Abstract
Background:
Methicillin-resistant Staphylococcus aureus (MRSA) is a major nosocomial pathogen worldwide, which has emerged over the past 30 years as a leading cause of both nosocomial and community-acquired infections. Accurate and rapid identification of MRSA in clinical specimens is essential for timely decision on effective antimicrobial chemotherapy.
Aim:
The present study was conducted to compare two conventional phenotypic methods, oxacillin disk diffusion (ODD) method and mannitol salt agar (MSA) with oxacillin, with polymerase chain reaction (PCR) for mecA gene (as standard).
Materials and Methods:
A total of 165 consecutive clinical isolates of S. aureus received at the Department of Microbiology in our tertiary care teaching hospital were included in the study. All the isolates were subjected to ODD (1 μg) method, culture in MSA with oxacillin, and PCR for mecA gene.
Results:
The sensitivity and specificity of ODD test were found to be 93.5% (86.4-97.3%) and 83.5% (79.2-85.8%), respectively, and that of MSA with oxacillin were found to be 87.1% (79.5-92.3%) and 89.3% (84.8-92.5%), respectively. The time taken for diagnosing MRSA by conventional methods is 48-72 h, which is more as compared to PCR which takes 18-24 h.
Conclusion:
This study recommends advocating PCR for mecA gene on a regular basis for detecting methicillin resistance in S. aureus isolates isolated from sterile body fluids or from special units such as intensive care units.
Keywords: MecA gene, methicillin resistance, polymerase chain reaction, Staphylococcus aureus
INTRODUCTION
Staphylococcus aureus is one of the major resistant pathogens in clinical practice. The reservoir of methicillin-resistant S. aureus (MRSA) is infected and colonized patients, and the major mode of transmission from patient to patient is through the contaminated hands of healthcare workers.[1] MRSA was first detected in Britain in 1961 and is now “quite common” in hospitals.[2] MRSA is defined as a strain of S. aureus that is resistant to a large group of antibiotics called β-lactams, that includes penicillins and cephalosporins.
MRSA is a major nosocomial pathogen worldwide which has emerged over the past 30 years as a leading cause of both nosocomial and community-acquired infections.[3,4] Methicillin resistance is caused by the presence of mecA gene, which encodes an additional 78 kDa low-affinity penicillin binding protein (PBP)-2a or PBP2' which has a low affinity for β-lactam antibiotics.[5] There has been a steady increase in the prevalence of MRSA all over the world.[6] In a meta-analysis of 31 studies, Cosgrove et al. reported that MRSA bacteremia is associated with increased mortality as compared with methicillin-sensitive S. aureus (MSSA) bacteremia.[7] The treatment of MRSA infections is entirely different from that of MSSA infections. Accurate and rapid detection of MRSA permits timely implementation of effective antimicrobial therapy, preventive infection control strategies like immediate patient isolation (and occasional ward closure), screening of the patient contacts and staff, and appropriate disinfection measures which in turn reduce the costs.[8,9] An additional concern is the emergence of vancomycin-intermediate S. aureus (VISA) and, more recently, vancomycin-resistant S. aureus (VRSA),[10,11] one of the main reasons assumed to be due to the injudicious use of vancomycin for wrongly diagnosed MRSA (false-positive MRSA).
Laboratory diagnosis and susceptibility testing are crucial steps in treating, controlling, and preventing MRSA infections. Discrepancies in detection have an adverse effect on patient management, thereby highlighting the importance of accuracy in detection. Hence, methods used to detect MRSA in clinical samples should have high sensitivity and specificity and, most importantly, the result should be available within a short time. Various methods have evolved for rapid detection of methicillin-resistant staphylococci, but the optimal method for the detection remains controversial. The most commonly used method in the laboratories is culture and antibiotic sensitivity test (AST) [oxacillin disk diffusion (ODD)]. Other methods available for diagnosing MRSA include mannitol salt agar (MSA) with oxacillin (agar screening method), minimum inhibitory concentration (MIC) tests, agar dilution tests etc., All these tests are the conventional phenotypic methods of MRSA identification.[12,13] Genotypic (molecular) method is the polymerase chain reaction (PCR) based method for detecting mecA gene, which remains the “gold standard” for diagnosing MRSA.[8,13,14]
The present study was conducted to compare two conventional phenotypic methods
ODD method (culture and sensitivity method) and
MSA with oxacillin with PCR for mecA gene.
This study would give us a definitive idea or would recommend the most appropriate test for diagnosing methicillin resistance in S. aureus with utmost accuracy and speed which is very much needed to avoid false positives and false negatives, as well as for early diagnosis. This in turn helps to prevent the emergence of new antibiotic-resistant strains as well as the spread of MRSA.
MATERIALS AND METHOD
It is a hospital-based observational study. Duration of the study was 1 year and 6 months (January 2009-June 2010). The study was started after getting the ethical clearance from the Scientific Research Committee of the institution. An informed written consent was obtained from all the subjects. A total of 165 consecutive, non-duplicated, clinically significant MRSA isolates were collected in Department of Microbiology at our tertiary care hospital, between January 2009 and June 2010.
Bacterial identification and antimicrobial susceptibility testing
The clinical specimens were inoculated on 5% sheep blood agar and MacConkey's agar (HiMedia, New Delhi, India), incubated at 37°C for 24-48 h, and examined for bacterial growth. S. aureus was identified using standard methods based on colony morphology, Gram's stain, catalase test, mannitol fermentation, and coagulase test. A total of 165 isolates were confirmed as S. aureus. They were tested for methicillin resistance based on modified Kirby-Bauer disk diffusion method using oxacillin disks (1 μg) on Mueller-Hinton agar in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines using the criteria of standard zone sizes of inhibition to define sensitivity or resistance.[15] The S. aureus strains were processed by the following three techniques for diagnosing MRSA:
(1) ODD method, (2) Culture in MSA medium containing oxacillin (at a concentration of 6 μg/ml of media) and 4% NaCl, and (3) PCR for the detection of mecA gene. Two standard strains, one MRSA ATCC (43300) and one MSSA ATCC (29213), were included in each batch of testing by all methods
Polymerase chain reaction for detection of mecA gene
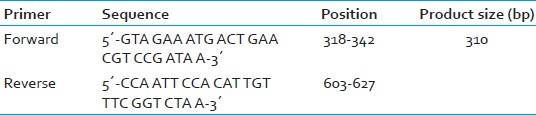
A crude extract of bacteria (DNA template) was prepared by microwave lysis method,[16] where a large colony of the organism was suspended in 10 μl of TE buffer (pH 8) in a microfuge tube. Bacteria were lysed by irradiation in a microwave oven using seven pulses of 60 s each with a 60 s interval in between. Then snap cooling at 4°C followed by centrifugation at 10,000 rpm for 30 s was done. The crude extract thus obtained was used for PCR as the DNA template. No further DNA extraction was carried out. The mecA gene was amplified using the primers, as described by Geha et al.,[17] and is given in Table 1.
Table 1.
Primer and its sequence[17]
The DNA template (crude extract) was amplified by PCR in 100 μl of a reaction mixture containing 200 μM (each) deoxynucleoside triphosphates, 2.5 μM (each) primers, 2.5 U of Taq DNA polymerase (Bangalore Genei), 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, and 0.01% gelatin. The procedural steps were as follows: Pre-denaturation for 4 min at 94°C, denaturation for 45 s at 94°C; annealing for 45 s at 55°C; and primer extension for 1 min at 72°C. Each step was repeated 30 times. Twenty microliters of the reaction mixture was loaded onto a 1.0% agarose gel with ethidium bromide. The band of amplified DNA was visualized under UV transilluminator. A 310 bp amplicon corresponds to mecA gene which is shown in Figure 1.
Figure 1.
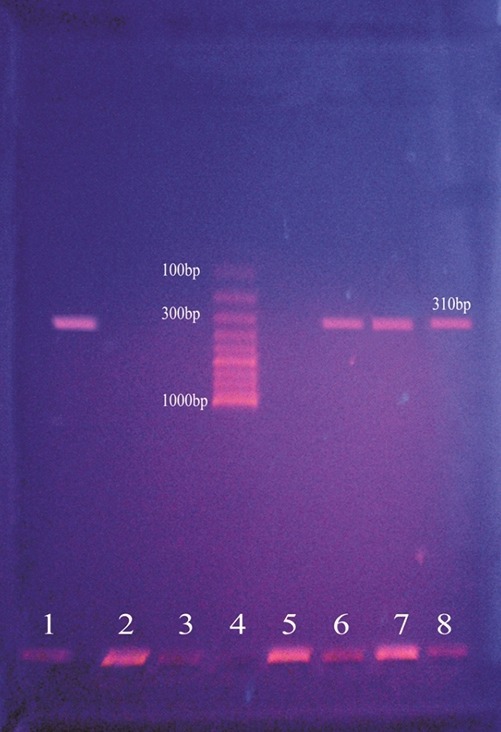
Results of mecA polymerase chain reaction
Oxacillin disk diff usion test
Disk diffusion test was performed on all isolates of S. aureus with 1 mg of oxacillin per disk on 25 ml of Mueller-Hinton agar without NaCl 2incubation at 37°C. The zone size was interpreted according to the CLSI as follows:[15] Susceptible, 13 mm; intermediate, 11-12 mm; and resistant, 10 mm [Figure 2].
Figure 2.
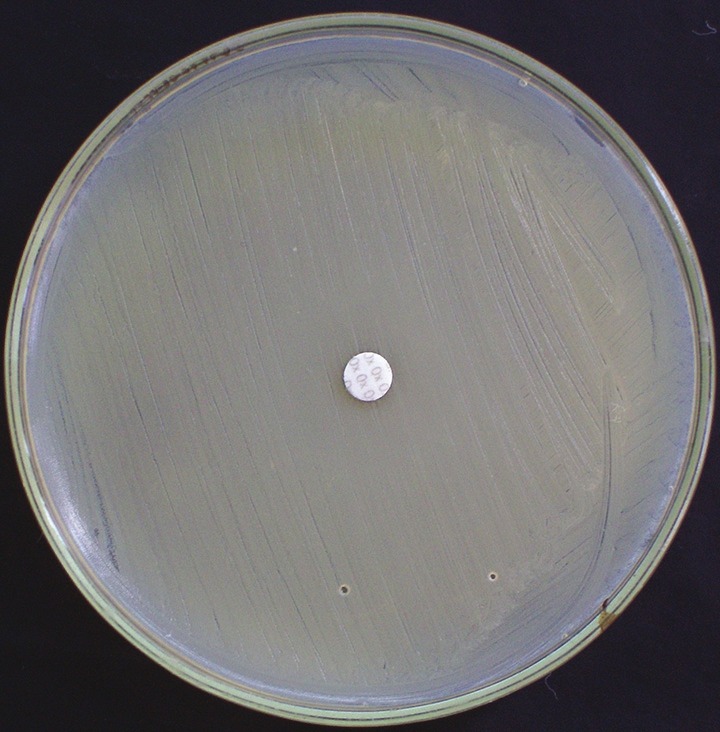
Oxacillin disk diffusion plate showing methicillin-resistant Staphylococcus aureus
Culture in mannitol salt agar containing oxacillin (oxacillin agar screen)
Weigh 11.1 g of MSA base into 100 ml of distilled water in a conical flask and autoclave it. After autoclaving, when the temperature reaches around 50°C, add oxacillin solution into the autoclaved medium so that the final concentration of oxacillin becomes 6 μg/ml of medium. Allow the medium to set. Streak the MSA plate with the sample and incubate at 37°C overnight (24 h). Any growth in the medium is taken as MRSA [Figure 3].
Figure 3.
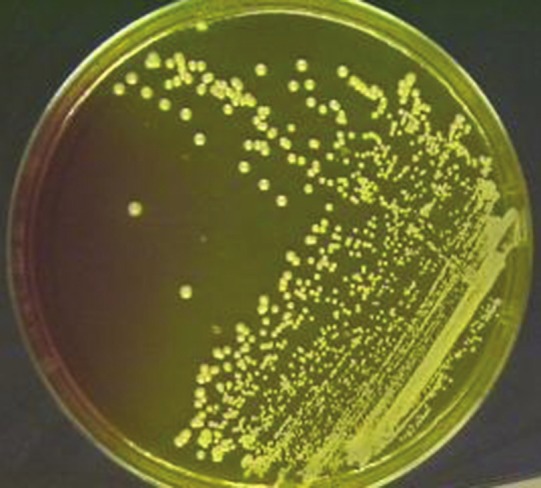
Mannitol salt agar plate with oxacillin showing methicillin-resistant Staphylococcus aureus
Statistical analysis
Comparative statistical analysis was done using R commander software. The ODD and the MSA method results were compared with the mecA PCR results using two-way contingency table analysis.
RESULTS
One hundred and sixty-five S. aureus isolates were included in the study. Out of the total 165 S. aureus isolates, 131 (79.39%) were pus samples, 23 (13.93%) were urine samples, 7 (4.24%) were blood samples, 3 (1.81%) were ear swab samples, and 1 (0.6%) was umbilical swab. The percentage of MRSA in the S. aureus isolates was found to be 37.5% in the present study. Of the total 165 samples tested, mecA PCR detected 62 (37.57%) MRSA and 103 (62.43%) MSSA. Out of the 62 true MRSA samples, 49 (79%) were pus samples, 10 (16.12%) were urine samples, 2 (3.23%) were blood samples, and 1 (1.6%) was an umbilical swab sample. The conventional microbiological method, culture and sensitivity using oxacillin antibiotic disk (ODD), detected 75 (45.45%) MRSA. MSA with oxacillin (oxacillin screen agar method) detected 65 (39.39%) MRSA. Four of the PCR-positive samples were detected as MSSA (not detected as MRSA) by ODD method. Of the 103 PCR-negative samples, 17 samples were detected as MRSA by this method. Eight of the PCR-positive samples were detected as MSSA (not detected as MRSA) by oxacillin screen agar method. Of the 103 PCR-negative samples, 11 samples were detected as MRSA by oxacillin screen agar method.
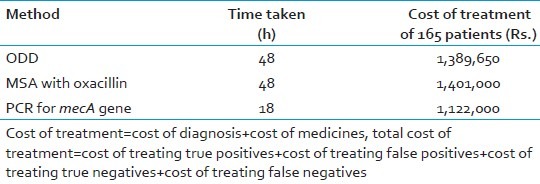
The ODD and the MSA method results were compared using two-way contingency table analysis. All the tests were compared for different parameters like speed, cost of treatment, sensitivity, and specificity with PCR for mecA gene, which is considered gold standard. The sensitivity and specificity for ODD method was found to be 93.5% and 83.5%, respectively, and that of MSA with oxacillin was 87.1% and 89.3%, respectively. PCR was found to be rapid as compared to the other methods [Tables 2 and 3].
Table 2.
Comparison of various parameters among oxacillin disk diffusion, mannitol salt agar with oxacillin, and polymerase chain reaction for mecA gene methods for detection of methicillin resistance
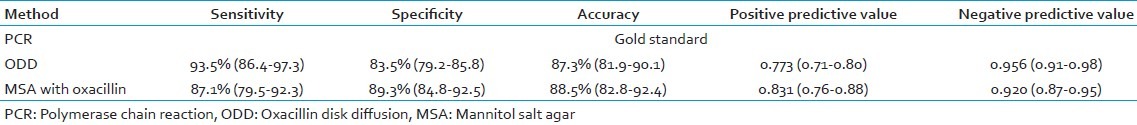
Table 3.
Comparison of speed and cost effectiveness among three methods
DISCUSSION
The present study revealed that ODD or AST was found to have sensitivity and specificity of 93.5% (86.4-97.3%) and 83.5% (79.2-85.85), respectively, and that of MSA with oxacillin was found to be 87.1% (79.5-92.3%) and 89.3% (84.8-92.5%), respectively. These findings revealed by our study are similar to those of many studies conducted in different parts of the world. Abu Hujier et al. in their study reported a sensitivity and specificity of 90% and 95.8%, respectively, for ODD.[18] Similar results were reported by Swenson et al.,[19] Skov et al.,[20] and Anila et al.[11]
The specificity is very low as compared to the PCR for mecA gene which is considered the “gold standard.” Both the sensitivity and specificity of AST are not acceptable. The lower levels of the confidence intervals of both sensitivity and specificity are far below 90%, which cannot be accepted for a method used for diagnosing methicillin resistance in S. aureus. The sensitivity of 93.5% means out of 100 true positives, only 93.5 will be diagnosed as positives and the remaining 7 will be misdiagnosed. Misdiagnosing seven MRSA isolates is not acceptable because if these isolates are not diagnosed as MRSA, the treatment pattern changes. The patient, instead of receiving vancomycin, will be prescribed other line of treatment normally given for MSSA. So, the patient does not get cured. More alarming situation is that by this time, MRSA would have been spread to other patients or health personnel. Finally, the patient will be forced to be given vancomycin. Ultimately, the cost of the whole treatment increases and the spread of MRSA in the hospital as well as the community will result. So, on comparison, the expenses due to the wrong diagnosis (missing MRSA) will be far more than the cost of PCR. Similarly, the specificity and accuracy of ODD were found to be 83.5% and 87.3%, respectively, which is far below the acceptable limits.
In our study, out of the 62 mecA PCR-positive isolates, only 58 were found positive by ODD method and 54 were found positive by MSA with oxacillin method. Four isolates were detected as MSSA by ODD test (phenotypically methicillin-susceptible strains) and eight isolates by MSA with oxacillin method.
The reason for these four to eight MRSA isolates detected as MSSA by the conventional methods can be attributed to the fact that accurate determination of methicillin resistance in staphylococci by conventional tests is subject to variations in inoculum size, incubation time, medium pH, medium salt concentration, etc.[16] These factors emphasize the need for a rapid, standardized, accurate, and sensitive method for detection of methicillin resistance in staphylococci, which is not dependent on growth conditions. Studies have reported that the heterogeneous nature of methicillin resistance in S. aureus limits the accuracy and reliability of phenotypic methods such as disk diffusion, broth and agar dilution tests.[21] Another important reason for these few MRSA isolates being detected phenotypically as MSSA is the over-expression of mecR and mecI genes which are co-repressors of mecA gene.[22,23] Of the 103 mecA PCR-negative isolates, only 86 were found to be negative by ODD and the rest of the 17 isolates was diagnosed as MRSA by ODD method. MSA with oxacillin method detected 92 MSSA and 11 MRSA.
The reason for this phenomenon (17 false positives) has been ascribed to the heterogeneous expression of methicillin resistance in many strains. Most of these isolates expressed resistance at the borderline of the inhibition zone and were thus termed “moderately resistant S. aureus” (MODSA). Under some test conditions, low-level resistance may also be seen in isolates which produce large amounts of penicillinase (penicillinase hyper producers), and these isolates have been referred to as “borderline oxacillin- resistant S. aureus” (BORSA).[24,25] It may be difficult to distinguish them from true resistant strains that carry the mecA gene, by routine tests.
MRSA isolates are either heterogeneous or homogenous in their expression of resistance. With homogenous expression, virtually all cells express resistance when tested by standard in vitro tests. However, testing of hetero-resistant isolates may result in some cells that appear susceptible and others resistant. Often only 1 in 104 to 1 in 108 mecA positive cells in the test population express resistance and heterogeneous expression results in isolates that appear to be borderline. The clinical problem posed by such isolates is that during chemotherapy with β-lactam antibiotics, production of PBP-2a may be induced, converting them into oxacillin-resistant strains. Hence detection of mecA gene is indispensable for precise differentiation of MRSA, and thus the PCR is a useful technique in clinical laboratories.
The PCR technique has many added advantages over the conventional techniques. The false-negative results of the conventional methods can be picked up by the PCR at a very early stage of the disease. PCR could accurately demonstrate the presence of mecA gene within 5 h of bacterial isolation and is a useful tool for rapid and unequivocal identification of MRSA. The expenses and workload of single PCR exceed the demand of testing one clinical specimen for the presence of MRSA. But if the daily number of MRSA screening tests increases, the workload per PCR decreases and finally outweighs the expenses for molecular reagents The time taken for diagnosing MRSA by conventional methods is 48-72 h, which is more as compared to PCR which takes 18-24 h. The cost of PCR is high as compared to the conventional phenotypic methods. But statistical analysis of the cost effectiveness revealed that PCR is the best option.
Our study concludes that the commonly used phenotypic tests are not completely reliable for the detection of methicillin resistance in S. aureus owing to their low sensitivity and specificity which are below the acceptable limits. Hence, PCR for mecA gene is the best method for detecting methicillin resistance in S. aureus with respect to accuracy, sensitivity, specificity, speed, and cost effectiveness. Increasing the number of samples for PCR on a regular basis will decrease the cost of a single PCR. This study advocates PCR for mecA gene on a regular basis for detecting methicillin resistance in S. aureus isolates from sterile body fluids or from special units such as intensive care units.
ACKNOWLEDGMENTS
Dr. Nilotpal Chowdhury, Associate Professor, Department of Pathology, is acknowledged for helping us with statistical analysis.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- 1.Eveillard M, Martin Y, Hidri N, Boussougant Y, Joly-Guillou ML. Carriage of methicillin-resistant Staphylococcus aureus among hospital employees: Prevalence, duration, and transmission to households. Infect Control Hosp Epidemiol. 2004;25:114–20. doi: 10.1086/502360. [DOI] [PubMed] [Google Scholar]
- 2.Ayliffe GA. The progressive international spread of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 1997;24:74–9. doi: 10.1093/clinids/24.supplement_1.s74. [DOI] [PubMed] [Google Scholar]
- 3.Chambers HF. The changing epidemiology of Staphylococcus aureus. Emerg Infect Dis. 2001;7:178–82. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timothy R. Peters, Dudley E. Hammon, Rima J. Jarrah, Elizabeth L. Palavecino, Elizabeth S. Blakeney, Katherine A. Poehling. Staphylococcal Toxic Shock Syndrome Complicating Influenza A Infection in a Young Child. ISRN Pulmonology, 2011. 2011:1–3. [Google Scholar]
- 5.Feudal C, Suvorov M, Vakulenko SB, Mobashery S. The basis for resistance to β-lactam antibiotics by penicillin-binding protein 2a of methicillin-resistant staphylococcus aureus. J Biol Chem. 2004;279:40802–6. doi: 10.1074/jbc.M403589200. [DOI] [PubMed] [Google Scholar]
- 6.Shopsin B, Kreiswirth B. Molecular epidemiology of methicillin-resistant Staphylococcus aureus. Emerg Infect Dis. 2001;7:322–6. doi: 10.3201/eid0702.010236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosgrove SE, Qi Y, Kaye KS, Harbarth S, Karchmer AW, Carmeli Y, et al. The impact of Methicillin Resistance in Staphylococcus aureus Bacteremia on Patient Outcomes: Mortality, Length of Stay, and Hospital Charges. Infect Control Hosp Epidemiol. 2005;26:166–74. doi: 10.1086/502522. [DOI] [PubMed] [Google Scholar]
- 8.Cooper BS, Stone SP, Kibbler CC, Cookson BD, Roberts JA, Medley GF, et al. Isolation measures in the hospital management of methicillin resistant Staphylococcus aureus (MRSA): Systematic review of the literature. BMJ. 2004;329:533. doi: 10.1136/bmj.329.7465.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Towner KJ, Talbot DC, Curran R, Webster CA, Humphreys H. Development and Evaluation of a PCR based immunoassay for the rapid detection of methicillin resistant Staphylococcus aureus. J Med Microbiol. 1998;47:607–13. doi: 10.1099/00222615-47-7-607. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention (CDC) Staphylococcus aureus resistant to vancomycin. JAMA. 2002;288:825. [Google Scholar]
- 11.Mathews AA, Thomas M, Appalaraju B, Jayalakshmi J. Evaluation and comparison of tests to detect methicillin resistant S.aureus. Indian J Pathol Microbiol. 2010;53:79–82. doi: 10.4103/0377-4929.59189. [DOI] [PubMed] [Google Scholar]
- 12.Brown DF, Edwards DI, Hawkey PM, Morrison D, Ridgway GL, Towner KJ, et al. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA) J Antimicrob Chemother. 2005;56:1000–18. doi: 10.1093/jac/dki372. [DOI] [PubMed] [Google Scholar]
- 13.Anand KB, Agrawal P, Kumar S, Kapila K. Comparison of cefoxitin disc diffusion test, oxacillin screen agar, and PCR for mecA gene for detection of MRSA. Indian J Med Microbiol. 2009;27:27–9. [PubMed] [Google Scholar]
- 14.Van Leeuwen WB, van Pelt C, Luijendijk A, Verbrugh HA, Goessens WH. Rapid Detection of Methicillin Resistance in Staphylococcus aureus Isolates by the MRSA-Screen Latex Agglutination Test. J Clin Microbiol. 1999;37:3029–30. doi: 10.1128/jcm.37.9.3029-3030.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CLSI 2011. Wayna, pa: Performance Standards for antimicrobial susceptibility testing, 21st Informational Supplement. Approved Standard M100-S21. [Google Scholar]
- 16.Menon P K, Wg Cdr, Nagendra A., Col Comparison of rapid method of DNA extraction using microwave irradiation with conventional phenol chloroform technique for use in multiplex PCR for mecA and femB genes. MJAFI. 2001;57:194–96. doi: 10.1016/S0377-1237(01)80041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geha DJ, Uhl JR, Gustaferro CA, Persing DH. Multiplex PCR for the identification of methicillin-resistant Staphylococci in the clinical laboratory. J Clin Microbiol. 1994;32:1768–72. doi: 10.1128/jcm.32.7.1768-1772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abu Hujier NS, Sharif FA. Detection of methicillin-resistant Staphylococcus aureus in nosocomial infections in Gaza Strip. Afr J Microbiol Res. 2008;2:235–41. [Google Scholar]
- 19.Swenson JM, Tenover FC. Cefoxitin disk study group.Results of Disk diffusion testing with Cefoxitin correlate with presence of mecA in Staphylococcus sp. J Clin Microbiol. 2005;43:3818, 28. doi: 10.1128/JCM.43.8.3818-3823.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skov R, Smyth R, Clausen M, Larsen AR, Frimodt-Møller N, Olsson-Liljequist B, et al. Evaluation of a cefoxitin30μg disk on Iso-Sensitest agar for detection of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2003;52:204–7. doi: 10.1093/jac/dkg325. [DOI] [PubMed] [Google Scholar]
- 21.Prasad KN, Kumar R, Tiwari DP, Mishra KK, Ayyagari A. Comparison of various conventional methods with a polymerase chain reaction assay for detecting methicillin resistant and susceptible Staphylococcus aureus strains. Indian J Med Res. 2000;112:198–202. [PubMed] [Google Scholar]
- 22.Khan AU, Sultan A, Tyagi A, Zahoor S, Akram M, Kaur S. Amplification of mecA gene in multi-drug resistant Staphylococcus aureus strains from hospital personnel. J Infect Dev Ctries. 2007;3:289–95. [PubMed] [Google Scholar]
- 23.Lewis RA, Dyke KG. MecI represses synthesis from the beta-lactamase operon of Staphylococcus aureus. J Antimicrob Chemother. 2000;45:139–44. doi: 10.1093/jac/45.2.139. [DOI] [PubMed] [Google Scholar]
- 24.Fluit AC, Wielders CL, Verhoef J, Schmitz FJ. Epidemiology and susceptibility of 3,051 Staphylococcus aureus isolates from 25 university hospitals participating in the European SENTRY study. J Clin Microbiol. 2001;39:3727–32. doi: 10.1128/JCM.39.10.3727-3732.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohanasoundaram KM, Lalitha MK. Comparison of phenotypic versus genotypic methods in the detection of methicillin resistance in Staphylococcus aureus. Indian J Med Res. 2008;127:78–84. [PubMed] [Google Scholar]


