Abstract
Although tumor molecular-profile-directed therapy appears promising in early clinical studies, there are many practical challenges to its successful clinical application in non-small-cell lung cancer (NSCLC). These challenges may be broadly classified as those relating to tumor (heterogeneity), tissue (acquisition and processing), testing (assays for molecular profiling) and trials (clinical evaluation of molecular markers and drugs). Strategies to overcome these challenges include (i) understanding the biological basis of tumor heterogeneity and of carcinogenesis in the large subset of patients with no currently evident driver events; (ii) technological advances in minimally invasive acquisition of tumor and next-generation sequencing (NGS) which would enable single-platform analysis of molecular alterations in limited tissue at a reasonable turnaround time (TAT); (iii) deliberation in early stages of drug development as well as clinical trial design to identify, validate and assess the clinical utility of biomarkers in conjunction with drugs and (iv) collaboration to improve understanding of and accrual to trials enrolling patients with rare molecular alterations.
Keywords: molecular profiling, next-generation sequencing, non-small-cell lung cancer, tumor heterogeneity
introduction
Improved understanding of the molecular mechanisms of non-small-cell lung cancer (NSCLC) which are essential for carcinogenesis and tumor progression has led to the development of drugs targeting these malignant-cell-specific vulnerabilities. However, these drugs are most efficacious in patients whose tumors harbor specific molecular alterations and their effectiveness may go undetected in unselected study groups. Clinical features alone have proven insufficient to predict the presence or absence of these genetic alterations.
Molecular profiling, the prospective analysis of tumor genetic expression, proteomic profile, deregulated cellular pathways and/or somatic mutations [1] could identify patients who are most likely to benefit from a specific drug and thereby, potentially improve outcomes, minimize toxic effects and abbreviate drug development. However, the increasing number of driver mutations in ever smaller subsets of patients and the availability of an array of candidate drugs (Table 1) have made clinical application of this paradigm challenging. In this review, we will discuss molecular profiling in NSCLC—the opportunities, challenges and potential strategies to overcome them.
Table 1.
Frequency of common genetic alterations in advanced non-small-cell lung cancer (NSCLC), their clinico-pathologic correlates and the drugs targeting them
| Genetic alteration | Gene | Frequency (%) | Major clinico-pathological correlates | Selected drugs targeting the gene/s (additional targets) (phase of clinical trial evaluation in NSCLC) |
|---|---|---|---|---|
| Mutation | EGFR | 10–35 | Asian, female, never smoker, adenocarcinoma | Erlotinib (approved) |
| Gefitinib (approved) | ||||
| Afatinib (EGFR/HER2) (phase III) | ||||
| Dacomitinib (Pan HER) (phase III) | ||||
| HER2 | 2–4 | Never smoker, female, adenocarcinoma | Lapatinib (EGFR/HER2) (phase III) | |
| Dacomitinib (Pan HER) (phase III) | ||||
| Afatinib (EGFR/HER2) (phase III) | ||||
| PI3K | 1–3 | Squamous cell carcinoma | BKM120 (phase II)Pictilisib (phase II) | |
| PX866 (phase II) | ||||
| XL147 (phase II) | ||||
| XL765 (PI3K/MTOR) (phase II) | ||||
| BEZ235 (PI3K/MTOR) (phase II) | ||||
| BYL719 (phase II) | ||||
| Perifosine (PI3K/AKT) (phase II) | ||||
| PF04691502 (phase II) | ||||
| PKI587 (PI3K/MTOR) (phase I) | ||||
| AKT1 | 1–2 | Not described | MK2206 (phase I) | |
| KRAS | 15–25 | Former/current smokers | None | |
| BRAF | 2–3 | Former/current smokers | Pazopanib (multiple kinases) (phase III) | |
| Dabrafenib (phase II) | ||||
| MEK | 1 | Adenocarcinoma | Selumetinib (phase II) | |
| Trametinib (phase II) | ||||
| DDR2 | Squamous cell carcinoma | Dasatinib (multiple kinases) (phase II) | ||
| Translocation | ALK | 3–7 | Younger age, never/light-smokers, adenocarcinoma | Crizotinib (MET/ALK/ROS) (approved) |
| AP26113 (ALK/EGFR) (phase II) | ||||
| LDK 378 (phase 1) | ||||
| ROS1 | 1 | Younger age, never/light smokers, adenocarcinoma | Crizotinib (MET/ALK/ROS) (phase II) | |
| KIF5B-RET | 1 | Younger age, never/light-smokers, adenocarcinoma | Sunitinib (multiple kinases) (not in clinical trials) | |
| Amplification | MET | 3 | EGFR mutant tumors following prior treatment with EGFR TKI | Onartuzumab (phase III) |
| Tivantinib (phase III)Cabozantinib (multiple kinases) (phase II) | ||||
| FGFR1 | 2 | Squamous cell carcinoma | Lenvatinib (multiple kinases) (phase II) | |
| brivanib alaninate (multiple kinases) (phase II) | ||||
| BGJ398 (pan-FGFR) (phase I) |
NSCLC, non-small-cell lung cancer; TKI, tyrosine kinase inhibitor.
clinical application of molecular profiling
Molecular profiling has been found feasible and of potential clinical benefit in patients with refractory metastatic solid tumors (Table 2) [2,3]. A pilot trial in refractory metastatic cancers demonstrated clinical benefit, defined as progression-free survival (PFS) ratio (PFS on molecular-profile-directed treatment/PFS on prior treatment) of ≥1.3 in 27% (18 out of 66) of patients who received molecular-profile-directed therapy (95% CI, 17%–38%; one-sided P = 0.007) [2]. A phase I program reported longer time to treatment failure compared with prior therapy (median 5.3 versus 3.2 months, P = 0.0003) and higher overall response rate (ORR) (29% versus 8%; P = 0.0001) in 161 patients with advanced malignancies and one genetic alteration who received molecular-profile-directed therapy compared with patients who received treatment not directed by the molecular profile [3].
Table 2.
Summary of reports of clinical application of molecular profiling
| Author, year of publication | N | Type of study | Patient characteristics | Tissue used | Targets interrogated | Technology used | Median turnaround time (TAT) in weeks | Success rate (all proposed tests carried out) (%) | Frequency of at least one genetic alteration (%) | Treated with a matched- targeted agent (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Metastatic malignancies | ||||||||||
| Von Hoff et al. 2010 [2] | 86 | Prospective multi-institutional trial with central laboratory | Refractory metastatic cancers | Mandatory fresh biopsy | 11 proteins 51 genes | IHC, FISH, Gene expression microarray | NR | 98 | 98 | 77 |
| Tsimberidou et al. 2011 [3]* | 955 | Prospective single institution trial | Refractory metastatic cancers | NR | NR | PCR, IHC, FISH | NR | 89 | 41.5 | 19 |
| Non-small-cell lung cancer | ||||||||||
| Kris, 2010 [5]* | 301 | Prospective single institution trial | Adenocarcinoma | Previously obtained tissue | EGFR, KRAS, BRAF, HER2, PIK3CA, MEK1 AKT1, ALK | PCR-based direct sequencing and Sequenoma, FISH | NR | 92 | 58 | 17 |
| Ortiz et al. 2011 [6]* | 226 | Prospective single institution trial | Non-squamous NSCLC | NR | EGFR, KRAS, BRAF, PIK3CA, HER2, ALK | PCR-based direct sequencing, FISH | 4 | 89 | 54 | 14 |
| Sequist et al. 2011 [4] | 589 | Retrospective single institution experience | NSCLC | NR | AKT1, APC, BRAF, CTNNB1, EGFR, ERBB2, FLT3, IDH1, JAK2, KIT, KRAS, NOTCH1, NRAS, PIK3CA, PTEN, TP53, ALK | SNaPshotb, FISH | 2.8 (range 1.0–8.9 weeks) | 95 | 51 | 22c |
| Kim et al. 2011 [7] | 255 | Prospective single-institution-randomized trial | Advanced pre-treated NSCLC | Mandatory fresh biopsy | EGFR,KRAS, BRAF, Cyclin D1, VEGF, VEGFR-2, RXRs α, β, γ | PCR-based direct sequencing, FISH, IHC | <2 | NR | 84 | 93 |
*Abstract only.
aSequenom: a multiplexed mass spectrometry-based assay.
bSNaPshot: multiplexed PCR-based assay.
c78 (22%) of the 353 patients with advanced disease were candidates for targeted therapy.
N, number of patients; PCR, polymerase chain reaction; NSCLC, non-small-cell carcinoma; NR, not reported.
In NSCLC, several reports have demonstrated the feasibility of molecular analyses in a majority of patients (Table 2) [4–7]. These studies used either archival tissue or fresh biopsies and multiple assays-fluorescence in situ hybridization (FISH), polymerase chain reaction (PCR)-based direct sequencing and/or multiplexed genotyping platforms. The median turnaround time (TAT) for results was 2 to 4 weeks and at least one genetic alteration was identified in 51%–84% of patients. The Biomarker-integrated Approaches of Targeted Therapy for Lung Cancer Elimination (BATTLE-1) trial was the first completed prospective, molecular-profile driven trial in NSCLC [7]. Chemo-refractory patients underwent mandatory pretreatment biopsies. Based on the tumor molecular profile, they were assigned to five biomarker groups (EGFR, KRAS/BRAF, VEGF/VEGFR2, RXR/Cyclin D1, None) and randomly assigned initially equally and later adaptively to four treatment groups (erlotinib, sorafenib, vandetanib or erlotinib plus bexarotene). The overall 8-week disease control rate (DCR) was 46% and eight of the 20 biomarker-treatment matches met the predefined criterion for efficacy, i.e. >80% probability of achieving a >30% 8-week DCR.
Drawing from these and our own experiences of a pilot trial of molecular profiling in lung cancer, we will discuss the challenges to clinical application of molecular profiling under four broad categories: tumor (heterogeneity), tissue (acquisition and processing), testing (assays for molecular profiling) and trials (clinical evaluation of molecular markers and drugs).
tissue: acquisition and processing of malignant tissue
Obtaining tissue with an adequate tumor fraction with minimal contamination of normal and necrotic cells for morphological confirmation and molecular analyses in a minimally invasive manner is perhaps the greatest challenge. The common methods of tumor acquisition include bronchoscopic and image-guided percutaneous transthoracic needle biopsy (TTNB). Bronchoscopy is suitable for central lesions, but typically yields limited tissue compared with TTNB, which is employed for more peripheral and mediastinal lesions. The reported rates of successful molecular profiling with TTNB vary depending on the fixation method and the extent of analysis carried out (Table 3) [8–11].
Table 3.
Selected reports evaluating common primary tumor acquisition methods in NSCLC
| Author, year of publication | Type of study | N | Procedure used | Type of tissue obtained | Molecular analyses carried out | Complications | Successful molecular analysis |
|---|---|---|---|---|---|---|---|
| Gill et al. 2012 [11]a | Single- institution retrospective review | 81 | CT-guided TTNB | Formalin-fixed paraffin embedded (FFPE) | PCR-sanger sequencing for hotspot mutations: EGFR, KRAS, BRAF, PIK3CA HER2; FISH: ALK | Pneumothorax: 23 (28.3%) | Sequencing: 64 (79%) |
| Chest tube: 6 (7%) | FISH: 60 (71%) | ||||||
| Hospitalization: 9 (11%) | |||||||
| Intra-parenchymal hemorrhage: 19 (23%) | |||||||
| 18 grade1; 1 grade 2 | |||||||
| Solomon et al. 2010 [9] | Selected consecutive patients from a phase II single-institution trial | 18 | CT or fluoroscopy guided TTNB | FFPE | EGFR by direct sequencing or PCR; KRAS by direct sequencing | Pneumothorax: 3 (17%) | 16 (89%) |
| No chest tube placement or hospitalization | |||||||
| Cheung et al. 2010 [10] | Retrospective review | 47 | CT-guided TTNB | Fresh frozen | EGFR by PCR | Pneumothorax: 6 (13%) | 47 (100%) |
| Chest tube: 2% | |||||||
| Hospitalization NR | |||||||
| Hemoptysis: 3 (6%) | |||||||
| Reck et al. 2011 [8] | Multicenter phase II trial | 255 | Bronchoscopic biopsy | Fresh frozen | Gene expression profiling | NR | 122 (48%) |
aAbstract only.
NSCLC, non-small-cell lung cancer; N, number of patients; CT, computed tomography; TTNB, transthoracic needle biopsy; FFPE, formalin-fixed paraffin embedded; PCR, polymerase chain reaction; FISH, fluorescence in situ hybridization; NR, not reported.
The need to minimize biopsy-related complications has prompted interest in the use of less invasive sources of tissue like cytological specimens, circulating tumor cells (CTCs) and free serum DNA [12–14]. Although molecular profiling of cytological samples (for e.g. aspirates like those obtained using endobronchial ultrasound-guided transbronchial needle aspiration and pleural effusions) was discouraged in the past, recent studies demonstrate the feasibility of using an adequately cellular tumor cytology sample [12, 15, 16]. Depending on the frequency of mutations and tumor cell proportion, cytological material can provide results comparable with surgical specimens [16]. CTCs captured using a microfluidic-based device containing epithelial-cell adhesion molecule-coated microposts successfully identified the expected EGFR activating and resistant mutations in 92% and 55% of patients with EGFR-mutated NSCLC [13]. In 12 patients for whom primary tumor samples, CTC, and plasma were all available, CTC-based detection had a sensitivity of 92%. Further studies are needed to clarify whether this potential source of cancer cells is representative of the primary tumor.
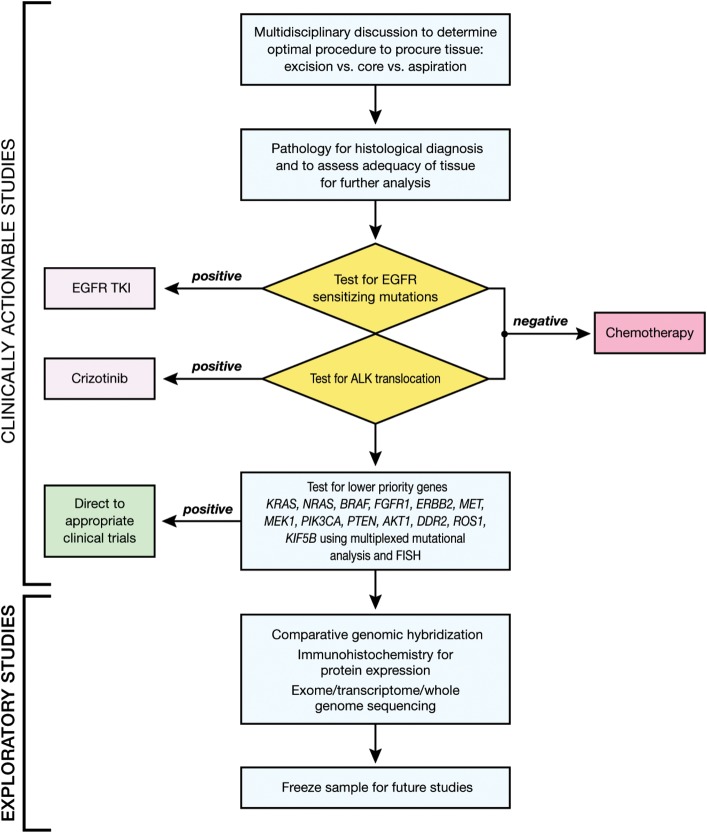
Once tissue is obtained, its strategic management, i.e. processing to preserve as much as possible for molecular testing, is essential. This involves planning and coordination between members of the thoracic oncology multidisciplinary team to choose the optimal tissue acquisition procedure as well as allocation of tissue for morphological diagnosis and prioritization of molecular studies [17]. Based on the assay used, amount and type of tissue available and the specific needs in individual cases, each laboratory must determine its own priority of tests. Figure 1 shows a schema which is based on the available data to facilitate optimal use of available tissue for treatment decisions and exploratory studies. It is conceivable that this model would be simplified in the near future with the clinical use of comprehensive clinical genomic analyses.
Figure 1.
An algorithm for tissue allocation to facilitate prioritization of testing for actionable molecular alterations in newly diagnosed patients with advanced non-small-cell lung cancer (NSCLC). A multidisciplinary discussion involving thoracic oncologists, surgeons, molecular pathologists, cytopathologists and interventional radiologists determines the optimal procedure to obtain tissue for molecular profiling. Following pathology review, the sample is tested for genetic alterations with approved therapies, EGFR mutations and ALK translocations. Patients with one of these alterations are directed to the corresponding therapies, while the rest are directed to standard first-line chemotherapy. The remaining tissue, if available, is used for analysis of lower priority genes followed by (or concurrently using the same platform) other exploratory studies to facilitate enrollment in molecular-profile-matched therapies at progression of disease.
Other important considerations after tissue acquisition include standardization of fixation and processing methods, TAT and quality control. Formalin-fixed paraffin-embedded (FFPE) specimens, the most common source of tissue, may yield poor-quality DNA due to cross-linking and degradation which leads to decreased amplicon length and artifactual mutations [18]. Cytological specimens may afford better preservation of nucleic acids and nuclear structure due to the use of alcohol-based fixation and direct smearing of cells as opposed to formalin fixation and tissue sections in FFPE specimens [19]. TAT, the interval from genotype requisition to result finalization, is an important consideration in advanced NSCLC due to the rapid clinical course. TAT may be prolonged, for example in cases where an alternative sample is requested from an outside institution when it is not feasible to obtain a new biopsy or when the initial specimen was of poor quality and retesting is needed. Guidelines have been issued regarding timelines for delivery of archived tissue (both from in-house and outside institutions) for testing of EGFR and ALK [20]. The Clinical Laboratory Improvement Amendments (CLIA) serve as a regulatory standard for tests which will be used for clinical decision making to ensure the accuracy, reliability and timeliness.
Alterations in signaling pathways following successive lines of treatment warrant molecular analysis on biopsies obtained at the time of progression rather than using the original diagnostic biopsy. The importance of assessing cancers throughout the disease course with repeated biopsies is exemplified by studies of patients with EGFR-mutant NSCLC with acquired resistance to EGFR tyrosine kinase inhibitors (TKIs) [21]. The mechanisms of resistance included genetic (e.g. EGFR T790M mutation, PIK3CA mutation) and histologic [e.g. transformation of NSCLC to small-cell lung cancer (SCLC)] alterations with significant impact on the choice of further treatment.
The pursuit of potentially improved outcomes should be balanced against the risks associated with the use of invasive procedures for tissue acquisition. Moreover, ethical concerns apply to mandatory biopsies for research participation [22]. Voluntary informed consent and close monitoring to minimize and manage procedure-related complications are imperative.
testing: assays for molecular profiling
Most laboratories use direct sequencing of PCR-amplified exon sequences for identification and confirmation of mutations, which are currently relevant to treatment (e.g. EGFR and KRAS). Although commonly used in many clinical settings, capillary-based DNA sequencing is limited by the small number of genes that can be profiled and its insufficient sensitivity to detect low frequency mutations in clinical samples where there is a substantial admixture of malignant and non-malignant cells.
Multiplexed panels (e.g. Sequenom and SNaPshot) aim to rapidly and simultaneously interrogate multiple common hotspot mutations in DNA from FFPE specimens. Sequenom's MassARRAY system is based on multiplexed PCR, multiplexed single-base primer extension and analysis of primer-extension products using matrix-assisted laser desorption/ionization time of flight mass spectrometry [23]. SNaPshot analyzes fluorescently labeled primer-extension products by conventional capillary electrophoresis [24, 25]. Multiplexed platforms interrogate only a limited number of loci and are unable to detect chromosomal rearrangements or determine gene copy number.
Next-generation sequencing (NGS) (e.g. single-nucleotide fluorescent base extension with reversible terminators and ligation-based sequencing) is based on the simultaneous detection of nucleotides in arrayed amplified DNA products originating from single-DNA molecules [26, 27]. NGS uses over-sampling, i.e. extensive repeated coverage (deep sequencing) and matched germline DNA sequencing to overcome experimental noise resulting from tumor heterogeneity, detect low-abundance mutations in samples with low tumor content and filter out single-nucleotide polymorphisms to more readily identify true somatic alterations. NGS can perform comprehensive whole genome sequencing, targeted sequencing of nucleic acid compartment of interest [e.g. transcriptomes (expressed genes) and exomes (coding and non-coding exons)] and detection of chromosomal rearrangements and copy number alterations at very high resolution. However at this time, clinical application of NGS is hampered by the large amount of data generated and the resultant statistical and computational challenges, long TAT, need for secondary verification, and cost.
FISH identifies gene amplifications and translocations and is the current standard for detecting ALK rearrangements (using ALK break-apart probe), independent of the specific fusion partner. However, FISH assays can be technically challenging, costly and may be difficult to perform in FFPE specimens due to destruction of tissue morphology [28]. Chromogenic in situ hybridization (CISH), a modification of FISH which uses peroxidase reaction instead of fluorescent dye, abrogates the need for fluorescence microscopy, but is not widely available [28]. Immunohistochemistry (IHC) is a sensitive, easy and cost-effective surrogate for genetic testing and a potential alternative to FISH [29]. The limitations of IHC include the generally low expression of the ALK fusion protein in NSCLC, need for standardization of pre-analytic conditions and antibody, lack of quantitation and inter-observer variability [28]. Based on its high sensitivity and moderate specificity, ALK IHC-based screening has been proposed: an initial screening followed by FISH evaluation of 2+ or 1+ and 2+ IHC-positive cases [30, 31]. Further studies are needed to validate the concordance between ALK IHC and FISH. Reverse transcriptase-PCR-based ALK testing offers advantages of extreme sensitivity, but may yield false-positive results due to contamination, are difficult to perform on poor-quality RNA extracted from FFPE specimens, need multiplexed assays to detect all the known ALK variants and does not detect previously uncharacterized ALK fusion partners [28]. In contrast to FISH and CISH which detect solitary amplifications and deletions in specific genes, comparative genomic hybridization (CGH) uses large-scale copy number assessment to identify chromosomal amplifications and deletions, but is not routinely used in a clinical setting.
Currently, clinical application of molecular profiling is limited to a few oncogenic point mutations and amplifications in NSCLC. Multiple assays are used to detect different molecular alterations (e.g. gene fusions by FISH; point mutations by sequencing) as a single platform is unable to detect all of them. The existing approach to drug development also involves development of a companion diagnostic assay in conjunction with a drug which entails optimization of the assay and its analytical and clinical validation, i.e. evidence that the result of the analytically validated assay correlates with the clinical outcome of interest and assessment of clinical utility for the intended use [32]. This approach is costly, especially when the molecular alterations are rare and large numbers of patients have to be screened as exemplified by RET fusions and DDR2 mutations which occur in <1% of NSCLC patients. Moreover, it is time consuming and requires a large amount of tissues to perform all the different assays. Clinical application of NGS-based platforms may offer a potential solution to this quandary by providing faster, cheaper, yet comprehensive assessment of tumor molecular landscape using limited tissue. Although the feasibility of targeted NGS of FFPE specimens to detect actionable mutations has been demonstrated, [33, 34] at this time, it may be neither feasible nor affordable to use NGS in a clinical setting.
tumor: heterogeneity
Heterogeneity exists between primary and metastatic NSCLC, within an individual tumor and following successive lines of therapy [21, 35]. Tumor heterogeneity assessments in NSCLC to date are limited by the number of genes evaluated (only EGFR and KRAS in most cases) and inconsistent techniques, often with limited sensitivity. The frequencies of heterogeneity reported in these studies have ranged between 0% and 29% for EGFR and 9% and 25% for KRAS mutations [36–43]. Moreover, biopsy specimens contain a mixture of malignant cells, adjacent normal cells and stroma, and infiltrating normal cells. While small core biopsies may not be representative of clonal heterogeneity of the entire tumor, conventional sequencing of a large sample may miss heterogeneity by representing only the dominant clone [44].
Intratumor heterogeneity may have implications on the choice of treatment, prognosis and emergence of resistance. The presence of a mixed population of EGFR-mutated and wild-type cells has been reported to result in reduced response to gefitinib [45]. Heterogeneity in the EGFR mutation status between the primary lung tumors and their metastases may explain the mixed response to EGFR TKIs in some patients [43]. The presence of low frequency of pretreatment MET-amplified and EGFR T790M-mutated cells could potentially predict acquired resistance and shorter PFS with EGFR TKI [46, 47]. Since it is not practical to understand the full extent of tumor heterogeneity by deep sequencing multiple simultaneous core biopsies from primary and metastatic sites, in practice, we attempt biopsy of the most rapidly growing tumors, which presumably contain the most biologically aggressive genetic alterations.
trial: clinical evaluation of molecular markers and drugs
Many anticancer therapies benefit only a subset of patients and the benefit may be overlooked by the traditional broad eligibility approach in clinical trials. Hence, the paradigm has shifted towards restricting study enrollment based on the presence of a biomarker which potentially identifies a population that is likely to respond to a given drug. Negative results of several phase III trials of EGFR TKI in unselected patients [48–52] and the rapid clinical development of crizotinib in patients with ALK-rearranged NSCLC [53, 54] provide contrasting examples of the importance of this strategy. Considering the increasing number of drugs and potentially druggable genetic alterations, newer clinical trial designs are needed to simultaneously develop multiple drugs and drug combinations in molecular-profile-defined subsets of patients [55]. There are several ongoing trials which employ novel clinical trial designs to evaluate in parallel multiple-targeted therapies in selected NSCLC patients.
An ongoing pilot trial at the National Cancer Institute is evaluating the feasibility of carrying out fresh biopsies for MP and the efficacy of multiple molecular-profile-directed therapies in 600 patients with advanced thoracic malignancies (NCT01306045). Real-time molecular analysis using multiple platforms (pyrosequencing, NGS, CGH and FISH) identifies oncogenic mutations, insertions, deletions, gene amplifications and translocations of 12 genes to guide treatment allocation while analysis of >190 cancer-related genes is used for the discovery of new biomarkers. Patients with ALK-rearranged NSCLC receive crizotinib, whereas the remaining patients are assigned to one of five experimental arms based on biomarker assessment (EGFR mutation or indel: erlotinib; KRAS, NRAS, HRAS or BRAF mutations: selumetinib; PIK3CA, AKT or PTEN mutations, PIK3CA amplification: MK2206; ERBB2 mutation/amplification: lapatinib; KIT mutation, PDGFRA mutation/amplification: sunitinib) or standard treatment if not eligible for any of the experimental arms. For each of the 15 possible treatment arms (three disease types––NSCLC, SCLC and thymic malignancies; five drugs), the study uses the optimal two-stage phase II design [56].
The BATTLE-1 trial randomized an initial cohort (n = 97; 40%) equally to each of the four treatment arms [7]. Subsequent patients (n = 158; 60%) were randomly assigned according to the Bayesian adaptive algorithm [57], which used the prior probability and 8-week DCR of the initial cohort to generate a ‘posterior’ probability of DCR for a given treatment which was, in turn, used to increasingly assign patients to treatment arms with the greatest efficacy. The Lung Cancer Mutation Consortium Protocol (LCMC), a collaborative effort which involves 14 cancer centers in the United States, aims to determine the frequency of oncogenic mutations in 1000 patients with advanced lung adenocarcinoma (NCT01014286) [58] The linked clinical and mutational analyses are used to determine the frequency of each mutation, its association with clinical features and outcome and its association with other mutations. As therapeutic protocols specific for these mutations are developed, patients are notified of their eligibility for these studies. A secondary goal of LCMC is to establish a consortium of sites that have the capability of conducting multiple mutation testing in a CLIA-certified lab.
There are several hurdles to molecular-profile-driven patient selection in early-phase clinical trials. This approach runs the risk of discounting the efficacy of a drug when an incorrect biomarker is used for patient selection, which often results from the complexity of signaling pathways, especially in early drug development. A molecular-profile-driven approach also raises issues of validating assays, especially when the assay is used to guide treatment decisions [32]. The efficacy of a drug may be overlooked in biomarker ‘negative’ population. For example, crizotinib resulted in ORR of 50% and 61%, respectively, in two multicenter single-arm studies in patients with ALK-translocated (defined as >15% cells with ALK rearrangement) locally advanced or metastatic NSCLC which led to its Food and Drug Administration (FDA) approval [53, 54]. However, in an expansion cohort of 19 ALK negative patients, crizotinib resulted in five partial responses (ORR 26%, 95% CI, 9%–51%) [59]. It is unclear whether the responses to crizotinib in ALK-negative patients are a result of its activity in other genetic alterations (e.g. MET amplification or ROS rearrangement) [60] or due to issues with validation of assay performance. To address these questions, FDA has recommended a clinical trial to explore the activity of crizotinib in ALK-negative patients, adequacy of current assay cut-off and the role of additional biomarkers. Alternative clinical trial designs which do not require biomarker selection before initiation of study may address some of these issues [61, 62]. Accrual to trials enrolling rare subgroups is also a challenge in molecular-profile-driven patient selection. LCMC is an example of a nationwide initiative, which could identify and maximize accrual to trials of rare molecular subtypes.
The remarkable responses seen in early-phase clinical trials of some molecular-profile-directed therapies such as crizotinib have also raised questions regarding the necessity and feasibility of randomized phase III trials before regulatory approval [63]. Foregoing phase III trials would expedite drug development, improve patient access and mitigate cost, but may yield less definitive safety and efficacy data [64]. Despite its potential risks, early efficacy results of selected drugs in a molecular-profile-defined population may potentially be used to forego pre-marketing phase III trials [63, 64].
conclusion and future directions
Among the several molecular targets that are being investigated in NSCLC, inhibition of only a few in selected patients has led to meaningful clinical benefit (e.g. EGFR, ALK), whereas many others have not proven useful (e.g. IGF1R). A lack of validated biomarkers and patient selection has led to many drugs showing no efficacy or even detrimental effects in clinical studies, despite strong rationale and pre-clinical activity. Molecular-profile-driven patient selection has demonstrated the potential to improve outcomes in NSCLC. The practical challenges to clinical application of molecular profiling may be conceptualized as those relating to ‘tumor’ (heterogeneity), ‘tissue’ (acquisition and processing), ‘testing’ (assays for molecular profiling) and ‘trials’ (clinical evaluation of molecular markers and drugs).
Multidisciplinary determination of the least invasive procedure to obtain adequate specimen, standardization of sample collection, processing and strategic management of available tissue can increase the success rate of molecular profiling. CTC and free serum DNA may be potential alternatives to invasive tissue acquisition in the future. Further studies are needed to understand the biological basis and implications of tumor heterogeneity as well as the role of tumor suppressor genes and epigenetic events in the large subset of NSCLC patients (∼ 40%) with no known driver mutations. Clinical use of NGS-based platforms could provide faster, cheaper and comprehensive assessment of tumor molecular landscape using limited tissue. Regulatory mechanisms should adapt to the need for such platforms. Beginning from the early stages of drug development and clinical trial design, efforts should focus on identifying, validating and assessing the clinical utility of biomarkers in conjunction with drugs. Collaborative efforts are needed to improve patient accrual to trials of drugs targeting less common genetic alterations. It is anticipated that comprehensive clinical genomic analysis will in the near future enable molecular profiling of the vast majority of patients who are cared for in a general oncology setting.
funding
This work was supported by the Intramural program, National Cancer Institute, National Institutes of Health.
disclosure
The authors have declared no conflicts of interest.
references
- 1.Macconaill LE, Van Hummelen P, Meyerson M, et al. Clinical implementation of comprehensive strategies to characterize cancer genomes: opportunities and challenges. Cancer Discov. 2011;1:297. doi: 10.1158/2159-8290.CD-11-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Von Hoff DD, Stephenson JJ, Jr, Rosen P, et al. Pilot study using molecular profiling of patients’ tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol. 2010;28:4877–4883. doi: 10.1200/JCO.2009.26.5983. [DOI] [PubMed] [Google Scholar]
- 3.Tsimberidou AM, Iskander NG, Hong DS, et al. Personalized medicine in a phase I clinical trials program: The M. D. Anderson Cancer Center Initiative. J Clin Oncol. 2011;29(suppl) doi: 10.1158/1078-0432.CCR-12-1627. (Abstr CRA2500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sequist LV, Heist RS, Shaw AT, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol. 2011;22:2616–2624. doi: 10.1093/annonc/mdr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kris MG, Lau C, Ang D, et al. Initial results of LC-MAP: An institutional program to routinely profile tumor specimens for the presence of mutations in targetable pathways in all patients with lung adenocarcinoma. J Clin Oncol. 2010;28(suppl) (Abstr 7009) [Google Scholar]
- 6.Ortiz TM, Joshi VA, Heon S, et al. The introduction of systematic genomic testing for patients with non-small cell lung cancer (NSCLC) at Dana-Farber Cancer Institute (DFCI) J Clin Oncol. 2011;29(suppl) doi: 10.1097/JTO.0b013e3182745bcb. (Abstr 7517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim ES, Herbst RS, Wistuba II, et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov. 2011;1:44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reck M, Hermes A, Tan EH, et al. Tissue sampling in lung cancer: a review in light of the MERIT experience. Lung Cancer. 2011;74:1–6. doi: 10.1016/j.lungcan.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Solomon SB, Zakowski MF, Pao W, et al. Core needle lung biopsy specimens: adequacy for EGFR and KRAS mutational analysis. Am J Roentgenol. 2010;194:266–269. doi: 10.2214/AJR.09.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung YC, Chang JW, Hsieh JJ, et al. Adequacy and complications of computed tomography-guided core needle biopsy on non-small cell lung cancers for epidermal growth factor receptor mutations demonstration: 18-gauge or 20-gauge biopsy needle. Lung Cancer. 2010;67:166–169. doi: 10.1016/j.lungcan.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Gill RR, Heon S, Yeap BY, et al. Genomic profiling of non-small cell lung cancer (NSCLC) for personalized targeted therapy using CT-guided transthoracic needle biopsy (TTNB) J Clin Oncol. 2012;30(suppl) (Abstr 10592) [Google Scholar]
- 12.Garcia-Olive I, Monso E, Andreo F, et al. Endobronchial ultrasound-guided transbronchial needle aspiration for identifying EGFR mutations. Eur Respir J. 2010;35:391–395. doi: 10.1183/09031936.00028109. [DOI] [PubMed] [Google Scholar]
- 13.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leary RJ, Kinde I, Diehl F, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2(20ra4) doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rekhtman N, Brandt SM, Sigel CS, et al. Suitability of thoracic cytology for new therapeutic paradigms in non-small cell lung carcinoma: high accuracy of tumor subtyping and feasibility of EGFR and KRAS molecular testing. J Thorac Oncol. 2011;6:451–458. doi: 10.1097/JTO.0b013e31820517a3. [DOI] [PubMed] [Google Scholar]
- 16.Smouse JH, Cibas ES, Janne PA, et al. EGFR mutations are detected comparably in cytologic and surgical pathology specimens of nonsmall cell lung cancer. Cancer. 2009;117:67–72. doi: 10.1002/cncy.20011. [DOI] [PubMed] [Google Scholar]
- 17.Travis WD, Rekhtman N. Pathological diagnosis and classification of lung cancer in small biopsies and cytology: strategic management of tissue for molecular testing. Semin Respir Crit Care Med. 2011;32:22–31. doi: 10.1055/s-0031-1272866. [DOI] [PubMed] [Google Scholar]
- 18.Williams C, Ponten F, Moberg C, et al. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol. 1999;155:1467–1471. doi: 10.1016/S0002-9440(10)65461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer AH, Cibas ES, Howell LP, et al. Role of cytology in the management of non-small-cell lung cancer. J Clin Oncol. 2011;29:3331–3332. doi: 10.1200/JCO.2011.35.2534. [DOI] [PubMed] [Google Scholar]
- 20.CAP/IASLC/AMP Lung Cancer Biomarkers Guideline draft recommendations. http://www.cap.org/apps/docs/membership/transformation/new/lung_public_comment_supporting_materials.pdf (11 June 2012, date last accessed) [Google Scholar]
- 21.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002003. 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peppercorn J, Shapira I, Collyar D, et al. Ethics of mandatory research biopsy for correlative end points within clinical trials in oncology. J Clin Oncol. 2010;28:2635–2640. doi: 10.1200/JCO.2009.27.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 24.Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. Embo Mol Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su Z, Dias-Santagata D, Duke M, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13:74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 27.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 28.Shaw AT, Solomon B, Kenudson MM. Crizotinib and testing for ALK. J Natl Compr Canc Netw. 2011;9:1335–1341. doi: 10.6004/jnccn.2011.0115. [DOI] [PubMed] [Google Scholar]
- 29.Mino-Kenudson M, Chirieac LR, Law K, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res. 2010;16:1561–1571. doi: 10.1158/1078-0432.CCR-09-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paik JH, Choe G, Kim H, et al. Screening of anaplastic lymphoma kinase rearrangement by immunohistochemistry in non-small cell lung cancer correlation with fluorescence in situ hybridization. J Thorac Oncol. 2011;6:466–472. doi: 10.1097/JTO.0b013e31820b82e8. [DOI] [PubMed] [Google Scholar]
- 31.Yi ES, Boland JM, Maleszewski JJ, et al. Correlation of IHC and FISH for ALK gene rearrangement in non-small cell lung carcinoma: IHC score algorithm for FISH. J Thorac Oncol. 2011;6:459–465. doi: 10.1097/JTO.0b013e318209edb9. [DOI] [PubMed] [Google Scholar]
- 32.Williams PM, Lively TG, Jessup JM, et al. Bridging the gap: moving predictive and prognostic assays from research to clinical use. Clin Cancer Res. 2012;18:1531–1539. doi: 10.1158/1078-0432.CCR-11-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagle N, Berger MF, Davis MJ, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen Y-Y, Fang E, Li Y, et al. The efficacy of targeted next-generation sequencing for detection of clinically actionable mutations in cancer. J Clin Oncol. 2012;30(suppl) (Abstr 10598) [Google Scholar]
- 35.Taillade L, Penault-Llorca F, Boulet T, et al. Immunohistochemichal expression of biomarkers: a comparative study between diagnostic bronchial biopsies and surgical specimens of non-small-cell lung cancer. Ann Oncol. 2007;18:1043–1050. doi: 10.1093/annonc/mdm072. [DOI] [PubMed] [Google Scholar]
- 36.Yatabe Y, Matsuo K, Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol. 2011;29:2972–2977. doi: 10.1200/JCO.2010.33.3906. [DOI] [PubMed] [Google Scholar]
- 37.Schmid K, Oehl N, Wrba F, et al. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res. 2009;15:4554–4560. doi: 10.1158/1078-0432.CCR-09-0089. [DOI] [PubMed] [Google Scholar]
- 38.Park S, Holmes-Tisch AJ, Cho EY, et al. Discordance of molecular biomarkers associated with epidermal growth factor receptor pathway between primary tumors and lymph node metastasis in non-small cell lung cancer. J Thorac Oncol. 2009;4:809–815. doi: 10.1097/JTO.0b013e3181a94af4. [DOI] [PubMed] [Google Scholar]
- 39.Sun LN, Zhang QA, Luan HL, et al. Comparison of KRAS and EGFR gene status between primary non-small cell lung cancer and local lymph node metastases: implications for clinical practice. J Exp Clin Canc Res. 2011;30:30. doi: 10.1186/1756-9966-30-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalikaki A, Koutsopoulos A, Trypaki M, et al. Comparison of EGFR and K-RAS gene status between primary tumours and corresponding metastases in NSCLC. Br J Cancer. 2008;99:923–929. doi: 10.1038/sj.bjc.6604629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gow CH, Chang YL, Hsu YC, et al. Comparison of epidermal growth factor receptor mutations between primary and corresponding metastatic tumors in tyrosine kinase inhibitor-naive non-small-cell lung cancer. Ann Oncol. 2009;20:696–702. doi: 10.1093/annonc/mdn679. [DOI] [PubMed] [Google Scholar]
- 42.Cortot AB, Italiano A, Burel-Vandenbos F, et al. KRAS mutation status in primary nonsmall cell lung cancer and matched metastases. Cancer. 2010;116:2682–2687. doi: 10.1002/cncr.25014. [DOI] [PubMed] [Google Scholar]
- 43.Chena ZY, Zhonga WZ, Zhanga XC, et al. EGFR mutation heterogeneity and the mixed response to EGFR tyrosine kinase inhibitors of lung adenocarcinomas. Oncologist. 2012;17:978–985. doi: 10.1634/theoncologist.2011-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polyak K, Shipitsin M, Campbell-Marrotta L, et al. Breast tumor heterogeneity: causes and consequences. Breast Cancer Res. 2009;11(Suppl 1):S18. [Google Scholar]
- 45.Taniguchi K, Okami J, Kodama K, et al. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer Sci. 2008;99:929–935. doi: 10.1111/j.1349-7006.2008.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turke AB, Zejnullahu K, Wu YL, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su KY, Chen HY, Li KC, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol. 2012;30:433–440. doi: 10.1200/JCO.2011.38.3224. [DOI] [PubMed] [Google Scholar]
- 48.Giaccone G, Herbst RS, Manegold C, et al. Gefitinib in combination with gemcitabine and cisplatin in advanced non-small-cell lung cancer: a phase III trial–INTACT 1. J Clin Oncol. 2004;22:777–784. doi: 10.1200/JCO.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 49.Herbst RS, Giaccone G, Schiller JH, et al. Gefitinib in combination with paclitaxel and carboplatin in advanced non-small-cell lung cancer: a phase III trial–INTACT 2. J Clin Oncol. 2004;22:785–794. doi: 10.1200/JCO.2004.07.215. [DOI] [PubMed] [Google Scholar]
- 50.Herbst RS, Prager D, Hermann R, et al. TRIBUTE: a phase III trial of erlotinib hydrochloride (OSI-774) combined with carboplatin and paclitaxel chemotherapy in advanced non-small-cell lung cancer. J Clin Oncol. 2005;23:5892–5899. doi: 10.1200/JCO.2005.02.840. [DOI] [PubMed] [Google Scholar]
- 51.Thatcher N, Chang A, Parikh P, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer) Lancet. 2005;366:1527–1537. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 52.Gatzemeier U, Pluzanska A, Szczesna A, et al. Phase III study of erlotinib in combination with cisplatin and gemcitabine in advanced non-small-cell lung cancer: the Tarceva Lung Cancer Investigation Trial. J Clin Oncol. 2007;25:1545–1552. doi: 10.1200/JCO.2005.05.1474. [DOI] [PubMed] [Google Scholar]
- 53.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crinò L, Kim D, Riely GJ, et al. Initial phase II results with crizotinib in advanced ALK-positive non-small cell lung cancer (NSCLC): PROFILE 1005. J Clin Oncol. 2011;29(suppl) (Abstr 7514) [Google Scholar]
- 55.Yap TA, Sandhu SK, Workman P. Envisioning the future of early anticancer drug development. Nat Rev Cancer. 2010;10:514–523. doi: 10.1038/nrc2870. [DOI] [PubMed] [Google Scholar]
- 56.Simon R. Optimal two-stage designs for phase II clinical trials. Control Clin Trials. 1989;10:1–10. doi: 10.1016/0197-2456(89)90015-9. [DOI] [PubMed] [Google Scholar]
- 57.Zhou X, Liu S, Kim ES, et al. Bayesian adaptive design for targeted therapy development in lung cancer––a step toward personalized medicine. Clin Trials. 2008;5:181–193. doi: 10.1177/1740774508091815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kris MG, Johnson BE, Kwiatkowski DJ, et al. Identification of driver mutations in tumor specimens from 1,000 patients with lung adenocarcinoma: The NCI's Lung Cancer Mutation Consortium (LCMC) J Clin Oncol. 2011;29 (abstr) (Abstr CRA7506) [Google Scholar]
- 59.FDA prescribing information Xalkori (crizotinib) http://www.accessdata.fda.gov/drugsatfda_docs/label/2011/202570s000lbl.pdf. Accessed June 11, 2012.
- 60.Bergethon K, Shaw AT, Ignatius Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Freidlin B, Jiang WY, Simon R. The cross-validated adaptive signature design. Clin Cancer Res. 2010;16:691–698. doi: 10.1158/1078-0432.CCR-09-1357. [DOI] [PubMed] [Google Scholar]
- 62.Freidlin B, Simon R. Adaptive signature design: an adaptive clinical trial design for generating and prospectively testing a gene expression signature for sensitive patients. Clin Canc Res. 2005;11:7872–7878. doi: 10.1158/1078-0432.CCR-05-0605. [DOI] [PubMed] [Google Scholar]
- 63.Miller FG, Joffe S. Equipoise and the dilemma of randomized clinical trials. N Engl J Med. 2011;364:476–80. doi: 10.1056/NEJMsb1011301. [DOI] [PubMed] [Google Scholar]
- 64.Sharma MR, Schilsky RL. Role of randomized phase III trials in an era of effective targeted therapies. Nat Rev Clin Oncol. 2011;9:208–214. doi: 10.1038/nrclinonc.2011.190. [DOI] [PubMed] [Google Scholar]