Abstract
Understanding the relative importance of heterosis and outbreeding depression over multiple generations is a key question in evolutionary biology and is essential for identifying appropriate genetic sources for population and ecosystem restoration. Here we use 2455 experimental crosses between 12 population pairs of the rare perennial plant Rutidosis leptorrhynchoides (Asteraceae) to investigate the multi-generational (F1, F2, F3) fitness outcomes of inter-population hybridization. We detected no evidence of outbreeding depression, with inter-population hybrids and backcrosses showing either similar fitness or significant heterosis for fitness components across the three generations. Variation in heterosis among population pairs was best explained by characteristics of the foreign source or home population, and was greatest when the source population was large, with high genetic diversity and low inbreeding, and the home population was small and inbred. Our results indicate that the primary consideration for maximizing progeny fitness following population augmentation or restoration is the use of seed from large, genetically diverse populations.
Keywords: outbreeding depression, inbreeding, heterosis, population size, conservation, genetic rescue
1. Introduction
Habitat loss and fragmentation have led to the increasing need for population augmentation and habitat restoration to conserve biodiversity. Determining the most appropriate sources of genetic material for genetic rescue [1,2] is critical to ensure the long-term viability of restored or augmented populations [3–5]. Owing to concerns about the loss of local adaptation and reduced progeny fitness through outbreeding depression [6–8], recommendations for sourcing genetic material for genetic rescue and restoration often advocate the use of local genotypes, genetically similar individuals or those from environments that match the transplant site [4,8–11]. Although these recommendations provide an important basis undertaking genetic rescue, there is currently little known about the relative contribution of genetic quality to restoration success. For example, non-local genotypes from large populations with high genetic diversity may provide more appropriate genetic sources for restoration than local genotypes from small, genetically depauperate populations. The potential benefits of genetic rescue may also be greater for small, inbred populations with lower genetic diversity [12,13]. Accordingly, introducing new genotypes may result in a trade-off between the benefits of counteracting inbreeding depression via heterosis, and the loss of fitness through outbreeding depression [8,11]. The dynamics of this trade-off may also change over generations, with greatest heterosis, as a consequence of maximum heterozygosity, expected in the first generation (F1). In comparison, for outbreeding depression, the dilution of locally adapted genomes may result in reduced fitness of F1 progeny, but any loss of fitness due to the break-up of favourable epistatic interactions (coadapted gene complexes) will not occur until the second (F2) and later generations [14]. Consequently, understanding the relative importance of heterosis versus outbreeding depression over multiple generations are fundamental questions that need to be addressed empirically in conservation.
Inbreeding depression and heterosis have been studied extensively since Darwin ([15], for reviews see [16,17]), whereas the importance of outbreeding depression for the fitness of hybrid progeny is poorly understood [11]. Most studies of outbreeding depression have assessed the fitness consequences of inter-population hybridization for F1 progeny and only a few have examined these issues over multiple generations [18–23]. Where outbreeding depression has been observed, it has been associated with large spatial scales [24], high genetic divergence [21] or hybridization of distinct sub-species or ecotypes [5]. Understanding correlates of heterosis and outbreeding depression can therefore provide an insight into the mechanisms underlying these processes and thus can help establish useful guidelines when considering genetic rescue.
Spatial scale, environmental variables and population characteristics may predict hybrid fitness, but their explanatory power depends on the underlying mechanisms and relative importance of hybrid vigour (heterosis) and/or hybrid breakdown (outbreeding depression). Geographical distance may correlate with outbreeding depression if genetic divergence and ecological differences among sites (environmental distance) increase with spatial scale [25]. By contrast, in heterogeneous environments with mosaic patterns of environmental variation, environmental distance or genetic divergence based on quantitative traits (QST) may more accurately predict the outcomes of inter-population hybridization than geographical distance [23,25]. Even in ecologically similar environments, populations may reach different adaptive peaks or accumulate different coadapted gene complexes [26,27], leading to hybrid breakdown in later generations following inter-population crossing. Thus, genetic differentiation may not follow an isolation-by-distance model nor correlate with environmental variation. Here, molecular genetic distance (FST) may more effectively predict hybrid fitness than geographical or environmental distance measures if it reflects differentiation in coadapted gene complexes. Characteristics of the local and foreign source population may also contribute to the trade-off between heterosis and outbreeding depression. Elevated inbreeding and the loss of genetic diversity in small populations can result in lower population fitness [28] and greater heterosis when augmenting small compared with large populations [12]. While sourcing from large populations may introduce greater genetic diversity, to our knowledge no studies have examined the importance of population size, genetic diversity or level of inbreeding in the foreign source population for the outcomes of inter-population hybridization.
Here, we examine the multi-generational implications of inter-population hybridization for plant fitness in 12 population pairs of the rare perennial plant Rutidosis leptorrhynchoides. We use controlled crosses within and between the home and foreign populations for the 12 pairs to produce control (within home population), F1, F2, F3 and backcrossed offspring and compare their fitness in an environment representative of the home population. Specifically, we ask: (i) do inter-population hybrids show heterosis or outbreeding depression? (ii) is this consistent over three generations and for backcrosses to the home and foreign populations? and (iii) can geographical distance (0.7 to approx. 600 km), environmental distance, FST, QST and characteristics of the home and foreign populations (reproductive population size, allelic diversity (AE) and inbreeding coefficient (FIS)) predict heterosis or outbreeding depression? Our results demonstrate the importance of source population characteristics for seed sourcing in genetic rescue and also provide some novel insights into the processes responsible for heterosis in fragmented plant populations.
2. Material and methods
(a). Species and population pairs
Rutidosis leptorrhynchoides F. Muell. (Asteraceae) is a multi-stemmed, herbaceous, insect-pollinated perennial with a sporophytic self-incompatibility system [29] that is endemic to highly fragmented temperate grasslands and grassy woodland communities of southeastern Australia. The 15 remaining diploid populations are distributed in two broad geographical zones; a northern zone in New South Wales and the Australian Capital Territory (less than 35°30′ S, greater than 148°30′ E), and a southern zone that extends through central Victoria (greater than 37° S, greater than 145°30′ E; see fig. 1 in [30]). The majority of R. leptorrhynchoides populations are less than 1000 plants and the largest consists of approximately 100 000 reproductive individuals. To assess the relative fitness of inter-population hybrids compared with progeny from crosses between plants from the home population, we chose 12 population pairs representing a range of spatial scales from less than 1 to 600 km (the first population is the home and the second the foreign population (distance between populations, kilometres); LW-QB (0.7), SR-CC (1.5), MA-BA (4.0), HH-MA (8.0), QB-RH (9.6), CR-LW (15.2), RH-CF (34.8), MJ-GB (71.9), GB-PO (78.9), SR-TR (506.2), CF-SA (516.0), GB-SA (575.1); see also the electronic supplementary material, table S1). Owing to the limited number of remaining R. leptorrhynchoides populations, site access restrictions and their uneven distribution in different geographical distance classes, some populations were used in multiple pairs. Population pairs were, however, selected to ensure replication at each distance class while minimizing the number of times a population was included (only GB and SR are used twice as a home population). While the use of populations in multiple pairs does raise issues associated with the assumption of independence, each pair is treated as independent and unique maternal families were used in each pair. We also correct the alpha level for the use of populations in multiple pairs (see §§3 and 4).
(b). Seed collection and crossing experiment
To produce the control (within home population progeny) and inter-population hybrids (F1, F2, F3 and backcrosses) for the 12 population pairs, we undertook a multi-generational crossing experiment from November 2002 until January 2005 (figure 1a). Parental material for each population pair was obtained by collecting seed for the home and foreign populations from one to three open-pollinated inflorescences of 30–60 maternal plants. Twelve to eighteen maternal plants were randomly chosen for each population in a pair. Four seeds from all families were then germinated on a moist filter paper in a growth cabinet maintained at 20°C with a 12 L : 12 D cycle. We then transplanted a randomly chosen seedling from each family into 20 cm 1 l capacity pot containing a soil mix of equal parts potting mix, sand and peat moss, and these were grown in a glasshouse maintained at 15–28°C. We supplemented with artificial light to ensure a year-round 14 hr photoperiod.
Figure 1.
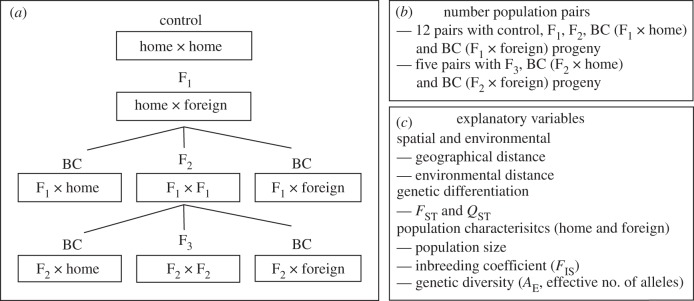
(a) The design of the crossing experiment used to generate the eight progeny types; control (home × home), F1 (home × foreign), F2 (F1 × F1), backcross (BC) (F1 × home), backcross (BC) (F1 × foreign), F3 (F2 × F2), backcross (BC) (F2 × home) and backcross (BC) (F2 × foreign). (b) The number of population pairs with each crossing treatment. (c) The predictor variables used to explain the among population pair variation in progeny fitness.
For the 12 population pairs, we produced control (home × home), F1 (home × foreign), F2 (F1 × F1) and backcrosses to the home ( F1 × home) and foreign (
F1 × home) and foreign ( F1 × foreign) populations. In addition, for five pairs (SR-CC, HH-MA, MJ-CF, GB-PO and SR-TR), F3 (F2 × F2), backcrosses to the home (
F1 × foreign) populations. In addition, for five pairs (SR-CC, HH-MA, MJ-CF, GB-PO and SR-TR), F3 (F2 × F2), backcrosses to the home (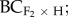 F2 × home) and foreign (
F2 × home) and foreign (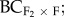 F2 × foreign) populations were produced (figure 1a,b, see also the electronic supplementary material, table S2). Each population pair contained 10–15 maternal crossing lines (crosses that originated from a single maternal family). We used a sub-sample of six maternal crossing lines for the five population pairs with F3 and backcrossed progeny.
F2 × foreign) populations were produced (figure 1a,b, see also the electronic supplementary material, table S2). Each population pair contained 10–15 maternal crossing lines (crosses that originated from a single maternal family). We used a sub-sample of six maternal crossing lines for the five population pairs with F3 and backcrossed progeny.
We used the following pollination protocol to produce the control and hybrid progeny. We randomly paired plants for all cross types in each population pair. Crosses were initiated on the day the first florets opened and were repeated three to four times, every second day. We gently brushed together a single inflorescence from each of the two randomly chosen plants to transfer pollen. The inflorescences were bagged on opening and between pollinations, and remained bagged until the seed had matured and dehisced (4 to 5 weeks). All cross-pollinations were reciprocal, where each inflorescence served as a pollen donor and recipient. We collected and counted seed for each reciprocal cross-pollination. Some crosses were unsuccessful owing to matching of self-incompatibility alleles (S-alleles) [31]. For these crosses, plants were randomly re-paired and the cross repeated. To produce each subsequent generation (F2, F3 and backcrosses), we randomly chose two seed from each side of the reciprocal cross for the 10–15 maternal lines in each population pair and germinated them using the same protocols as the parental generation. We then randomly chose one of these plants at reproductive maturity (approx. 12–16 weeks). The number of reciprocal crosses within each population pair ranged from 150 to 240, so that across all population pairs this experiment involved 2455 cross-pollinations.
(c). Fitness experiment
To examine the consequences of inter-population hybridization for progeny fitness, we compared their fitness with control (within home population) progeny in an environment representative of the home population (see below). From multivariate analysis of bioclimatic and soil (edaphic) variables, we found that populations of R. leptorrhynchoides divide into two climatic zones (north and south), but within each climate zone, edaphic variation was the primary driver of environmental heterogeneity [30]. Consequently, our experimental approach involved growing progeny from each population pair in soil collected from the home population and in an outdoor enclosure at CSIRO Plant Industry in Canberra, Australian Capital Territory (35°16′23″ S, 149°06′42″ E) so that plants experienced daily and seasonal climatic conditions (variation in temperature and moisture) that were representative of the home site (northern climate zone). The soil (100–312 l) was collected from multiple locations within each home population. The overall climate at this location is representative of the local population for all population pairs assessed because all local populations were from the northern climate zone [30]. Consequently, differences in soil were the main driver of environmental differences among the nine population pairs where the home and foreign populations were from the northern climate zone. For the three pairs where the foreign population was from the southern climate zone, both soil and climate differences contributed to environmental differences among sites. Previous studies of this species [30] found a correlation between QST and environmental distance based on climate and soils. This suggests that that these ecological variables are related to the environmental selective pressures in each population. Consequently, although we were unable to assess the importance of other biotic (e.g. competition, pathogens) or abiotic factors, our experimental framework enabled us to compare progeny performance in an environment that captured the local soil and climatic conditions. The soil samples from each population were mixed and used in an 80 : 20 ratio with river sand (to improve drainage) in 10 cm diameter 0.5 l capacity pots.
We used 12 maternal lines for each population pair (except CF-SA, n = 10). For each maternal line and cross type, we randomly selected one side of the reciprocal cross and counted and bulk weighted 12 seed. Seeds were then cold treated in a refrigerator set at approximately 5°C for 72 h. We planted one to three seeds into each of four pots for each cross type and maternal line in April–May 2005. For each population pair, n = 200–312 pots and for all population pairs n = 3200 pots and 9397 planted seed. We grouped together a single pot of each cross type for each maternal line, and each of the four replicated sets were positioned in a complete randomized block design with four blocks. The position of the replicate in each block was arranged using a randomized row and column structure. We supplemented natural precipitation with hand-watering every 1–2 days as required.
(d). Fitness components
To compare the relative performance of inter-population hybrids and progeny from the home population, we measured seedling survival as well as two adult fitness components (at 12 months); number of inflorescences (reproduction) and biomass. We recorded survival weekly for the first 3 months. For pots where multiple seedlings had germinated, all seedlings except the one closest to the geometric centre of the pot were removed before competition ensued. At 12 months, we harvested and obtained dried weights for above- and below-ground biomass by drying samples at 70°C for 3 days before weighing them on a four decimal place gram balance. The root mass was washed prior to drying. Together, these provided an estimate of total biomass (hereafter biomass). Previous demographic work for R. leptorrhynchoides [32] found that seedling and adult survivorship, as well as adult reproductive characteristics had the highest elasticity values and therefore have a high contribution to population growth rate. Survival from seedling to adult was much higher in our experiment (greater than 95%) compared with observations of plants in the field. Thus, in our experiment, we use biomass as a surrogate for adult survival, as plant size has been shown to be associated with survival in natural populations (A. G. Young 2000, unpublished data).
3. Statistical analysis
(a). The effect of crossing treatment on fitness components
To examine the effect of cross type (control, F1, F2, F3 and backcrosses) and population pair (and their interaction) on progeny fitness, we used; (i) a generalized linear model (logistic regression) for seedling survival (pooled across families), (ii) generalized linear mixed models for number of inflorescences (normal distribution, log link function) and (iii) REML linear mixed models for biomass. For all analyses, seed weight was fitted as a covariate, but where it was non-significant was removed from the final model. For all mixed models, cross type and population pair (and their interaction) were fitted as the main effect in the fixed model, while maternal line (nested within population pair), row and column position (in each block) and block were fitted in the random model. Differences between cross types were assessed using least significant difference (LSD; at α = 0.05).
(b). Linear regression analysis
We analysed the relation between the difference in fitness between control (within home population progeny) and hybrid progeny (F1, F2, F3 and backcrosses) and variables: (i) log home reproductive population size (logHPS), (ii) log foreign reproductive population size (logFPS), (iii) effective number of alleles in the foreign population (AEF), (iv) inbreeding coefficient of the home (FISH) and foreign (FISF) populations, (v) log geographical distance (logGD), (iv) environmental distance (EnvD), (v) QST, and (vi) FST (figure 1c), for each fitness component using multiple (and single) linear regressions. Details on the derivation of each of these explanatory variables can be found in [30], while the values for each population pair are listed in the electronic supplementary material, table S1. We used stepwise ANOVA for model selection to identify the single variable, or combinations of variables that best explained the difference in fitness between control and hybrid progeny. We then analysed identified variables as simple or multiple linear regressions. A maximum of two explanatory variables were used in the multiple regression models for the F1, F2 and backcrosses (n = 12) and a single variable used for the F3 and backcrosses (n = 5). Regression analysis of biomass for the F1, F2 and backcrosses to the foreign population (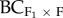 ) identified MJ-GB as a high leverage data point. Accordingly, we present the results of the analyses with and without this population pair. Significance was tested at α = 0.05; however where 0.05 < p < 0.1 we report results as marginally significant. To account for the non-independence of population pairs that shared a home population (i.e. SR-CC, SR-TR and GB-PO, GB-SA), we use a Bonferonni correction (α = 0.05/2) and test significance of these relations at α = 0.025. We used Genstat 13th edn (VSN International, Oxford, UK) for all analyses.
) identified MJ-GB as a high leverage data point. Accordingly, we present the results of the analyses with and without this population pair. Significance was tested at α = 0.05; however where 0.05 < p < 0.1 we report results as marginally significant. To account for the non-independence of population pairs that shared a home population (i.e. SR-CC, SR-TR and GB-PO, GB-SA), we use a Bonferonni correction (α = 0.05/2) and test significance of these relations at α = 0.025. We used Genstat 13th edn (VSN International, Oxford, UK) for all analyses.
4. Results
(a). Fitness of inter-population hybrids compared with progeny from the home population
The fitness of progeny from inter-population crosses over multiple generations (F1–F3 and backcrosses) was either no different or increased (heterosis) compared with progeny from the home population for the majority of population pairs and fitness components. Including all population pairs, significant heterosis was observed for seedling survival (p = 0.002), whereas inter-population hybrids had similar fitness to progeny from the home population for biomass (p = 0.058) and reproduction (number of inflorescences, p = 0.076; table 1). There was, however, variation in heterosis among population pairs for all fitness components. Here, we examine this variation among population pairs in relation to a range of distance matrices and characteristics of the home and foreign source populations (see [30]; figure 1c and electronic supplementary material, table S1). We found that the level of inbreeding (FIS), genetic diversity (AE) and size of the source and home population were the most consistent predictors of heterosis across a range of fitness components. Heterosis was greater when the foreign population was large and genetically diverse with low levels of inbreeding and the home population small or inbred. We now examine these results in more detail for three fitness components identified as high elasticity traits, seedling survival, biomass and reproduction (number of inflorescences).
Table 1.
Statistical analyses and per cent difference between the control (within home population cross) and each crossing treatment (treatment - control, as % of control) for a number of fitness components of R. leptorrhynchoides including all 12 population pairs. (Positive (or negative) values represent the per cent increase (or decrease) in the crossing treatment (F1, F2, F3 and backcrosses) compared with the control. Positive (or negative) values in bold indicate significant heterosis (or outbreeding depression; LSD at α = 0.05). H is the home (local) population and F is the foreign (source) population.)
| crossing treatment |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| fitness component | F | p-value | F1 (H × F) | BC (F1 × F) | F2 (F1 × F1) | BC (F1 × H) | BC (F2 × F) | F3 (F2 × F2) | BC (F2 × H) |
| seedling survival | |||||||||
| cross type | 3.26 | 0.002 | 1.0 | 0.3 | 0.5 | −0.2 | 1.5 | 2.7 | 3.9 |
| population pair | 4.48 | <0.001 | |||||||
| cross type × population pair | 1.24 | 0.120 | |||||||
| biomass | |||||||||
| cross type | 1.95 | 0.058 | 2.1 | −0.7 | 4.4 | 2.1 | −1.0 | 0.3 | 1.0 |
| population pair | 99.60 | <0.001 | |||||||
| cross type × population pair | 0.89 | 0.698 | |||||||
| number inflorescences | |||||||||
| cross type | 1.84 | 0.076 | 8.4 | 11.9 | 11.7 | 6.1 | −5.0 | 3.0 | −0.2 |
| population pair | 31.71 | <0.001 | |||||||
| cross type × population pair | 2.18 | <0.001 | |||||||
(b). Seedling survival
We found that the population characteristics and distance matrices only explained the among population pair variance in seedling survival for F2 progeny. Here, the inbreeding coefficient of the foreign population (FISF) and environmental distance explained 39 per cent of the variance in seedling survival (p = 0.043, see the electronic supplementary material, figure S1), with greater heterosis in the F2 when the foreign source population had low levels of inbreeding (low FIS) and populations came from similar environments (smaller environmental distance).
(c). Biomass
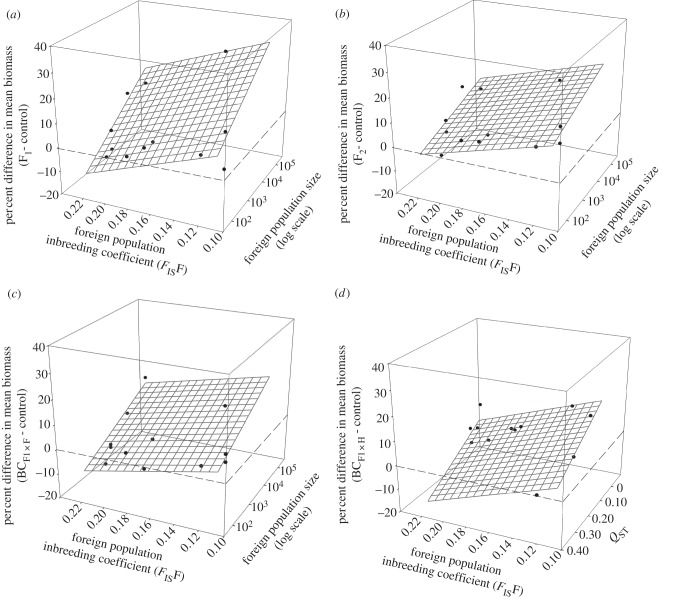
We found that the inbreeding coefficient (FISF) and reproductive size of the foreign source population explained 32–69% of the variance in biomass of the F1 (figure 2a; R2 = 0.32, p = 0.071, without MJ-GB R2 = 0.46, p = 0.034), F2 (figure 2b; R2 = 0.36, p = 0.053, without MJ-GB R2 = 0.55, p = 0.017) and backcross progeny (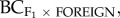 figure 2c; R2 = 0.69, p = 0.002, without MJ-GB R2 = 0.59, p = 0.012), with greater heterosis when the source population was large with low levels of inbreeding. For backcrosses to the home population (
figure 2c; R2 = 0.69, p = 0.002, without MJ-GB R2 = 0.59, p = 0.012), with greater heterosis when the source population was large with low levels of inbreeding. For backcrosses to the home population (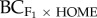 ), the inbreeding coefficient (FISF) of the source population and QST explained 60 per cent of the variance in progeny fitness (figure 2d; R2 = 0.60, p = 0.006). Here, heterosis was greater in population pairs where the source population had lower levels of inbreeding (low FISF) and where quantitative genetic divergence (QST) was low. For the F3 generation, FIS of the source population (FISF) explained 60 per cent of the variance in progeny fitness (p = 0.077; table 2), with greater heterosis when the source population had low levels of inbreeding. By contrast, for backcrosses to the home population (
), the inbreeding coefficient (FISF) of the source population and QST explained 60 per cent of the variance in progeny fitness (figure 2d; R2 = 0.60, p = 0.006). Here, heterosis was greater in population pairs where the source population had lower levels of inbreeding (low FISF) and where quantitative genetic divergence (QST) was low. For the F3 generation, FIS of the source population (FISF) explained 60 per cent of the variance in progeny fitness (p = 0.077; table 2), with greater heterosis when the source population had low levels of inbreeding. By contrast, for backcrosses to the home population (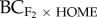 ), heterosis for biomass was greater when the home population was inbred (table 2; R2 = 0.74, p = 0.038).
), heterosis for biomass was greater when the home population was inbred (table 2; R2 = 0.74, p = 0.038).
Figure 2.
The difference in mean biomass between the control (within home population progeny) and (a) F1, (b), F2, (c) backcrosses to the foreign population (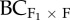 ) and (d) backcrosses to the home population (
) and (d) backcrosses to the home population (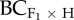 ), as a function of the inbreeding coefficient of the foreign population (FISF) and foreign population size (FPS) (log scale) for (a), (b) and (c) and quantitative genetic divergence (QST) for (d). Values above the dashed horizontal (zero) line represent heterosis and below represent outbreeding depression. The equations for these relations are: (a) R2 = 0.32, p = 0.071, biomass = 3.7–141.6 × FISF + 8.2 × LogFPS, (without MJ-GB: R2 = 0.46, p = 0.034), (b) R2 = 0.36, p = 0.053, biomass = 14.9–130.5 × FISF + 4.4 × LogFPS (without MJ-GB: R2 = 0.55, p = 0.017), (c) R2 = 0.69, p = 0.002, biomass = 1.1–107.4 × FISF + 5.9 × LogFPS (without MJ-GB: R2 = 0.59, p = 0.012), (d) R2 = 0.60, p = 0.006, biomass = 46.9–219.1 × FISF –80.0 × QST.
), as a function of the inbreeding coefficient of the foreign population (FISF) and foreign population size (FPS) (log scale) for (a), (b) and (c) and quantitative genetic divergence (QST) for (d). Values above the dashed horizontal (zero) line represent heterosis and below represent outbreeding depression. The equations for these relations are: (a) R2 = 0.32, p = 0.071, biomass = 3.7–141.6 × FISF + 8.2 × LogFPS, (without MJ-GB: R2 = 0.46, p = 0.034), (b) R2 = 0.36, p = 0.053, biomass = 14.9–130.5 × FISF + 4.4 × LogFPS (without MJ-GB: R2 = 0.55, p = 0.017), (c) R2 = 0.69, p = 0.002, biomass = 1.1–107.4 × FISF + 5.9 × LogFPS (without MJ-GB: R2 = 0.59, p = 0.012), (d) R2 = 0.60, p = 0.006, biomass = 46.9–219.1 × FISF –80.0 × QST.
Table 2.
Results of the linear regression analysis examining the effect of the inbreeding coefficient of the home (FISH) and foreign (FISF) populations on the difference in biomass for F3 and backcrossed progeny compared to the control (within home population cross) for five population pairs of R. leptorrhynchoides.
| fitness component | cross type | R2 | p-value | explanatory variable | slope | model equation |
|---|---|---|---|---|---|---|
| biomass | F3 (F2 × F2) | 0.60 | 0.077 | FISF | − | y = 16.65 − 99.6 × FISF |
| BC (F2 × home) | 0.74 | 0.038 | FISH | + | y = −44.4 + 252.9 × FISH |
(d). Reproduction
We found that the genetic diversity of the foreign source population (measured by the effective number of alleles, AE) was the most consistent predictor of heterosis for the number of inflorescences in the F1, F2 and backcross generations. For the F1 (figure 3a; R2 = 0.36, p = 0.023) and F2 (figure 3b; R2 = 0.43, p = 0.012), the effective number of alleles (AE) of the source population explained 36–43% of the variance in fitness, with greater heterosis in population pairs where the source population was more diverse. Similarly, the effective number of alleles in the source population explained 33 per cent of the variance in the number of inflorescences for backcross progeny to the foreign population in the second generation (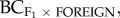 R2 = 0.33, p = 0.029; figure 3c). However, for backcrossed progeny to the home population (
R2 = 0.33, p = 0.029; figure 3c). However, for backcrossed progeny to the home population (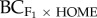 ), heterosis was greater when environmental distance was large and FST small (R2 = 0.58, p = 0.008, electronic supplementary material, figure S2).
), heterosis was greater when environmental distance was large and FST small (R2 = 0.58, p = 0.008, electronic supplementary material, figure S2).
Figure 3.
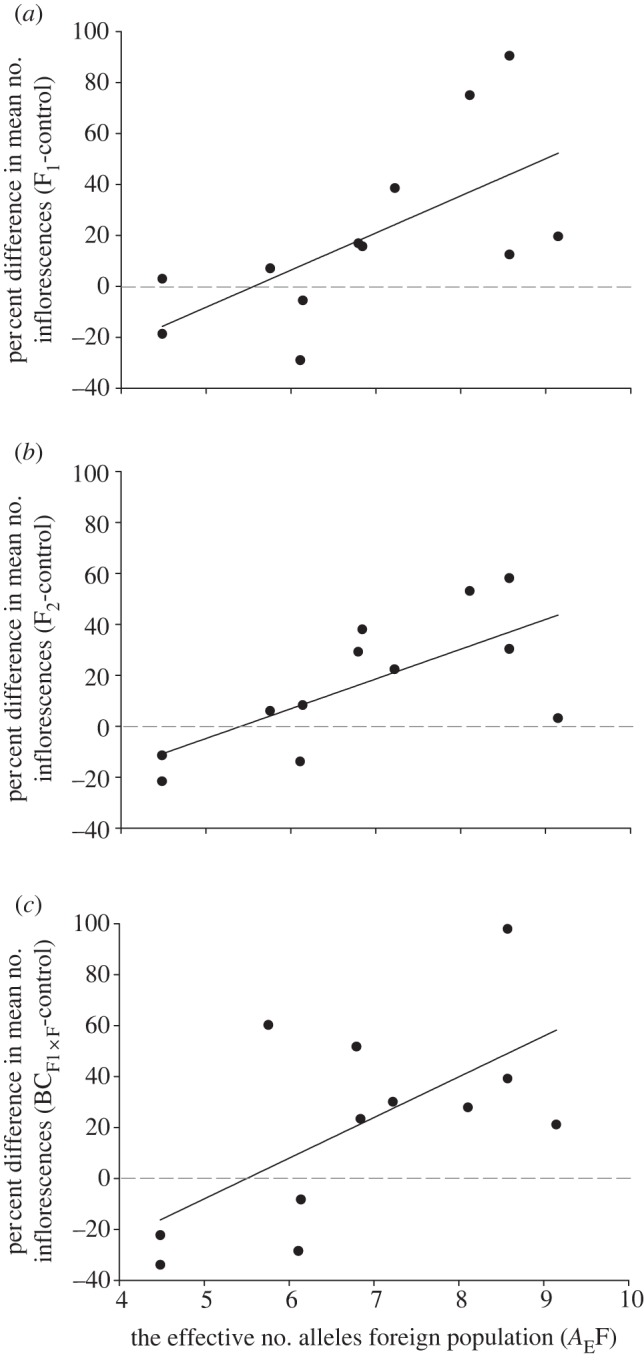
The difference in the mean number of inflorescences (no. inflor.) between the control (within home population progeny) and (a) F1, (b), F2 and (c) backcrosses to the foreign population (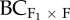 ) as a function of the effective number of alleles in the foreign population (AEF). Values above the dashed horizontal (zero) line represent heterosis and below represent outbreeding depression. The equations for these relations are: (a) R2 = 0.36, p = 0.023, no. inflor. = –81.0 + 14.6 × AEF, (b) R2 = 0.43, p = 0.012, no. inflor. = –63.2 + 11.7 × AEF and (c) R2 = 0.33, p = 0.029, no. inflor. = –87.9 + 16.0 × AEF.
) as a function of the effective number of alleles in the foreign population (AEF). Values above the dashed horizontal (zero) line represent heterosis and below represent outbreeding depression. The equations for these relations are: (a) R2 = 0.36, p = 0.023, no. inflor. = –81.0 + 14.6 × AEF, (b) R2 = 0.43, p = 0.012, no. inflor. = –63.2 + 11.7 × AEF and (c) R2 = 0.33, p = 0.029, no. inflor. = –87.9 + 16.0 × AEF.
5. Discussion
Our study demonstrates the importance of source population characteristics for heterosis following hybridization among fragmented populations of a rare perennial plant. For a range of fitness components and across multiple generations, hybrid progeny fitness was greatest when the foreign source population was large, with high allelic diversity and low levels of inbreeding (low FIS). These results have important implications for understanding potential trade-offs between heterosis and outbreeding depression following genetic rescue. Currently, guidelines regarding the movement of genotypes for population augmentation or ecological restoration advocate the use of local or genetically similar genotypes owing to the risk of outbreeding depression [8]. Our results suggest that for small, inbred populations inter-population crossing results in heterosis, rather than outbreeding depression and that progeny fitness is maximized by sourcing genetic material from large, outbred populations with high genetic diversity. The maintenance of heterosis beyond the first generation also suggests that these effects may be beneficial for population growth rate for several generations. Compared with characteristics of the source population, geographical distance and environmental distance had little power to predict hybrid progeny fitness. Our results highlight the central role of small population processes in the trade-off between heterosis and outbreeding depression following inter-population hybridization.
Genetic drift and inbreeding in small populations result in reduced population fitness owing to loss of genetic diversity and an increase in the expression of mildly deleterious alleles [12,33]. Inter-population hybridization can counter this loss of fitness through either the masking of deleterious recessive alleles or overdominance (heterozygote advantage) [34,35]. For R. leptorrhynchoides, inbreeding occurs via mating among close relatives (bi-parental inbreeding) and the loss of mating types (S-alleles) in small populations can further reduce effective population size [36]. As expected from theory [37] and recent studies of inbreeding and outbreeding depression [23,38,39], small, inbred populations of R. leptorrhynchoides experienced greater heterosis than large, outbred populations. The novel finding of our study is the importance of these processes in the foreign source population for hybrid progeny fitness. The increase in heterosis when genotypes were sourced from large, genetically diverse populations may result from a number of processes. First, the increased efficiency of selection in large populations can result in a lower frequency of deleterious recessive alleles, leading to greater heterosis via dominance. While there is less evidence for overdominance [35], this may also contribute to progeny fitness when genotypes are sourced from larger populations with high genetic diversity. Sourcing from populations with higher diversity may provide more variation for the evolution of novel, highly fit genotypes through recombination in the F2 and subsequent generations [18]. However, this is more likely to effect the variance in fitness rather than the mean. Given the low number of genetic clusters in this species (K = 3, see [30] for Structure analysis), heterosis may be reduced when matings occur between populations belonging to the same genetic cluster. However, we found that heterosis was not limited to pairs where the populations had very different average membership to the three genetic clusters (see the electronic supplementary material, figure S3). This suggests that high genetic diversity or low inbreeding in the source population are the more likely drivers of the observed heterosis, rather than genetic dissimilarity of the home and source populations. Our results are intriguing and although the exact mechanism(s) underlying these patterns are at this stage unclear, they indicate the importance of considering source population characteristics for genetic rescue.
In combination, population size, genetic diversity and the level of inbreeding of the source population consistently explained variation in hybrid performance across a range of fitness components. Population size is often correlated with genetic diversity and fitness [28,40], but not necessarily the level of inbreeding (FIS) [40]. Similarly, we found that population size was related to genetic diversity (AE; R2 = 0.28, p = 0.025) but not the inbreeding coefficient (FIS; p > 0.05) for R. leptorrhynchoides (see the electronic supplementary material, appendix S1). High fine-scale spatial genetic structure in this species [41] may contribute to higher rates of inbreeding in larger populations, so that these two population characteristics explain different components of heterosis. Taken together, these results suggest that population size may not always be an effective surrogate for source population quality and that additional information on inbreeding and the level of genetic diversity can assist in the choice of the most appropriate source populations for restoration.
Geographical distance had very little power to predict hybrid progeny fitness for any of the fitness components or generations. Spatial scale is often used to delineate seed sourcing zones in conservation with emphasis on the use of local genotypes. Along with previous studies where geographical distance had limited value for predicting hybrid progeny fitness [5,24,42], our results suggest that other criteria such as the size and diversity of the source populations are more important considerations for genetic rescue. The importance of environmental distance for progeny fitness in R. leptorrhynchoides was generally low, but varied among traits and generations. Consequently, along with characteristics of the source population, ecological differences among sites remain an important consideration when selecting genetic sources for restoration [5,42]. Previous work on adaptive differentiation in R. leptorrhynchoides found that local adaptation was generally low, except for some local genotype advantage for biomass when QST was high [30]. Here, along with FIS of the source population, we found low QST was associated with greater heterosis for biomass in backcrosses to the home population. This suggests that for traits where local adaptation is related to quantitative genetic divergence, heterosis can be maximized by sourcing from outbred populations that are less phenotypically divergent.
While the importance of genetic divergence for hybrid vigour is well established for crop breeding [43], our results challenge the current paradigm of avoiding the movement of genotypes among divergent natural plant populations [8]. Similar to Willi et al. [23], we found that FST and QST had little power to predict either heterosis or outbreeding depression, and that there was no negative effect of gene flow among populations with FST values of 0.03–0.11 and QST values 0.01–0.33. For R. leptorrhynchoides, these two matrices were only marginally correlated and (r = 0.44, p = 0.064), indicating that they reflect different measures of population differentiation. The lack of association between hybrid progeny fitness and FST over a number of generations suggests that this measure of genetic distance does not reflect patterns of genetic coadaptation. We found that QST was not related to any loss of fitness through the dilution of local genes in the F1 or backcrosses to the foreign population (where progeny had a reduced average proportion of local genes), even though QST was associated with ecological differences among populations (QST and environmental distance, r = 0.64, p = 0.006) and local adaptation for some traits [30]. Gene flow among moderately divergent populations of this species is therefore likely to have either no effect or result in hybrid vigour, especially for inbred populations and when the genetic and demographic characteristics of the source populations are considered.
Genetic rescue and the choice of appropriate source populations is a major challenge in conservation and restoration [7,8]. Our results demonstrate that, instead of detrimental effects, mixing genetic material from divergent populations resulted in hybrid vigour when genotypes are sourced from large populations with high genetic diversity and low levels of inbreeding, and that this fitness benefit is maintained over generations. This highlights the importance of genetic and demographic characteristics of both the recipient and source population when considering artificial gene flow among populations. In line with both theory [37,44] and empirical studies [18,23], our study supports the potential role of genetic rescue as a means of increasing the probability of local population persistence. This may be especially important for species where there is a frequency-dependent selection for rare alleles (e.g. S-alleles). Consequently, rather than a trade-off between inbreeding and outbreeding, genetic rescue may provide a twofold fitness benefit for self-incompatible species. The first benefit is owing to increased fertilization success [31,39] and, the second is attributable to the multi-generational fitness benefits of heterosis, particularly when immigrants are sourced from large, outbred and genetically diverse populations.
Acknowledgements
We thank Graham Pickup for technical assistance. The New South Wales Department of Environment and Climate Change, ACT Parks, Conservation and Lands and the Department of Sustainability and Environment in Victoria provided permits for seed and soil collection. We thank Spencer C. H. Barrett and two anonymous reviewers for comments that improved the quality of the manuscript. This research was by supported by an Australian Research Council PhD stipend and CSIRO Plant Industry ‘top-up’ scholarship to M.P.
References
- 1.Richards CM. 2000. Inbreeding depression and genetic rescue in a plant metapopulation. Am. Nat. 155, 383–394 10.1086/303324 (doi:10.1086/303324) [DOI] [PubMed] [Google Scholar]
- 2.Ingvarsson PK. 2001. Restoration of genetic variation lost: the genetic rescue hypothesis. Trends Ecol. Evol. 16, 62–63 10.1016/S0169-5347(00)02065-6 (doi:10.1016/S0169-5347(00)02065-6) [DOI] [PubMed] [Google Scholar]
- 3.Fenster CB, Dudash MR. 1994. Genetic considerations for plant population restoration and conservation. In Restoration of endangered species: conceptual issues, planning and implementation (eds Bowles ML, Whelan CJ.), pp. 34–62 Cambridge, UK: Cambridge University Press [Google Scholar]
- 4.Hufford KM, Mazer SJ. 2003. Plant ecotypes: genetic differentiation in the age of ecological restoration. Trends Ecol. Evol. 18, 147–155 10.1016/S0169-5347(03)00002-8 (doi:10.1016/S0169-5347(03)00002-8) [DOI] [Google Scholar]
- 5.Montalvo AM, Ellstrand NC. 2001. Nonlocal transplantation and outbreeding depression in the subshrub Lotus scoparius (Fabaceae). Am. J. Bot. 88, 258–269 10.2307/2657017 (doi:10.2307/2657017) [DOI] [PubMed] [Google Scholar]
- 6.Templeton AR. 1986. Coadaptation and outbreeding depression. In Conservation biology: the science of scarcity and diversity (ed. Soule ME.), pp. 105–116 Sunderland, MA: Sinauer [Google Scholar]
- 7.Tallmon DA, Luikart G, Waples RS. 2004. The alluring simplicity and complex reality of genetic rescue. Trends Ecol. Evol. 19, 489–496 10.1016/j.tree.2004.07.003 (doi:10.1016/j.tree.2004.07.003) [DOI] [PubMed] [Google Scholar]
- 8.Edmands S. 2007. Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol. Ecol. 16, 463–475 10.1111/j.1365-294X.2006.03148.x (doi:10.1111/j.1365-294X.2006.03148.x) [DOI] [PubMed] [Google Scholar]
- 9.Lesica P, Allendorf FW. 1999. Ecological genetics and the restoration of plant communities: mix or match? Restor. Ecol. 7, 42–50 10.1046/j.1526-100X.1999.07105.x (doi:10.1046/j.1526-100X.1999.07105.x) [DOI] [Google Scholar]
- 10.Knapp EE, Rice KJ. 1994. Starting from seed: genetic issues in using native grasses for restoration. Ecol. Restor. 12, 40–45 [Google Scholar]
- 11.Dudash MR, Fenster CB. 2000. Inbreeding and outbreeding depression in fragmented populations. In Genetics, demography and viability of fragmented populations (eds Young AG, Clarke GM.), pp. 35–53 Cambridge, UK: Cambridge University Press [Google Scholar]
- 12.Ellstrand NC, Elam DR. 1993. Population genetic consequences of small population size: implications for plant conservation. Annu. Rev. Ecol. Syst. 24, 217–242 10.1146/annurev.es.24.110193.001245 (doi:10.1146/annurev.es.24.110193.001245) [DOI] [Google Scholar]
- 13.Hedrick PW, Kalinowski ST. 2000. Inbreeding depression in conservation biology. Annu. Rev. Ecol. Syst. 31, 139–162 10.1146/annurev.ecolsys.31.1.139 (doi:10.1146/annurev.ecolsys.31.1.139) [DOI] [Google Scholar]
- 14.Lynch M. 1991. The genetic interpretation of inbreeding depression and outbreeding depression. Evolution 45, 622–629 10.2307/2409915 (doi:10.2307/2409915) [DOI] [PubMed] [Google Scholar]
- 15.Darwin C. 1876. The effects of cross and self-fertilization in the vegetable kingdom. London, UK: Murray [Google Scholar]
- 16.Keller LF, Waller DM. 2002. Inbreeding effects in wild populations. Trends Ecol. Evol. 17, 230–241 10.1016/S0169-5347(02)02489-8 (doi:10.1016/S0169-5347(02)02489-8) [DOI] [Google Scholar]
- 17.Charlesworth D, Charlesworth B. 1987. Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Syst. 18, 237–268 10.1146/annurev.es.18.110187.001321 (doi:10.1146/annurev.es.18.110187.001321) [DOI] [Google Scholar]
- 18.Erickson DL, Fenster CB. 2006. Intraspecific hybridization and the recovery of fitness in the native legume Chamaecrista fasciculata. Evolution 60, 225–233 [PubMed] [Google Scholar]
- 19.Fenster CB, Galloway LF. 2000. Inbreeding and outbreeding depression in natural populations of Chamaecrista fasciculata (Fabaceae). Conserv. Biol. 14, 1406–1412 10.1046/j.1523-1739.2000.99234.x (doi:10.1046/j.1523-1739.2000.99234.x) [DOI] [Google Scholar]
- 20.Keller M, Kollmann J, Edwards PJ. 2000. Genetic introgression from distant provenances reduces fitness in local weed populations. J. Appl. Ecol. 37, 647–659 10.1046/j.1365-2664.2000.00517.x (doi:10.1046/j.1365-2664.2000.00517.x) [DOI] [Google Scholar]
- 21.Edmands S. 1999. Heterosis and outbreeding depression in interpopulation crosses spanning a wide range of divergence. Evolution 53, 1757–1768 10.2307/2640438 (doi:10.2307/2640438) [DOI] [PubMed] [Google Scholar]
- 22.Gharrett AJ, Smoker WW. 1991. Two generations of hybrids between even- and odd-year pink salmon (Oncorhynchus gorbuscha): a test for outbreeding depression? Can. J. Fish Aquat. Sci. 48, 1744–1749 10.1139/f91-206 (doi:10.1139/f91-206) [DOI] [Google Scholar]
- 23.Willi Y, Kleunen MV, Dietrich S, Fischer M. 2007. Genetic rescue persists beyond first-generation outbreeding in small populations of a rare plant. Proc. R. Soc. B 274, 2357–2364 10.1098/rspb.2007.0768 (doi:10.1098/rspb.2007.0768) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fenster CB, Galloway LF. 2000. Population differentiation in an annual legume: genetic architecture. Evolution 54, 1157–1172 [DOI] [PubMed] [Google Scholar]
- 25.Edmands S. 2002. Does parental divergence predict reproductive compatibility? Trends Ecol. Evol. 17, 520–527 10.1016/S0169-5347(02)02585-5 (doi:10.1016/S0169-5347(02)02585-5) [DOI] [Google Scholar]
- 26.Wade MJ, Goodnight CJ. 1998. Perspective: the theories of Fisher and Wright in the context of metapopulations: when nature does many small experiments. Evolution 52, 1537–1553 10.2307/2411328 (doi:10.2307/2411328) [DOI] [PubMed] [Google Scholar]
- 27.Wright S. 1931. Evolution in Mendelian populations. Genetics 16, 97–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reed DH, Frankham R. 2003. Correlation between fitness and genetic diversity. Conserv. Biol. 17, 230–237 10.1046/j.1523-1739.2003.01236.x (doi:10.1046/j.1523-1739.2003.01236.x) [DOI] [Google Scholar]
- 29.Young AG, Miller C, Gregory E, Langston A. 2000. Sporophytic self-incompatibility in diploid and tetraploid races of Rutidosis leptorrhynchoides (Asteraceae). Aust. J. Bot. 48, 667–672 10.1071/BT99024 (doi:10.1071/BT99024) [DOI] [Google Scholar]
- 30.Pickup M, Field DL, Rowell DM, Young AG. 2012. Predicting local adaptation in fragmented plant populations: implications for restoration genetics. Evol. Appl. (doi:10.1111/j.1752-4571.2012.00284.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pickup M, Young AG. 2008. Population size, self-incompatibility and genetic rescue in diploid and tetraploid races of Rutidosis leptorrhynchoides (Asteraceae). Heredity 100, 268–274 10.1038/sj.hdy.6801070 (doi:10.1038/sj.hdy.6801070) [DOI] [PubMed] [Google Scholar]
- 32.Young AG, Brown AHD, Murray BG, Thrall PH, Miller CH. 2000. Genetic erosion, restricted mating and reduced viability in fragmented populations of the endangered grassland herb Rutidosis leptorrhynchoides. In Genetics, demography and viability of fragmented populations (eds Young AG, Clarke GM.), pp. 335–359 Cambridge, UK: Cambridge University Press [Google Scholar]
- 33.Young A, Boyle T, Brown T. 1996. The population genetic consequences of habitat fragmentation for plants. Trends Ecol. Evol. 11, 413–418 10.1016/0169-5347(96)10045-8 (doi:10.1016/0169-5347(96)10045-8) [DOI] [PubMed] [Google Scholar]
- 34.Lynch M, Walsh B. 1998. Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer Association [Google Scholar]
- 35.Charlesworth B, Charlesworth D. 1999. The genetic basis of inbreeding depression. Genet. Res. 74, 329–340 10.1017/S0016672399004152 (doi:10.1017/S0016672399004152) [DOI] [PubMed] [Google Scholar]
- 36.Young AG, Pickup M. 2010. Low S-allele numbers limit mate availability, reduce seed set and skew fitness in small populations of a self-incompatible plant. J. Appl. Ecol. 47, 541–548 10.1111/j.1365-2664.2010.01798.x (doi:10.1111/j.1365-2664.2010.01798.x) [DOI] [Google Scholar]
- 37.Ingvarsson PK, Whitlock MC. 2000. Heterosis increases the effective migration rate. Proc. R. Soc. Lond. B 267, 1321–1326 10.1098/rspb.2000.1145 (doi:10.1098/rspb.2000.1145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Escobar JS, Nicot A, David P. 2008. The different sources of variation in inbreeding depression, heterosis and outbreeding depression in a metapopulation of Physa acuta. Genetics 180, 1593–1608 10.1534/genetics.108.092718 (doi:10.1534/genetics.108.092718) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willi Y, Fischer M. 2005. Genetic rescue in interconnected populations of small and large size of the self-incompatible Ranunculus reptans. Heredity 95, 437–443 10.1038/sj.hdy.6800732 (doi:10.1038/sj.hdy.6800732) [DOI] [PubMed] [Google Scholar]
- 40.Leimu R, Mutikainen P, Koricheva J, Fischer M. 2006. How general are positive relationships between plant population size, fitness and genetic variation? J. Ecol. 94, 942–952 10.1111/j.1365-2745.2006.01150.x (doi:10.1111/j.1365-2745.2006.01150.x) [DOI] [Google Scholar]
- 41.Wells GP, Young AG. 2002. Effects of seed dispersal on spatial genetic structure in populations of Rutidosis leptorrhynchoides with different levels of correlated paternity. Genet. Res. 79, 219–226 10.1017/s0016672302005591 (doi:10.1017/s0016672302005591) [DOI] [PubMed] [Google Scholar]
- 42.Raabová J, Münzbergová Z, Fischer M. 2007. Ecological rather than geographic or genetic distance affects local adaptation of the rare perennial herb, Aster amellus. Biol. Conserv. 139, 348–357 10.1016/j.biocon.2007.07.007 (doi:10.1016/j.biocon.2007.07.007) [DOI] [Google Scholar]
- 43.Moll RH, Lonnquist JH, Fortuno JV, Johnson EC. 1965. The relationship of heterosis and genetic divergence in maize. Genetics 52, 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitlock MC, Ingvarsson PK, Hatfield T. 2000. Local drift load and the heterosis of interconnected populations. Heredity 84, 452–457 10.1046/j.1365-2540.2000.00693.x (doi:10.1046/j.1365-2540.2000.00693.x) [DOI] [PubMed] [Google Scholar]