Abstract
Despite a burgeoning science of cultural evolution, relatively little work has focused on the population structure of human cultural variation. By contrast, studies in human population genetics use a suite of tools to quantify and analyse spatial and temporal patterns of genetic variation within and between populations. Human genetic diversity can be explained largely as a result of migration and drift giving rise to gradual genetic clines, together with some discontinuities arising from geographical and cultural barriers to gene flow. Here, we adapt theory and methods from population genetics to quantify the influence of geography and ethnolinguistic boundaries on the distribution of 700 variants of a folktale in 31 European ethnolinguistic populations. We find that geographical distance and ethnolinguistic affiliation exert significant independent effects on folktale diversity and that variation between populations supports a clustering concordant with European geography. This pattern of geographical clines and clusters parallels the pattern of human genetic diversity in Europe, although the effects of geographical distance and ethnolinguistic boundaries are stronger for folktales than genes. Our findings highlight the importance of geography and population boundaries in models of human cultural variation and point to key similarities and differences between evolutionary processes operating on human genes and culture.
Keywords: cultural evolution, cultural FST, folktale, population genetics
1. Introduction
Parallels between processes of genetic and cultural evolution [1,2] mean that method and theory developed to analyse biological data can be used to study language, culture and the archaeological record [3–7]. A major focus of empirical research on cultural variation and change has been the analysis of data coding for the presence or absence of population-level cultural traits across ethnolinguistic groups (groups defined along ethnic and/or linguistic lines). It has been argued that these traits are frequently transmitted with a high degree of fidelity down ethnolinguistic lineages, analogous to genetic inheritance in biological species, supporting what has been dubbed the ‘cultures as species’ model [8]. Language change may be a paradigm example of such ‘species-like’ cultural evolution [9], and language family trees inferred using phylogenetic methods are now routinely used as lineages on which to model the evolution of a wide variety of population-level cultural traits [10–12].
However, cultures do not always behave like species [13–15]. Characterizing ethnolinguistic groups as having population-level cultural traits can be problematic when there is significant heterogeneity within groups. Furthermore, while horizontal transmission of genes between species is rare, the exchange of cultural traits between ethnolinguistic groups is not. As Boyd et al. [14] argue, there exists a spectrum of possibilities for the degree of coherence of culture within ethnolinguistic groups, ranging from core cultural traditions with less cohesive peripheral aspects, to assemblages of bounded cultural packages lacking core traditions, to mere collections of ephemeral and unbounded cultural traits. Where within-population variation and horizontal transmission are high, a macro-evolutionary ‘cultures as species’ model provides, at best, an incomplete picture that ignores internal diversity and the micro-evolutionary processes shaping patterns of variation. There is thus a need for research methods that quantify, rather than ignore, within-population and spatial variation in culture.
Population geneticists have developed a suite of tools for characterizing patterns and processes of genetic variation within species owing to mutation, selection, gene flow and drift [16,17]. Wright's F-statistic (FST) [18] and associated metrics such as the ΦST statistic [19] are routinely used to measure how variance in genetic diversity is partitioned within and between populations. The FST quantifies the relative variation of traits within versus between populations and is calculated as the correlation of randomly chosen variants within a population relative to a similar correlation across the meta-population [20]. An FST or ΦST value of 0 indicates no differentiation between populations, whereas a value of 1 indicates complete differentiation.
Analyses of autosomal single nucleotide polymorphisms (SNPs) in human populations around the world have yielded average FST estimates of between 0.052 and 0.130 [21], indicating that, on a global scale, roughly 5–13% of human autosomal genetic variation occurs between populations. Between-population variation can be much lower when examining genetic diversity within continents, particularly in Europe [22,23]. Recently, high-resolution studies of SNP data from European populations have found extremely low average FST estimates of 0.0025–0.004 between populations [24,25]. Low levels of genetic diversity between human populations have been used to argue against the validity of the biological concept of race [26,27] and against the feasibility of genetic group-level selection in humans [28].
Another important line of inquiry in population genetics uses spatial analysis of human genetic diversity to shed light on the processes shaping our gene pool. Although human genetic variation falls into a number of regional clusters, the predominant pattern is clinal, with much of the apparent regional clustering attributable to discontinuous spatial sampling [29]. Genetic distance between human populations increases with geographical distance at both continental and global scales, and across a variety of markers [22]. A smooth clinal pattern of genetic variation is often taken to support an ‘isolation by distance’ (IBD) model [18,29] in which individuals tend to migrate short distances between neighbouring populations, taking their genes with them, resulting in gradual diffusion of genetic variants across the landscape. In Europe, an IBD model is supported by a remarkable fit between genes and geography: a recent study of high-resolution autosomal SNP data found that the first and second principal components of genetic variation recreated a map of the continent, albeit explaining only a small percentage of the overall variation (0.30% and 0.15%, respectively) [25]. Conversely, departures from a clinal pattern of human genetic variation expected under the IBD ‘null’ model have been used to identify population boundaries, prehistoric migrations and ancient selection pressures [30–32].
Research on human population structure and spatial variation has allowed population geneticists to gain insights into human prehistory and the processes operating within populations that give rise to global patterns of genetic diversity [23,29]. It has long been argued by anthropologists and archaeologists that research on cultural evolution also needs to take ‘population thinking’ seriously [5,33,34]. By quantifying population structure and spatial variation in cultural diversity, we can learn how micro-scale processes operating within populations act to shape macro-scale between-population variation in human culture.
Recently, scholars have begun to borrow theory and analytical tools from population genetics to study cultural variation within populations. Random copying models analogous to Kimura's [16] neutral genetic drift model have been used to predict variation and change in the archaeological record [34–37] and in contemporary culture [5]. Bell et al. [28] used cross-cultural data from the World Values Survey to calculate pairwise cultural FST values for 150 neighbouring countries, which they compared with previously published genetic FST values (like the genetic FST, cultural FST is a measure of the relative variation of traits within versus between populations). They found that the average cultural FST value between neighbouring countries (mean = 0.080) was an order of magnitude larger than the average SNP genetic FST value between the same countries (mean = 0.0053), which they argued demonstrates a greater potential for group selection on culture than genes. Rzeszutek et al. [38] examined cross-cultural variation in song characteristics across 16 Formosan-speaking ethnolinguistic groups and found an overall ΦST of 0.02, indicating that approximately 2 per cent of variation was between populations. In addition, debates in experimental economics have begun to focus on within- versus between-population variation in strategies employed in economic games [39–41].
While these studies make important first steps towards quantifying cultural variation within and between populations, none of them investigated how this variation is patterned spatially. This renders estimates of population structure (such as FST and ΦST values) difficult to interpret because, as research on human genetic diversity has repeatedly demonstrated, apparent population structure can be an artefact of discontinuous spatial sampling, rather than group boundaries [29]. There is therefore a need for research that quantifies the independent effects of group boundaries and geography on patterns of cultural diversity and examines when and why these patterns vary across different elements of culture.
Here, we adapt tools from population genetics to quantify the influence of both population structure and geography on 700 variants of the folktale ‘The tale of the kind and the unkind girls’ [42], drawn from 31 ethnolinguistic populations across Europe (see the electronic supplementary material, figure S1). Described by the folklorist Thompson [43] as ‘one of the most popular of oral tales’ (p. 126), versions of this folktale are found all over Europe. Two variants appear in the Brothers Grimm fairy tale collection (Die drei Männlein im Walde and Frau Holle), and a motif was used by Shakespeare in The merchant of Venice. Variants of the folktale typically tell a moralistic story of a kind girl who is rewarded for her generosity and an unkind girl who is punished for her selfishness (see the electronic supplementary material).
There are a number of features of this folktale dataset that make it particularly attractive for studying the influence of population structure and geography on cultural variation. First, the dataset includes multiple samples of folktale variants drawn from the same ethnolinguistic group, allowing the quantification of within- versus between-group variation. Second, the dataset includes geographical information for 84 per cent of the folktale variants, which affords an opportunity to disentangle effects of group membership and geography. Third, most of the folktale variants included in the dataset were collected during the late nineteenth and early twentieth centuries, before communication technology and air travel transformed how ideas and people spread. Fourth, given that variation in this folktale was likely to have been predominantly selectively neutral (i.e. not ‘functional’ in the sense of being tested against the natural environment [34,44]), it may provide a plausible ‘null’ model of cultural diffusion, akin to IBD in population genetics, against which the effects of selection, population boundaries and cultural ancestry can be tested. Finally, the folktale variants in the dataset were independently coded for narrative content by a noted folklore scholar [42] according to the well-established historic–geographic method of folklore analysis [43].
We examine the independent effects of population structure and geography on variation in this folktale across Europe using three stages of analysis. First, we quantify individual folktale variation within versus between ethnolinguistic groups and examine whether between-population folktale variation is greater than between-population genetic variation, as has been found for other cultural traits [28]. Second, we investigate the processes underlying any between population differences. We test whether individual folktale variation shows a predominantly clinal pattern, like that observed in human population genetic variation [29], and quantify the independent effects of geography and ethnolinguistic affiliation. Third, we examine how the various folktale populations cluster in Europe, using pairwise population ΦST distances. We ask whether these populations show a hierarchical, tree-like pattern of branching, probably reflecting sequential colonization and vertical inheritance of coherent (perhaps linguistic) lineages, or a more reticulate pattern, aligned to geography, suggesting a process of local diffusion.
2. Material and methods
(a). Data
We sourced folktale data from Roberts’ study of the Tale of the kind and the unkind girls [42]—tale type 480 according to the Aarne–Thompson–Uther tale type index [45]. Roberts indicated the presence and absence of important narrative elements in each folktale variant using multistate character codings according to principles of the historic–geographic method of folklore analysis [43] (see the electronic supplementary material, table S1). For example, one coded narrative element is the location where the main protagonist meets some other key characters, with the location coded according to 12 character-states, including at the bottom of a well, by a river, in a field, on a mountain-side and in a cave.
We assigned folktale variants to populations using the ethnolinguistic assignments provided by the source dataset. We analysed only those folktale variants that were drawn from ethnolinguistic populations in Europe because many of the other geographical regions were poorly sampled and included folktale variants that might reflect more recent post-colonial movements rather than long-standing geographical and ethnolinguistic patterns [42]. In total, our analysis included 700 folktale variants drawn from 31 European ethnolinguistic populations, with a mean of 23 folktale variants per population (Armenian, 3; Basque, 2; Bulgarian, 8; Czech, 11; Danish, 48; English, 8; Estonian, 16; Finnish, 83; Swedish in Finland, 25; Flemish, 6; French, 16; German, 61; Greek, 11; Icelandic, 11; Irish, 22; Italian, 33; Latvian, 13; Norwegian, 48; Polish, 45; Portuguese, 2; Romanian, 4; Russian, 32; Finno-Ugric in Russia, 23; Scottish, 3; Slovenian, 6; Spanish, 11; Swedish, 101; Swiss German, 3; Turkish, 32; Walloon, 3; Yugoslavian, 13).
We recoded the presence or absence of narrative elements as ‘1’ or ‘0’, respectively, to produce a matrix of 700 folktale variants coded across 393 binary traits (traits coded as ‘other’ were excluded because it is a catchall category such that a shared presence of ‘other’ does not represent similarity). For analysis, this presence/absence matrix was converted to a Jaccard distance matrix reflecting pairwise distances between all folktale variants (see the electronic supplementary material, table S2). The Jaccard distance for each pair of folktale variants was calculated as the sum of the number of traits that are present in one variant but not the other, divided by the sum of the number of traits that are present in one or both of the variants. The Jaccard distance is particularly appropriate for analysing this cultural dataset because it standardizes for the number of traits observed for each pair and shared absences do not contribute to similarity [44].
The geographical locations of the folktale variants are shown in the electronic supplementary material, figure S1. They were estimated using locality information included in the source dataset. Sixteen per cent of the folktale variants did not include locality information beyond ethnolinguistic affiliation. For these folktale variants, geographical coordinates were assigned as the centroid location of the points sampled from the ethnolinguistic group to which they belonged. Removing these cases from the analysis did not qualitatively affect any of the results we report. Geographical coordinates were used to calculate pairwise geographical distance and logged geographical distance matrices between individual folktale variants, and between the 31 ethnolinguistic populations (using the centroid of geographical coordinates for each population). Pairwise distances were calculated using great circle distances in GenAlEx v. 6.4 [46] (see the electronic supplementary material, tables S3 and S4).
We used linguistic divergence between ethnolinguistic groups to index cultural ancestry. A language dissimilarity matrix was calculated using patristic distances between Indo-European languages inferred from Gray & Atkinson's [47] phylogenetic analysis of the Indo-European language family. All Indo-European ethnolinguistic populations included in the folktale dataset were represented in Gray and Atkinson's analysis, with the exception of Scottish. Nevertheless, Scottish can be reliably placed as a close sister language to Irish in the Indo-European tree [48] so we assigned Scottish the same distance as Irish to all languages (except to Irish itself, which was assigned a distance equivalent to the minimum distance between languages observed in the initial data). Assigning distances to languages outside the Indo-European family is more problematic. Higher-level language family groupings have been proposed, but they remain highly controversial [49], making precise estimates of distances between languages from different language families unfeasible. To generate approximate values, we set distances between languages from different language families (Indo-European, Turkic and Finno-Ugric) to 1.25 times the maximum observed distance between Indo-European languages. The ethnolinguistic category ‘Finno-Ugric in Russia’ was also problematic, because the particular Finno-Ugric languages were not recorded. Because Finno-Ugric shows a comparable level of internal diversity to Indo-European [50], we set a distance for languages within the Finno-Ugric language family (Finnish, Estonian and ‘Finno-Ugric speakers in Russia’) to the average distance between languages in the Indo-European language family (see the electronic supplementary material, table S5). We found our results were robust across a range of between-family distance multipliers from 1 to 3 (with values higher than 1.25 explaining less of the variance; see the electronic supplementary material, tables S6 and S7).
An ethnolinguistic identity matrix for individual folktale variants was created by scoring the distance between folktale variants as 0 if they came from the same ethnolinguistic group and 1 if they came from a different group. These usually correspond to language speaker populations (e.g. Spanish), but twice to subpopulations within a language (Swiss German, Swedish speakers in Finland) and once to a group of related languages (Finno-Ugric speakers in Russia).
(b). Analysis
Cultural population structure across ethnolinguistic groups was investigated using the analysis of molecular variance (AMOVA) [19] technique as implemented in Arlequin v. 3.5.1.2 [51]. AMOVA provides a measure of the proportion of variance within versus between populations using between-population ΦST values—a value of 0 indicates no differentiation between populations, whereas a value of 1 indicates complete differentiation. Unlike the FST statistic, which is based on variant frequencies, the ΦST statistic extracts additional information from the data by accounting for distances between variants. The method takes as input a pairwise matrix of distances between sampled variants, together with information on the population each variant was sampled from. Because AMOVA makes no assumptions about the units of analysis or the mechanisms generating diversity, it is equally suited to analysing cultural data from ethnolinguistic groups or genetic data from biological populations. Although geneticists use a measure of genetic distances between sequences, here we use our Jaccard distance matrix of distances between folktale variants. By calculating pairwise population ΦST values across ethnolinguistic groups, it is possible to quantify the average level of within- versus between-group variation, as well as population pairwise distances. Negative ΦST values have no interpretation and, following standard practice, were set to zero. Statistical significance of ΦST values was tested using 1000 random permutations (see the electronic supplementary material, table S5).
Spatial autocorrelations among (i) individual folktale variants and (ii) pairwise ΦST values for ethnolinguistic populations were calculated using the method implemented in GenAlEx v. 6.4 [46]. This autocorrelation method uses a pairwise geographical distance matrix and a pairwise folktale distance matrix to calculate an autocorrelation coefficient r across a specified range of geographical distance classes. The autocorrelation coefficient provides a measure of the similarity between pairs of folktales whose geographical separation falls within each distance class. Tests for statistical significance were performed using two methods, calculating r across 1000 random permutations and 1000 bootstrap estimates [52].
In order to investigate the independent effects of geography, ethnolinguistic affiliation and cultural ancestry on variation in individual folktale variants, we calculated correlations and partial correlations between the folktale, geographical, linguistic and ethnolinguistic identity distance matrices using Mantel and partial Mantel [53,54] tests in Arlequin v. 3.5.1.2 [51], with significance assessed using 1000 random permutations. We used the same approach to test for correlations between geographical distance, linguistic distance and pairwise ΦST values between ethnolinguistic populations.
In order to visualize the pattern of relationships between populations and identify population clusters, we constructed a NeighbourNet [55] in SplitsTree v. 4.11.3 from the folktale pairwise population ΦST values. The NeighbourNet algorithm is useful for identifying complex transmission histories of population divergence and convergence [56,57]. The method does not assume a simple tree-like model of evolution; instead, evidence for such a model appears as bifurcating ‘tree-like’ splits in the graph. Conversely, evidence for convergence or horizontal transmission owing to cultural borrowing will appear as reticulate, ‘box-like’ structures representing conflicting population subdivisions.
3. Results and discussion
(a). Population structure
Our AMOVA reveals moderate but highly significant population structure in folktale variation across the sampled ethnolinguistic groups, with 9.1 per cent of the variation among individual folktales occurring between populations (average ΦST = 0.091, p < 0.001). Some of the ethnolinguistic groups in our dataset had small sample sizes, which can result in unreliable ΦST values. To investigate whether they may have biased our results, we repeated the AMOVA with small populations (less than five variants) removed. Consistent with the full analysis, we again found 9 per cent between-population variation (ΦST = 0.090, p < 0.001). This value is comparable to levels of variation observed in attitudes and values between neighbouring nations (8%) [28] and to between-population behavioural variation in economic games (4–38%) [39,40].
A value of 9.1 per cent is also within the range of between-population variation in global human autosomal genetic diversity, which range from 5 per cent to 13 per cent [21]. However, estimates of between-population genetic variation in comparable European populations range from 0.25 per cent to 0.40 per cent [24,25]. This order of magnitude difference in Europe fits with the finding that cultural FST scores calculated using variation in attitudes and values between neighbouring nations (FST = 0.08 or 8%) are higher than genetic FST scores for the same populations (FST = 0.005 or 0.5%) [28].
When comparing our results with estimates of human genetic diversity, it is important to note that, while each sampled genotype can be tied to an individual person, here we are not tracking characteristics (behavioural or genetic) of individual people—that is, we do not have information about which individuals in a population know which folktale variant(s). Although tracking the characteristics of individual people is appropriate for some cultural traits [28], it makes little sense for traits such as folktales because, unlike genes, one person can know many folktales and folktales can move without people. Instead, our approach tracks the cultural entities themselves, in effect treating individual folktale variants in ethnolinguistic groups like population geneticists treat genetically distinct haploid organisms in biological populations. Rzeszutek et al. [38] used a similar approach in their analysis of Formosan song variants, although, interestingly, our estimate of between-population variation is closer to Bell et al.'s 8 per cent than Rzeszutek et al.'s 2 per cent (see §3d for a possible explanation for this).
While AMOVA allows us to quantify variation between ethnolinguistic groups, it does not tell us whether the differences we observe are the result of measurable ethnolinguistic boundaries and divergence along cultural lineages, or purely clinal patterns of geographical variation, or some combination of the two. In order to determine how the between-population differences we observe arose, we first consider the effects of geography and then test for departures from a purely clinal model based on ethnolinguistic affiliation.
(b). Geographical clines
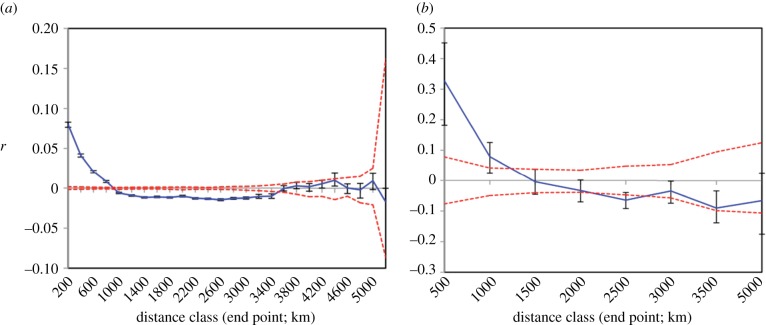
Mantel tests on individual folktale data show clear clinal patterning (table 1). Logged geographical distance is the best single predictor of folktale similarity, explaining 8.9 per cent of the variance (r2 = 0.089, p < 0.001; unlogged geographical distance explains 6.4 per cent of the variance (r2 = 0.064, p < 0.001)). By comparison, ethnolinguistic identity and language distance explain 6.8 per cent (r2 = 0.0683, p < 0.001) and 2.6 per cent (r2 = 0.0262, p < 0.001) of the variance in folktale similarity, respectively. Spatial autocorrelation analysis also shows a highly significant relationship between individual folktale distance and geographical distance (figure 1a). Although the correlation is small, it is roughly an order of magnitude greater than observed in similar analyses of autosomal genetic distances between individuals across Europe [24,25].
Table 1.
Results of Mantel and partial Mantel tests [53,54] of correlations between individual folktale Jaccard distance values, geographical distance, logged geographical distance, linguistic distance and ethnolinguistic group.
| variance explained (%) | p-value | |
|---|---|---|
| individual folktale distance values (n = 700) predicted by | ||
| geography | 6.38 | <0.001 |
| log(geography) | 8.92 | <0.001 |
| ethnolinguistic group | 6.83 | <0.001 |
| language | 2.62 | <0.001 |
| log(geography) controlling for ethnolinguistic group | 6.64 | <0.001 |
| log(geography) controlling for language | 8.50 | <0.001 |
| ethnolinguistic group controlling for log(geography) | 3.68 | <0.001 |
| ethnolinguistic group controlling for language | 6.14 | <0.001 |
| language controlling for log(geography) | 0.54 | 0.006 |
| language controlling for ethnolinguistic group | 0.99 | <0.001 |
Figure 1.
Folktale spatial autocorrelation analysis [46]. Spatial correlogram plot showing correlation coefficient (r) as a function of distance for (a) individual-level data from 700 folktales using pairwise Jaccard distances and (b) population-level data from 31 ethnolinguistic groups using pairwise ΦST values. The permuted 95% CI (dashed lines) and the bootstrapped 95% confidence error bars are also shown. Variation in error estimates is influenced by the number of pairwise comparisons within each distance class. (Online version in colour.)
Our population-level analyses also show clear clinal spatial structure (table 2). Unlike the individual folktale analyses, geographical distance explains more of the variance in pairwise population ΦST values (14.8%, r2 = 0.148, p < 0.001) than does logged geographical distance (13.0%, r2 = 0.130, p < 0.001). By comparison, language distance explains 7.5 per cent of the variance (r2 = 0.0751, p < 0.014). These findings hold when populations with small sample sizes are excluded (table 2). The shape and magnitude of spatial autocorrelation at the population level (figure 1b) is similar to that found in analyses of human genetic variation between populations in Europe [58].
Table 2.
Results of Mantel and partial Mantel tests [53,54] of correlations between population pairwise matrices of folktale ΦST values, geographical distance, logged geographical distance and linguistic distance.
| variance explained (%) | p-value | |
|---|---|---|
| population folktale ΦST values (n = 31) predicted by | ||
| geography | 14.75 | <0.001 |
| log(geography) | 12.99 | <0.001 |
| language | 7.51 | 0.014 |
| geography controlling for language | 12.61 | 0.005 |
| language controlling for geography | 4.33 | 0.099 |
| population folktale ΦST values (minus groups with < 5 folktales; n = 23) predicted by | ||
| geography | 16.78 | 0.001 |
| language | 8.23 | 0.035 |
| geography controlling for language | 14.40 | 0.013 |
| language controlling for geography | 4.40 | 0.140 |
Partial Mantel tests provide insights into the processes driving these spatial patterns of folktale variation at the individual level (table 1). Logged geographical distance remains a significant predictor of individual folktale variation, even after controlling for ethnolinguistic identity (r2 = 0.085, p < 0.001) and language distance (r2 = 0.066, p < 0.001), explaining 8.5 per cent and 6.6 per cent of the variance, respectively. This indicates that spatial patterning is not simply the result of cultural divisions (as measured by ethnolinguistic affiliation) or cultural ancestry (as measured by language distance). In fact, the strongest individual folktale correlations occur at distances of less than 200 km, suggesting highly localized within-group effects of geography on folktale variation (figure 1a).
The importance of geography is reinforced at the population level (table 2). Geographical distance explains 12.6 per cent of the variance in between-population ΦST values when controlling for language distance (r2 = 0.126, p < 0.001), but language distance is not a significant predictor of ΦST values when controlling for geographical distance (r2 = 0.043, p < 0.106). This suggests the folktale and language histories are decoupled, either because the folktales spread much later than the spread of languages across Europe or because any legacy of deep cultural ancestry inherited down language lineages has been obscured by subsequent folktale evolution and geographical diffusion.
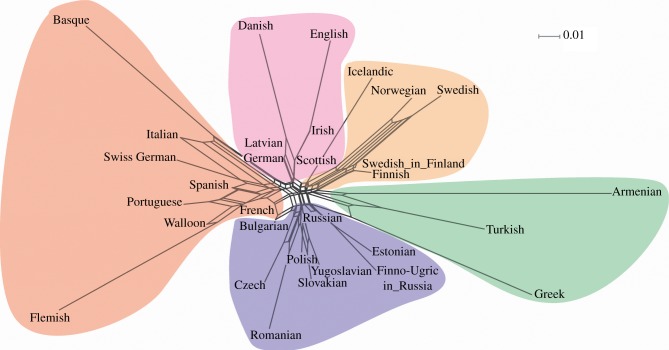
The NeighbourNet constructed from pairwise ΦST values between ethnolinguistic groups reveals a highly reticulate network and regional clustering of populations (figure 2). This does not support the idea that current folktale variation is the result of a sequential colonization of the landscape by vertically inherited, coherent cultural lineages (linguistic or otherwise). Convergent evolution of traits and/or trait reversals could account for some reticulation in the graph, but they would not be expected to generate the regional clustering we observe. Together, then, our individual and population-level results point to the primacy of local cultural diffusion processes between neighbouring folktale variants.
Figure 2.
NeighbourNet [55] of European folktale populations. The relationship between folktale populations across Europe, based on population folktale ΦST values. Populations that are closer together tend to have more similar folktales. Box-like structures show the reticulate nature of folktale similarity, indicating extensive horizontal transmission (as opposed to vertical transmission down cultural lineages). Shaded polygons show the five clusters discussed in the main text. (Online version in colour.)
(c). Ethnolinguistic boundaries
Measureable differences between groups do not necessarily point towards population structure since they could be the result of clinal variation that is masked by discontinuities in spatial sampling [29]. On Boyd et al.'s [14] spectrum of cultural descent types, this would suggest that folktales are simply diffusing across the landscape and are not part of coherent cultural traditions. By testing for departures from a purely clinal model, we can determine whether ethnolinguistic boundaries act as a barrier to the spread of folktales.
A partial Mantel test that uses ethnolinguistic identity to predict folktale variation while controlling for geography shows that ethnolinguistic identity explains a significant proportion of the variation in individual folktales, even after controlling for geographical distance (r2 = 0.037, p < 0.001). Ethnolinguistic identity therefore represents a barrier to folktale transmission. Based on the regression coefficients from our model incorporating geographical distance and ethnolinguistic identity, we can infer that the magnitude of this cultural barrier effect is equivalent to multiplying geographical distance between folktale variants by a factor of 10 (the relationship is multiplicative, rather than additive, because we are using logged geographical distance). In other words, folktales from the same culture found 100 km apart are, on average, as similar as folktales found 10 km apart in different cultures.
Studies of human genetic diversity have likewise identified barriers to gene flow that may be related to ethnolinguistic identity [21,30]. In both the folktale and genetic case, barriers could arise if there is a reduced probability of transmission across ethnolinguistic boundaries. If folktales cross ethnolinguistic boundaries less easily than genes, this could partly explain higher folktale ΦST values. However, in the case of folktales, another possibility is that cultural transmission biases operating within, but not across, ethnolinguistic groups may differentially impact which folktale elements are successfully copied. Content-dependent biases, such as favouring certain motifs for their meaning in certain cultures, or context-dependent biases, such as conformist or prestige bias [33,59], could lead to highly successful variants that are particular to each group.
(d). Patterns and processes of human cultural evolution
Our findings highlight key similarities and differences between patterns and processes of folktale and genetic variation in Europe. Like genetic variation, most folktale variation occurs within ethnolinguistic groups. However, across Europe, the folktales in our study show an order of magnitude more between-population variation than genes. Three factors are likely to be at work here. First, faster rates of cultural evolution could increase the likelihood of between-population differences arising [3]—although this also increases within-population variation. Second, the ethnolinguistic barrier effect we identify suggests that content- and/or context-dependent cultural transmission biases [33] are acting to limit information flow across group borders, suppress internal variation and/or accentuate group differences. Third, the stronger spatial autocorrelation in culture than genes (itself possibly a result of faster rates of cultural evolution) means that, in addition to any population boundary effects, for a given geographical scale, we expect greater between-population differences in culture than genes. If so, cultural FST or ΦST values may be particularly sensitive to the geographical scale of the population being sampled. This may help to explain why the cultural ΦST values from this study, drawn from large European language groups, and cultural FST values from countries around the globe [28] are four times larger than the cultural ΦST values from the considerably more localized Formosan-speaking groups [38].
Recently, empirical data on cultural and genetic FST values have been applied to debates about the units of selection in human evolution [28]. The folktale variants we examine here are unlikely to affect the survival of the individuals or groups that carry them and so are essentially selectively neutral traits. Nevertheless, our findings highlight an important caveat when interpreting FST or ΦST values more generally. Bell et al. [28] argue that higher cultural than genetic FST values between neighbouring groups suggests greater potential for cultural group selection. Yet, our partial Mantel tests on individual folktales show that variation is more strongly related to geographical distance (6.6% of the variation) than ethnolinguistic identity (3.7% of the variation). Hence, while populations differ and significant cultural barriers exist, geographical distance appears to be the most important factor. If this pattern generalizes to other elements of culture then, because much cultural competition is likely to have played out on a local valley-to-valley or village-to-village scale, actual differences between competing groups may be much less than is indicated by FST or ΦST values calculated on the basis of large-scale ethnolinguistic identities—the same is true for genetic variation. This highlights the importance of considering the spatial dimension of cultural and genetic variation when evaluating theoretical models of competition between groups.
(e). The cultural landscape of Europe
The NeighbourNet in figure 2 represents graphically the pattern of regional clustering in folktale variation. The five clusters we identify provide insights into possible cultural spheres of influence in Europe since the folktale's inception. Cluster (i) includes the western European Romance-speaking populations (excluding Romanian) as well as other non-Romance-speaking western European populations (Basque, Flemish and Swiss German). Cluster (ii) includes the eastern European Slavic-speaking populations, plus other non-Slavic-speaking eastern European populations (Romanian, and Finno-Ugric speakers from Russia). Cluster (iii) includes the southeastern European populations (Turkish, Greek and Armenian). Cluster (iv) includes northern European North Germanic-speaking populations (excluding Danish), plus Finnish. Interestingly, Swedes in Finland are placed alongside Finnish, not Swedish, reinforcing the importance of geography over cultural ancestry. The remaining cluster (v) is less obviously a geographical grouping, comprising German, Danish and Latvian in mainland northern Europe plus English, Irish and Scottish from the British Isles. The British Isles have met with waves of immigration and trade from the ancestors of these northern European groups, from Viking expansion beginning in the ninth century AD to trade networks such as the Hanseatic League, which linked the Baltic to Northern Europe and Britain from the thirteenth century AD. If this grouping is preserving the traces of early contact then the folktale stretches back beyond the earliest attested variants, which do not appear until the fourteenth century [42].
4. Conclusion
Much has been made of analogies between processes of biological and cultural evolution and the potential for interdisciplinary cross-fertilization [1–7]. While there exist important disanalogies between cultural and biological processes, particularly with regard to micro-evolutionary transmission mechanisms [33,59], our findings suggest that methods and theory from population genetics can nonetheless be usefully applied to characterize population structure and variation in cultural packages such as folktales. Our comparisons of the broad patterns that emerge on a continental scale in folktale and genetic diversity point to some key similarities and differences in the forces shaping the two. In addition, the location information from individual folktale variants allowed us to tease apart the relative effects of population structure and geography on cultural diversity. Future work using the approach we describe here could examine how these patterns differ across other aspects of human culture, such as variation in material culture assemblages through time in the archaeological record [6], providing important insights into processes of cultural transmission and the interplay between human genetic and cultural evolution.
Acknowledgements
We thank three anonymous reviewers for helpful comments on the manuscript. The research leading to these results was supported by a Rutherford Discovery Fellowship (Q.D.A.), ARC Discovery Fellowship (S.J.G.) and John Templeton Foundation grant no. 28745 (S.J.G. and Q.D.A.) and an ESRC Large Grant (REF RES-060-25-0085) entitled ‘Ritual, Community, and Conflict’.
References
- 1.Atkinson QD, Gray RD. 2005. Curious parallels and curious connections: phylogenetic thinking in biology and historical linguistics. Syst. Biol. 54, 513–526 10.1080/10635150590950317 (doi:10.1080/10635150590950317) [DOI] [PubMed] [Google Scholar]
- 2.Mesoudi A, Whiten A. 2004. Is human cultural evolution Darwinian? Evidence reviewed from the perspective of the Origin of Species. Evolution 51, 1–11 10.1111/j.0014-3820.2004.tb01568.x (doi:10.1111/j.0014-3820.2004.tb01568.x) [DOI] [PubMed] [Google Scholar]
- 3.Gray RD, Greenhill SJ, Ross RM. 2007. The pleasures and perils of Darwinizing culture (with phylogenies). Biol. Theory 2, 360–375 10.1162/biot.2007.2.4.360 (doi:10.1162/biot.2007.2.4.360) [DOI] [Google Scholar]
- 4.Mesoudi A, Whiten A, Laland KN. 2006. Towards a unified science of cultural evolution. Behav. Brain Sci. 29, 329–347 10.1017/S0140525X06009083 (doi:10.1017/S0140525X06009083) [DOI] [PubMed] [Google Scholar]
- 5.Bentley RA, Hahn MW, Shennan SJ. 2004. Random drift and culture change. Proc. R. Soc. Lond. B 271, 1443–1450 10.1098/rspb.2004.2746 (doi:10.1098/rspb.2004.2746) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eerkens JW, Lipo CP. 2007. Cultural transmission theory and the archaeological record: providing context to understanding variation and temporal changes in material culture. J. Archaeol. Res. 15, 239–274 10.1007/s10814-007-9013-z (doi:10.1007/s10814-007-9013-z) [DOI] [Google Scholar]
- 7.O'Brien MJ, Lyman RL. 2003. Cladistics and archaeology. Salt Lake City, UT: University of Utah Press [Google Scholar]
- 8.Pagel M, Mace R. 2004. The cultural wealth of nations. Nature 428, 275–278 10.1038/428275a (doi:10.1038/428275a) [DOI] [PubMed] [Google Scholar]
- 9.Pagel M. 2009. Human language as a culturally transmitted replicator. Nat. Rev. Genet. 10, 405–415 10.1038/nrg2560 (doi:10.1038/nrg2560) [DOI] [PubMed] [Google Scholar]
- 10.Fortunato L, Holden C, Mace R. 2006. From bridewealth to dowry? A Bayesian estimation of ancestral states of marriage transfers in Indo-European groups. Hum. Nat. 17, 355–376 10.1007/s12110-006-1000-4 (doi:10.1007/s12110-006-1000-4) [DOI] [PubMed] [Google Scholar]
- 11.Currie TE, Greenhill SJ, Gray RD, Hasegawa T, Mace R. 2010. Rise and fall of political complexity in island South-East Asia and the Pacific. Nature 467, 801–804 10.1038/nature09461 (doi:10.1038/nature09461) [DOI] [PubMed] [Google Scholar]
- 12.Jordan FM, Gray RD, Greenhill SJ, Mace R. 2009. Matrilocal residence is ancestral in Austronesian societies. Proc. R. Soc. B 276, 1957–1964 10.1098/rspb.2009.0088 (doi:10.1098/rspb.2009.0088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore JH. 1994. Putting anthropology back together again: the ethnogenetic critique of cladistic theory. Am. Anthropol. 96, 925–948 10.1525/aa.1994.96.4.02a00110 (doi:10.1525/aa.1994.96.4.02a00110) [DOI] [Google Scholar]
- 14.Boyd R, Richerson PJ, Borgerhoff-Mulder M, Durham WH. 1997. Are cultural phylogenies possible? In Human by nature, between biology and the social sciences (eds Weingart P, Richerson PJ, Mitchell SD, Maasen S.), pp. 355–386 Mahwah, NJ: Lawrence Erlbaum Associates [Google Scholar]
- 15.Borgerhoff-Mulder M, Nunn CL, Towner MC. 2006. Cultural macroevolution and the transmission of traits. Evol. Anthropol. 15, 52–64 10.1002/evan.1020 (doi:10.1002/evan.1020) [DOI] [Google Scholar]
- 16.Kimura M. 1983. The neutral theory of molecular evolution. Cambridge, UK: Cambridge University Press [Google Scholar]
- 17.Maynard Smith J. 1998. Evolutionary genetics, 2nd edn Oxford, UK: Oxford University Press [Google Scholar]
- 18.Wright S. 1943. Isolation by distance. Genetics 28, 114–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Excoffier LG, Smouse PE, Quattro JM. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holsinger KE, Weir BS. 2009. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat. Rev. Genet. 10, 639–650 10.1038/nrg2611 (doi:10.1038/nrg2611) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbujani B, Colonna V. 2011. Genetic basis of human biodiversity: an update. In Biodiversity hotspots: distribution and protection of conservation priority areas (eds Zachos FE, Habel JC.), pp. 97–119 Berlin, Germany: Springer [Google Scholar]
- 22.Cavalli-Sforza LL, Menozzi P, Piazza A. 1994. The history and geography of human genes. Princeton, NJ: Princeton University Press [Google Scholar]
- 23.Li JZ, et al. 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319, 1100–1104 10.1126/science.1153717 (doi:10.1126/science.1153717) [DOI] [PubMed] [Google Scholar]
- 24.Lao O, et al. 2008. Correlation between genetic and geographic structure in Europe. Curr. Biol. 18, 1241–1248 10.1016/j.cub.2008.07.049 (doi:10.1016/j.cub.2008.07.049) [DOI] [PubMed] [Google Scholar]
- 25.Novembre J, et al. 2008. Genes mirror geography within Europe. Nature 456, 98–101 10.1038/nature07331 (doi:10.1038/nature07331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewontin RC. 1972. The apportionment of human diversity. Evol. Biol. 6, 381–398 10.1007/978-1-4684-9063-3_14 (doi:10.1007/978-1-4684-9063-3_14) [DOI] [Google Scholar]
- 27.Marks J. 2010. Ten facts about human variation. In Human evolutionary biology (ed. Muehlenbein MP.), pp. 265–276 Cambridge, UK: Cambridge University Press [Google Scholar]
- 28.Bell AV, Richerson PJ, McElreath R. 2009. Culture rather than genes provides greater scope for the evolution of large-scale human prosociality. Proc. Natl Acad. Sci. USA 106, 17 671–17 674 10.1073/pnas.0903232106 (doi:10.1073/pnas.0903232106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Handley LJL, Manica A, Goudet J, Balloux F. 2007. Going the distance: human population genetics in a clinal world. Trends Genet. 23, 432–439 10.1016/j.tig.2007.07.002 (doi:10.1016/j.tig.2007.07.002) [DOI] [PubMed] [Google Scholar]
- 30.Barbujani G, Sokal RR. 1990. Zones of sharp genetic change in Europe are also linguistic boundaries. Proc. Natl Acad. Sci. USA 87, 1816–1819 10.1073/pnas.87.5.1816 (doi:10.1073/pnas.87.5.1816) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novembre J, Stephens M. 2008. Interpreting principal component analysis of spatial population genetic variation. Nat. Genet. 40, 646–649 10.1038/ng.139 (doi:10.1038/ng.139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itan Y, Powell A, Beaumont MA, Burger J, Thomas M. 2009. The origin of lactase persistence in Europe. PLoS Comput. Biol. 5, e10000491. 10.1371/journal.pcbi.1000491 (doi:10.1371/journal.pcbi.1000491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyd R, Richerson PJ. 1985. Culture and the evolutionary process. Chicago, IL: University of Chicago Press [Google Scholar]
- 34.Neiman FD. 1995. Stylistic variation in evolutionary perspective. Am. Antiquity 60, 7–36 10.2307/282074 (doi:10.2307/282074) [DOI] [Google Scholar]
- 35.Kempe M, Lycett SJ, Mesoudi A. 2012. An experimental test of the accumulated copying error model of cultural mutation for Acheulean handaxe size. PLoS ONE 7, e48333. 10.1371/journal.pone.0048333 (doi:10.1371/journal.pone.0048333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eerkens JW, Lipo CP. 2005. Cultural transmission, copying errors, and the generation of variation in material culture and the archaeological record. J. Anthropol. Archaeol. 24, 316–334 10.1016/j.jaa.2005.08.001 (doi:10.1016/j.jaa.2005.08.001) [DOI] [Google Scholar]
- 37.Lycett SJ. 2008. Acheulean variation and selection: does handaxe symmetry fit neutral expectations? J. Archaeol. Sci. 35, 2640–2648 10.1016/j.jas.2008.05.002 (doi:10.1016/j.jas.2008.05.002) [DOI] [Google Scholar]
- 38.Rzeszutek T, Savage PE, Brown S. 2011. The structure of cross-cultural musical diversity. Proc. R. Soc. B 279, 1606–1612 10.1098/rspb.2011.1750 (doi:10.1098/rspb.2011.1750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gachter S, Herrmann B, Thoni C. 2010. Culture and cooperation. Phil. Trans. R. Soc. B 365, 2651–2661 10.1098/rstb.2010.0135 (doi:10.1098/rstb.2010.0135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henrich J, et al. 2012. Culture does account for variation in game behavior. Proc. Natl Acad. Sci. USA 109, E32–E33 10.1073/pnas.1118607109 (doi:10.1073/pnas.1118607109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamba S, Mace R. 2012. Demography and ecology drive variation in cooperation across human populations. Proc. Natl Acad. Sci. USA 108, 14 426–14 430 10.1073/pnas.1105186108 (doi:10.1073/pnas.1105186108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts WE. 1958. The tale of the kind and the unkind girls: Aa-Th 480 and related tales. Berlin, Germany: De Gruyter [Google Scholar]
- 43.Thompson S. 1946. The folktale. New York, NY: Holt, Rinehart and Winston, Inc [Google Scholar]
- 44.Rogers DS, Ehrlich PR. 2008. Natural selection and cultural rates of change. Proc. Natl Acad. Sci. USA 105, 3416–3420 10.1073/pnas.0711802105 (doi:10.1073/pnas.0711802105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uther H-J. 2004. The types of international folktales: a classification and bibliography, based on the system of Antti Aarne and Stith Thompson. Helsinki, Finland: Suomalainen Tiedeakatemia, Academia Scientiarum Fennica [Google Scholar]
- 46.Peakall R, Smouse PW. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295 10.1111/j.1471-8286.2005.01155.x (doi:10.1111/j.1471-8286.2005.01155.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gray RD, Atkinson QD. 2003. Language-tree divergence times support the Anatolian theory of Indo-European origin. Nature 426, 435–439 10.1038/nature02029 (doi:10.1038/nature02029) [DOI] [PubMed] [Google Scholar]
- 48.Gillies W. 1993. Scottish Gaelic. In The Celtic languages (ed Ball MJ.), pp. 145–227 London, UK: Routledge [Google Scholar]
- 49.Campbell L, Poser WJ. 2008. Language classification: history and method. Cambridge, UK: Cambridge University Press [Google Scholar]
- 50.Marcantonio A. 2002. The Uralic language family: facts, myths and statistics. Oxford, UK: Blackwell Publishing [Google Scholar]
- 51.Excoffier LG, Laval G, Schneider S. 2005. Arlequin ver 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50 [PMC free article] [PubMed] [Google Scholar]
- 52.Peakall R, Ruibal M, Lindenmayer DB. 2003. Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes. Evolution 57, 1182–1195 10.1111/j.0014-3820.2003.tb00327.x (doi:10.1111/j.0014-3820.2003.tb00327.x) [DOI] [PubMed] [Google Scholar]
- 53.Mantel N. 1967. The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220 [PubMed] [Google Scholar]
- 54.Smouse PE, Long JC, Sokal RR. 1986. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Biol. 35, 627–632 [Google Scholar]
- 55.Bryant D, Moulton V. 2004. Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21, 255–265 10.1093/molbev/msh018 (doi:10.1093/molbev/msh018) [DOI] [PubMed] [Google Scholar]
- 56.Bryant D, Filimon F, Gray RD. 2005. Untangling our past: languages, trees, splits and networks. In The evolution of cultural diversity: phylogenetic approaches (eds Mace R, Holden CJ, Shennan S.), pp. 69–85 London, UK: UCL Press [Google Scholar]
- 57.Gray RD, Bryant D, Greenhill SJ. 2010. On the shape and fabric of human history. Phil. Trans. R. Soc. B 365, 3923–3933 10.1098/rstb.2010.0162 (doi:10.1098/rstb.2010.0162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosser ZH, et al. 2000. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am. J. Hum. Genet. 67, 1526–1543 10.1086/316890 (doi:10.1086/316890) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Henrich J, Gil-White F. 2001. The evolution of prestige: freely conferred deference as a mechanism for enhancing the benefits of cultural transmission. Evol. Hum. Behav. 22, 165–196 10.1016/S1090-5138(00)00071-4 (doi:10.1016/S1090-5138(00)00071-4) [DOI] [PubMed] [Google Scholar]