Abstract
Keratocan is an extracellular matrix protein that belongs to the small leucine-rich proteoglycan family which also includes the lumican, biglycan, decorin, mimecan and fibromodulin. Members of this family are known to play a role in regulating cellular processes such as proliferation and modulation of osteoprogenitor lineage differentiation. The aims of this study were to evaluate the expression pattern of the keratocan within the osteoprogenitor lineage and assess its role in regulating osteoblast maturation and function. Results from gene expression analyses of cells at different maturation stages within the osteoblast lineage indicate that keratocan is differentially expressed by osteoblasts and shows little or no expression by osteocytes. During primary osteoblast cultures, high keratocan mRNA expression was observed on day 14, while lower expression was detected at days 7 and 21. To assess the effects of keratocan on osteoprogenitor cell differentiation, we evaluated primary calvarial cell cultures from keratocan deficient mice. The mineralization of calvarial osteoblast cultures derived from keratocan null (kera−/−) mice was lower than in wild type osteoblast cultures. Furthermore, analysis of RNA derived from kera−/− calvarial cell cultures showed a reduction in the mature osteoblast differentiation markers, i.e., bone sialoprotein (BSP) and osteocalcin (OC). In addition, we have evaluated the bone formation in keratocan deficient mice. Histomorphometric analysis indicated that homozygous knockout mice have a significantly decreased rates of bone formation rate and mineral apposition. Taken together our results demonstrate the expression of keratocan by osteoblast lineage cells and its ability to modulate osteoblast function.
Keywords: keratocan, osteoblast, osteocyte, differentiation
INTRODUCTION
Proteoglycans are among the non-collagen, extracellular matrix proteins present in the bone tissue, and participate in collagen fibrillogenesis [1; 2; 3]. Studies of the gene expression profile of proteoglycans in bone have generated insights into their specific functions during osteoblast maturation [4; 5]. Keratocan is a member of the small, leucine-rich, proteoglycan family (SLRP) [6; 7]. SLRPs are extracellular matrix (ECM) molecules that contain a tandem array of leucine-rich repeat (LRR) motifs flanked by conserved cysteine residues in N-and C-terminal domains of the core proteins [8]. SLRPs are divided into three classes based on the spacing of their N-terminal cysteines, the number of LRRs they contain, and their corresponding gene structures. Class I (decorin, biglycan, and asporin) and class II SLRPs (fibromodulin, proline/arginine-rich end leucine-rich repeat protein (PRELP), keratocan, lumican, and osteoadherin/osteomodulin) contain 10–12 LRRs, whereas class III SLRPs (EPN/PG-Lb/DSPG3, opticin, and osteoglycin/mimecan) contain 6–8 LRRs [9]. SLRPs members, biglycan and decorin have been found in many skeletal regions including articular and epiphyseal cartilage, and the periosteum [10]. Mice lacking biglycan and decorin demonstrated the importance of the effects of microenvironment on stem cells. The phenotype of mice doubly deficient in both biglycan and decorin can be attributed to deregulation of transforming growth factor beta (TGFβ) sequestration, resulting in the increased apoptosis of osteoprogenitors [11]. In contrast, mice deficient in fibromodulin have enhanced alveolar bone formation at postnatal days 0 and 7, but normal levels at day 21 [12].
The expression of keratocan has been reported in the cornea and thinner corneal stroma was observed in keratocan deficient mice in comparison to wild-type littermates. As demonstrated by transmission electron microscopy, Kera−/− mice have larger stromal fibril diameters and less organized packing of collagen fibrils in stroma than those of the wild type animals [6]. These studies indicate that keratocan in cornea acts as a vital component in collagen fibrillogenesis [13; 14]. Using immunohistochemistry keratocan was also detected in tendon where it is likely to play a role in collagen fibrillogenesis [15].
Recently, using visual reporters of osteoblast lineage differentiation markers and microarray analysis, we have observed the expression of keratocan in the population of cells expressing the osteoblastic marker (Col2.3GFP) [5]. In this study, we investigated the role of keratocan in osteoblast differentiation and function using keratocan deficient mice. Our result shows a reduction in bone markers and decreased mineralization in primary calvarial osteoblast cultures derived from kera−/− mice. Similarly in vivo evaluation of 3-month-old kera−/− mice demonstrated decreases in both bone formation and mineral apposition rates. This result suggests a role for keratocan in the normal expression of differentiation markers, i.e., bone sialoprotein (BSP) and osteocalcin (OC) by osteoblasts.
MATERIALS AND METHOD
Mouse Models
The 2.3 kb of rat type I collagen promoter fragment-cyan fluorescent reporter (pOBCol2.3CFP) and dentin matrix protein 1- topaz fluorescent reporter (DMP-1-GFP) mice have been previously developed and characterized [16; 17]. Mice deficient for Keratocan (kera−/−) have been developed and characterized by Dr. Winston Kao [6]. Experimental mice were generated by breeding Kera+/− mice to produce all three genotypes (kera−/−, kera+/−, and kera+/+). Different genotypes were identified using the following primers kera-1, 5′-gatggcctagtcggccatcactgcaaagag-3′; kera-2, 5′-cacccagagtattaaacagtttggggttgc-3′; neo(+), 5′-cgcttcctcgtgctttacggtatcgccgctc-3′. [6].
Calvarial Osteoblast Culture
Calvarial cells were isolated from 5–7 day-old neonatal wild type and kera−/− mice using a modified version of the protocol developed by Wong and Cohn [18; 19]. Calvaria were subjected to four, sequential, 30-min digestions in an enzyme mixture containing 0.05% trypsin and 1.5 U/ml collagenase P at 37 °C. Cell fractions 2–4 were pooled and enzyme activity was terminated by addition of culture media (Dulbecco’s modified Eagle’s medium (DMEM) Life Technologies) supplemented with 10% FBS (Hyclone). Cells were plated at a density of 1.5 ×105 cells/well in 6-well culture plates. Differentiation medium (αMEM, 10% FBS, 50 μg/ml ascorbic acid, 4 mM β-glycerophosphate) was added when cells reached confluence, usually on day 7 after plating. Cells were harvested on days 7, 14 and 21 for RNA or stained for alkaline phosphatase expression or mineralization.
Cell Separation Using Flow Cytometry Cell Sorting
Neonatal calvaria derived from 5–8 day old dual reporter transgenic mice (Col2.3CFP/DMP-1GFP) were enzymatically digested using a procedure described above [5]. Cells were sorted using a FACS Vantage (Becton Dickinson) cell sorter with appropriate lasers to excite GFP at 488nm and CFP at 413nm. Details of the cell sorting procedure and isolation of cells for RNA extraction have been previously described [5].
Histochemical Analysis of Cell Cultures
Histochemical staining for ALP activity was performed using a commercially available kit (86-R, Sigma Diagnostics, Inc., MO, USA) according to the manufacturer’s instructions. Mineralization was assessed using the von Kossa silver nitrate staining method. The results of staining procedures were recorded using a scanner (UMax Astra 4000U), and composite images were generated using Adobe Photoshop.
RNA extraction
Total RNA was isolated from cell populations using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Measurement of RNA yield was performed using a NanoDrop 1000A Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
RT-PCR
Following the extraction and quantification procedure, RNA was subjected to DNase treatment (DNase I, Invitrogen) to eliminate genomic DNA contamination. cDNA was synthesized using an Invitrogen Superscript First-strand Synthesis System for RT-PCR. TaqMan® Gene Expression Assays were purchased from ABI and real time PCR was performed on the 7500 Real-Time PCR System (assay ID: Kera, Mm00515230_m1, Osteocalcin, Mm03413826_mH, BSP, Mm00492555_m1, and ALP, Mm01187117). GAPDH was used as internal control (Mm99999915_g1). Before using the ΔΔCT method for quantification, validation experiments were performed to demonstrate that the amplification efficiencies of target genes and the reference gene were approximately equal.
Northern blot
Total RNA (10 ug) was separated on a 2.2 M formaldehyde/1% agarose gel and transferred onto a nylon membrane (Maximum Strength Nytran Plus, Schleicher & Schuell). Membranes were probed with a 440 bp mouse OC fragment (p923) [20] and mouse BSP [21]. Probes were radiolabeled by the random primer method using (α-32P) dCTP (New England Nuclear, 3000 Ci/mmol) to obtain specific activities of approximately 1 × 109 cpm/μg. Filters were hybridized with 3 × 106 cpm/ml 32P-labeled probe at 42 °C in 50% formamide, 5x SSPE (1x SSPE = 0.149 M NaCl, 10 mM NaH2PO4, 1 mM EDTA, pH 7.4), 1.2x Denhardt’s and 0.5% sodium dodecyl sulfate and washed according to published procedures [22].
Bone histomorphorphometric analysis
To perform dynamic histomorphometry, 3-month-old mice were injected intraperitoneally with 10 mg/kg of calcein and 90 mg/kg of xylenol orange. Calcein was injected 7 days before animals were sacrificed, while xylenol orange was injected2 days before sacrifice. The femurs were fixed in 10% formalin at 4 °C, and immobilized in embedding media (Cryomatrix, Thermo Shandon, Pittsburgh). The embedded bones were longitudinally sectioned in 5 μm thick sections using CryoJane tape system (Instrumedics, NJ USA). Sections were covered in 50%glycerol/PBS and dynamic histomorphometry was completed using the Osteomeasure system (OsteoMetrics, Inc., Atlanta, GA). Measurements were completed within the trabecular region region 400μm away from growth plate and 225μm away from both sides of the endocortical suface. The measurements, terminology, and unitsused for histomorphometry were basedon the convention of standardized nomenclature [23].
Immunohistochemistry
Calvaria from 6 weeks old mice were fixed for 3 days in 4% paraformaldehyde/PBS (pH 7.4) at 4°C, decalcified for 3 days, placed in 30% sucrose/PBS overnight, and embedded. For immunohistochemistry, 5 μm cryosections were briefly rinsed in PBS, blocked with 5% normal serum in 1% BSA/PBS for 1 hour, and then incubated overnight with rabbit polyclonal anti-keratocan (SC-66941, Santa Cruz Biotech, CA, USA). Signal was detected with Envision System HRP (DAB) (Dakoctomation, Carpinteria, CA), sections were counter stained with hematoxylin and imaged with a microscope equipped with a CCD camera.
RESULTS
Keratocan expression in cells of the osteoblast lineage
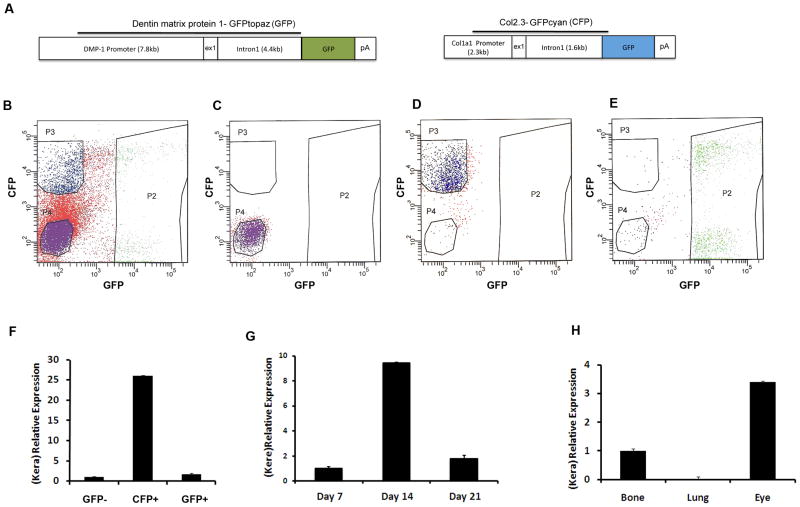
To demonstrate the expression of keratocan by osteoblast lineage cells, we performed real time PCR analysis of RNA derived from freshly isolated osteoblasts and osteocytes obtained from neonatal calvaria at postnatal days 5–8. We utilized previously established GFP transgenic mice in which defined stages of osteoblast differentiation were marked by expression of specific variants of GFP protein (Fig. 1A). The Col2.3 promoter directs the expression to osteoblasts and osteocytes, while DMP-1 promoter can be used to identify preosteocytes and osteocytes. Using a combination of different variants of GFP stage specific cell population can be separated [5; 22]. Osteoblasts and osteocytes were separated using flow cytometry cell sorting as described previously [5]. Gates utilized for cell sorting are shown in Figure 1B, and purity of the sorted population was confirmed by re-analysis (Fig. 1C–E). Analysis of gene expression using real time PCR demonstrated the strong expression of keratocan by mature osteoblasts (Fig. 1F, CFP+ group), while a weak signal was observed in the group that corresponds to osteocytes (Figure 1F, GFP+ group). This finding was confirmed during differentiation of primary calvarial osteoblast cultures, i.e. we detected peak mRNA levels of keratocan mRNA at day 14 of culture, while lower expression was observed at days 7 and 21 respectively (Fig. 1G). By day 14 osteoprogenitor cells differentiate into mature osteoblasts that express established markers characteristic of matrix producing bone cells. We compared the level of keratocan expression in bone to other tissues that have been shown to express high keratocan levels. The expression of keratocan in bone was three-fold lower than in corneal-containing eye tissue. Keratocan expression was not detected in the sample of lung that we have utilized as negative control (Fig. 1H).
Figure 1. Detection of Keratocan mRNA in osteoblast lineage cells.
(A) Transgenic constructs used to generate Dmp1-GFPtopaz (GFP) and Col2.3GFPcyan (CFP). (B) Isolation of cell populations based on visual transgene expression was completed using flow cytometry with sorting gates that clearly define populations (P2, DMP-1GFP+; P3, Col2.3CFP+; P4, Dmp-1GFP-/Col2.3CFP-). Reanalysis of the isolated population of GFP- cells (C), CFP+ (D) and GFP+ (E).
(F) Keratocan expression in freshly isolated calvarial osteoblasts and osteocytes obtained from dual transgenic mice (DMP-1topaz/Col2.3cyan). Higher expression of keratocan was observed in the Col2.3CFP positive population than in the DMP-1GFP positive cells. Four biological replicates were analyzed by real time PCR. (G) Real time PCR detection of keratocan mRNA in primary mouse calvarial osteoblast cultures. Keratocan was highly expressed on day 14 compared to earlier (day 7) or later (day 21) time points. (H) Real time PCR detection of keratocan mRNA in samples derived from bone, lung and eye.
Having determined that keratocan mRNA is present in osteoblasts, we analyzed keratocan expression at the protein level. Consistent with the mRNA results, keratocan protein was detected in osteoblasts as demonstrated by immunohistochemistry (Figure 2), Keratocan expression was observed in the osteoblast lining the periosteal and endosteal surfaces.
Figure 2. Detection of keratocan protein by immunohistochemistry.
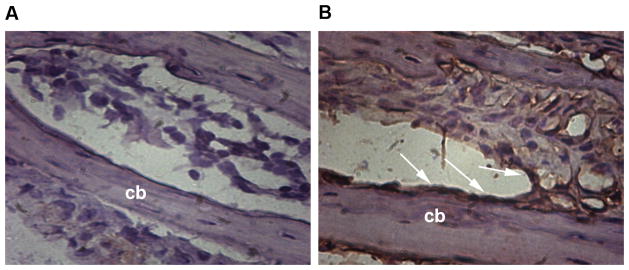
(A–B) Cryosections of calvarial bone harvested from 4-week-old mice were immunostained for keratocan and counterstained with hematoxylin. The left panel (A) shows a negative control without primary antibody. Right panel (B) shows alternate section stained with primary anti keratocan antibody. Keratocan expression was observed in osteoblasts lining bone surfaces (B, arrows indicate osteoblast layer; cb, cortical bone).
Analysis of bone formation in Kera deficient Mice
We performed histomorphormetric analysis on bone tissues derived from 3-month-old wild type and keratocan knockout mice. Our results demonstrate a significant decrease in the mineral apposition rate (MAR) (Fig. 3A, P=0.05) in femurs derived from keratocan deficient mice compared to samples derived from wild type control littermates. Furthermore, the bone formation rate was significantly decreased in kera−/− mice when compared to the wild type (Fig. 3B, p= 0.007). Mice heterozygous for keratocan also had both MAR and BFR closer to the levels of the complete knockout. Mineralizing surface also showed a decreasing trend in Kera−/− mice compared to wild type mice.
Figure 3. Dynamic histomorphometric analysis of Kera−/− mice.
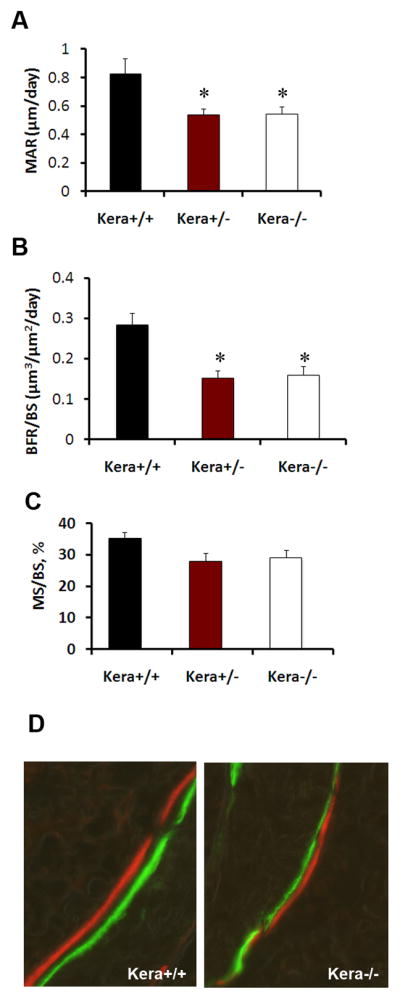
Frozen sections were obtained from 3 months old male kera−/− (n=5), kera+/− (n=7) and wild type littermates (n=8). Mice received an i.p. injection of calcein and following 5 days interval they were injected i.p. with xylenol orange. Sections were analyzed for histomorphometric parameters (A) Mineral apposition rate (MAR, μm/day), (B) Bone formation rate/bone surface (BFR/BS, μm3/μm2/day) and (C) Mineralizing surface, %. Values represent mean ± SD, *p≤0.05. (D) Representative images of Calcein-Xylenol orange in the trabecular region of kera+/+ and Kera−/−.
Effects of Keratocan deficiency on osteoprogenitor differentiation In vitro
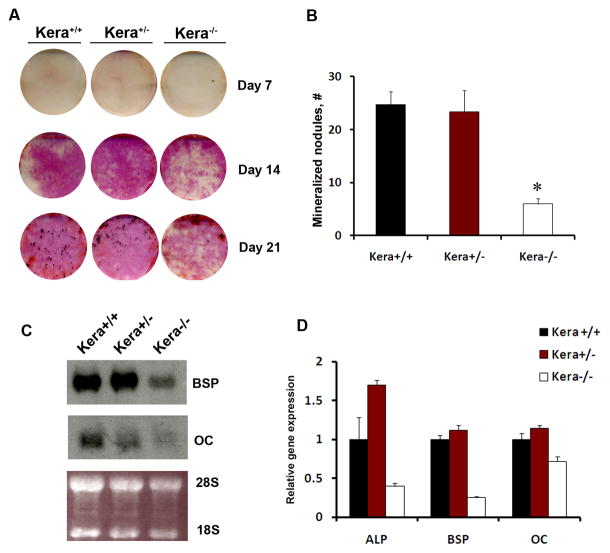
The histomorphometric observation prompted us to evaluate whether we could explain the results by studying the phenotype of osteoblast cells isolated from the calvaria (parietal bones) of kera−/− mice. Primary calvarial osteoblasts cultures from kera−/− mice showed detectable differences in the expression of ALP compared to wild type (Figure 4A, D). Decreased mineral deposition, as detected by von Kossa staining, was observed on day 21 of culture (Figure 4B). In addition, Northern blot analysis of RNA demonstrated decreased expression of mature osteoblast markers like bone sialoprotein and osteocalcin at 21 days of culture in cells obtained from kera−/− mice (Figure 4C, D). Results obtained in vitro are in line with the observation that keratocan is preferentially expressed in osteoblasts. Therefore, we did not expect to find significant changes in osteogenic parameters until the lineage maturation reached the osteoblast stage.
Figure 4. Decreased differentiation of primary osteoblasts derived from kera−/−mice.
A) Alkaline phosphatase expression (days 7–21) and mineralization (day 14–21) were analyzed using Von Kossa method (black spots) during calvarial osteoprogenitor cell differentiation in vitro. A representative of three biological experiments is shown. (B) mCOB cells derived from kera−/− show decreased mineralization while heterozygous mice did not show significant change in the mineralization, * p≤0.05). C) Following osteogenic induction osteocalcin and bone sialoprotein expression in mCOB cultures derived from kera−/− mice were analyzed by Northern blot analysis. A decrease in both bone sialoprotein and osteocalcin is observed on day 21 in kera−/−, compared to wild type (kera+/+) mice. D) Real time PCR analysis of mRNA expression for ALP, BSP and OC (results presented in figure 4D are technical replicates derived from one of two experiments;
DISCUSSION
This study confirms that keratocan is expressed in bone, as demonstrated by the presence of keratocan mRNA and protein in mature osteoblasts. Keratocan is a member of the small, leucine-rich, proteoglycan family (SLRP). This family includes lumican, decorin, and biglycan[24]. During embryonic development members of this family have been reported to participate in bone development [10]. Interestingly, we observed the highest expression of keratocan in mature osteoblasts as compared to osteoprogenitors and osteocytes. Mature osteoblasts are very active in the bone formation process, synthesizing and secreting collagenous and non-collagenous proteins. The expression of SLRPs in osteoblast lineage cells in culture tends to follow a pattern corresponding to their reported function or role in bone development. For instance, while biglycan was detected in forming growth plates, fibromodulin, a closely related homologue to keratocan, was detected at the primary ossification center [25]. Also, biglycan and fibromodulin were highly expressed in the disc and articular cartilage of the temporomandibular joint (TMJ) [26], and therefore, mice deficient in both biglycan and fibromodulin developed accelerated osteoarthritis with visible small vertical clefts in the condylar cartilage and partial disruption of the disk [26; 27]. It has been previously demonstrated that ablation of the Kera gene resulted in subtle structural alterations of the collagenous matrix in the mouse cornea, suggesting that keratocan might play a role in maintaining the appropriate corneal shape to ensure normal vision [6]. We have investigated whether keratocan plays any role in the bone tissue.
We observed decreased expression of osteoblast differentiation markers in day 21 calvarial osteoblasts derived from a keratocan−/− mice. It is intriguing that at day 21 of culture, osteoblasts from kera−/− mice demonstrated decreased mineral deposition. This observation suggests that keratocan is important at the mature osteoblast stage. In addition, In vivo results obtained by dynamic histomorphometry showed decreased MAR and BFR in kera−/− mice compared to wild type. Mice heterozygous for the keratocan mutation exhibited MAR and BFR changes similar to the complete knockout. We could speculate that this finding may suggest that there is a critical amount of the keratocan necessary for proper osteoid formation. Indeed, during in vitro differentiation markers of osteogenic differentiation were not different between kera+/+ and kera+/−. This difference of the effects of keratocan deficiency In vivo and In vitro, could be due to the lack of effect of microenvironment or other cell lineages in an In vitro system. During bone formation, mature osteoblasts are known to synthesize and secrete high levels of collagen, which is critical step preceding collagen fibrillogenesis. Previously, the role of keratocan in collagen fiber formation has been documented [28]. Based on our observation of decreased mineral apposition rate in the keratocan deficient mouse, it can be hypothesized that keratocan participates in the formation of osteoid. Collagen fibers are well ordered within the osteiod, and disruption of this order may lead to alterations in the subsequent mineralization of osteoid. Further studies will be required to characterize the molecular mechanism of keratocan in osteoblast function and activity.
Acknowledgments
Supported by: This work has been supported by grants from the National Institutes of Health, NIAMS (R03-AR053275) and Institutional support to IK through NIH grant (UDEO16495A) and National Eye Institute (EY011845 to WWK).
Literature
- 1.Tasheva E, Koester A, Paulsen A, Garrett A, Boyle D, Davidson H, Song M, Fox N, Conrad G. Mimecan/osteoglycin-deficient mice have collagen fibril abnormalities. Mol Vis. 2002;8:407–15. [PubMed] [Google Scholar]
- 2.Kalamajski S, Oldberg A. The role of small leucine-rich proteoglycans in collagen fibrillogenesis. Matrix Biol. 2010;29:248–53. doi: 10.1016/j.matbio.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Raouf A, Ganss B, McMahon C, Vary C, Roughley P, Seth A. Lumican is a major proteoglycan component of the bone matrix. Matrix Biol. 2002;21:361–7. doi: 10.1016/s0945-053x(02)00027-6. [DOI] [PubMed] [Google Scholar]
- 4.Sinha R, Tuan R. Regulation of human osteoblast integrin expression by orthopedic implant materials. Bone. 1996;18:451–7. doi: 10.1016/8756-3282(96)00044-0. [DOI] [PubMed] [Google Scholar]
- 5.Paic F, Igwe J, Nori R, Kronenberg M, Franceschetti T, Harrington P, Kuo L, Shin D, Rowe D, Harris S, Kalajzic I. Identification of differentially expressed genes between osteoblasts and osteocytes. Bone. 2009;45:682–92. doi: 10.1016/j.bone.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu C, Birk D, Hassell J, Kane B, Kao W. Keratocan-deficient mice display alterations in corneal structure. J Biol Chem. 2003;278:21672–7. doi: 10.1074/jbc.M301169200. [DOI] [PubMed] [Google Scholar]
- 7.Kao W, Liu C. The use of transgenic and knock-out mice in the investigation of ocular surface cell biology. Ocul Surf. 2003;1:5–19. doi: 10.1016/s1542-0124(12)70003-4. [DOI] [PubMed] [Google Scholar]
- 8.Hocking A, Shinomura T, McQuillan D. Leucine-rich repeat glycoproteins of the extracellular matrix. Matrix Biol. 1998;17:1–19. doi: 10.1016/s0945-053x(98)90121-4. [DOI] [PubMed] [Google Scholar]
- 9.Ameye L, Young M. Mice deficient in small leucine-rich proteoglycans: novel in vivo models for osteoporosis, osteoarthritis, Ehlers-Danlos syndrome, muscular dystrophy, and corneal diseases. Glycobiology. 2002;12:107R–16R. doi: 10.1093/glycob/cwf065. [DOI] [PubMed] [Google Scholar]
- 10.Bianco P, Fisher L, Young M, Termine J, Robey P. Expression and localization of the two small proteoglycans biglycan and decorin in developing human skeletal and non-skeletal tissues. J Histochem Cytochem. 1990;38:1549–63. doi: 10.1177/38.11.2212616. [DOI] [PubMed] [Google Scholar]
- 11.Bi Y, Stuelten C, Kilts T, Wadhwa S, Iozzo R, Robey P, Chen X, Young M. Extracellular matrix proteoglycans control the fate of bone marrow stromal cells. J Biol Chem. 2005;280:30481–9. doi: 10.1074/jbc.M500573200. [DOI] [PubMed] [Google Scholar]
- 12.Goldberg M, Ono M, Septier D, Bonnefoix M, Kilts T, Bi Y, Embree M, Ameye L, Young M. Fibromodulin-deficient mice reveal dual functions for fibromodulin in regulating dental tissue and alveolar bone formation. Cells Tissues Organs. 2009;189:198–202. doi: 10.1159/000151370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlson E, Wang I, Liu C, Brannan P, Kao C, Kao W. Altered KSPG expression by keratocytes following corneal injury. Mol Vis. 2003;9:615–23. [PubMed] [Google Scholar]
- 14.Michelacci Y. Collagens and proteoglycans of the corneal extracellular matrix. Braz J Med Biol Res. 2003;36:1037–46. doi: 10.1590/s0100-879x2003000800009. [DOI] [PubMed] [Google Scholar]
- 15.Rees SG, Waggett AD, Kerr BC, Probert J, Gealy EC, Dent CM, Caterson B, Hughes CE. Immunolocalisation and expression of keratocan in tendon. Osteoarthritis Cartilage. 2009;17:276–9. doi: 10.1016/j.joca.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Kalajzic I, Braut A, Guo D, Jiang X, Kronenberg MS, Mina M, Harris MA, Harris SE, Rowe DW. Dentin matrix protein 1 expression during osteoblastic differentiation, generation of an osteocyte GFP-transgene. Bone. 2004;35:74–82. doi: 10.1016/j.bone.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Kalajzic I, Kalajzic Z, Kaliterna M, Gronowicz G, Clark SH, Lichtler AC, Rowe D. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17:15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 18.Kalajzic I, Terzic J, Rumboldt Z, Mack K, Naprta A, Ledgard F, Gronowicz G, Clark S, Rowe D. Osteoblastic response to the defective matrix in the osteogenesis imperfecta murine (oim) mouse. Endocrinology. 2002;143:1594–601. doi: 10.1210/endo.143.5.8807. [DOI] [PubMed] [Google Scholar]
- 19.Wong G, Cohn D. Target cells in bone for parathormone and calcitonin are different: enrichment for each cell type by sequential digestion of mouse calvaria and selective adhesion to polymeric surfaces. Proc Natl Acad Sci U S A. 1975;72:3167–71. doi: 10.1073/pnas.72.8.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Celeste A, Rosen V, Buecker J, Kriz R, Wang E, Wozney J. Isolation of the human gene for bone gla protein utilizing mouse and rat cDNA clones. EMBO J. 1986;5:1885–90. doi: 10.1002/j.1460-2075.1986.tb04440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young M, Ibaraki K, Kerr J, Lyu M, Kozak C. Murine bone sialoprotein (BSP): cDNA cloning, mRNA expression, and genetic mapping. Mamm Genome. 1994;5:108–11. doi: 10.1007/BF00292337. [DOI] [PubMed] [Google Scholar]
- 22.Kalajzic I, Kalajzic Z, Kaliterna M, Gronowicz G, Clark S, Lichtler A, Rowe D. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone MinerRes. 2002;17:15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 23.Parfitt A, Villanueva A, Foldes J, Rao D. Relations between histologic indices of bone formation: implications for the pathogenesis of spinal osteoporosis. J Bone Miner Res. 1995;10:466–73. doi: 10.1002/jbmr.5650100319. [DOI] [PubMed] [Google Scholar]
- 24.Rees S, Dent C, Caterson B. Metabolism of proteoglycans in tendon. Scand J Med Sci Sports. 2009;19:470–8. doi: 10.1111/j.1600-0838.2009.00938.x. [DOI] [PubMed] [Google Scholar]
- 25.Wilda M, Bächner D, Just W, Geerkens C, Kraus P, Vogel W, Hameister H. A comparison of the expression pattern of five genes of the family of small leucine-rich proteoglycans during mouse development. J Bone Miner Res. 2000;15:2187–96. doi: 10.1359/jbmr.2000.15.11.2187. [DOI] [PubMed] [Google Scholar]
- 26.Wadhwa S, Embree M, Ameye L, Young M. Mice deficient in biglycan and fibromodulin as a model for temporomandibular joint osteoarthritis. Cells Tissues Organs. 2005;181:136–43. doi: 10.1159/000091375. [DOI] [PubMed] [Google Scholar]
- 27.Wadhwa S, Embree M, Kilts T, Young M, Ameye L. Accelerated osteoarthritis in the temporomandibular joint of biglycan/fibromodulin double-deficient mice. Osteoarthritis Cartilage. 2005;13:817–27. doi: 10.1016/j.joca.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 28.Douglas T, Heinemann S, Bierbaum S, Scharnweber D, Worch H. Fibrillogenesis of collagen types I, II, and III with small leucine-rich proteoglycans decorin and biglycan. Biomacromolecules. 2006;7:2388–93. doi: 10.1021/bm0603746. [DOI] [PubMed] [Google Scholar]