse lineage diversification [2,3]. However, empirical tests explicitly linking symbioses to both niche expansion and species diversification remain scarce [5,6].
Symbiotic interactions are particularly conspicuous and important in mediating interactions between plant-feeding (phytophagous) insects and their host plants [4,7]. Symbioses between insects and fungi have evolved in a variety of taxa [8–14]. A diverse insect group known to mediate plant–insect interaction through symbiotic association with fungi is the ambrosia gall midges (Diptera: Cecidomyiidae) [9–11,15–28]. Phylogenetic analysis of the fungal symbiont places it within a monophyletic clade of Botryosphaeria dothidea fungi [29]. Botryosphaeria dothidea are cosmopolitan, typically free-living, ubiquitous plant pathogens capable of feeding on a wide array of plant families world wide [29,30].
Fungal-symbiotic gall midge females from six genera within four morphologically well-separated taxonomic tribes transport conidia (fungal spores) and oviposit them along with their eggs into plant tissues. The gall structures induced by species of ambrosia gall midges on a variety of different plant tissue types (root, stem, bud, leaf, flower or seed) are lined internally with fungal hyphae which the developing larval gall midges use as food [10,11,15,31–35], but also for defence against natural enemies [29,30,36,37].
The genera of ambrosia gall midges display different adaptations for transportation of the fungal spores, including specialized mycangial structures or modified hairs that wrap around fungal spores [10,11,34]. As with other phytophagous insects [38–40], gall midge host-plant shifts occur most often between closely related plants relative to distantly related plants, because shifts among distantly related plants require substantially more adaptation [6,39]. Thus, gall midges are constrained in the range of possible host-plant shifts by the genetic distance between plant taxa [41,42].
Host-plant preferences are well characterized for gall midge species in 351 genera [41,43] which also may be categorized by the presence or the absence of fungal symbionts [15,30,43]. Both molecular and morphological analyses have also revealed that a small proportion (53 of the approx. 6100+gall midge species) [43] are polyphagous (capable of using multiple host-plant species; electronic supplementary material, table S3 and figure S4). Here, using time calibrated phylogenies of gall midge host-plants, I test two hypotheses derived from evolutionary theories of symbiosis [2–4,44] and diversification [1,45] concerning the role of symbiotic interaction in niche expansion and phytophagous insect diversification: (i) plant-feeding insects engaged in symbiosis should exhibit niche expansion (display greater genetic distance between the host plants they use), when compared with non-symbiotic lineages; and (ii) such niche expansion should be concomitant with elevated diversification (figure 1).
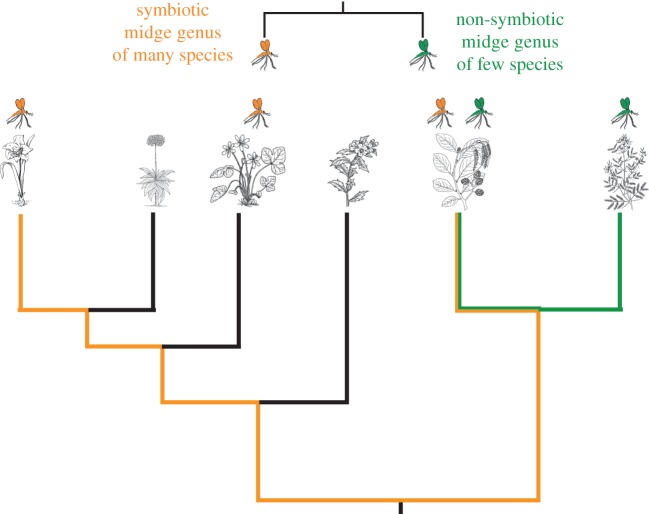
Figure 1.
Predicted effects of symbiotic association between plant-feeding insects and fungi on niche expansion and diversification. In this example, a genus of gall midges (orange) that has evolved a symbiotic association with fungi is 50 per cent more diverse and uses four times as much of the host-plant phylogeny as its strictly plant-feeding sister lineage (green). Branch lengths are proportional to time, the maximum phylogenetic distance used by symbiotic gall midges is traced in orange, maximum phylogenetic distance used by strictly plant-feeding gall midges is traced in green, blank branches are not included in maximum path calculation. (Online version in colour.)
2. Material and methods
(a). Host-plant data
Host-plant preferences for species from 351 genera of gall-inducing midges using 141 plant families were assembled and each midge taxa scored for the presence or the absence of symbiotic fungi from the literature [43]. Details of host-plant preferences and use of fungi by gall-inducing midge genera and polyphagous species is presented in electronic supplementary material, tables S1 and S2. The species richness of each gall midge genus was obtained from published sources [43] (see the electronic supplementary material, table S2).
(b). Host-plant phylogenetics
Relationships among host-plant taxa were reconstructed for gall midge genera and polyphagous gall midge species using Phylomatic [46], a tool which compiles published angiosperm phylogenies yielding a working hypothesis about their phylogenetic relationships. Where necessary host-plant phylogenies were further resolved using published phylogenies. Studies based on multiple genes were preferred and support values greater than 80 per cent were required to resolve relationships. Branch lengths of phylogenetic trees were scaled to time using the BLADJ function within Phylocom [46] and fossil calibration points from the literature [47]. The phylogenies of gall midge host-plants (see the electronic supplementary material, figures S1 and S2) are deposited in TreeBase (www.treebase.org). The maximum phylogenetic range (maximum patristic distance) of the host-plant phylogeny used by each gall midge taxa was calculated using functions in the R package APE.
(c). Statistical methods
To test the hypothesis that plant-feeding insects engaged in symbiosis should exhibit niche expansion, comparisons between fungal-symbiotic and non-fungal-symbiotic gall midges were performed at both the generic and specific taxonomic levels. Comparisons of both observed and expected numbers of polyphagous fungal-symbiotic and non-fungal-symbiotic gall-inducing midges and the observed versus expected numbers of plant families used by fungal-symbiotic and non-fungal-symbiotic gall-inducing midges were performed using χ2 goodness of fit tests. The expected numbers of gall midges were derived from the total numbers of fungal-symbiotic and non-fungal-symbiotic gall midges and total numbers of plant families used by fungal-symbiotic and non-fungal-symbiotic gall midge species. For example, the expected number of fungal-symbiotic polyphagous gall midges is given by ((total number of fungal−symbiotic gall midges/total number of gall midges) × total number of polyphagous gall midges). It is conceivable that further investigation of some of the described cases of polyphagy using molecular methods will reveal some of them to be cryptic species associated with a single host plant, however, it is unlikely that these cases would be biased towards fungal-symbiotic gall midges relative to those species which do not employ fungal symbionts.
Many species of gall midges are narrowly host-specific leading to a host-phylogenetic distance of zero. An overabundance of zeros may prove problematic for traditional data modelling techniques, such as generalized linear models (GLM), these situations arise when dependent variables are comprised of more zeros than expected under a Poisson distribution [48–51]. An over abundance of zeros in dependent variables is often generated when one process gives rise to zero and another to non-zero data. In such cases, restricting-dependent variables to non-zero data may result in biased parameter estimates [48–51]. Such zero weighted datasets are well modelled with a two part zero-inflated generalized linear model termed a hurdle model [49] which accounts for the over dispersion resulting from zero-inflated data [48–51]. Host-plant phylogenetic distance (maximum patristic distance) used by non-fungal-symbiotic gall midge genera was compared with that of fungal-symbiotic gall midge genera using hurdle zero-inflated generalized linear models [48] with species richness of gall midge genera included as a model covariate as implemented in the R statistical packages pscl [52] and mgcv [53]. Host-plant phylogenetic distance used by symbiotic and non-symbiotic polyphagous gall midge species was compared via a Wilcoxon signed-rank test.
Morphologically defined taxonomic tribes [43] correspond well with a molecular phylogeny (see the electronic supplementary material, figure S3). However, the lack of a fully resolved phylogeny for Cecidomyiidae precludes the use of a traditional sister group based approach, thus comparisons of the species richness of fungal-symbiotic gall midge genera to non-fungal-symbiotic genera were performed in two ways: (i) mean species richness of fungal-symbiotic genera were compared with all genera without symbiotic fungi within their taxonomic tribes using a paired t-test and (ii) to account for remaining uncertainty in taxonomic tribes fungal-symbiotic genera were compared with all non-fungal-symbiotic plant-feeding genera in family cecidomyiidae using a Wilcoxon signed-rank test.
That increased species richness in fungal-symbiotic lineages is a result of increased diversification rather than an effect of clade age was tested in two ways. First, the lengths of the pendant edges from the cecidomyiid phylogeny (electronic supplementary material) of fungal-symbiotic genera were compared with those of non-fungal-symbiotic genera using the R package APE [54] and a Wilcoxon signed-rank test. Second, I developed and employed a novel diversification metric integrating niche breadth, species richness and taxon age. diversification (D) = species richness (SR) × niche breadth (NB)/taxon age (T) so, D = (SR × NB)/T. Thus, applied here as: D = (gall midge genus species richness × maximum patristic distance of plant phylogeny used by gall midge genus)/pendant edge length of gall midge genus from the gall midge phylogeny. D for fungal-symbiotic gall midge genera was then compared with D of non-fungal-symbiotic gall midge genera using a Wilcoxon signed-rank test. All statistical analyses were performed within the R language statistical framework [55].
3. Results
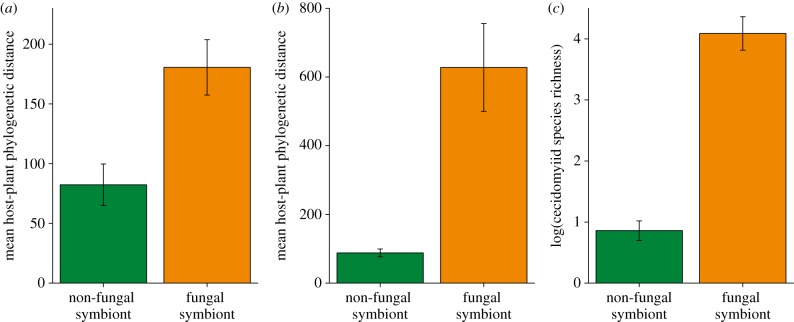
I tested hypothesis (i) that plant-feeding insects with symbiotic associations should display niche expansion in two ways. First, I performed two comparisons among polyphagous gall midge species (see the electronic supplementary material, table S3) using χ2-tests; (i) the observed versus expected numbers of polyphagous gall midge species that do and do not use fungal symbionts; and (ii) the observed versus expected number of plant families used by polyphagous gall midge species with and without fungal symbionts. Results indicate that there are significantly more polyphagous gall midges that are symbiotic than expected (χ2 = 21.62, p < 0.001) and that symbiotic gall midge species feed upon a greater number of plant families than expected (χ2 = 38.82, p < 0.001). Second, the phylogenetic range of host plants used by gall midges with and without fungal symbionts was compared using both a zero-inflated generalized linear model [48] and Wilcoxon signed-rank tests. I performed this comparison both between gall midge genera with and without fungal symbionts while accounting for the species richness of gall midge genera and between known cases of polyphagous gall midge species also with and without fungal symbionts. Gall midges employing fungal symbionts to mediate interactions with host-plant species consistently use a significantly expanded range of host plants relative to non-symbiotic lineages (table 1, figures 2a,b and 3a,b).
Table 1.
(a) Results of zero-inflated generalized linear model (hurdle model) comparing phylogenetic range (maximum patristic distance) of host plants (HP) used by symbiotic and non-symbiotic gall midge genera. The sample size n, β, β s.e., z-score and p-value are provided for both the Poisson and binomial parts of the model for each independent variable. (b) Wilcoxon comparison of the breadth of the host plant phylogeny (maximum patristic distance) used by polyphagous gall midge species using a fungal symbiont with the breadth of the phylogeny used by polyphagous gall midge species without fungal symbionts.
| model and response variables | independent variables | n | β | β s.e. | z | p-value |
|---|---|---|---|---|---|---|
| (a) gall midge genera: (i) Poisson part | ||||||
| HP phylogenetic range | (intercept) | 350 | 6.17516 | 0.009 | 654.416 | <0.001 |
| HP phylogenetic range | symbiont | 350 | 0.188 | 0.019 | 9.652 | <0.001 |
| HP phylogenetic range | gall midge genera species richness | 350 | 0.059 | 0.003 | 15.886 | <0.001 |
| gall midge genera: (ii) binomial part | ||||||
| HP phylogenetic range | (intercept) | 350 | −1.645 | 0.146 | −11.267 | <0.001 |
| HP phylogenetic range | symbiont | 350 | 3.254 | 1.105 | 2.944 | 0.003 |
| (b) polyphagous gall midge species | n | test | estimate | p-value | ||
| HP phylogenetic range non-symbiotic | HP phylogenetic range symbiotic | 44 | Wilcoxon | W = 114 | 0.003 | |
Figure 2.
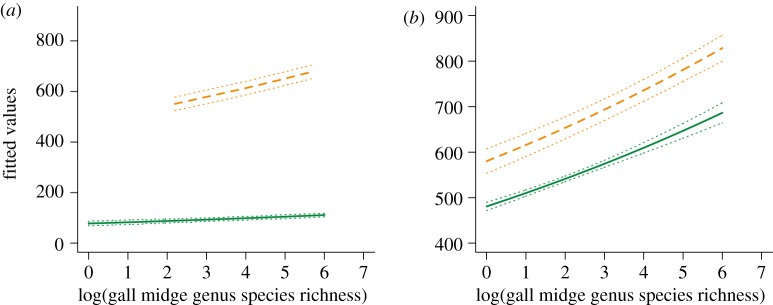
Plot of zero-inflated (hurdle) generalized linear model comparing phylogenetic range used by symbiotic (orange dashed lines) and non-symbiotic (green solid lines) gall midge genera, in which the response variable is phylogenetic breadth of the host-plant phylogeny used (host-plant phylogenetic distance ∼ log(gall midge species richness)+symbiotic status | symbiotic status). For symbiotic and non-symbiotic gall midge genera of similar species richness the symbiotic genera consistently use a greater proportion of the host-plant phylogeny. (a) Fitted values for binomial portion of hurdle model. (b) Fitted values for Poisson portion of hurdle model. In both (a) and (b) CIs are plus or minus twice the s.e. from model fit. (Online version in colour.)
Figure 3.
Maximum patristic distance of host-plant taxa used by (a) polyphagous gall midge species and (b) gall midge genera with and without fungal symbionts. Gall midge taxa with fungal associations use a greater breadth of host plant taxa relative to species lacking fungal symbionts as defined by genetic distance between hosts. (c) Comparison of the species richness of plant-feeding gall midge genera with symbiotic fungal associations to relatives within their taxonomic tribes that do not have fungal associations. Means with s.e. bars are indicated on each plot. (Online version in colour.)
To test hypothesis two, that fungal-symbiotic gall midge lineages are more diverse relative to lineages without such symbionts, I first compared the species richness of gall midge genera using symbiotic fungi to all related non-fungal-symbiotic lineages within their taxonomic tribes using a paired t-test. Secondly, as it is possible that a robust molecular phylogeny of family Cecidomyiidae would reveal phylogenetic rearrangements of genera among tribes the species richness of fungal-symbiotic gall midge genera was compared with all other plant-feeding genera within the family. Consistent with prediction (2), gall midge lineages using symbiotic fungi to mediate insect–plant interactions are significantly more diverse than both their tribal relatives lacking fungal symbionts (t = 21.71, d.f. = 3, p < 0.0003; figure 3c, electronic supplementary material, figure S4) and all plant-feeding genera within the family (W = 97.5, p < 0.0001). As judged by the available sequence data and a Wilcoxon test fungal-symbiotic gall midge genera are not significantly older when compared with their available non-fungal-symbiotic relatives (W = 76.5, p = 0.75). Comparison of diversification (D) of fungal-symbiotic gall midge genera with D of non-fungal-symbiotic gall midge genera suggests that symbiotic genera are diversifying faster (W = 132, p = 0.0008; figure 4).
Figure 4.
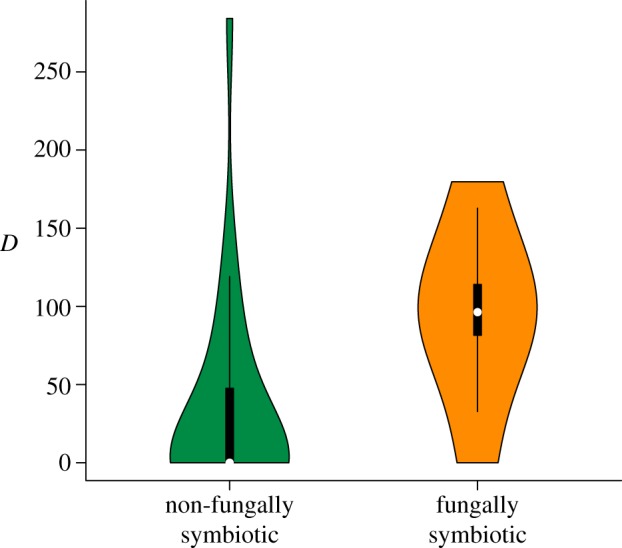
Violin plots [56] showing comparison of the diversification (D) of non-fungally symbiotic gall midge genera with fungal-symbiotic gall midge genera suggesting fungal-symbiotic gall midge genera are diversifying at a significantly faster rate relative to their non-symbiotic relatives. Whereby, D = SR × NB/T. (Online version in colour.)
4. Discussion
In accord with both predictions these results consistently support a role for symbiosis in niche expansion and diversification of phytophagous insects. Phytophagous insects which have evolved symbiotic association with fungi exhibit niche expansion, in the form of use of a broader range of host-plant taxa, because their fungal symbionts are cosmopolitan plant pathogens capable of digesting dozens of plant families worldwide [30,33]. Buffering interactions with plant taxa through symbiotic interaction with generalist fungi may thus relax phylogenetic constraints on the host-plant preferences of phytophagous insects imposed by characteristics of their host-plant lineages such as host-plant chemistry [57], natural enemies [36,58] or plant defences [59].
Key innovations facilitate the rapid migration from one peak in the adaptive landscape [60,61] to another perhaps higher peak [62] catalysing diversification [63]. For plant-feeding insects, such as gall-inducing midges, higher peaks in the adaptive landscape constitute those with more ecological opportunity in the form of more available niches (unexploited host-plant taxa, plant parts or time periods of plant growth) [41], fewer competitors or fewer natural enemies [36]. Incorporation of fungal symbionts into the life cycle of plant-feeding insects is thus a key innovation allowing transitions across a deep valley in the adaptive landscape, a shift to a genetically distant host-plant taxon, to be similarly tractable to shifts between genetically similar host-plant taxa. Thus, by facilitating the colonization of distantly related host-plant taxa, symbioses may allow insect lineages to simultaneously escape the limitations present at smaller scales, such as local competition for limited niches, and take advantage of the ecological opportunities that accompany shifts to novel adaptive zones [64], catalysing diversification at larger scales. Colonization of such novel adaptive zones is likely accompanied by the opportunity to diversify sensu Simpson [65] ‘…more or less simultaneous divergence of numerous lines all from much the same ancestral type into different, also diverging adaptive zones’. [65, p. 223], predicting elevated diversification of fungal-symbiotic insect lineages relative to lineages without fungal associations. Thus, among phytophagous insects, colonization of disparate adaptive zones may facilitate both phenotypic divergence [40] and diversification through division of available niche space such as closely related host-plant species, novel plant parts and time periods [36,41].
The 548 fungal-symbiotic gall midge species with well-described host-plant preferences are roughly evenly divided between tropical (190 species), temperate (215 species) and arid (143 species) biogeographical regions. Furthermore, only 24 fungal-symbiotic gall midge species use plant families that are not also used by non-fungal-symbiotic gall midge species suggesting the effect of symbiosis on niche expansion and diversification is not simply a correlate of particular host-plant families or biogeographical regions.
While good taxonomic characters delineate the fungal-symbiotic genera, different fungal-symbiotic genera have unique adaptations associated with symbiosis, and the preliminary molecular phylogeny matches well with taxonomy it is impossible to say for certain that the genera are monophyletic without a completely resolved molecular phylogeny. Furthermore, comparison of the richness of genera is not ideal nor is the estimation of lineage age without a complete phylogeny. Thus, a complete molecular phylogeny for the group would provide a more robust framework for testing the hypotheses here thereby substantially strengthening the inference that symbiosis catalysed diversification of this group.
Taken together these results support predictions derived from evolutionary theories of symbiosis and diversification that symbiotic interaction catalyses niche expansion and diversification. The importance of symbiotic interaction in diversification is unlikely to be exclusive to gall midge–fungal associations or to plant-feeding insects. For instance, it is likely that microbial mutualists also promote diversification through expansion of ecological opportunities available to their hosts in other taxa [4,6]. Thus, studies of the consequences of symbiosis for diversification in other taxonomic groups and other contexts of symbiosis would further illuminate the role played by symbiosis in diversification.
Acknowledgements
I am grateful to Bernard Crespi, Laura Weir, Patrik Nosil, Christine Parent, Arne Mooers and Will Stein for comments that greatly improved this manuscript. I thank Ruth Joy for helpful statistical advice and Suzanne Kuhnholz for translation of German manuscripts into English. I am greatly indebted to the SFU FAB* lab for helpful discussion. This work was supported by an NSERC discovery grant to Arne Mooers and a CIHR operating grant to Jamie Scott. The Interdisciplinary Research in Mathematics and Computing Sciences Centre at Simon Fraser University provided facilities.
References
- 1.Schluter D. 2000. The ecology of adaptive radiation, viii, 288 p. Oxford, UK: Oxford University Press [Google Scholar]
- 2.Moran NA. 2007. Symbiosis as an adaptive process and source of phenotypic complexity. Proc. Natl Acad. Sci. USA 104(Suppl. 1), 8627–8633 10.1073/pnas.0611659104 (doi:10.1073/pnas.0611659104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moran NA. 2006. Symbiosis. Curr. Biol. 16, R866–R871 10.1016/j.cub.2006.09.019 (doi:10.1016/j.cub.2006.09.019) [DOI] [PubMed] [Google Scholar]
- 4.Takiya DM, Tran P, Dietrich CH, Moran NA. 2006. Co-cladogenesis spanning three phyla: leafhoppers (Insecta: Hemiptera: Cicadellidae) and their dual bacterial symbionts. Mol. Ecol. 15, 4175–4191 10.1111/j.1365-294X.2006.03071.x (doi:10.1111/j.1365-294X.2006.03071.x) [DOI] [PubMed] [Google Scholar]
- 5.Futuyma DJ. 2003. Accounting for biological diversity. Evolution 57, 1216–1220 [Google Scholar]
- 6.Janson EM, Stireman JO, Singer MS, Abbot P. 2008. Phytophagous insect-microbe mutualisms and adaptive evolutionary diversification. Evolution 62, 997–1012 10.1111/j.1558-5646.2008.00348.x (doi:10.1111/j.1558-5646.2008.00348.x) [DOI] [PubMed] [Google Scholar]
- 7.Buchner P. 1965. Endosymbioses of animals with plant microorganisms. New York, NY: Wiley Interscience [Google Scholar]
- 8.Bentz BJ, Six DL. 2006. Ergosterol content of fungi associated with Dendroctonus ponderosae and Dendroconus rufipennis (Coleoptera: Curculionidae, Scolytinae). Ann. Entomol. Soc. Am. 99, 189–194 10.1603/0013-8746(2006)099[0189:ECOFAW]2.0.CO;2 (doi:10.1603/0013-8746(2006)099[0189:ECOFAW]2.0.CO;2) [DOI] [Google Scholar]
- 9.Gagne RJ. 1989. The plant-feeding gall midges of North America, 356 p Ithaca, FL: Cornell University Press [Google Scholar]
- 10.Bissett J, Borkent A. 1988. Ambrosia galls: the significance of fungal nutrition in the evolution of the Cecidomyiidae. San Diego, CA: Academic Press [Google Scholar]
- 11.Borkent A, Bissett J. 1985. Gall midges (Diptera: Cecidomyiidae) are vectors of their fungal symbionts. Symbiosis 1, 185–194 [Google Scholar]
- 12.Krishnamurthy K. 1984. Plant–insect–fungus association in some plant galls. Proc. Indian Acad. Sci. 93, 265–273 10.1007/BF03186245 (doi:10.1007/BF03186245) [DOI] [Google Scholar]
- 13.Farrell BD, Sequeira AS, O'Meara BC, Normark BB, Chung JH, Jordal BH. 2001. The evolution of agriculture in beetles (Curculionidae: Scolytinae and Platypodinae). Evolution 55, 2011–2037 [DOI] [PubMed] [Google Scholar]
- 14.Mikheyev AS, Mueller UG, Abbot P. 2006. Cryptic sex and many-to-one coevolution in the fungus-growing ant symbiosis. Proc. Natl Acad. Sci. USA 103, 10 702–10 706 10.1073/pnas.0601441103 (doi:10.1073/pnas.0601441103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohfritsch O. 2008. Plants, gall midges, and fungi: a three-component system. Entomol. Exp. Appl. 128, 208–216 10.1111/j.1570-7458.2008.00726.x (doi:10.1111/j.1570-7458.2008.00726.x) [DOI] [Google Scholar]
- 16.Trelease W. 1884. Notes on the relations of two cecidomyians to fungi. Psyche 4, 195–200 10.1155/1884/86407 (doi:10.1155/1884/86407) [DOI] [Google Scholar]
- 17.Docters Van Leeuwen WM. 1939. An ambrosia-gall on Symplocos fasciculata Zoll. Annal. Jardin Botanique Buitenzorg 49, 27–42 [Google Scholar]
- 18.Docters Van Leeuwen WM. 1929. Ueber eine galle auf Symplocus fasiculata Zoll., verursacht durch eine gallmucke: Asphondylia bursaria Felt, die mit einem fungus zusammen lebt. Marcellia 25, 61–66 [Google Scholar]
- 19.Camp RR. 1981. Insect–fungus blister galls on Solidago graminifolia and S. rugosa. I. A macroscopic and light microscopic study of the host–parasite relationship. Can. J. Bot. 59, 2466–2477 10.1139/b81-298 (doi:10.1139/b81-298) [DOI] [Google Scholar]
- 20.Highland HA. 1964. Life-history of Asphondylia ilicicola (Diptera: Cecidomyiidae), a pest of American Holly. J. Econ. Entomol. 57, 81–83 [Google Scholar]
- 21.Batra LR. 1964. Insect-fungus blister galls of Solidago and Aster. J. Kans. Entomol. Soc. 37, 227–234 [Google Scholar]
- 22.Batra LR, Lichtwardt RW. 1963. Association of fungi with some insect galls. J. Kans. Entomol. Soc. 36, 262–278 [Google Scholar]
- 23.Batra SWT, Batra LR. 1967. The fungus gardens of insects. Sci. Am. 217, 112–120 10.1038/scientificamerican1167-112 (doi:10.1038/scientificamerican1167-112) [DOI] [Google Scholar]
- 24.Meyer J. 1952. Cecidogenese de la galle de Lasioptera rubi Heeger et role nourricier d'un mycelium symbiotique. Seances Academie des Sciences 234, 2556–2558 [Google Scholar]
- 25.Mendes de Sá CE, Silveira FA, Santos JC, dos Santos Isaias RM, Fernandes GW. 2009. Anatomical and developmental aspects of leaf galls induced by Schizomyia macrocapillata Maia (Diptera: Cecidomyiidae) on Bauhiniabrevipes Vogel (Fabaceae). Revista Brasileira de Botânica 32, 319–327 10.1590/S0100-84042009000200011 (doi:10.1590/S0100-84042009000200011) [DOI] [Google Scholar]
- 26.Neger FW. 1910. Ambrosiapilze III. Weitere Beobachtungen an Ambrosiagallen. Berichte der Deutschen Botanischen Gesellschaft 28, 455–480 [Google Scholar]
- 27.Yukawa J, Rohfritsch O. 2005. Biology and ecology of gall-inducing Cecidomyiidae (Diptera). In Biology, ecology and evolution of gall-inducing arthropods (eds Raman A, Schaefer CW, Withers TM.), pp. 273–304 Gainesville, FL: Science Publishers Inc [Google Scholar]
- 28.Goidanich A. 1941. Interpretazione simbiotica di una associazione mico-entomatica gallare. Atti della Reale Accademia delle scienze di Torino 76, 208–221 [Google Scholar]
- 29.Janson EM, Peeden ER, Stireman JO, Abbot P. 2010. Symbiont-mediated phenotypic variation without co-evolution in an insect–fungus association. J. Evol. Biol. 23, 2212–2228 10.1111/j.1420-9101.2010.02082.x (doi:10.1111/j.1420-9101.2010.02082.x) [DOI] [PubMed] [Google Scholar]
- 30.Heath JJ, Stireman JO. 2010. Dissecting the association between a gall midge, Asteromyia carbonifera, and its symbiotic fungus, Botryosphaeria dothidea. Entomologia Experimentalis Et Applicata 137, 36–49 10.1111/j.1570-7458.2010.01040.x (doi:10.1111/j.1570-7458.2010.01040.x) [DOI] [Google Scholar]
- 31.Adair RJ, Burgess T, Serdani M, Barber P. 2009. Fungal associations in Asphondylia (Diptera: Cecidomyiidae) galls from Australia and South Africa: implications for biological control of invasive acacias. Fungal Ecol. 2, 121–134 10.1016/j.funeco.2009.02.003 (doi:10.1016/j.funeco.2009.02.003) [DOI] [Google Scholar]
- 32.Haridass ET. 1987. Midge–fungus interactions in a cucurbit stem gall. Phytophaga 1, 57–74 [Google Scholar]
- 33.Janson EM, Grebenok RJ, Behmer ST, Abbot P. 2009. Same host-plant, different sterols: variation in sterol metabolism in an insect herbivore community. J. Chem. Ecol. 35, 1309–1319 10.1007/s10886-009-9713-6 (doi:10.1007/s10886-009-9713-6) [DOI] [PubMed] [Google Scholar]
- 34.Rohfritsch O. 1997. Morphological and behavioural adaptations of the gall midge Lasioptera arundinis (Shiner) (Diptera: Cecidomyiidae) to collect and transport conidia of it's fungal symbiont. Tijdschrift voor Entomologie 140, 59–66 [Google Scholar]
- 35.Mamaev BM, Krivosheina NP. 1993. The larvae of the gall midges (Diptera, Cecidomyiidae). Rotterdam, The Netherlands: A. A. Balkema [Google Scholar]
- 36.Stireman JO, Janson EM, Carr TG, Devlin H, Abbot P. 2008. Evolutionary radiation of Asteromyia carbonifera (Diptera: Cecidomyiidae) gall morphotypes on the goldenrod Solidago altissima (Asteraceae). Biol. J. Linnean Soc. 95, 840–858 10.1111/j.1095-8312.2008.01101.x (doi:10.1111/j.1095-8312.2008.01101.x) [DOI] [Google Scholar]
- 37.Weis AE. 1982. Use of a symbiotic fungus by the gall maker Asteromyia carbonifera to inhibit attack by the parasitoid Torymus capite. Ecology 63, 1602–1605 10.2307/1938883 (doi:10.2307/1938883) [DOI] [Google Scholar]
- 38.Janz N, Nylin S, Wahlberg N. 2006. Diversity begets diversity: host expansions and the diversification of plant-feeding insects. BMC Evol. Biol. 6, 1–10 10.1186/1471-2148-6-1 (doi:10.1186/1471-2148-6-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Futuyma DJ, Agrawal AA. 2009. Macroevolution and the biological diversity of plants and herbivores. Proc. Natl Acad. Sci. USA 106, 18 054–18 061 10.1073/pnas.0904106106 (doi:10.1073/pnas.0904106106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nyman T, Farrell BD, Zinovjev AG, Vikberg V. 2006. Larval habits, host-plant associations, and speciation in nematine sawflies (Hymenoptera: Tenthredinidae). Evolution 60, 1622–1637 [PubMed] [Google Scholar]
- 41.Joy JB, Crespi B. 2012. Island phytophagy: explaining the remarkable diversity of phytophagous insects. Proc. R. Soc. B 279, 3250–3255 10.1098/rspb.2012.0397 (doi:10.1098/rspb.2012.0397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roskam JC. 1985. Evolutionary patterns in gall midge–host plant associations (Diptera: Cecidomyiidae). Tijdschrift voor Entomologie 128, 193–213 [Google Scholar]
- 43.Gagne RJ. 2010. A catalog of the Cecidomyiidae (Diptera) of the world. Digital version 1. See http://www.ars.usda.gov/SP2UserFiles/Place/12754100/Gagne_2010_World_Catalog_Cecidomyiidae.pdf
- 44.Moran NA, Tran P, Gerardo NM. 2005. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteriodetes. Appl. Environ. Microbiol. 71, 8802–8810 10.1128/AEM.71.12.8802-8810.2005 (doi:10.1128/AEM.71.12.8802-8810.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nosil P. 2012. Ecological speciation, 280 p. Oxford, UK: Oxford University Press [Google Scholar]
- 46.Webb CO, Ackerly DD, Kembel SW. 2006. Phylocom: software for the analysis of community phylogenetic structure and trait evolution, 3.4 edn See http://www.phylodiversity.net/phylocom/ [DOI] [PubMed] [Google Scholar]
- 47.Bell CD, Soltis DE, Soltis PS. 2010. The age and diversification of angiosperms revisited. Am. J. Bot. 97, 1296–1303 10.3732/ajb.0900346 (doi:10.3732/ajb.0900346) [DOI] [PubMed] [Google Scholar]
- 48.Zuur AF, Ieno EN, Walker N, Saveliev AA, Smith GM. 2009. Mixed effects models and extensions in ecology with R. New York, NY: Springer [Google Scholar]
- 49.Yesilova A, Kaydan MB, Kaya Y. 2010. Modeling insect-egg data with excess zeros using zero-inflated regression models. Hacettepe J. Math. Stat. 39, 273–282 [Google Scholar]
- 50.Baker-Méio B, Marquis RJ. 2012. Context-dependent benefits from ant-plant mutualism in three sympatric varieties of Chamaecrista desvauxii. J. Ecol. 100, 242–252 10.1111/j.1365-2745.2011.01892.x (doi:10.1111/j.1365-2745.2011.01892.x) [DOI] [Google Scholar]
- 51.Aldstadt J, Koenraadt JM, Fansiri T, Kijchalao U, Richardson J, Jones JW, Scott TW. 2011. Ecological modeling of Aedes aegypti (L.) pupal production in rural Kamphaeng Phet, Thailand. PLoS Neglected Trop. Dis. 5, 3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeileis A, Kleiber C, Jackman S. 2008. Regression models for count data in R. J. Stat. Softw. 27, 1–25 [Google Scholar]
- 53.Wood SN. 2011. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J. R. Stat. Soc. 73, 3–36 10.1111/j.1467-9868.2010.00749.x (doi:10.1111/j.1467-9868.2010.00749.x) [DOI] [Google Scholar]
- 54.Paradis E. 2006. Analysis of phylogenetics and evolution with R. New York, NY: Springer [Google Scholar]
- 55.Team RDC. 2012. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 56.Hinze J, Nelson R. 1998. Violin plots: a box plot-density trace synergism. Am. Stat. 52, 181–184 [Google Scholar]
- 57.Becerra JX, Noge K, Venable DL. 2009. Macroevolutionary chemical escalation in an ancient plant–herbivore arms race. Proc. Natl Acad. Sci. USA 106, 18 062–18 066 10.1073/pnas.0904456106 (doi:10.1073/pnas.0904456106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stone GN, Schonrogge K, Atkinson RJ, Bellido D, Pujade-Villar J. 2002. The population biology of oak gall wasps (Hymenoptera: Cynipidae). Annu. Rev. Entomol. 47, 633–668 10.1146/annurev.ento.47.091201.145247 (doi:10.1146/annurev.ento.47.091201.145247) [DOI] [PubMed] [Google Scholar]
- 59.Armbruster WS. 2009. Macroevolutionary patterns of defense and pollination in Dalechampia vines: adaptation, exaptation, and evolutionary novelty. Proc. Natl Acad. Sci. USA 106, 18 085–18 090 10.1073/pnas.0907051106 (doi:10.1073/pnas.0907051106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wright S. 1932. The roles of mutation, inbreeding, crossbreeding, and selection in evolution. In International congress of genetics (ed. Jones DF.), pp. 356–366 Ithaca, NY: Genetics Society of America [Google Scholar]
- 61.Vamosi JC, Vamosi SM. 2010. Key innovations within a geographical context in flowering plants: towards resolving Darwin's abominable mystery. Ecol. Lett. 13, 1270–1279 10.1111/j.1461-0248.2010.01521.x (doi:10.1111/j.1461-0248.2010.01521.x) [DOI] [PubMed] [Google Scholar]
- 62.Thompson JN. 2012. The role of coevolution. Science 335, 410–411 10.1126/science.1217807 (doi:10.1126/science.1217807) [DOI] [PubMed] [Google Scholar]
- 63.Hodges SA, Arnold ML. 1995. Spurring plant diversification: are floral nectar spurs a key innovation? Proc. R. Soc. Lond. B 262, 343–348 10.1098/rspb.1995.0215 (doi:10.1098/rspb.1995.0215) [DOI] [Google Scholar]
- 64.Parent CE, Crespi BJ. 2009. Ecological opportunity in adaptive radiation of Gálapagos endemic land snails. Am. Nat. 174, 898–905 10.1086/646604 (doi:10.1086/646604) [DOI] [PubMed] [Google Scholar]
- 65.Simpson GG. 1953. The major features of evolution. New York, NY: Columbia University Press [Google Scholar]