Abstract
A comparative genomic analysis predicted that many members of the under-characterized COG0523 subfamily of putative P-loop GTPases function in metal metabolism. In this work we focused on the uncharacterized Escherichia coli protein YeiR by studying both the physiology of a yeiR mutant and the in vitro biochemical properties of YeiR expressed as a fusion with the maltose-binding protein (YeiR-MBP). Our results demonstrate that deletion of yeiR increases the sensitivity of E. coli to EDTA or cadmium and this phenotype in linked to zinc depletion. In vitro, the tagged protein binds several Zn2+ ions with nanomolar affinity and oligomerizes in the presence of metal. The GTPase activity of YeiR is similar to that measured for other members of the group, but GTP hydrolysis is enhanced by Zn2+ binding. These results support the predicted connection between the COG0523 P-loop GTPases and roles in metal homeostasis.
Keywords: GTPase, zinc homeostasis, metal-binding, COG0523, cadmium
Introduction
GTPases are often referred to as molecular switches. The binding and hydrolysis of GTP cause conformational changes that effectively turn “on” or “off” interactions with other macromolecules.1 As can be predicted by the commonality of the fold, a myriad of functions are carried out by GTPases, such as regulatory roles in translation,2, 3 signal transduction,4 cell motility,5 intracellular trafficking,6 cell cycle regulation,7 and membrane transport.8
The GTPase superclass can be divided based on shared structural and sequence features.9 One resulting subdivision is the G3E family of GTPases, which is defined by specific G protein motifs: a glutamate residue that substitutes for the conserved aspartate in the Walker B motif and an intact base recognition motif NKXD.9 The G3E family is further divided into four subfamilies, three of which are represented by HypB, UreG, and MeaB. These GTPases are involved in metallocenter assembly, a process that often requires multiple accessory proteins to generate catalytically active metalloenzymes.10–12 The production of the Ni2+-containing enzymes [NiFe]-hydrogenase and urease depends on NTP hydrolysis by HypB13–15 and UreG,16, 17 respectively, and MeaB contributes to the assembly of B12-dependent methylmalonyl-CoA mutase.18 The fourth subfamily, composed of proteins containing the COG0523 domain (referred to as COG0523), is far more ubiquitous than the other subfamilies but as of yet remains predominately uncharacterized.
Only sparse reports are found on COG0523 proteins. The first member of this family to be identified was found in Pseudomonas denitrificans and named CobW. Disruption of cobW, which is located in a gene cluster with cobalamin synthesis genes, leads to a defect in the production of cobalamin.19 Cobalamins are composed of a Co(III) bound in an octahedral configuration to a corrin macrocycle.20 The proposed role for CobW is in presentation of cobalt to the cobaltchetelase component of the pathway, which is responsible for inserting cobalt into the corrin ring.21 However, 20 years after its discovery, there is as of yet no experimental evidence for this function. Since “cobalamin biosynthesis protein” was the first role assigned to a member of COG0523, this annotation has been propagated by sequence similarity (e.g. BLAST) to nearly all COG0523 genes in current databases.
Other partially characterized members of COG0523 include the nitrile hydratase activators, which are required for iron-type nitrile hydratase (NHase) activity.22 NHases employ either an octahedral non-heme iron or non-corrin cobalt in the hydration of nitriles to amides.23 The active site residues of both NHase enzymes are highly conserved and the same coordination geometry was determined for both metals.24 The metal specificity is thought to be due to activator proteins.22, 25–28 In the case of iron-type NHase, the corresponding activator protein is a COG0523 protein and is referred to as Nha3. As with CobW, however, even though the involvement of Nha3 in iron-type NHase activation is documented, its exact role is not known.29
The most recently studied COG0523 protein is YciC from Bacillus subtilis. Several observations led to the initial hypothesis that yciC codes for a component of a low-affinity zinc transporter,30 but subsequent work has suggested that this protein may play a chaperone role.31 The gene, yciC, is repressed by Zur, a zinc-responsive transcription factor,30 and deletion of yciC in combination with deletion of the high-affinity zinc transporter genes results in an EDTA-sensitive growth defect and perturbed growth in zinc-deficient medium.30, 32 As with CobW and Nha3, the mechanism responsible for these phenotypes remains elusive.
Although experimental evidence has linked a few COG0523 proteins to various metal-dependent processes, metal-binding and GTPase activities have not been confirmed. All COG0523 proteins contain a CXCC sequence located between the Walker A and the Walker B motifs of the GTPase domain (Figure S1). This sequence is proposed to contribute to a metal-binding site because it aligns with the known metal-binding residues of HypB33, 34 and a similar motif is found in COX17, which uses this motif for transporting copper to mitochondrial cytochrome oxidase in eukaryotic cells.35 The conservation of this putative metal-binding motif suggests that metal binding could be a defining characteristic of this family of putative P-loop GTPases and thus inform their function.
To test the hypothesis that proteins in the COG0523 family function in metal homeostasis, this study focused on the physiology and metal-binding properties of Escherichia coli YeiR, an uncharacterized member of COG0523. YeiR was previously assigned to subgroup 10 of COG0523, which has an enrichment of genes that are downstream from putative Zur-binding sites.36 However, unlike other members of its subgroup, the E. coli yeiR gene was not predicted to be regulated by Zur. Its inclusion in this COG0523 subgroup led us to analyze the effect of metal depletion on the growth of an E. coli yeiR mutant, while its homology to other metal-binding GTPases led us to characterize its ability to hydrolyze GTP and bind metals.
We found that deletion of yeiR resulted in enhanced sensitivity of E. coli to the metal chelator ethylenediaminetetraacetate (EDTA). This growth defect can be suppressed by the addition of Zn2+ and is exacerbated by deletion of the genes encoding the high-affinity Zn2+ transporter, ZnuABC. Our studies also show that purified YeiR-MBP binds Zn2+ and that metal binding modulates the quaternary structure of the protein. Finally, the GTPase activity of YeiR is similar to that found for other members of the G3E group such as HypB and UreG but it is accelerated by metal loading, in contrast to the characterized impact of metal on the activity of the other proteins in this family.
Results
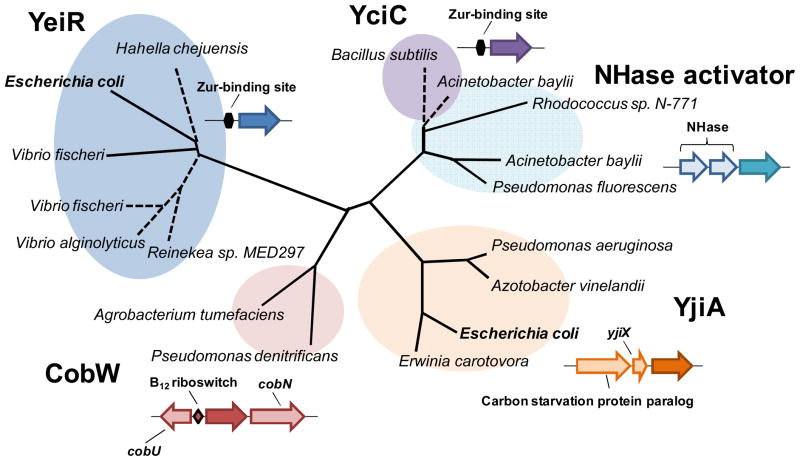
Phylogenetically, YeiR is a member of a COG0523 clade that is distinct from groups containing previously investigated COG0523 proteins.36 YeiR, however, is closely related to putatively zinc-regulated COG0523 proteins from two Vibrio species, Reinekea sp. MED297 and Hahella chejuensis (Figure 1). Therefore like YciC from B. subtilis, YeiR could be involved in a response to zinc depletion. On the other hand, YjiA, a second COG0523-domain containing protein encoded in the E. coli genome (Figure 1), is not predicted to be involved in zinc homeostasis because this protein belongs to a COG0523 subgroup that does not contain zinc-regulated members.36
Figure 1. Phylogenetic tree of selected COG0523 proteins including the two hypothetical members from E. coli, YeiR and YjiA.
Protein sequences were downloaded from the SEED database (http://theseed.uchicago.edu/FIG/) and aligned with the MUSCLE program.73 The neighbor joining tree was constructed using the PHYLIP package.74 Predicted repression of the corresponding gene by Zur36 is indicated by a dotted line. Typical gene organization involving COG0523 from each of the 5 subgroups is shown. CobW refers the COG0523 proteins proposed to be involved in cobalamin biosynthesis. NHase activators are COG0523 proteins involved in Fe-type nitrile hydratase activation and YciC refers to COG0523 proteins shown to be involved in survival during zinc depletion.75
EDTA sensitivity of the yeiR mutant
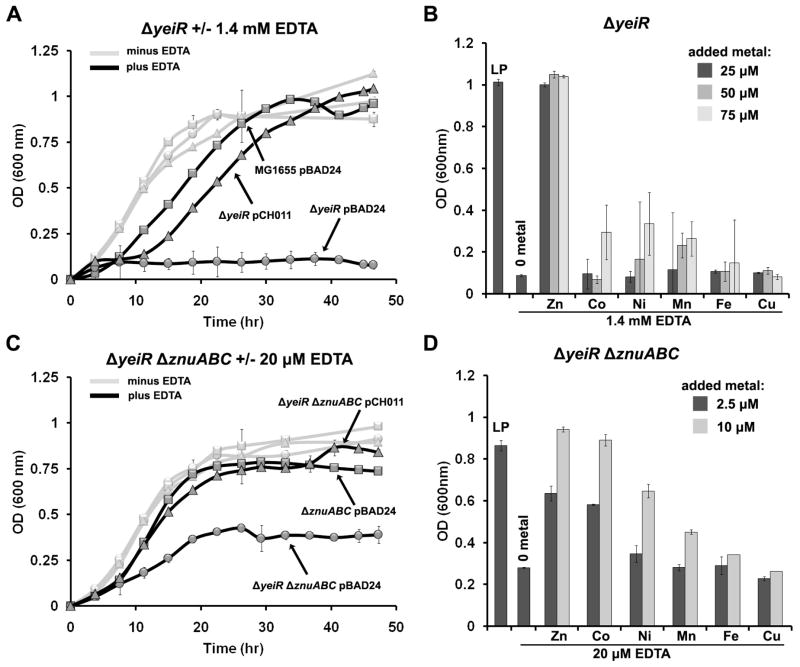
The parent MG1655 (WT) and yeiR mutant E. coli strains were grown in a low-phosphate (LP) minimal medium that is commonly used for the characterization of metal-related physiology and was previously used to study low-affinity Zn2+ transporters in E. coli.37 In order to create conditions of metal depletion, the non-specific metal ion chelator EDTA was added to the medium at various concentrations (Figure S2).37, 38 Compared to MG1655, a drastic growth defect was observed for the yeiR mutant when grown in the presence of 1.4 mM EDTA (Figure 2A). Deletion of yeiR was complemented by expressing yeiR in trans from the PBAD promoter of pCH011 (Figure 2A), whereas yeiR carrying a mutation of each cysteine in the conserved CXCC motif were unable to rescue growth (Figure S2). A similar growth defect was not observed with the WT strain until concentrations of EDTA above 2 mM were used (Figure S2). As EDTA is a non-specific chelator, various metal salts were added one at a time to the growth medium supplemented with 1.4 mM EDTA. The ΔyeiR strain growth defect was suppressed only by the addition of Zn2+ whereas Ni2+, Cu2+, Co2+, Mn2+ and Fe3+ did not rescue cell growth (Figure 2B and S2).
Figure 2. EDTA sensitivity phenotypes.
Overnight cultures of E. coli derivatives grown in LB at 37°C were washed once in LP medium and normalized to an OD600nm of 1.0. Cells were then diluted 1/500 into fresh LP medium. Growth curves were measured using a Bioscreen C at 37°C. For each panel, error bars represent ± the standard deviation of 3 replicates. (A) E. coli MG1655 (WT) pBAD24 (squares), ΔyeiR pBAD24 (circles) and ΔyeiR pCH011 (PBAD::yeiR) (triangles) in LP medium were grown in the absence (grey line) or presence (black line) of 1.4 mM EDTA. (B) Growth of the ΔyeiR strain in LP medium in the absence of EDTA (LP) or presence of 1.4 mM EDTA +/− the indicated metals at either 25 – 75 μM metal. (C) ΔznuABC::cam pBAD24 (squares), ΔznuABC::cam ΔyeiR pBAD24 (circles) and ΔznuABC::cam ΔyeiR pCH011 (PBAD::yeiR) (triangles) were grown in the absence (grey line) or presence (black lines) of 20 μM EDTA. Error bars represent ± the standard deviation of 3 replicates. (D) Growth of the ΔznuABC::cam ΔyeiR mutant in LP medium in the absence of EDTA (LP) or presence of 20 μM EDTA +/− the indicated metals at either 2.5 μM or 10 μM metal.
As previously noted,32, 38 it is difficult to prepare zinc-limited medium for bacterial growth because of efficient scavenging systems. Instead, zinc deficiency can be specifically introduced by inactivating high-affinity zinc uptake, which aggravates growth defects caused by Zn2+ depletion.32, 37 Therefore, to further evaluate whether the observed growth phenotype of the yeiR mutant was due specifically to disruption of zinc homeostasis, the znuABC transporter genes were deleted in the ΔyeiR strain background and the strains were grown in the presence of EDTA (Figure S3). As shown in Figure 2C, the phenotype is exacerbated in this strain and only 20 μM EDTA was required to detect a growth defect upon deletion of yeiR, which was rescued by expressing yeiR in trans from PBAD. Higher concentrations of EDTA were required to detect the same EDTA sensitivity in the ΔznuABC::cam strain as compared to the ΔyeiR ΔznuABC::cam strain (Figure S3). In contrast to yeiR, deletion of yjiA, the second COG0523-encoding gene in E. coli, had no effect on the growth of either the ΔznuABC::cam strain or the ΔyeiR ΔznuABC::cam strain in the presence of EDTA (Figure S3).
The first characterization of E. coli znuABC was performed in the strain MC4100, which contains a deletion in yeiR.39 The authors found that growth of znuA::MudX and znuB::MudX on rich solid medium was inhibited in the presence of 400 μM EDTA. A repeat of those experiments with the MG1655-derived strains, ΔznuABC::cam and ΔznuABC::cam ΔyeiR, revealed that a 10-fold increase in EDTA concentration is required to inhibit the growth of the ΔznuABC::cam strain with yeiR present (data not shown).
Rescue of the ΔznuABC::cam ΔyeiR mutant growth defect in LP medium with exogenously added metal was also performed. A distinct pattern of metal rescue was observed. As shown above (Figure 2B), a molar ratio of 56:1 EDTA to Zn was sufficient to suppress the ΔyeiR mutant phenotype and no other metals at that same concentration had a noticeable effect on growth. However, with the genes encoding the high-affinity zinc transporter also deleted, a molar ratio of 2:1 EDTA to Zn (10 μM zinc) was required to detect full suppression of the phenotype (Figure 2D). In addition, 10 μM cobalt appeared to suppress the growth defect of the ΔznuABC::cam ΔyeiR mutant in the presence of 20 μM EDTA to the same extent as 10 μM zinc. Nickel at 10 μM was able to restore growth to 65% of the strain’s growth in LP medium without EDTA.
Cadmium sensitivity
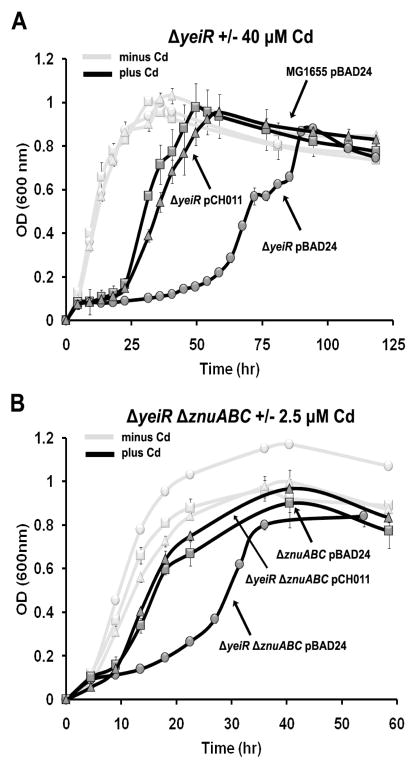
To further probe the growth defect induced by deleting yeiR, WT and ΔyeiR strains were grown in the presence of Cd2+ (Figure S4). Cadmium is an environmental toxin and although the exact mechanism of cytotoxicity is not understood it is thought to disrupt the homeostasis of essential transition metals.38, 40 The ΔyeiR strain exhibited a clear growth defect in the presence of 40 μM Cd2+ as compared to WT (Figure 3A and S4). As was observed with EDTA, less Cd2+ (2.5 μM) was required to detect the ΔyeiR strain phenotype if the znuABC genes were also deleted (Figure 3B and Figure S5).
Figure 3. Cadmium sensitivity phenotypes.
E. coli derivatives were grown as described for Figure 2 except Cd2+ was added to the appropriate cultures instead of EDTA. (A) E. coli MG1655 (WT) pBAD24 (squares), ΔyeiR pBAD24 (circles) and ΔyeiR pCH011 (PBAD::yeiR) (triangles) in LP medium were grown in the absence (grey line) or presence (black line) of 40μM Cd2+. (B) ΔznuABC::cam pBAD24 (squares), ΔznuABC::cam ΔyeiR pBAD24 (circles) and ΔznuABC::cam ΔyeiR pCH011 (PBAD::yeiR) (triangles) were grown in the absence (grey line) or in the presence (black line) of 2.5 μM Cd2+. Error bars represent ± the standard deviation of 3 replicates.
The effect of supplementation with various metals in equimolar concentration to Cd2+ on the growth of the MG1655 and ΔyeiR strains was also tested. The presence of Zn2+ was able to partially suppress the effect of Cd2+ on the growth of the ΔyeiR mutant, as was Mn2+ to a lesser extent (Figure S6). In contrast to the mutant, the growth defect that was observed with the WT strain was not affected by the presence of Zn2+ or Mn2+ (Figure S6). The presence of Co2+, Ni2+ or Fe3+ did not lead to suppression or further exacerbation of the Cd2+-induced growth defect of the mutant or WT strain, while the addition of Cu2+ further exacerbated growth of both strains (Figure S6).
In vitro metal binding to YeiR
Initially, E. coli YeiR was overexpressed without a tag, purified and used for metal-binding experiments. However, the wild type protein (YeiR) tended to aggregate and the addition of metal to the protein exacerbated precipitation. To increase the solubility and expression levels of YeiR, an N-terminal maltose-binding protein (MBP) fusion was constructed and purified (Figure S7), and all subsequent experiments were performed with YeiR-MBP. As isolated, YeiR-MBP contained trace amounts of metal bound to the protein (~ 10%) as determined by metal analysis, so all protein preparations were treated with 10 mM EDTA, along with 1 mM TCEP in an anaerobic glovebox to minimize disulfide bond formation. Following gel filtration chromatography to remove the small molecules, no metal bound to the protein was detected.
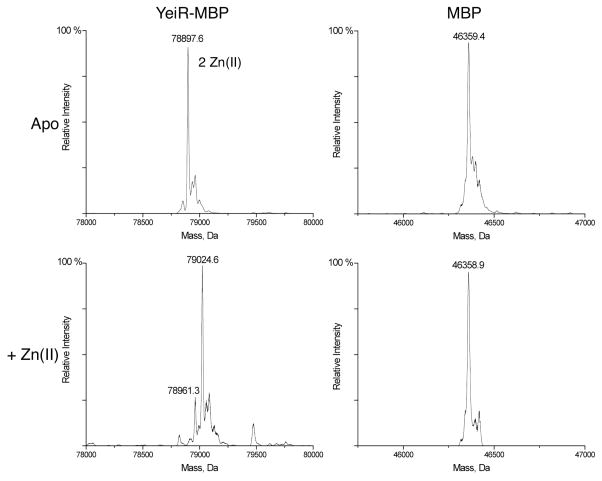
To determine if Zn2+ can bind to the protein, apo-YeiR-MBP was incubated with excess metal overnight inside an anaerobic glovebox followed by passage through a PD10 gel filtration column to eliminate unbound metal. The samples were then analyzed by using inductively coupled plasma atomic emission spectroscopy (ICP-AES) or buffer exchanged into ammonium acetate and examined via electrospray ionization mass spectrometry (ESI-MS). In the latter method, most of the protein detected was loaded with two Zn2+ ions (Figure 4 and S8), but 3.3 ± 0.6 Zn2+ ions were detected per protein by using ICP-AES. These data suggest that YeiR can bind up to three zinc ions, but only two remain bound to the protein following the ionization process in the ESI-MS. As a control, this experiment was also performed with isolated MBP. Only the apo-MBP form was detected via ESI-MS (Figure 4), indicating that the tag does not bind Zn2+ under these experimental conditions, which was confirmed via ICP-AES.
Figure 4. Zinc binding to YeiR-MBP and MBP.
YeiR-MBP or MBP was incubated with EDTA and TCEP for 48 hours at 4°C in an anaerobic glovebox followed by gel filtration chromatography to yield apo-YeiR-MBP. Samples of 150 μM protein were incubated with at least four molar equivalents of zinc, after which they were passed through a gel filtration column and buffer exchanged into 10 mM ammonium acetate, pH 7.5 before analysis by ESI-MS. The predicted masses of YeiR-MBP and MBP are 78,896.9 and 46359.8 Da, respectively.
As an alternative method to monitor zinc binding to YeiR-MBP, the metal was titrated into a protein solution containing the fluorometric metal chelator Mag-Fura-2 (MF-2), and the formation of the Zn(II)-MF-2 complex was monitored by the decrease in emission at 500 nm following excitation at 366 nm. At least 15 μM Zn2+ was needed to quench the fluorescence of 0.5 μM MF-2 when 5 μM YeiR-MBP was present, consistent with a stoichiometry of three Zn2+ ions per YeiR-MBP (Figure 5A). Furthermore, an initial plateau region was detected in which the signal of apo-MF-2 did not change until 5 μM Zn2+ was added to the solution (Figure 5A), suggesting that there is one site on the protein that is significantly tighter than the ≈ 100 nM binding affinity exhibited by the competitor MF-2.41, 42 To determine the binding affinities for the remaining two Zn2+ sites, the curve starting after the addition of 5 μM metal was analyzed with the program DYNAFIT43 by using a two binding site model. This experiment yielded Zn2+ affinities of Kd2 43 ± 6 and Kd3 408 ± 18 nM (Figure 5B). As a control, zinc was titrated into an MF-2 solution containing MBP, and the change in signal was the same as when the protein was absent, confirming no strong zinc binding by the tag (data not shown).
Figure 5. MF-2 competitions of zinc binding to YeiR-MBP.
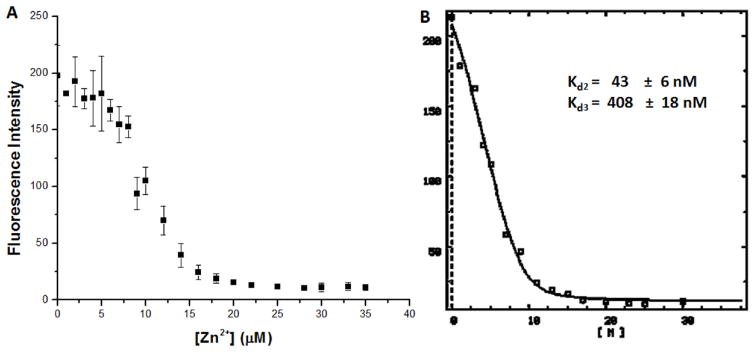
(A) Zn2+(0 to 35 μM) was titrated into samples containing 5 μM protein and 0.5 μM Mag-fura-2, after which the samples were incubated at 4°C inside a glovebox. Mag-fura-2 was excited at 366 nm, and the decreasing fluorescence upon metal addition was monitored at 500 nm. The data points are averages from three experiments, and the error bars represent 1 standard deviation. (B) The data were re-zeroed at 5 μM metal added, and then the modified data set was analyzed by using DYNAFIT 43 using a script describing the competition between the protein and the Mag-fura-2 for Zn2+, assuming two metal sites on YeiR-MBP.
Given that several other G3E P-loop GTPases are implicated in nickel delivery during metallocenter assembly of nickel-containing enzymes,11 we also examined nickel binding to YeiR. Upon addition of Ni2+ to YeiR-MBP a broad peak with a maximum around 325 nm became apparent in the electronic absorption spectrum (Figure S7B, ε325 = (3.43 ± 0.3) × 103 M−1cm−1). Both the intensity and position of the peak suggest that it originates from a Cys-S−→Ni2+ ligand-to-metal charge transfer (LMCT).44 Control Ni2+ titrations were conducted with the MBP protein and no changes in the electronic absorption spectrum of the apo-protein were detected (data not shown), suggesting that the LMCT arises from nickel binding to YeiR. This interpretation is supported by experiments with a mutant YeiR-MBP protein in which the three cysteines in the C63XCC66 motif were mutated to alanine, serine and serine, respectively (referred to as the ASS mutant). Following incubation of this mutant protein with Ni2+ no change in the electronic absorption spectrum compared with apo-protein was observed (Figure S7B), suggesting that the nickel-binding ligands include cysteines from that motif. To determine if zinc also binds to the amino acids that make up this motif, Zn2+ was titrated into the Ni2+-loaded WT protein, resulting in a decrease in the electronic absorption signal, indicating that Zn2+ also binds to YeiR-MBP in a site that at least partially overlaps that of a Ni2+ site (Figure S7C). However, when the ASS mutant was incubated with excess Zn2+ followed by gel filtration chromatography, the amount of zinc ions detected by ICP-AES only decreased to 2.6 ± 0.4, suggesting that a zinc binding site has not been completely disrupted by the mutations.
GTPase activity
The sequence of YeiR includes the four canonical motifs of G-proteins: the phosphate binding Walker A motif, the guanine recognition motif NKXD, the Walker B motif, and the invariant N38 and E39 residues in the switch I region that bind the γ-phosphate of the NTP.45–47
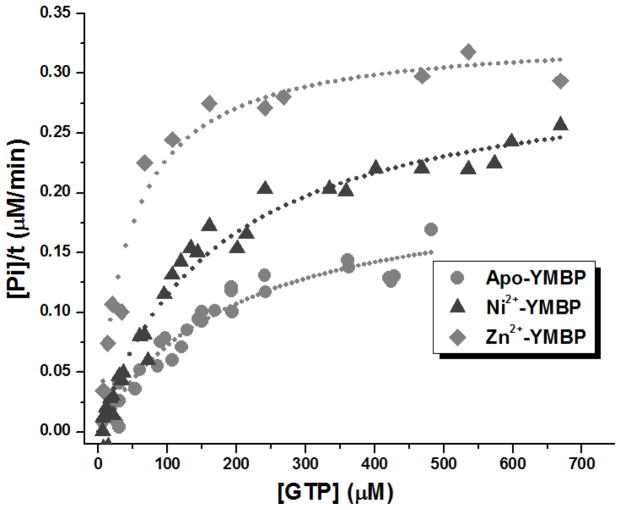
The GTPase activity of the YeiR-MBP was monitored by using the Malachite Green colorimetric assay for free phosphate at varying GTP concentrations, revealing saturation behavior (Figure 6). The experiment was carried out with either 1 μM apo-protein inside the anaerobic glovebox or with protein first incubated overnight with either Ni2+ or Zn2+. The GTPase activity of the apo-protein was low, displaying a Km of 220 ± 81 μM and a kcat of 0.19 ± 0.03 min−1 (Table 1). This activity is similar to that observed for other P-loop GTPases such as E. coli HypB (kcat = 0.17 min−1, KM = 4 μM),48 Bacillus pasteurii UreG (kcat = 0.04 min−1),49, Methlobacterium extorquens MeaB (kcat = 0.039 min−1)18 and even the human P-loop GTPase protein hMMAA (kcat = 0.15 min−1, KM = 24 μM).50 Nickel-bound protein exhibits a slight improvement in GTPase activity over the apo-protein, with a KM of 177 ± 50 μM and a kcat of 0.33 ± 0.05 min−1. The catalytic efficiency of the protein increased further when Zn2+ was added to apo-YeiR-MBP, yielding a reaction turnover number only slightly higher than that of the apo-protein with a kcat of 0.32 ± 0.03 min−1, but displaying a KM (49 ± 22 μM) four times lower than that of the apo-protein (Table 1), resulting in an 8-fold increase in kcat/KM.
Figure 6. GTPase Activity of YeiR-MBP.
Apo-protein (1 μM) or protein first incubated with nickel or zinc was incubated with the indicated concentrations of GTP for 2.5 h at 37°C, followed by quantification of the released phosphate. Representative data sets and fits to the Michaelis-Menten equation are shown and the calculated kinetic constants are listed in Table 1.
Table 1.
GTPase Activity of YeiR-MBP
| Protein | KM (μM) | kcat (min−1) | kcat/KM (sec−1 M−1) |
|---|---|---|---|
| Apo-YeiR | 220 ± 81 | 0.19 ± 0.03 | 14 ± 6 |
| YeiR + Ni2+ | 177 ± 50 | 0.33 ± 0.05 | 31 ± 10 |
| YeiR + Zn2+ | 49 ± 22 | 0.32 ± 0.03 | 110 ± 21 |
The GTPase activity was determined for 1 μM protein by using the Malachite Green (MG) assay for free phosphate. Holo protein samples contained a 3-fold excess of either Zn2+ or Ni2+. Samples with varying concentrations of GTP were incubated at 37° C for 2.5 hours prior to analysis. Control samples without protein were analyzed to determine the rate of background hydrolysis. The data were fit to the Michaelis-Menten equation. Experiments were performed at least in triplicate and errors represent one standard deviation.
Metal-dependent quaternary structure
A property of many SIMIBI (signal recognition particle, MinD, BioD) NTPases is a dynamic equilibrium between monomeric and dimeric states.9 To investigate the oligomeric state of YeiR-MBP in solution, protein samples were analyzed by size exclusion chromatography (Table S1 and Figure S9). These experiments revealed a variable amount of protein eluting after the monomer protein peak (Figure S9), which was attributed to a small fraction of protein degradation. YeiR-MBP eluted as a monomer and the addition of GTP did not appear to have any significant effect on the quaternary structure of YeiR-MBP (Figure S9). On the other hand, when either Ni2+ or Zn2+ was added to the samples in the absence of nucleotide, peaks corresponding to larger oligomeric species were apparent. This type of metal-dependent dimerization has been observed in other members of the family such as H. pylori UreG, which dimerizes in the presence of Zn2+ but not Ni2+ ions.51 Protein samples incubated with a metal as well as nucleotide elute as oligomers that cannot be resolved.
Discussion
YeiR belongs to the G3E family of P-loop GTPases,9, 36 and experimental information collected for other subfamilies including HypB,42, 48 UreG,14, 16, 17 and MeaB,18, 52 indicate that they are GTPases involved in cofactor loading. By analogy, we propose that YeiR and other members of the COG0523 group could have similar properties. However, while all HypB and UreG proteins studied are involved in nickel homeostasis and all MeaB proteins studied are involved in cobalamin homeostasis, the members of the COG0523 subfamily are linked to different metals: CobW19 to cobalamin (cobalt-containing cofactor), the NHase activator to iron22, 28 and YciC to zinc.32 The phenotypes caused by deleting yeiR from the E. coli genome have led us to propose that like YciC of B. subitlis, the role of YeiR in E. coli is also linked to zinc.
The yeiR mutant has an EDTA-sensitive phenotype, which we suggest was due to competition for zinc by the chelator. The growth defect was specifically rescued by the addition of exogenous zinc and deletion of the high-affinity, highly-specific zinc transporter (ZnuABC) increased the efficacy of EDTA to cause the phenotype. Several other transporters have been shown to participate in the uptake of zinc under growth conditions of zinc sufficiency,37, 53 but the high-affinity zinc transport system is expressed in response to zinc limitation.39 ZnuA structural changes induced specifically by zinc are thought to be responsible for the high selectivity of this transporter.54 Not only was the yeiR mutant more sensitive to EDTA with this transporter missing, but zinc no longer rescued the growth defect to the same extent as when the transporter was present.
As suggested previously,55 the results presented here imply that ZnuA is able to compete with EDTA for binding to zinc in the medium. As such, approximately 60-fold less EDTA is needed to observe a growth defect of the ΔyeiR mutant if the transporter is absent. The different metal suppression profiles collected for the ΔyeiR and ΔznuABC::cam ΔyeiR mutants could also be a direct result of the competition between the transporter and EDTA. With znuABC present on the chromosome, only 25 μM zinc was needed to suppress the impact of 1.4 mM EDTA (Figure 2B). However, with znuABC deleted, 10 μM zinc was required to suppress the effects of 20 μM EDTA (Figure 2D), a much higher ratio of metal to chelator.
Additionally, at such a high ratio of metal to EDTA, cobalt and to a lesser extent nickel were able to partially suppress the EDTA-dependent growth defect exhibited by the ΔznuABC::cam ΔyeiR mutant. At this ratio of EDTA:metal (2:1), it is possible that cobalt and nickel are displacing zinc from EDTA, which is then available for uptake through low-affinity transporters.39 The stability constants between EDTA and Co2+ and EDTA and Zn2+ are roughly the same (log K equals 18.1 and 18.3, respectively).56 The stability constant between EDTA and Ni2+ is roughly two orders of magnitude higher (log K equal to 20.4).56 A remote possibility is that cobalt and nickel are also able to directly rescue the ΔznuABC::cam ΔyeiR mutant growth defect. However, this prospect remains to be explored.
The yeiR mutant also has a Cd2+-sensitive phenotype. Intracellular zinc limitation induced by cadmium has been postulated to be responsible for cadmium toxicity.38, 57 Accordingly, we observed that the ΔznuABC::cam mutant, which is unable to transport zinc in a high-affinity capacity,39 is more sensitive to cadmium than the isogenic parent and the ΔznuABC::cam ΔyeiR mutant is more sensitive still. Zinc was able to partially rescue the cadmium-phenotype of the ΔyeiR strain, although not as specifically as with the EDTA-experiments.
These results point to a link between YeiR and zinc as has been found for YciC from B. subtilis, but YeiR and YciC are members of distinct COG0523 clades of the COG0523 superfamily.36 The YeiR clade, however, does contain putatively zinc-regulated proteobacterial COG0523 proteins (Figure 1). As observed for yeiR, deletion of yciC also leads to an EDTA-dependent growth defect when znuA is deleted,30, 58 and the yciC phenotype was later confirmed to be due specifically to zinc depletion.32 While the involvement of yciC in zinc homeostasis in supported by its regulation by Zn2+ through Zur31, Zur regulatory sites are not predicted to exist upstream of yeiR.36, 59 Furthermore, yeiR has not been identified in any of the studies that examined the response of E. coli to zinc depletion.38, 60 Thus, it is unclear whether expression of yeiR is influenced by the metal requirements of the cell.
In this study, we also sought to characterize metal binding to YeiR, which is predicted from its sequence similarity to the known nickel-binding proteins UreG and HypB, and to confirm GTPase activity, a characteristic of G3E family proteins that has not been investigated for the COG0523 subgroup. Several different methods were used to establish that YeiR-MBP binds multiple zinc ions. Although we can not rule out a contribution from the MBP tag to metal binding, the observation that zinc impacts the biochemical activities of this protein supports at least one specific metal-binding site on YeiR. Zinc influences the GTPase activity of YeiR-MBP, which could provide a mechanism for linking the activity of the protein to zinc homeostasis. In contrast, a physiological connection between Ni2+ and YeiR in vivo was not established and less stimulation of the GTPase activity of YeiR-MBP was observed in the presence of Ni2+. It was previously shown that the activity of several HypB homologs are also affected by metal binding,42, 61 but in those cases metal inhibits GTPase hydrolysis, so YeiR is the first example to display GTP hydrolysis acceleration upon metal loading. Mutation of the cysteine residues of the CXCC motif embedded in the GTPase domain abrogates the ability of the protein to complement the EDTA sensitivity of the yeiR strain of E. coli, supporting a functional role, but whether this is due to direct zinc binding at this site is not clear. Our experiments suggest that Zn2+ is able to compete with Ni2+ bound to the cysteine ligands in this motif, but mutation of these cysteines does not dramatically diminish zinc binding in vitro. Furthermore, it is clear that there are multiple metal-binding sites on the protein, including the poly-histidine motif (HxHxH) found in the C-terminal section of the protein (Figure S1). Further work will be necessary to establish the function of this CXCC motif and the detailed coordination environments of the metals. Nevertheless, taken together these results support a functional link between the GTPase and metal-binding activities of this family of proteins.
Ensuring proper metal allocation to the active sites of metalloenzymes is critical for the survival of any organism. Studies involving the maturation of [NiFe]-hydrogenase and of urease have provided detailed pictures of metallocenter assembly.11, 62 The mechanism and function of each accessory factor required for maturation of these enzymes is still not fully understood, but in both cases the G3E P-loop GTPase is involved in the incorporation of the Ni(II) ion into the enzyme’s active site. By analogy, YeiR could have similar roles in the activation of a target enzyme(s) whose function becomes essential when the cell is faced with metal stress such as that induced by either EDTA or cadmium. Whether the affect of zinc binding on GTPase activity is an indication that the target protein requires a zinc cofactor remains to be determined.
Materials and Methods
Materials
Restriction endonucleases and T4 DNA ligase were obtained from New England Biolabs. Pfu DNA polymerase was purchased from Stratagene. Kanamaycin, TCEP and IPTG were obtained from BioShop (Toronto, ON). Phusion High-Fidelity DNA polymerase was purchased from Finnzymes. High Flow Amylose resin was acquired from New England Biolabs and all other chromatography media were purchased from GE Healthcare. Pure metal salts and analytical grade metal solutions were purchased from Sigma-Aldrich. All studies were prepared with Milli-Q water, 18.2 MΩ-cm resistance. Buffers for all metal assays were treated with Chelex-100 (Bio-Rad) to minimize trace metal contamination. Primers (Table S2) were purchased from Sigma Genosys or Integrated DNA Technologies. Metal salts used for suppression of the EDTA-dependent growth defect (ZnSO4, CoCl2, CuCl2, MnCl2, NiCl2, FeCl3) were Puratronic® grade from Alfa Aesar.
Bacterial strains and growth conditions
E. coli strains were routinely grown at 37°C in LB-Lennox medium (LB) or a low-phosphate Tris-buffered minimal medium (LP)63 that contained 0.2% glycerol and 0.3% casamino acids. The ferric citrate in this medium was replaced with FeSO4 at 3 mg/L. When required, growth media were solidified with 15 g of agar/liter for preparation of plates. Kanamycin (kan, 50 μg/ml), ampicillin (amp, 100μg/ml) and chloramphenicol (cam, 30 μg/ml) were added as required. E. coli K-12 MG1655 was used as the wild type strain in all experiments. The yeiR::kan allele from the Keio collection (JW2161)64 was transferred to MG1655 by P1 transduction.65 The kan marker was excised from the chromosome by FLP-recombinase-mediated recombination.66 The ΔznuABC::cam allele from E. coli GR35237 was transferred to MG1655 and ΔyeiR by P1 transduction. For EDTA and CdSO4 sensitivity experiments, overnight cultures of strains grown in LB were washed once with LP medium then normalized to an optical density at 600 nm of 1.0. Normalized cultures were then diluted 1/500 into fresh LP medium. Growth curves were generated with a Bioscreen C (Growth Curves USA, Piscataway, NJ) at 37°C with intensive shaking. All growth experiments were repeated independently at least three times. Strains were routinely checked by PCR amplification with primers external and internal to the deleted gene (Table S2).
Construction of Expression Vectors and Mutants
For yeiR rescue experiments, yeiR was amplified by polymerase chain reaction (PCR) using Phusion High-Fidelity DNA polymerase according to the manufacturer’s guidelines from MG1655 genomic DNA using the YeiR_pBAD24fw and YeiR_pBAD24rev (Table S2) that were designed with restriction sites for NcoI (forward) and XbaI (reverse). PCR-amplified yeiR was then cloned into pBAD24 resulting in pCH011. For site-specific mutagenesis of yeiR used in rescue experiments, the overlap extension method67 was employed with primers listed in Table S2 and pCH011 as template.
For overexpression, the coding sequence for YeiR was amplified from E. coli DH5α and cloned into a pET24b(+) vector. The yeiR gene was then amplified from this vector using the primers YeiRfw and YeiRrev (Table S2) that were designed with restriction sites for NdeI (forward) and XhoI (reverse), cut with those enzymes, and ligated into the pIADL-16 vector to produce the plasmid expressing YeiR-MBP with an N-terminal MBP tag.68 The triple mutant (ASS) was generated from the YeiR-MBP parent vector by using QuikChange PCR mutagenesis with Pfu polymerase followed by DpnI digestion. Plasmids were transformed into XL-2 Blue competent cells and isolated using the Qiagen plasmid mini-prep kit. All plasmids were sequenced in the forward and reverse directions (ACGT, Toronto, ON or the UF ICBR sequencing core, Gainesville, FL) to verify the cloned sequences and mutations.
Protein Expression
For expression of wild type and mutant YeiR-MBP, the plasmids were transformed into BL21(DE3) cells (Novagen). An overnight culture of 50 ml of the transformed cells was grown at 37° C and 15 ml of it was used to inoculate 1.5 L of LB medium, supplemented with 50 μg/ml kan. The cells were grown aerobically at 37° C until the OD measured at 600 nm reached 0.6, at this point the temperature was set to 15° C and the cells were induced with 0.25 mM IPTG. After 3–4 hours of incubation, the cells were harvested by centrifugation and resuspended in 50 ml of 20 mM Tris, pH 7.5, containing 100 mM NaCl. The cells were frozen with liquid nitrogen and stored at −80° C until needed.
Protein Purification
Two chromatography steps were required to produce protein that was >95% pure, as evidenced by a band at approximately 79 kDa in reducing SDS-PAGE analysis (Figure S7A). Thawed cells were treated with 5 mM TCEP, 1 mM PMSF and supplemented with a Complete Mini EDTA-free protease inhibitor cocktail tablet (Roche Applied Sciences). The cells were then sonicated on ice, followed by centrifugation at 4° C and 18,000 rpm (RC5B PLUS SORVALL, using an SS-34 rotor) for 40 min. The supernatant was filtered through a 0.45 μm syringe filter before being loaded onto an Amylose column. The column was washed with 50 column volumes of Buffer A (20 mM Tris, pH 7.5) containing 0.2 M NaCl at 4o C. The protein was then eluted with column buffer plus 10 mM maltose. Fractions were screened by SDS-PAGE (12.5%), and fractions containing the protein of interest were pooled together and dialyzed against Buffer A. Samples were loaded onto a MonoQ 10/100 GL (GE Healthcare) column equilibrated with buffer A and eluted with a NaCl gradient, with the protein eluting at ~150 mM NaCl. Following SDS-PAGE, protein fractions were pooled and concentrated using Amicon Ultra 3K MWCO centrifuge concentrators (Millipore) to final concentrations of at least 250 μM. Protein concentration was determined by using the calculated extinction coefficient of 113,580 M−1cm−1 for YeiR-MBP at 280 nm. The molecular mass of the purified proteins was determined by ESI-MS (Department of Chemistry, University of Toronto); the predicted molecular weight for the wild type fusion protein is 78,896.9 Da, assuming loss of the N-terminal methionine, and the observed MW was 78,898.0 Da. Protein samples were stored at −80° C until use. All experiments described in this report were performed with the fusion protein YeiR-MBP.
Apo-proteins were generated by incubating the purified protein with 10 mM EDTA and 1 mM TCEP for at least 48 hours in an anaerobic glovebox at 4° C, after which the samples were passed through two sequential PD10 gel filtration columns equilibrated with 25 mM HEPES, pH 7.6,100 mM NaCl in the glovebox. The free thiol content of the fractions was quantified by monitoring the absorption signal at 412 nm after reaction with DTNB in the presence of 6 M guanidinium hydrochloride; β-mercaptoethanol was used as a standard.
CD Spectroscopy
Mutant and wild type YeiR-MBP samples (30 μM) were analyzed on a Jasco J-170 spectropolarimeter using a cuvette with a 1 mm path length over a wavelength range of 190–350 nm at room temperature. Each experiment was the average of five scans.
Metal Titrations
Ni2+ binding was assessed by electronic absorption spectroscopy using the observed signal at 325 nm. Ni2+ titrations were prepared by incubating 400 nM apo-protein with varying amounts of Ni2+ and monitoring the metal binding by using a GBC Cintra 404 spectrometer and a 1-cm path length cuvette.
Mag-Fura-2 Competitions
To confirm Zn2+ binding to the protein, the metal-binding competitor Mag-Fura-2 was used, which has a reported mid-nMKd.69 The zinc-binding affinity of Mag-Fura-2 to Zn2+ was previously determined to be 101 nM using the same buffer system used in these studies.42 Stocks of Mag-Fura-2 (Invitrogen) were prepared in MilliQ water and quantified by using the reported extinction coefficient of 22,000 M−1cm−1 at 369 nm.69 Stoichiometry was determined by incubating 5 μM protein with 0.5 μM Mag-fura-2, after which the samples were titrated with 0 to 40 μM Zn2+ and left overnight at 4°C inside a glovebox. Mag-fura-2 was excited at 366 nm, and the decreasing fluorescence upon metal addition was monitored at 500 nm. The data were analyzed by using DYNAFIT43 using a script describing the competition between the protein and the Mag-fura-2 for Zn2+. All fluorescence experiments were conducted on a JY HORIBA Fluorolog-3 spectrofluorimeter.
Mass Spectroscopy
Apo-protein was incubated overnight with excess nickel or zinc acetate. The samples were then passed through a PD10 column equilibrated with 25 mM HEPES, pH 7.6, 100 mM NaCl in the glovebox to remove unbound metal. The most concentrated protein sample was then passed through a second PD10 column equilibrated with 10 mM ammonium acetate (pH 7.5). The samples were capped under anaerobic conditions to minimize oxidation prior to ESI-MS measurements. The mass spectra were acquired on an AB/SciexQStarXL mass spectrometer equipped with an ion spray source in the positive mode. Ions were scanned from 1000–3000 m/z with accumulations of up to 10 min per spectrum. The instrument parameters are as follows: ion source gas 50.0 psi, curtain gas 45.0 psi, ion spray voltage 4,200.0 V, declustering potential 150.0 V, focusing potential 210.0 V, collision gas 3.0, and MPC (detector) 2,200 V. The spectra were deconvoluted using the Bayesian protein reconstruction program over a mass range of 78,000 – 80,000 Da.
Analytical Superdex
Samples of 150 μl of 100 μM YeiR-MBP were incubated in an anaerobic glovebox at 4° C overnight with 3 equivalents of Ni2+ or Zn2+. Samples that included GTP were incubated an hour before injection with 600 μM nucleotide. The samples were loaded on to an Analytical Superdex 200 10/300 column (GE Healthcare) equilibrated with 25 mM HEPES, pH 7.5, 200 mM NaCl and 1 mM TCEP. The column was calibrated with thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.4 kDa) (BioRad).
GTPase Activity
The GTPase activity was determined using the Malachite Green (MG) assay for free phosphate.70, 71 A variation of this assay uses Tween 20 to stabilize the dye complex so smaller concentrations (up to 20 μM) of phosphate are detected.72 The MG reagent was prepared with 10 ml 0.045% malachite green in water with 0.1 N HCl, 2.5 ml of 7.5% ammonium molybdate in water and 200 μl of 11% Tween 20; the mix was allowed to stand for 30 min. The reaction mixture consisted of 1 μM protein, 50 mM Tris, pH 7.5, 50 mM KCl, and 5 mM MgCl2. Samples with varying concentrations of GTP were incubated at 37° C for 2.5 hours. After this, 160 μl of each reaction was plated on a 96 well plate and 40 μl of the MG reagent was added to each sample. After 3 minutes of gentle shaking, sodium citrate was added to a final concentration of 3.5%. The plate was mixed again by shaking and then incubated for 30 minutes at room temperature. The absorbance was measured at 630 nm with an EL808 Ultra microplate reader (Bio-Tek Instruments). Buffer and GTP only controls were run to determine background signals. The amount of phosphate released was calculated on the basis of a standard curve from phosphate standards (Molecular Probes). The data were analyzed using OriginPro 7.5 to fit the data to the Michaelis-Menten equation. To determine the effects of metal, 10 μM protein samples were incubated overnight with 30 μM of either Zn2+ or Ni2+ before dilution to 1 μM, and control samples in the absence of protein contained the same amounts of metal ion.
Supplementary Material
Acknowledgments
This work was supported the National Institutes of Health (Grant R01GM70641-01 to VdC-L), as well as funding from the Natural Sciences and Engineering Research Council of Canada and the Canada Research Chairs Program. The authors thank members of the Zamble lab for helpful discussions, N. Khanam for preparation of the ASS mutant vector, Dr. T. Ngu for preparation of the ESI-MS figure, and Prof. C. Rensing for the znuABC::cam strain of E. coli.
Works Cited
- 1.Vetter IR, Wittinghofer A. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 2.Hauryliuk VV. Mol Biol (Mosk) 2006;40:769–783. [PubMed] [Google Scholar]
- 3.Rodnina MV, Stark H, Savelsbergh A, Wieden HJ, Mohr D, Matassova NB, Peske F, Daviter T, Gualerzi CO, Wintermeyer W. Biol Chem. 2000;381:377–387. doi: 10.1515/BC.2000.050. [DOI] [PubMed] [Google Scholar]
- 4.Narumiya S. J Biochem. 1996;120:215–228. doi: 10.1093/oxfordjournals.jbchem.a021401. [DOI] [PubMed] [Google Scholar]
- 5.Wittmann T, Waterman-Storer CM. J Cell Sci. 2001;114:3795–3803. doi: 10.1242/jcs.114.21.3795. [DOI] [PubMed] [Google Scholar]
- 6.Segev N. Semin Cell Dev Biol. 2011;22:1–2. doi: 10.1016/j.semcdb.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meier TI, Peery RB, McAllister KA, Zhao G. Microbiology. 2000;146(Pt 5):1071–1083. doi: 10.1099/00221287-146-5-1071. [DOI] [PubMed] [Google Scholar]
- 8.Molendijk AJ, Ruperti B, Palme K. Curr Opin Plant Biol. 2004;7:694–700. doi: 10.1016/j.pbi.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 9.Leipe DD, Wolf YI, Koonin EV, Aravind L. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 10.Rosenzweig AC. Chem Biol. 2002;9:673–677. doi: 10.1016/s1074-5521(02)00156-4. [DOI] [PubMed] [Google Scholar]
- 11.Kaluarachchi H, Chan Chung KC, Zamble DB. Nat Prod Rep. 2010;27:681–694. doi: 10.1039/b906688h. [DOI] [PubMed] [Google Scholar]
- 12.Kuchar J, Hausinger RP. Chem Rev. 2004;104:509–525. doi: 10.1021/cr020613p. [DOI] [PubMed] [Google Scholar]
- 13.Maier T, Lottspeich F, Böck A. Eur J Biochem. 1995;230:133–138. [PubMed] [Google Scholar]
- 14.Mehta N, Benoit S, Maier RJ. Microb Pathog. 2003;35:229–234. doi: 10.1016/s0882-4010(03)00151-7. [DOI] [PubMed] [Google Scholar]
- 15.Olson JW, Maier RJ. J Bacteriol. 2000;182:1702–1705. doi: 10.1128/jb.182.6.1702-1705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moncrief MB, Hausinger RP. J Bacteriol. 1997;179:4081–4086. doi: 10.1128/jb.179.13.4081-4086.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soriano A, Hausinger RP. Proc Natl Acad Sci U S A. 1999;96:11140–11144. doi: 10.1073/pnas.96.20.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Padovani D, Labunska T, Banerjee R. J Biol Chem. 2006;281:17838–17844. doi: 10.1074/jbc.M600047200. [DOI] [PubMed] [Google Scholar]
- 19.Crouzet J, Levy-Schil S, Cameron B, Cauchois L, Rigault S, Rouyez MC, Blanche F, Debussche L, Thibaut D. J Bacteriol. 1991;173:6074–6087. doi: 10.1128/jb.173.19.6074-6087.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hodgkin DG, Pickworth J, Robertson JH, Trueblood KN, Prosen RJ, White JG. Nature. 1955;176:325–328. doi: 10.1038/176325a0. [DOI] [PubMed] [Google Scholar]
- 21.Heldt D, Lawrence AD, Lindenmeyer M, Deery E, Heathcote P, Rigby SE, Warren MJ. Biochem Soc Trans. 2005;33:815–819. doi: 10.1042/BST0330815. [DOI] [PubMed] [Google Scholar]
- 22.Nojiri M, Yohda M, Odaka M, Matsushita Y, Tsujimura M, Yoshida T, Dohmae N, Takio K, Endo I. J Biochem. 1999;125:696–704. doi: 10.1093/oxfordjournals.jbchem.a022339. [DOI] [PubMed] [Google Scholar]
- 23.Yamada H, Kobayashi M. Biosci Biotechnol Biochem. 1996;60:1391–1400. doi: 10.1271/bbb.60.1391. [DOI] [PubMed] [Google Scholar]
- 24.Endo I, Nojiri M, Tsujimura M, Nakasako M, Nagashima S, Yohda M, Odaka M. J Inorg Biochem. 2001;83:247–253. doi: 10.1016/s0162-0134(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 25.Nojiri M, Nakayama H, Odaka M, Yohda M, Takio K, Endo I. FEBS Lett. 2000;465:173–177. doi: 10.1016/s0014-5793(99)01746-9. [DOI] [PubMed] [Google Scholar]
- 26.Nishiyama M, Horinouchi S, Kobayashi M, Nagasawa T, Yamada H, Beppu T. J Bacteriol. 1991;173:2465–2472. doi: 10.1128/jb.173.8.2465-2472.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Z, Hashimoto Y, Shiraki K, Kobayashi M. Proc Natl Acad Sci U S A. 2008;105:14849–14854. doi: 10.1073/pnas.0803428105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashimoto Y, Nishiyama M, Horinouchi S, Beppu T. Biosci Biotechnol Biochem. 1994;58:1859–1865. doi: 10.1271/bbb.58.1859. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, Zheng Y, Yamagishi H, Odaka M, Tsujimura M, Maeda M, Endo I. FEBS Lett. 2003;553:391–396. doi: 10.1016/s0014-5793(03)01070-6. [DOI] [PubMed] [Google Scholar]
- 30.Gaballa A, Helmann JD. J Bacteriol. 1998;180:5815–5821. doi: 10.1128/jb.180.22.5815-5821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabriel SE, Miyagi F, Gaballa A, Helmann JD. J Bacteriol. 2008;190:3482–3488. doi: 10.1128/JB.01978-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gabriel SE, Helmann JD. J Bacteriol. 2009;191:6116–6122. doi: 10.1128/JB.00802-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gasper R, Scrima A, Wittinghofer A. J Biol Chem. 2006;281:27492–27502. doi: 10.1074/jbc.M600809200. [DOI] [PubMed] [Google Scholar]
- 34.Leach MR, Sandal S, Sun H, Zamble DB. Biochemistry. 2005;44:12229–12238. doi: 10.1021/bi050993j. [DOI] [PubMed] [Google Scholar]
- 35.Srinivasan C, Posewitz MC, George GN, Winge DR. Biochemistry. 1998;37:7572–7577. doi: 10.1021/bi980418y. [DOI] [PubMed] [Google Scholar]
- 36.Haas CE, Rodionov DA, Kropat J, Malasarn D, Merchant SS, de Crécy-Lagard V. BMC Genomics. 2009;10:470. doi: 10.1186/1471-2164-10-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grass G, Wong MD, Rosen BP, Smith RL, Rensing C. J Bacteriol. 2002;184:864–866. doi: 10.1128/JB.184.3.864-866.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graham AI, Hunt S, Stokes SL, Bramall N, Bunch J, Cox AG, McLeod CW, Poole RK. J Biol Chem. 2009;284:18377–18389. doi: 10.1074/jbc.M109.001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patzer SI, Hantke K. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 40.Verbruggen N, Hermans C, Schat H. Curr Opin Plant Biol. 2009;12:364–372. doi: 10.1016/j.pbi.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 41.de Seny D, Heinz U, Wommer S, Kiefer M, Meyer-Klaucke W, Galleni M, Frere JM, Bauer R, Adolph HW. J Biol Chem. 2001;276:45065–45078. doi: 10.1074/jbc.M106447200. [DOI] [PubMed] [Google Scholar]
- 42.Sydor AM, Liu J, Zamble DB. J Bacteriol. 2011;193:1359–1368. doi: 10.1128/JB.01333-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuzmic P. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 44.Solomon EI. Inorg Chem. 2006;45:8012–8025. doi: 10.1021/ic060450d. [DOI] [PubMed] [Google Scholar]
- 45.Bourne HR, Sanders DA, McCormick F. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 46.la Cour TF, Nyborg J, Thirup S, Clark BF. EMBO J. 1985;4:2385–2388. doi: 10.1002/j.1460-2075.1985.tb03943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pai EF, Krengel U, Petsko GA, Goody RS, Kabsch W, Wittinghofer A. EMBO J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maier T, Jacobi A, Sauter M, Böck A. J Bacteriol. 1993;175:630–635. doi: 10.1128/jb.175.3.630-635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zambelli B, Stola M, Musiani F, De Vriendt K, Samyn B, Devreese B, Van Beeumen J, Turano P, Dikiy A, Bryant DA, Ciurli S. J Biol Chem. 2005;280:4684–4695. doi: 10.1074/jbc.M408483200. [DOI] [PubMed] [Google Scholar]
- 50.Froese DS, Kochan G, Muniz JR, Wu X, Gileadi C, Ugochukwu E, Krysztofinska E, Gravel RA, Oppermann U, Yue WW. J Biol Chem. 2010;285:38204–38213. doi: 10.1074/jbc.M110.177717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zambelli B, Turano P, Musiani F, Neyroz P, Ciurli S. Proteins. 2009;74:222–239. doi: 10.1002/prot.22205. [DOI] [PubMed] [Google Scholar]
- 52.Korotkova N, Lidstrom ME. J Biol Chem. 2004;279:13652–13658. doi: 10.1074/jbc.M312852200. [DOI] [PubMed] [Google Scholar]
- 53.Beard S, Hashim R, Wu G, Binet M, Hughes M, Poole R. FEMS Microbiol Lett. 2000;184:231–235. doi: 10.1111/j.1574-6968.2000.tb09019.x. [DOI] [PubMed] [Google Scholar]
- 54.Yatsunyk LA, Easton JA, Kim LR, Sugarbaker SA, Bennett B, Breece RM, Vorontsov II, Tierney DL, Crowder MW, Rosenzweig AC. J Biol Inorg Chem. 2008;13:271–288. doi: 10.1007/s00775-007-0320-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berducci G, Mazzetti AP, Rotilio G, Battistoni A. FEBS Lett. 2004;569:289–292. doi: 10.1016/j.febslet.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 56.Stumm W, Morgan JJ. Aquatic chemistry. John Wiley and Sons, Inc; New york: 1996. [Google Scholar]
- 57.Zhang B, Georgiev O, Hagmann M, Günes C, Cramer M, Faller P, Vásak M, Schaffner W. Mol Cell Biol. 2003;23:8471–8485. doi: 10.1128/MCB.23.23.8471-8485.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaballa A, Wang T, Ye RW, Helmann JD. J Bacteriol. 2002;184:6508–6514. doi: 10.1128/JB.184.23.6508-6514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Panina EM, Mironov AA, Gelfand MS. Proc Natl Acad Sci U S A. 2003;100:9912–9917. doi: 10.1073/pnas.1733691100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sigdel T, Easton J, Crowder M. J Bacteriol. 2006;188:6709–6713. doi: 10.1128/JB.00680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cai F, Ngu TT, Kaluarachchi H, Zamble DB. J Biol Inorg Chem. 2011;16:857–868. doi: 10.1007/s00775-011-0782-y. [DOI] [PubMed] [Google Scholar]
- 62.Carter EL, Flugga N, Boer JL, Mulrooney SB, Hausinger RP. Metallomics. 2009;1:207–221. doi: 10.1039/b903311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mergeay M, Nies D, Schlegel HG, Gerits J, Charles P, Van Gijsegem F. J Bacteriol. 1985;162:328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratories; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 66.Cherepanov PP, Wackernagel W. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 67.Sambrook J, Russell D. Molecular Cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2001. [Google Scholar]
- 68.McCafferty DG, Lessard IA, Walsh CT. Biochemistry. 1997;36:10498–10505. doi: 10.1021/bi970543u. [DOI] [PubMed] [Google Scholar]
- 69.Golynskiy MV, Gunderson WA, Hendrich MP, Cohen SM. Biochemistry. 2006;45:15359–15372. doi: 10.1021/bi0607406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Motomizu S, Wakimoto T, Tôei K. Talanta. 1984;31:235–240. doi: 10.1016/0039-9140(84)80269-6. [DOI] [PubMed] [Google Scholar]
- 71.Attin T, Becker K, Hannig C, Buchalla W, Wiegand A. Clin Oral Investig. 2005;9:203–207. doi: 10.1007/s00784-005-0313-8. [DOI] [PubMed] [Google Scholar]
- 72.Itaya K, Ui M. Clin Chim Acta. 1966;14:361–366. doi: 10.1016/0009-8981(66)90114-8. [DOI] [PubMed] [Google Scholar]
- 73.Edgar R. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.PHYLIP FJ. (Phylogeny Inference Package) version 3.67. http://evolution.genetics.washington.edu/phylip.html.
- 75.Gabriel SE, Miyagi F, Gaballa A, Helmann JD. J Bacteriol. 2008;190:3482–3488. doi: 10.1128/JB.01978-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.