Abstract
A central issue in ecology is the understanding of the establishment of biotic interactions. We studied the factors that affect the assembly of the commensalistic interactions between vascular epiphytes and their host plants. We used an analytical approach that considers all individuals and species of epiphytic bromeliads and woody hosts and non-hosts at study plots. We built models of interaction probabilities among species to assess if host traits and abundance and spatial overlap of species predict the quantitative epiphyte–host network. Species abundance, species spatial overlap and host size largely predicted pairwise interactions and several network metrics. Wood density and bark texture of hosts also contributed to explain network structure. Epiphytes were more common on large hosts, on abundant woody species, with denser wood and/or rougher bark. The network had a low level of specialization, although several interactions were more frequent than expected by the models. We did not detect a phylogenetic signal on the network structure. The effect of host size on the establishment of epiphytes indicates that mature forests are necessary to preserve diverse bromeliad communities.
Keywords: Bromeliaceae, Chamela-Cuixmala biosphere reserve, ecological interactions, Mexico, Tillandsia, tropical dry forest
1. Introduction
An important aim in community ecology is to identify the factors that drive the establishment of species interactions. Mutualisms, antagonisms and more recently commensalisms have been depicted as complex networks [1–3], and have revealed patterns of community organization common in ecological systems, such as the tendency of species to interact with subsets of the interaction partners of more generalized species (nestedness, [4]). Quantitative differences in network properties among these interaction types have been identified [5–7], indicating differences in the underlying structuring factors. If interactions are neutral, the structure of the network is explained by the random interaction among individuals in the community and species abundance determines network patterns [8]. However, deterministic factors can also affect the structure of a network. There has been debate on whether neutral or biological factors such as complementarity in species phenotypes determine structural patterns of networks [9–12]. Phylogenetic analyses can help understand network topology because evolutionary history can influence ecological interactions [13,14]. Recent advances have been made in identifying the factors that influence network structure in mutualisms [12,15–17] and antagonisms [18–20], though there is a lag in the study of commensalistic interactions. Network structures may be the result of multiple, hierarchical, non-exclusive interactions among factors [21], but few studies have assessed several factors simultaneously [12,15,18]. Identifying the factors that structure interaction networks provides novel insights into the ecological and evolutionary processes at the community level that shape interactions, with possible implications for the management and conservation of species, particularly for groups with high vulnerability to extinction.
Vascular epiphytes (henceforth referred as epiphytes) are plants that establish obligate interactions with other plants (phorophytes) using them as a substrate without parasitizing them [22]. This makes them a vulnerable group to anthropogenic disturbance, together with their long life cycles, slow growth and the stressing environment in which they live [22]. Epiphytes represent 10 per cent of the diversity of living vascular plants [23] attaining their highest diversity in the Neotropics [24]. Studies of networks have found high values of nestedness [3,5,25]; no phylogenetic signal in species interaction patterns [26]; and the generalization of species partly or fully explained by species abundance [5,25]. However, no attempt has been made to incorporate other explanatory variables that might determine the structure of epiphyte–phorophyte networks. Certain host traits are associated with the presence of epiphytes. For example, bark ornamentation affects seed establishment [27,28]; bark porosity affects the humidity of the substrate [28,29]; and the production of secondary compounds can inhibit the germination of epiphytes [29,30]. Host tree size is related to epiphyte diversity and abundance [27,31,32]. Larger trees have more complex structures, providing more microhabitats and more substrate area for seeds to land. Tree size is also related to age, and older trees tend to have more epiphytes than younger trees because they have been exposed for a longer period of time to the seed rain of epiphytes. Additionally, bigger branches provide more stability, as these branches are less likely to fall [33,34]. Wood density is expected to affect epiphyte–phorophyte interactions because it influences branch stability, and because species with dense wood have slower growth rates [20] that affects the time of exposure to seed rain when tree size is considered. Thus, the combination of traits in a host species will influence the colonization, establishment and success of the interaction.
One of the main focuses in species interactions is their specialization. The observed specificity of epiphytes on host species may be owing to their specialization or to sampling effects posed by community attributes of the flora, such as species richness and abundance [21,22]. Low diversity forests have only few potential phorophyte species, and epiphytes may appear as highly specialized. On the other hand, species in forests with high alpha diversity have low densities, and rare species are seldom recorded, appearing more specialized than common species. Community studies have only considered common host, and sometimes epiphyte species even in network studies [3,25,31,35,36], and have used null models incorporating species abundance to overcome this problem when assessing specialization [3,25,31,36]. A further limitation to assess specialization (or neutrality) has been the exclusion of absence data, i.e. trees that do not bear epiphytes, because the real abundance of host species or species that lack epiphytes is not considered [22,37] (but see [31]).
Here, we assess which factors predict the structure of a quantitative epiphyte–phorophyte network in a tropical dry forest in western Mexico. We hypothesize that bark texture, size of woody individuals and wood density of woody species, together with the abundance and spatial overlap of species, determine network level patterns of species interactions and specialization. We evaluate the relative contribution of these factors to network structure, and whether patterns of interactions are conserved in the phylogeny of epiphytes and phorophytes.
2. Material and methods
Epiphyte–phorophyte networks in the tropical dry forest offer a great opportunity to investigate which factors contribute to emergent patterns of network organization because epiphyte and woody individuals can be easily recorded during the leafless season of this low canopy forest. This is an important advantage because excluding species and individuals that do not interact precludes the appropriate assessment of factors that may limit the establishment of interactions. We focus on the epiphytic bromeliads of a tropical dry forest in western Mexico. Rather than trying to document all possible interactions in the system, we focused on assessing if the factors considered can explain the network structure given the pool of individuals and species at the study sites.
(a). Study area and field data
The work was conducted in conserved tropical dry forest at the Chamela-Cuixmala Biosphere Reserve and its vicinity, in the central western coast of Mexico in the state of Jalisco (19°22′–19°35′ N, 104°56′–105°03′ W). The climate is very seasonal; the wet season occurs between July and October [38]. In this forest there are 421 woody species (potential phorophytes) reported and 29 vascular epiphyte species [39] distributed among the families Cactaceae (1 species), Orchidaceae (10 species) and Bromeliaceae (18 species). We focused on the Bromeliaceae, a monophyletic family of the monocots, which is the family with the highest epiphyte species richness and abundance in the region, and contributes importantly to the diversity of epiphytes in Neotropical forests [37].
We registered all interactions between bromeliad epiphytes and woody species at three 20 × 20 m plots (19°30.065′ N, 105°02.584′ W; 19°30.532′ N, 105°02.410′ W; 19°24.297′ N, 104°58.968′ W). We registered all epiphytes larger than approximately 3 cm in height, which can be confidently seen and identified from the ground using binoculars. Data on epiphytic bromeliads was collected during the dry season of 2007/2008 (from November to May) when the canopy is leafless and bromeliads are easy to observe. Two Tillandsia species have the potential of secondary dispersal by asexual means (Tillandsia intermedia, R. Sayago 2007, personal observation; Tillandsia usneoides, [37]) when ramets (clones) detach. We defined an individual as an epiphyte physically separated from other epiphytes.
All the woody plants at the plots with a diameter at 1.3 m above ground (trees and shrubs) or at the base (lianas) greater than 2.5 cm (DBH henceforth) were marked with an aluminium tag. For each marked individual we collected the following information: (i) DBH, (ii) species identity, and (iii) the abundance of each epiphytic bromeliad species. Leaf and reproductive samples were collected for each woody species. Plant identification was conducted by the authors and corroborated with herbarium specimens preserved at the Chamela Biological Station (Universidad Nacional Autonoma de Mexico). We refer to phorophytes only when referring to woody individuals bearing epiphytes, and to woody species to refer to all potential phorophytes present at the plots.
Bark texture for all woody species (see the electronic supplementary material, appendix S1) was recorded for at least one, and up to three of the largest individuals of each species based on four previously defined categories, from smooth (category 1) with lack of ornamentation (e.g. spines, lenticels, fissures) and including smooth exfoliating barks (e.g. some Bursera species), to rough (category 4) with coarse ornamentation (large lenticels and/or spines and/or deep fissures). Trees with rugose and ornamented bark are expected to have a greater load of epiphytes [22,29]. Data on wood density (specific gravity) of all tree species at the plots was obtained from the literature (see the electronic supplementary material, appendix S1).
(b). Observed interaction network
Ninety per cent of the bromeliad epiphytes were found on woody individuals with DBH greater than or equal to 5 cm, and we only considered these individuals for the network analyses. We constructed an interaction matrix Y describing the epiphyte–phorophyte network, pooling the data from the three sites. In this matrix the I rows correspond to woody species (all potential phorophytes) and the J columns to bromeliad species (epiphytes), and a cell yij is an integer that represents the number of interactions that occur between woody species i and bromeliad species j, i.e. the sum of all epiphyte individuals of species i at the three plots, recorded on individuals of woody species j. Any woody species i at the plots, whose individuals did not host bromeliads is present in this matrix with ∑i yi* = 0.
We use network statistics that characterize several aspects of network structure [40] to describe the observed network and to compare observed values with the values obtained from models of network determinants using functions in R [12,40–42] (see the electronic supplementary material, appendix S2): (i) connectance (C = number of links/IJ); (ii) nestedness [4], two metrics: N = 100−T, where T is the matrix temperature [43], and nestedness based on overlap and decreasing fill (NODF) [44]; (iii) interaction evenness [45]; (iv) 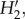 a quantitative metric of specialization that controls for the interaction frequencies expected from the total observations per species, i.e. the effect of the differences in the abundance of species, that results in abundant species interacting more frequently and with more partners, is removed [46]; (v) generality and vulnerability, the weighted mean number of phorophyte species per epiphyte species, and epiphyte species per phorophyte species, respectively [47]; and (vi) the average interaction strength asymmetry for phorophytes and for epiphytes [8]. The first two statistics are based on unweighted links, whereas the remaining are based on weighted links. To allow for comparison, we assessed the significance of nestedness [43,48] using the equivalent of null model 2 of Bascompte et al. [4].
a quantitative metric of specialization that controls for the interaction frequencies expected from the total observations per species, i.e. the effect of the differences in the abundance of species, that results in abundant species interacting more frequently and with more partners, is removed [46]; (v) generality and vulnerability, the weighted mean number of phorophyte species per epiphyte species, and epiphyte species per phorophyte species, respectively [47]; and (vi) the average interaction strength asymmetry for phorophytes and for epiphytes [8]. The first two statistics are based on unweighted links, whereas the remaining are based on weighted links. To allow for comparison, we assessed the significance of nestedness [43,48] using the equivalent of null model 2 of Bascompte et al. [4].
(c). Models of network determinants
We follow the conceptual and methodological framework proposed by Vázquez et al. [12], in which the observed matrix is a function of multiple interaction probability matrices determined by different factors. We built probability matrices from models that consider epiphyte and woody species abundance, bark texture and wood density of woody species, size (DBH) of woody individuals, and presence/absence of woody species and epiphytes species at each site (spatial overlap). In addition, we develop models that consider the joint effect of two or more factors, thus, all possible combinations (plant size is always assessed together with abundance, see below for details) of two, three and four factors, and a full model with the five factors are considered.
Consider a matrix X of the same size as Y, whose entries xij are given weights according to a particular model defined by the factors we are assessing. To calculate a probability matrix P from any matrix X, these weights are converted to probabilities of occurrence of pairwise interactions by normalizing the matrix so that all its elements sum to one. For the different models the calculation of xij is described in table 1. In the abundance model (A) interactions are determined by the abundance of species. The spatial overlap model (S) is based on the presence and absence of species on a local (site) scale, considering two species cannot interact if they do not co-occur at a site. The wood density model (W) weighs each species by its specific gravity. The model that considers bark texture (B) assigns higher probabilities of interaction to woody species with rougher bark texture. Model AD considers the joint influence of plant size and abundance. Because plant size is a trait of the individual and networks depict interactions among species, when considering the DBH of all individuals, the resulting probability matrix necessarily incorporates information on the abundance of each woody species. Models that consider the joint influence of spatial overlap and abundance are calculated so that not only presence/absence data are considered for each site, but the abundance of each species as well, thus incorporating the fine information on local abundance of this system. The performance of all models was compared to the performance of a null model that assumes that all interactions have the same probability to occur.
Table 1.
Model calculation of pairwise interaction weights (xij). (Letters composing the model name indicate the factors that it considers: A, abundance; B, bark texture of woody species; D, plant size (DBH); S, spatial overlap of species; W, wood density of woody species.)
| models | xij calculation | variables |
|---|---|---|
| null | xij = 1/IJ | I—total number of woody species |
| J—total number of epiphyte species | ||
| A | xij = aiaj | ai—number of individuals of species i |
| aj—number of individuals of species j | ||
| B | xij = P(presence of epiphytesi | bark texturei) | P(presence of epiphytes) = ez/(1+ez)a,b |
| S | xij = s | s—number of sites in which species i and j co-occur |
| W | xij = wi | wi—woody species specific gravity |
| ADc, ABD |  |
M—total number of individuals of species i |
| P(presence of epiphytes) = ez/(1+ez)a | ||
| nm = 100.777log(v1), expected number of epiphytes for individual md; v1—DBH category of m | ||
| AS, ADS, ABDS | xij is calculated as for A, AD and ABD, respectively, but calculated for each of the three sites separately, and summing the three weights of xij obtained for each site. | |
| AB, BS, ABS | The respective probability matrices are calculated as the element-wise multiplication of the probability matrices P of models A and B, B and S, AS and B, respectively. The resulting matrices are normalized again to obtain P. | |
| AW, BW, SW, ADW, ABW, ASW, BSW, ABSW, ABDW, ADSW, ABDSW | The respective probability matrices are calculated as the element-wise multiplication of each of the probability matrices P of all models above (except W), and the probability matrix of model W. The resulting matrices are normalized again to obtain P. | |
ae is the Napier's constant; z is calculated from a logistic regression equation that describes the log odds of presence of epiphytes on a tree, z = ln(odds(presence of epiphyes)), in which bark category and DBH are the explaining variables. z = b0 + b1 v1 + b2 v2, where b0 is the intercept, v1 is the DBH category of the tree, v2 is the bark category of the species to which the tree belongs, and b1 and b2 are the regression coefficients (see the electronic supplementary material, appendix S2).
bIn this model v1 is held constant to five, the minimum DBH category size of the individuals considered.
cIn this model v2 is held constant to the intermediate category (two), the bark category to which most tree species belong (see the electronic supplementary material, appendix S1).
dnm is the back-transformation of the log (expected number of epiphytes on a woody individual), calculated from the regression equation that describes the linear relation between log(DBH of phorophytes) and log(epiphyte abundance) (see the electronic supplementary material, appendix S2).
Two approaches are used to compare the performance of models in explaining the observed interaction network [12] (see the electronic supplementary material, appendix S2).
— The likelihood that a probability matrix explains the observed matrix is calculated assuming a multinomial distribution [12] and compared among models using the Akaike's information criterion (AIC) [49]. AIC = −2 ln(L) + 2k, where k is the number of parameters used to generate a probability matrix, which is the number of factors involved in the calculation of each probability matrix, except for plant size that involved two parameters (probability of presence and expected abundance). The model with the lowest ΔAIC, the difference in AIC between a given model and the AIC of matrix Y fitted to itself, indicates the model that better fits the data.
— A randomization algorithm [12] that assigns the total number of observed interactions (sum of elements in Y) to cells of a matrix of size Y with probabilities defined by a probability matrix is used to generate 1000 quantitative networks for each model; all network statistics are calculated for each generated network to obtain the 95% confidence intervals (CI) of the distribution of values of the statistics, against which the statistics of the observed network are compared. The only constraint of this algorithm [12] is to assign at least one interaction per species. To allow for empty rows, we did not use the constraint for woody species, though we left the constraint for epiphyte species. This simulates a scenario in which all woody species can potentially be colonized by epiphytes, and allows testing if the factors considered explain the absence of epiphytes on certain woody species. Network statistics are calculated excluding non-interacting woody species.
We identified the species with the predicted number of interactions that deviate the most from the observed values by assessing whether the observed value of each pairwise interaction falls outside the lower and upper 95% CI of 1000 runs of the algorithm.
(d). Phylogenetic signal in species associations
We evaluated the influence of evolutionary history on network patterns by assessing the presence of phylogenetic signal (i) on species degree (number of links per species); (ii) on species strength, a measure of the importance of a species to the partner's set that considers the relative frequency of the species on each partner [50]; and (iii) on the assemblage of interacting partners of the species (ecological similarity) [13]. Phylogenetic signal is assessed separately for epiphytes and for phorophytes, and was evaluated for the regional species pool (data from the three sites) and for the assemblages at each sampled plot. Phylogenetic hypotheses were constructed using Phylomatic software [51] for host species, and Mesquite v. 2.75 [52] for epiphyte assemblages, based on Barfuss et al. [53] and Chew et al. [54]. We transformed all branch lengths to 1, because branch length information is absent for the Tillandsia phylogeny. The amount of phylogenetic signal for (i) and (ii) was evaluated calculating the K-statistic [55], and for (iii) with Mantel tests comparing phylogenetic distance matrices with ecological distance matrices. Analyses were performed over 100 fully resolved trees. See full details in the electronic supplementary material, appendix S2.
3. Results
We recorded 363 woody individuals, of which 221 (DBH greater than or equal 5 cm) were included in the network analyses, belonging to 50 species and 20 plant families. The network included 1304 bromeliads recorded on 142 phorophytes (64% of woody individuals). We registered 151 links between 12 Tillandsia spp. and 36 (72%) woody species (figure 1). Five of these Tillandsia spp. are endemic to Mexico (see the electronic supplementary material, appendix S1). The observed network showed low levels of specialization, with 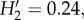 connectance of 0.35, and a significantly nested structure (N = 90.5, mean for null matrices = 62.21, p < 0.001; NODF = 62.89, mean for null matrices = 44.75, p < 0.01). Epiphyte species interact on average with 8.8 phorophyte species (generality), and phorophyte species interact with 4.9 epiphyte species (vulnerability). Interaction evenness is high (0.82), indicating no dominance of few interactions. The average strength of interactions between epiphytes and their partners is close to symmetry (mean asymmetry for epiphytes is very low = −0.023), whereas phorophytes tend to experience stronger effects from their interaction partners (−0.389).
connectance of 0.35, and a significantly nested structure (N = 90.5, mean for null matrices = 62.21, p < 0.001; NODF = 62.89, mean for null matrices = 44.75, p < 0.01). Epiphyte species interact on average with 8.8 phorophyte species (generality), and phorophyte species interact with 4.9 epiphyte species (vulnerability). Interaction evenness is high (0.82), indicating no dominance of few interactions. The average strength of interactions between epiphytes and their partners is close to symmetry (mean asymmetry for epiphytes is very low = −0.023), whereas phorophytes tend to experience stronger effects from their interaction partners (−0.389).
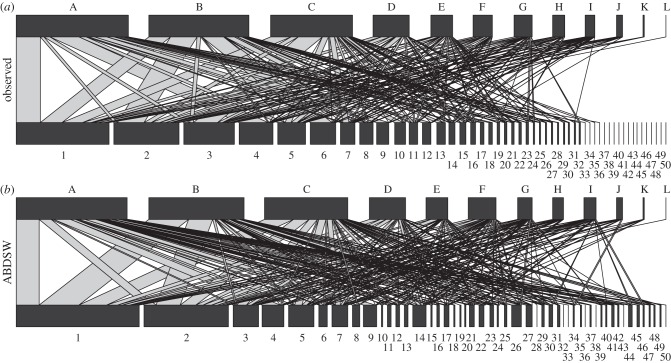
Figure 1.
(a) The bromeliad–phorophyte network and (b) the network yielded by one run of the best performing model of network determinants, ABDSW. Widths of rectangles represent the relative interaction frequencies of bromeliad (a) and phorophyte (b) species. Links among species and their relative frequency are represented by the lines connecting the rectangles and their width, respectively. Networks are drawn to the same scale. Species order and code is the same at both networks. Species identity is shown in the electronic supplementary material, appendix S1.
In the likelihood analysis all the models tested had a better performance than the null matrix, and partially explained the observed patterns of pairwise interactions (figure 2). The full model (ABDSW, figure 1) was the best performing model, with ΔAIC = 1171.41, which is 73.14 units away from the second lowest ΔAIC model (figure 2). Thus, the likelihood that the observed data are explained by model ABDSW is higher than for all other models. Abundance contributed the most to explain the observed patterns (figure 2, compare performance of models including one factor). The model that considers abundance and spatial overlap of woody species and epiphytes (i.e. local presence and abundance, model AS) is the model of two factors that better fits the observed data. Size of woody individuals is the third factor in importance for explaining interactions between epiphytes and phorophytes, being ADS the model of three factors with lowest ΔAIC. Finally, wood density followed by bark texture contributed the least to explain the observed data (see figure 2 dark bars).
Figure 2.
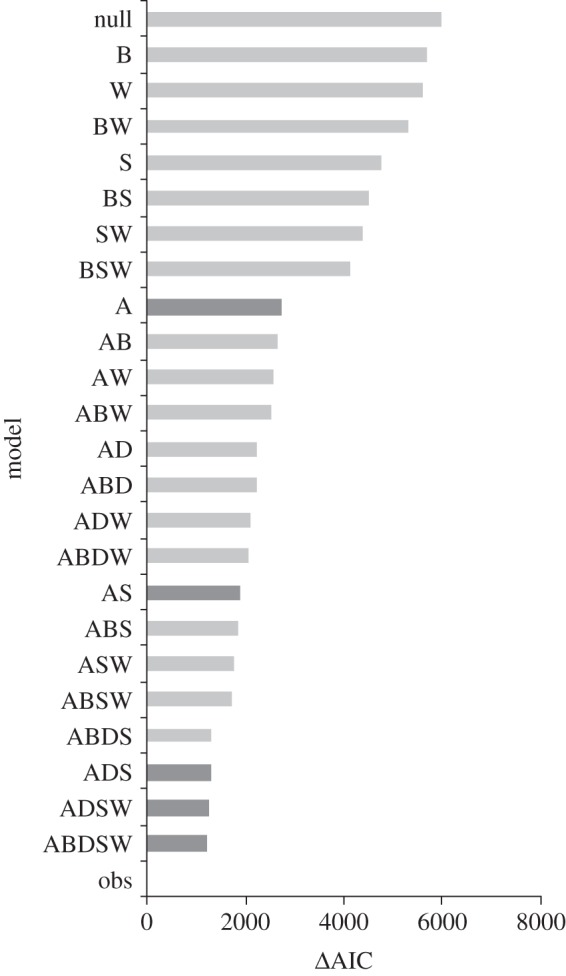
ΔAIC values for models of determinants of network structure that incorporate data on: A, species abundance; B, bark texture of woody species; D, plant size (DBH); S, spatial overlap; W, wood density and their combinations. obs, observed data; null, null model. The dark grey bars indicate the best performing models of one, two, three, four and five factors.
The models jointly including abundance, size of woody individuals and spatial overlap in general performed well for predicting network indices (figure 3 and electronic supplementary material, figure S1). In particular, connectance, interaction evenness, nestedness (NODF) and phorophyte interaction strength asymmetry were predicted by these models (figure 3 and electronic supplementary material, figure S1). Epiphyte interaction strength asymmetry was predicted or extremely close to the lower CI of models that incorporate abundance (figure 3). Similarly, nestedness (N) was predicted by models that incorporate abundance, and by model S (see the electronic supplementary material, figure S1).
Figure 3.
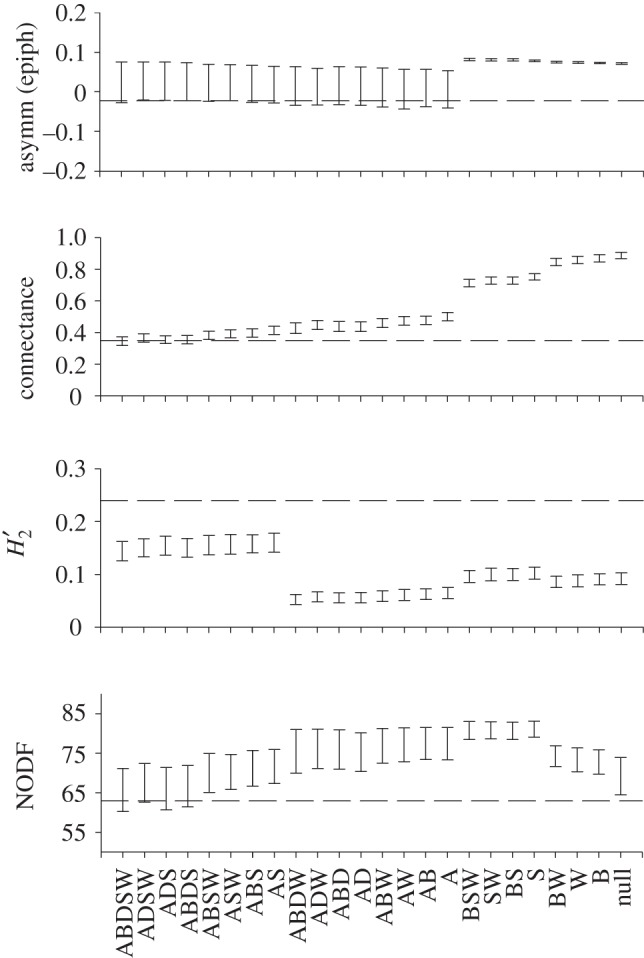
Network statistics for the observed epiphyte–phorophyte network and for networks yielded by models of determinants of network structure. Model names as in figure 2. The dashed line indicates the observed value. Interaction strength asymmetry of epiphytes, connectance, 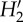 specialization and nestedness (NODF) are shown. Other statistics are reported in the electronic supplementary material, figure S1.
specialization and nestedness (NODF) are shown. Other statistics are reported in the electronic supplementary material, figure S1.
No probability matrix predicted the values observed for indexes related to specialization; model matrices were more generalized. Epiphyte and phorophyte species interact with less species than predicted by any model (generality and vulnerability, electronic supplementary material, figure S1). Abundance, and to a lesser extent spatial overlap, contribute to explain vulnerability, being the models that include these factors the closest to the observed value, while all factors contribute to explain generality. The observed 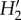 specialization is higher than predicted by any probability matrix. Incorporating information on the spatial heterogeneity of species abundance (models including AS) leads to higher values, yet lower than the observed (figure 3). The number of phorophyte species was overestimated by all models (see the electronic supplementary material, figure S1); ABDW (CI = 43–49) and ABDSW (CI = 44–49) yielded the matrices with the lowest number of phorophytes, yet more than the 36 observed.
specialization is higher than predicted by any probability matrix. Incorporating information on the spatial heterogeneity of species abundance (models including AS) leads to higher values, yet lower than the observed (figure 3). The number of phorophyte species was overestimated by all models (see the electronic supplementary material, figure S1); ABDW (CI = 43–49) and ABDSW (CI = 44–49) yielded the matrices with the lowest number of phorophytes, yet more than the 36 observed.
The frequency of most links fell within the 95% CI of the frequencies predicted by the best performing models, i.e. models including ADS (mean = 92.4% of the links; electronic supplementary material, table S1 and figure S2). When links fell outside the 95% CI, more often, observed links were stronger than predicted by these models (see the electronic supplementary material, table S1). An improvement in the performance of the models can be observed, as the frequency of a larger number of interactions get closer to the line of best fit between mean values of a model and observed frequencies, and to the observed network (see the electronic supplementary material, figures S3 and S2, respectively). Finally, no significant phylogenetic signal was detected for the traits considered (K-statistic range for species degree and strength: 0.32–0.86; p-values ≥ 0.14 for the z-statistic for ecological similarity; electronic supplementary material, table S2 and figure S4).
4. Discussion
We found that individual traits (woody individual's size) and species traits (bark texture and wood density), and the abundance and spatial overlap of species contributed differentially to predict network metrics and the frequency of pairwise interactions in epiphytic bromeliads—woody species commensalisms in a tropical dry forest. Consistent with the finding that these ecological factors largely determine species–species interactions, and with the low specialization of epiphytes on host species, we did not detect a phylogenetic signal in the network, i.e. specialization, species strength and interaction partners are not conserved, neither in the phylogeny of epiphytes nor in the phylogeny of phorophytes.
(a). Network structure
Our results are consistent with previous work which suggests that neutrality contributes to network structure [8,15,25]. Neutrality only partly accounted for the observed patterns (see performance of model A). Species abundance explains network patterns when neutrality determines the establishment of interactions, but this is a component of the community structure, which is influenced in a complex way by many biological factors (see causal model of Vazquez et al. [21]), among them, the spatial and temporal distribution of species. Once considering the constraints posed by the local spatial and temporal distribution of species (models including ADS, figure 3, and electronic supplementary material, figure S1), neutrality explained to a large extent network patterns. That is, network assembly, involving abundance, spatial overlap and phorophyte size (proxy for time of exposure to epiphyte seed rain) implies that networks are gradually built up by random encounters of individuals (regardless of the species to which they belong) concurring in space and time. However, it is clear that other biological factors also affect the establishment of interactions, including bark texture and wood density of hosts. The network specialization is comparable to ant-nectar plant (‘low intimacy’) and seed dispersal networks [56]. Ecological differences among epiphyte species may lead to differentiation in host use, e.g. if species differ in their susceptibility to nutrient, light or humidity levels and if substrate suitability varies among phorophyte species.
Clumped spatial distributions of woody species in tropical dry forests [57], and epiphyte communities (e.g. orchids and ferns [22]) may account for the unexplained network structure. Anemochory of Tillandsia can limit their dispersal kernel to a few metres resulting in spatial aggregation [28]. Asexual reproduction contributes for aggregated patterns when ramets detach and establish in the same or a neighbour host. Our models partly account for a clumped distribution: first, models that consider spatial overlap and abundance take into account differences in epiphyte abundances among plots. Second, when Tillandsia reproduces sexually or asexually, the probability of some progeny being established on the same host is large, influencing the epiphyte abundance–host size relation considered in the models (table 1).
Using a spatially explicit model, Morales & Vázquez [58] simulated the assembly of plant–frugivore networks using scenarios with varying levels of spatial autocorrelation of tree species and distances of bird foraging movements. Spatial structure and limited mobility affected the ‘degree of mixing’ in the system, imposing constraints to encounter probabilities [58]. A similar scenario could be found in our system with limited seed dispersal of Tillandsia and aggregated woody species. In Morales & Vázquez [58], a scenario analogous to our system (limited animal mobility and random individual tree spatial distribution), they show that with increasing spatial autocorrelation of tree identity, nestedness (N) varies slightly, connectance and evenness decrease, strength asymmetry increases, and unexpected presences/absences of interactions occur. Our findings agree with their results (figure 3 compare model A against AS, ADS and observed values which putatively increase in spatial structure), and the lower number of links, the many stronger interactions (see the electronic supplementary material, table S1, links above upper CI) and the absence of interactions in some woody species are evident in figure S2 (see the electronic supplementary material). This suggests that spatial processes contributed to structure the network.
We did not find evidence of a phylogenetic signal in the network structural patterns analysed, and the results were consistent across sites. A phylogenetic signal is not uncommon in mutualistic and antagonistic interactions [13,14,59]. Similar to our findings, the commensalistic networks between orchid epiphytes and their host trees do not show a phylogenetic signal [26]. The lack of phylogenetic signal could be related to the small size of the phylogenies [55]. Conversely, a phylogenetic signal may be present in other ways (e.g. in species roles [60], within or among compartments [61], within clades [59]). Evolutionary history may influence network structure at other scales and analysing a phylogenetically more diverse epiphyte community or a larger network might lead to the detection of a phylogenetic signal. Additionally, if traits important for species associations are phylogenetically conserved, then phylogeny will indirectly influence network structure [62,63]. Abundance does not show a phylogenetic signal in the epiphyte or phorophyte assemblage (see the electronic supplementary material, table S2 and figure S4). Wood density is highly conserved across the entire seed plant phylogeny [64]. Here our results showed a weak signal, and the contribution of wood density on the network structure is low.
(b). Pairwise interactions
Our results are consistent with previous epiphyte community reports that have found stronger observed interactions when compared with results from null models that consider phorophyte abundance [32,36]. When accounting for the effect of other factors, a higher percentage of the interactions is explained (see the electronic supplementary material, table S1; model ABDSW versus A). Nevertheless, several observed interactions remain stronger (more frequent) than expected by the full model. This is consistent with a higher observed than model yielded network specialization (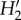 ). In our dataset, the most extreme case was that of Caesalpinia sclerocarpa, the only species in which all 12 Tillandsia species were registered. This is a timber species with dense wood (see the electronic supplementary material, appendix S1) and, therefore, more stable branches. A slower growth rate expected from dense wood also implies that size in this species represents older individuals than in other species. Despite considering all factors (ABDSW), five out of the 12 interactions were stronger than predicted. Caesalpinia sclerocarpa's architecture, with ramifications high on the trunk and fairly horizontal branches may favour the establishment of epiphytes. Species with dense wood are capable of growing taller [65], and this is one of the tallest species in the forest; Tillandsia are light demanding [66] and may be more successful high in the canopy. By contrast, the most abundant woody species, Apoplanesia paniculata, interacted with 10 Tillandsia spp., but was found interacting less frequently than expected by its abundance, bark texture, wood density, spatial overlap and tree sizes, with two Tillandsia species.
). In our dataset, the most extreme case was that of Caesalpinia sclerocarpa, the only species in which all 12 Tillandsia species were registered. This is a timber species with dense wood (see the electronic supplementary material, appendix S1) and, therefore, more stable branches. A slower growth rate expected from dense wood also implies that size in this species represents older individuals than in other species. Despite considering all factors (ABDSW), five out of the 12 interactions were stronger than predicted. Caesalpinia sclerocarpa's architecture, with ramifications high on the trunk and fairly horizontal branches may favour the establishment of epiphytes. Species with dense wood are capable of growing taller [65], and this is one of the tallest species in the forest; Tillandsia are light demanding [66] and may be more successful high in the canopy. By contrast, the most abundant woody species, Apoplanesia paniculata, interacted with 10 Tillandsia spp., but was found interacting less frequently than expected by its abundance, bark texture, wood density, spatial overlap and tree sizes, with two Tillandsia species.
Despite several weaker links than predicted by ABDSW (11 cases, electronic supplementary material, table S1), we found little evidence for axenic species (free of epiphytes). From the 14 species recorded with no epiphytes, only Guazuma ulmifolia and Jacquinia pungens were expected to interact with the most abundant epiphyte, T. usneoides. Overall, epiphytes tended to be absent or were less frequent on woody individuals that were young, on woody species with low abundance, present at a single site, and with smooth bark and/or soft wood. In the same way, less abundant epiphyte species tended to be hosted by a lower number of woody species.
(c). Conservation implications
Understanding community assembly will contribute to the conservation of interactions. Consequences of network structure for commensalistic interactions might be different than for other interactions where coevolution is involved. As other epiphyte–phorophyte networks [3,5,25], the bromeliad–phorophyte network showed high values of nestedness (N and NODF), higher than other types of interactions [5]. Several processes may lead to a nested pattern [63,67,68]. The specialization asymmetry implied in nestedness, suggested to arise through a coevolutionary process in other systems, with implications for extinction risk [4,63,69,70], seems to be here a consequence of the abundance and temporal and spatial distribution of species of a mostly generalized system. The one-way specialization of these bromeliads on an epiphytic life form but with low host specificity shows that many species can be adequate hosts, which in regard to host availability makes them less vulnerable to disturbance. However, hosts with suitable traits will give epiphytes higher probabilities of survival, with important implications for the conservation and management of this group.
Our analysis highlights the importance of time for the establishment of epiphyte communities. It takes at least one decade to the first reproduction of bromeliads [22,37], and the recruitment of species with different reproductive strategies (monocarpic versus polycarpic) may be affected differentially by disturbances. Epiphytes provide important resources for many taxa [22] and their disappearance will cascade through the ecosystem. Tillandsia is the richest genus of the Bromeliaceae, and Mexico is a centre of diversification for this genus, where 43 per cent of its species, most of which are endemic [37,71], occur. Our study contributes to understanding the processes that assemble ecological interaction networks in general and commensalistic networks in particular. Vascular epiphytes are the most vulnerable plant life form, and this study highlights ecological factors that shape their communities.
Acknowledgements
We are grateful to Andy Hector and two anonymous referees for their comments on the manuscript. We thank the Centro de Investigaciones en Ecosistemas and the Estación de Biología Chamela of the Universidad Nacional Autónoma de México (UNAM) for logistic support. Financial support was provided by grants to M.Q. from CONACyT-Mexico (grants nos. 2002-C01–0597, CB-2005-51043, 2009-C01-131008 and 2010-155016), Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT IN201011) and the Inter-American Institute for Global Change Research (IAI) Project CRN2-21. This paper constitutes a partial fulfilment of the Graduate Programme in Biological Sciences of UNAM. R.S. acknowledges the scholarship and financial support provided by CONACyT, and Posgrado en Ciencias Biológicas UNAM. CONACyT and UNAM financed M.L.'s postdoctoral position. M. Gargollo allowed us to work on her property. Technical assistance was provided by E. Castro, H. Ferreira, G. Sánchez-Montoya and A. Valencia-García.
References
- 1.Carvalheiro LG, Buckley YM, Ventim R, Fowler SV, Memmott J. 2008. Apparent competition can compromise the safety of highly specific biocontrol agents. Ecol. Lett. 11, 690–700 10.1111/j.1461-0248.2008.01184.x (doi:10.1111/j.1461-0248.2008.01184.x) [DOI] [PubMed] [Google Scholar]
- 2.Memmott J. 1999. The structure of a plant-pollinator food web. Ecol. Lett. 2, 276–280 10.1046/j.1461-0248.1999.00087.x (doi:10.1046/j.1461-0248.1999.00087.x) [DOI] [PubMed] [Google Scholar]
- 3.Burns KC, Zotz G. 2010. A hierarchical framework for investigating epiphyte assemblages: networks, meta-communities, and scale. Ecology 91, 377–385 10.1890/08-2004.1 (doi:10.1890/08-2004.1) [DOI] [PubMed] [Google Scholar]
- 4.Bascompte J, Jordano P, Melian CJ, Olesen JM. 2003. The nested assembly of plant-animal mutualistic networks. Proc. Natl Acad. Sci. USA 100, 9383–9387 10.1073/pnas.1633576100 (doi:10.1073/pnas.1633576100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piazzon M, Larrinaga AR, Santamaria L. 2011. Are nested networks more robust to disturbance? A test using epiphyte-tree, comensalistic networks. PLoS ONE 6, e19637. 10.1371/journal.pone.0019637 (doi:10.1371/journal.pone.0019637) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fontaine C, Thébault E, Dajoz I. 2009. Are insect pollinators more generalist than insect herbivores? Proc. R. Soc. B 276, 3027–3033 10.1098/rspb.2009.0635 (doi:10.1098/rspb.2009.0635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thébault E, Fontaine C. 2010. Stability of ecological communities and the architecture of mutualistic and trophic networks. Science 329, 853–856 10.1126/science.1188321 (doi:10.1126/science.1188321) [DOI] [PubMed] [Google Scholar]
- 8.Vázquez DP, Melian CJ, Williams NM, Blüthgen N, Krasnov BR, Poulin R. 2007. Species abundance and asymmetric interaction strength in ecological networks. Oikos 116, 1120–1127 10.1111/j.2007.0030-1299.15825.x (doi:10.1111/j.2007.0030-1299.15825.x) [DOI] [Google Scholar]
- 9.Jordano P, Bascompte J, Olesen JM. 2003. Invariant properties in coevolutionary networks of plant-animal interactions. Ecol. Lett. 6, 69–81 10.1046/j.1461-0248.2003.00403.x (doi:10.1046/j.1461-0248.2003.00403.x) [DOI] [Google Scholar]
- 10.Santamaría L, Rodríguez-Gironés MA. 2007. Linkage rules for plant–pollinator networks: trait complementarity or exploitation barriers?. PLoS Biol. 5, e31. 10.1371/journal.pbio.0050031 (doi:10.1371/journal.pbio.0050031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stang M, Klinkhamer PGL, van der Meijden E. 2007. Asymmetric specialization and extinction risk in plant–flower visitor webs: a matter of morphology or abundance? Oecologia 151, 442–453 10.1007/s00442-006-0585-y (doi:10.1007/s00442-006-0585-y) [DOI] [PubMed] [Google Scholar]
- 12.Vázquez DP, Chacoff NP, Cagnolo L. 2009. Evaluating multiple determinants of the structure of plant–animal mutualistic networks. Ecology 90, 2039–2046 10.1890/08-1837.1 (doi:10.1890/08-1837.1) [DOI] [PubMed] [Google Scholar]
- 13.Rezende EL, Lavabre JE, Guimaraes PR, Jordano P, Bascompte J. 2007. Non-random coextinctions in phylogenetically structured mutualistic networks. Nature 448, U925–U926 10.1038/nature05956 (doi:10.1038/nature05956) [DOI] [PubMed] [Google Scholar]
- 14.Eklöf A, Helmus MR, Moore M, Allesina S. 2012. Relevance of evolutionary history for food web structure. Proc. R. Soc. B 279, 1588–1596 10.1098/rspb.2011.2149 (doi:10.1098/rspb.2011.2149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stang M, Klinkhamer PGL, Waser NM, Stang I, van der Meijden E. 2009. Size-specific interaction patterns and size matching in a plant-pollinator interaction web. Ann. Bot. 103, 1459–1469 10.1093/aob/mcp027 (doi:10.1093/aob/mcp027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donatti CI, Guimaraes PR, Galetti M, Pizo MA, Marquitti FMD, Dirzo R. 2011. Analysis of a hyper-diverse seed dispersal network: modularity and underlying mechanisms. Ecol. Lett. 14, 773–781 10.1111/j.1461-0248.2011.01639.x (doi:10.1111/j.1461-0248.2011.01639.x) [DOI] [PubMed] [Google Scholar]
- 17.Olesen JM, Bascompte J, Dupont YL, Elberling H, Rasmussen C, Jordano P. 2011. Missing and forbidden links in mutualistic networks. Proc. R. Soc. B 278, 725–732 10.1098/rspb.2010.1371 (doi:10.1098/rspb.2010.1371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cagnolo L, Salvo A, Valladares G. 2011. Network topology: patterns and mechanisms in plant-herbivore and host-parasitoid food webs. J. Anim. Ecol. 80, 342–351 10.1111/j.1365-2656.2010.01778.x (doi:10.1111/j.1365-2656.2010.01778.x) [DOI] [PubMed] [Google Scholar]
- 19.Poisot T, Thrall PH, Hochberg ME. 2012. Trophic network structure emerges through antagonistic coevolution in temporally varying environments. Proc. R. Soc. B 279, 299–308 10.1098/rspb.2011.0826 (doi:10.1098/rspb.2011.0826) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enquist BJ, West GB, Charnov EL, Brown JH. 1999. Allometric scaling of production and life-history variation in vascular plants. Nature 401, 907–911 10.1038/44819 (doi:10.1038/44819) [DOI] [Google Scholar]
- 21.Vázquez DP, Blüthgen N, Cagnolo L, Chacoff NP. 2009. Uniting pattern and process in plant–animal mutualistic networks: a review. Ann. Bot. 103, 1445–1457 10.1093/aob/mcp057 (doi:10.1093/aob/mcp057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benzing DH. 1990. Vascular epiphytes. Cambridge, UK: Cambridge University Press [Google Scholar]
- 23.Kress WJ. 1986. The systematic distribution of vascular epiphytes: an update. Selbyana 9, 2–22 [Google Scholar]
- 24.Nieder J, Prosperi J, Michaloud G. 2001. Epiphytes and their contribution to canopy diversity. Plant Ecol. 153, 51–63 10.1023/a:1017517119305 (doi:10.1023/a:1017517119305) [DOI] [Google Scholar]
- 25.Burns KC. 2007. Network properties of an epiphyte metacommunity. J. Ecol. 95, 1142–1151 10.1111/j.1365-2745.2007.01267.x (doi:10.1111/j.1365-2745.2007.01267.x) [DOI] [Google Scholar]
- 26.Silva IA, Ferreira AWC, Lima MIS, Soares JJ. 2010. Networks of epiphytic orchids and host trees in Brazilian gallery forests. J. Trop. Ecol. 26, 127–137 10.1017/S0266467409990551 (doi:10.1017/S0266467409990551) [DOI] [Google Scholar]
- 27.Johansson D. 1974. Ecology of vascular epiphytes in West African rain forest. Acta Phytogeogr. Suecica 59, 1–59 [Google Scholar]
- 28.Cascante-Marin A, von Meijenfeldt N, de Leeuw HMH, Wolf JHD, Oostermeijer JGB, den Nijs JCM. 2009. Dispersal limitation in epiphytic bromeliad communities in a Costa Rican fragmented montane landscape. J. Trop. Ecol. 25, 63–73 10.1017/s0266467408005622 (doi:10.1017/s0266467408005622) [DOI] [Google Scholar]
- 29.Callaway RM, Reinhart KO, Moore GW, Moore DJ, Pennings SC. 2002. Epiphyte host preferences and host traits: mechanisms for species-specific interactions. Oecologia 132, 221–230 10.1007/s00442-002-0943-3 (doi:10.1007/s00442-002-0943-3) [DOI] [PubMed] [Google Scholar]
- 30.Frei JK, Dodson CH. 1972. The chemical effect of certain bark substrates on the germination and early growth of epiphytic orchids. Bull. Torrey Bot. Club 6, 301–307 10.2307/2997072 (doi:10.2307/2997072) [DOI] [Google Scholar]
- 31.Laube S, Zotz G. 2006. Neither host-specific nor random: vascular epiphytes on three tree species in a Panamanian lowland forest. Ann. Bot. 97, 1103–1114 10.1093/aob/mcl067 (doi:10.1093/aob/mcl067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zotz G, Schultz S. 2008. The vascular epiphytes of a lowland forest in Panama-species composition and spatial structure. Plant Ecol. 195, 131–141 10.1007/s11258-007-9310-0 (doi:10.1007/s11258-007-9310-0) [DOI] [Google Scholar]
- 33.Zimmerman JK, Olmsted IC. 1992. Host tree utilization by vascular epiphytes in a seasonally inundated forest (Tintal) in Mexico. Biotropica 24, 402–407 10.2307/2388610 (doi:10.2307/2388610) [DOI] [Google Scholar]
- 34.Tersteege H, Cornelissen JHC. 1989. Distribution and ecology of vascular epiphytes in lowland rain-forest of Guyana. Biotropica 21, 331–339 10.2307/2388283 (doi:10.2307/2388283) [DOI] [Google Scholar]
- 35.Vergara-Torres CA, Pacheco-Alvarez M, Flores-Palacios A. 2010. Host preference and host limitation of vascular epiphytes in a tropical dry forest of central Mexico. J. Trop. Ecol. 26, 563–570 10.1017/s0266467410000349 (doi:10.1017/s0266467410000349) [DOI] [Google Scholar]
- 36.Benavides AM, Vasco A, Duque AJ, Duivenvoorden JF. 2011. Association of vascular epiphytes with landscape units and phorophytes in humid lowland forests of Colombian Amazonia. J. Trop. Ecol. 27, 223–237 10.1017/s0266467410000726 (doi:10.1017/s0266467410000726) [DOI] [Google Scholar]
- 37.Benzing DH. 2000. Bromeliaceae profile of an adaptative radiation. Cambridge, UK: Cambridge. University Press [Google Scholar]
- 38.García-Oliva F, Camou A, Maass JM. 2002. El clima de la región central de la costa del Pacífico mexicano. In Historia Natural de Chamela (eds Noguera FA, Vega-Rivera JH, García-Aldrete AN, Quesada M.), pp. 3–10 México, DF: Universidad Nacional Autónoma de México [Google Scholar]
- 39.Lott JE. 2002. Lista anotada de las plantas vasculares de Chamela-Cuixmala. In Historia natural de Chamela (eds Noguera FA, Vega JH, García ANA, Quesada M.), pp. 99–153 México, DF: Universidad Nacional Autónoma de México [Google Scholar]
- 40.Dormann CF, Fründ J, Blüthgen N, Gruber B. 2009. Indices, graphs and null models: analyzing bipartite ecological networks. Open Ecol. J. 2, 7–24 10.2174/1874213000902010007 (doi:10.2174/1874213000902010007) [DOI] [Google Scholar]
- 41.Oksanen J, Blanchet FG, Kindt R, Legendre P, O'Hara RB, Simpson GL, Solymos PHSMH, Wagner H. 2011. Vegan: Community ecology package. R package version 1.17–8. Vienna, Austria: R Foundation for Statistical Computing. See http://CRANR-projectorg/package=vegan [Google Scholar]
- 42.R Development Core Team 2011. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org/ [Google Scholar]
- 43.Rodriguez-Girones MA, Santamaria L. 2006. A new algorithm to calculate the nestedness temperature of presence-absence matrices. J. Biogeogr. 33, 924–935 10.1111/j.1365-2699.2006.01444.x (doi:10.1111/j.1365-2699.2006.01444.x) [DOI] [Google Scholar]
- 44.Almeida-Neto M, Guimaraes P, Guimaraes PR, Jr, Loyola RD, Ulrich W. 2008. A consistent metric for nestedness analysis in ecological systems: reconciling concept and measurement. Oikos 117, 1227–1239 10.1111/j.0030-1299.2008.16644.x (doi:10.1111/j.0030-1299.2008.16644.x) [DOI] [Google Scholar]
- 45.Tylianakis JM, Tscharntke T, Lewis OT. 2007. Habitat modification alters the structure of tropical host-parasitoid food webs. Nature 445, 202–205 10.1038/nature05429 (doi:10.1038/nature05429) [DOI] [PubMed] [Google Scholar]
- 46.Blüthgen N, Menzel F, Blüthgen N. 2006. Measuring specialization in species interaction networks. BMC Ecol. 6, 9. 10.1186/1472-6785-6-9 (doi:10.1186/1472-6785-6-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bersier LF, Banasek-Richter C, Cattin MF. 2002. Quantitative descriptors of food-web matrices. Ecology 83, 2394–2407 10.2307/3071801 (doi:10.2307/3071801) [DOI] [Google Scholar]
- 48.Guimaraes PR, Jr, Guimaraes P. 2006. Improving the analyses of nestedness for large sets of matrices. Environ. Model Softw. 21, 1512–1513 10.1016/j.envsoft.2006.04.002 (doi:10.1016/j.envsoft.2006.04.002) [DOI] [Google Scholar]
- 49.Akaike H. 1973. Information theory and an extension of the maximum likelihood principle. In 2nd Int. Symp. on Information Theory (eds Petrov BN, Caskaki F.), pp. 267–281 Budapest, Hungary: Akademiai Kiado [Google Scholar]
- 50.Bascompte J, Jordano P, Olesen JM. 2006. Asymmetric coevolutionary networks facilitate biodiversity maintenance. Science 312, 431–433 10.1126/science.1123412 (doi:10.1126/science.1123412) [DOI] [PubMed] [Google Scholar]
- 51.Webb CO, Donoghue MJ. 2005. Phylomatic: tree assembly for applied phylogenetics. Mol. Ecol. Notes 5, 181–183 10.1111/j.1471-8286.2004.00829.x (doi:10.1111/j.1471-8286.2004.00829.x) [DOI] [Google Scholar]
- 52.Maddison WP, Maddison DR. 2011. Mesquite: a modular system for evolutionary analysis. Version 2.75 See http://mesquiteproject.org. [Google Scholar]
- 53.Barfuss MHJ, Samuel R, Till W, Stuessy TF. 2005. Phylogenetic relationships in subfamily Tillandsioideae (Bromeliaceae) based on DNA sequence data from seven plastid regions. Am. J. Bot. 92, 337–351 10.3732/ajb.92.2.337 (doi:10.3732/ajb.92.2.337) [DOI] [PubMed] [Google Scholar]
- 54.Chew T, De Luna E, Gonzalez D. 2010. Phylogenetic relationships of the pseudobulbous tillandsia species (Bromeliaceae) inferred from cladistic analyses of ITS 2, 5.8S ribosomal RNA gene, and ETS sequences. Syst. Bot 35, 86–95 10.1600/036364410790862632 (doi:10.1600/036364410790862632) [DOI] [Google Scholar]
- 55.Blomberg SP, Garland T, Ives AR. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745 10.1554/0014-3820(2003)057[0717:tfpsic]2.0.co;2 (doi:10.1554/0014-3820(2003)057[0717:tfpsic]2.0.co;2) [DOI] [PubMed] [Google Scholar]
- 56.Blüthgen N, Menzel F, Hovestadt T, Fiala B, Blüthgen N. 2007. Specialization, constraints, and conflicting interests in mutualistic networks. Curr. Biol. 17, 341–346 10.1016/j.cub.2006.12.039 (doi:10.1016/j.cub.2006.12.039) [DOI] [PubMed] [Google Scholar]
- 57.Hubbell SP. 1979. Tree dispersion, abundance and diversity in tropical dry forest. Science 203, 1299–1309 10.1126/science.203.4387.1299 (doi:10.1126/science.203.4387.1299) [DOI] [PubMed] [Google Scholar]
- 58.Morales JM, Vázquez DP. 2008. The effect of space in plant-animal mutualistic networks: insights from a simulation study. Oikos 117, 1362–1370 10.1111/j.0030-1299.2008.16737.x (doi:10.1111/j.0030-1299.2008.16737.x) [DOI] [Google Scholar]
- 59.Gómez JM, Verdú M, Perfectti F. 2010. Ecological interactions are evolutionarily conserved across the entire tree of life. Nature 465, 918–921 10.1038/nature09113 (doi:10.1038/nature09113) [DOI] [PubMed] [Google Scholar]
- 60.Stouffer DB, Sales-Pardo M, Sirer MI, Bascompte J. 2012. Evolutionary conservation of species’ roles in food webs. Science 335, 1489–1492 10.1126/science.1216556 (doi:10.1126/science.1216556) [DOI] [PubMed] [Google Scholar]
- 61.Rezende EL, Albert EM, Fortuna MA, Bascompte J. 2009. Compartments in a marine food web associated with phylogeny, body mass, and habitat structure. Ecol. Lett. 12, 779–788 10.1111/j.1461-0248.2009.01327.x (doi:10.1111/j.1461-0248.2009.01327.x) [DOI] [PubMed] [Google Scholar]
- 62.Naisbit RE, Rohr RP, Rossberg AG, Kehrli P, Bersier LF. 2012. Phylogeny versus body size as determinants of food web structure. Proc. R. Soc. B 279, 3291–3297 10.1098/rspb.2012.0327 (doi:10.1098/rspb.2012.0327) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rezende EL, Jordano P, Bascompte J. 2007. Effects of phenotypic complementarity and phylogeny on the nested structure of mutualistic networks. Oikos 116, 1919–1929 10.1111/j.2007.0030-1299.16029.x (doi:10.1111/j.2007.0030-1299.16029.x) [DOI] [Google Scholar]
- 64.Swenson NG, Enquist BJ. 2007. Ecological and evolutionary determinants of a key plant functional trait: wood density and its community-wide variation across latitude and elevation. Am. J. Bot. 94, 451–459 10.3732/ajb.94.3.451 (doi:10.3732/ajb.94.3.451) [DOI] [PubMed] [Google Scholar]
- 65.Niklas KJ. 1993. Influence of tissue density-specific mechanical-properties on the scaling of plant height. Ann. Bot. 72, 173–179 10.1006/anbo.1993.1096 (doi:10.1006/anbo.1993.1096) [DOI] [Google Scholar]
- 66.Reyes-García C, Griffiths H, Rincon E, Huante P. 2008. Niche differentiation in tank and atmospheric epiphytic bromeliads of a seasonally dry forest. Biotropica 40, 168–175 10.1111/j.1744-7429.2007.00359.x (doi:10.1111/j.1744-7429.2007.00359.x) [DOI] [Google Scholar]
- 67.Blüthgen N, Fründ J, Vázquez DP, Menzel F. 2008. What do interaction network metrics tell us about specialization and biological traits? Ecology 89, 3387–3399 10.1890/07-2121.1 (doi:10.1890/07-2121.1) [DOI] [PubMed] [Google Scholar]
- 68.Zhang F, Hui C, Terblanche JS. 2011. An interaction switch predicts the nested architecture of mutualistic networks. Ecol. Lett. 14, 797–803 10.1111/j.1461-0248.2011.01647.x (doi:10.1111/j.1461-0248.2011.01647.x) [DOI] [PubMed] [Google Scholar]
- 69.Memmott J, Waser NM, Price MV. 2004. Tolerance of pollination networks to species extinctions. Proc. R. Soc. Lond. B 271, 2605–2611 10.1098/rspb.2004.2909 (doi:10.1098/rspb.2004.2909) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ashworth L, Aguilar R, Galetto L, Aizen MA. 2004. Why do pollination generalist and specialist plant species show similar reproductive susceptibility to habitat fragmentation? J. Ecol. 92, 717–719 10.1111/j.0022-0477.2004.00910.x (doi:10.1111/j.0022-0477.2004.00910.x) [DOI] [Google Scholar]
- 71.Espejo-Serna A, Lopez-Ferrari AR, Ramirez-Morillo I, Holst BK, Luther HE, Till W. 2004. Checklist of Mexican Bromeliaceae with notes on species distribution and levels of endemism. Selbyana 25, 33–86 [Google Scholar]