Abstract
A microtubule (MT) sub-population that emanates from Golgi membrane has been recently shown to comprise a significant part of MT network in interphase cells. In this study, we address whether Golgi membrane, which is being extensively remodeled throughout the cell cycle, retains its ability to nucleate MTs at diverse cell cycle stages. Live cell imaging and immunofluorescence microscopy reveals that Golgi-derived MTs form at multiple stages of the cell cycle, including G1, G2 and distinct phases of mitosis. However, the capacity of Golgi to nucleate MTs in mitosis is strongly down-regulated as compared to interphase, indicating that this property is cell-cycle regulated. We demonstrate that Golgi-derived MTs are indispensable for efficient Golgi assembly in telophase, and speculate that these non-centrosomal MTs may hold specific functions at other cell cycle stages.
Keywords: microtubules, Golgi, Golgi-derived microtubules, mitosis, cell cycle
Introduction
Microtubules (MTs) are spatially and temporally regulated according to the stage of the cell cycle. In interphase, MTs form an extended array radiating throughout the cytoplasm; in mitosis, MTs organize in a dynamic bipolar array named the mitotic spindle that quickly disassembles as mitosis is completed (reviewed in (Desai and Mitchison, 1997)). To a large extent, MT organization relies on the activity and positioning of MT nucleation sites, the MT Organizing Centers (MTOCs). In mitosis, the duplicated centrosomes are the main MTOCs and nucleate two asters of MTs (Mazia, 1987). MT nucleation can also be promoted near chromosomes and organized in the vicinity of the kinetochore (Kalab et al., 2006; Maiato et al., 2004), so that assembly of the mitotic spindle occurs through a cooperative process involving parallel pathways (O'Connell and Khodjakov, 2007). Furthermore, peripheral non-centrosomal MTs of yet unclear origin can be recruited into the mitotic spindle (Moutinho-Pereira et al., 2009; Tulu et al., 2003).
Similar to the mitotic spindle, MTs comprising the interphase MT network can also be supplied from diverse sources. While the centrosome is thought to be the main interphase MTOC (Luders and Stearns, 2007), additional non-centrosomal MTs are required for multiple specific cellular tasks (Bartolini and Gundersen, 2006; Vinogradova et al., 2012). Importantly, the Golgi apparatus was recently identified as a significant MTOC producing up to 50% of MTs in interphase cells (Chabin-Brion et al., 2001; Efimov et al., 2007). MT nucleation at the Golgi requires a number of specific molecular factors, including MT dynamics regulators CLASPs (Akhmanova et al., 2001; Maiato et al., 2003), which are linked to the Golgi apparatus through the trans-Golgi protein GCC185 (Efimov et al., 2007) and a large multifunctional cis-Golgi protein AKAP450 (AKAP350, CG-NAP) (Rivero et al., 2009). Golgi-emanating MTs comprise an asymmetric MT population, which is essential for correct Golgi complex assembly and directional cell migration (Efimov et al., 2007; Miller et al., 2009; Rivero et al., 2009; Vinogradova et al., 2012).
Although it is clear that Golgi-derived MTs are essential and a significant component of interphase MT network, their role and existence in mitosis has not been addressed yet. At the same time, it is well established that the Golgi membranes persist throughout the cell cycle while their arrangement undergoes significant reorganization (Robbins and Gonatas, 1964). An interphase Golgi ribbon is a continuous system composed by interconnected membrane stacks. Starting late G2, it undergoes several tightly regulated stages of fragmentation (Lucocq and Warren, 1987), a process required for mitotic entry (Sutterlin et al., 2002). It is thought that Golgi fragmentation allows for the equal distribution of membranes into the daughter cells (Robbins and Gonatas, 1964). In telophase, Golgi membranes begin to reassemble in stacks of cisternae, which then cluster together and upon cytokinesis assemble into a single Golgi apparatus in the perinuclear region (Lucocq et al., 1989).
In this study, we address whether Golgi membranes, which are being extensively remodelled during cell cycle, retain the ability to support MT nucleation. We further discuss possible functional implications of these non-centrosomal MTs at distinct cell cycle stages.
Results
Golgi Organization in G1 and G2
In order to investigate how Golgi derived MTs are regulated throughout the cell cycle, RPE1 and LLC-PK1α cells were synchronized in early S-phase with Aphidicolin (Ikegami et al., 1978) or in G2 through the inhibition of Cdk1/cyclin B1 complex with the small molecule RO-3306 (Vassilev et al., 2006). Mitotic arrests were done by treating cells with 100 ng/mL of nocodazole (De Brabander et al., 1976; Vasquez et al., 1997).
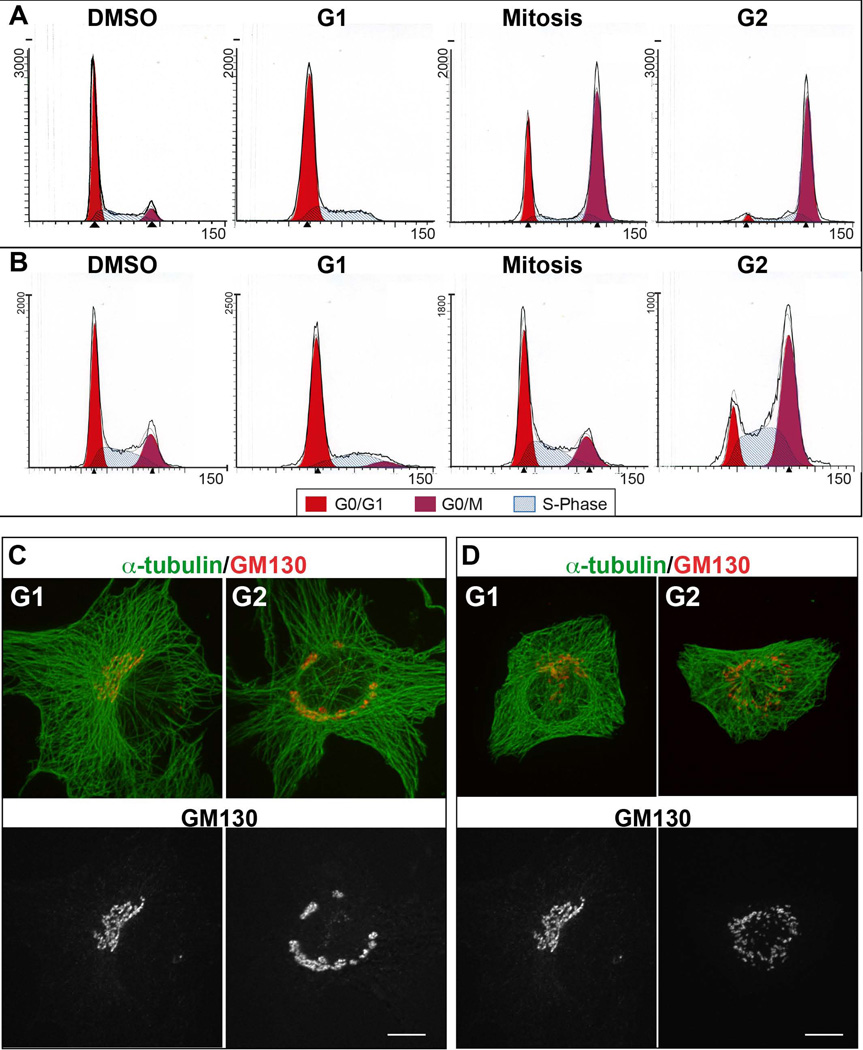
To determine synchronization efficiency, propidium iodide labeled cells were analyzed by FACS according to their DNA content (Figure 1A and B). In control (DMSO) normal cycling RPE1 and LLC-PK1 cells show the typical FACS profile, where around 50 to 60% of the population was in G0/G1, 30% in S phase and 8 to 20% of cells are in G2 and mitosis. Aphidicolin treatment arrested 70% of cells in G1, with the remaining 28% of cells in S phase, and small G2/Mitosis population. In RPE1, RO-3306 treatment increases G2/Mitosis population to 61%, keeping 12% of cells in S phase and decreasing G1 population to 27%. G2 synchronization was inefficient in LLC-PK1 cells since 48% of the population was in G1, 32% in S phase, and 20% in G2/Mitosis. RPE1 mitotic arrest with nocodazole led to 81% of cells in G2/Mitosis, leaving 3% of the population in G1 and 16% in S phase. In LLC-PK1, nocodazole treatment increased G2/M cells to 49%, followed by an increase to 39% of S phase population and 13% of G1 cells. These results indicate that the drug treatments are properly arresting RPE1 cells, however LLC-PK1 are more resistant to cell cycle synchronization. For this reason, in experiments described below LLC-PK1 cells were chosen for G2 analysis based on the centrosome duplication detected by GFP-centrin visualization or by the number of major MT asters.
Figure 1. Golgi and MT Organization in G1 versus G2 Synchronized Cells.
(A, B) Propidium Iodide fluorescence profiles of RPE (A) and LLC-PK1 (B) cells in DMSO treated, G1, G2 or mitotic arrest. The cells synchronized as noted were analyzed by flow cytometry according to their DNA content. (C, D) Representative images of RPE (C) or LLC-PK1 (D) cells in G1 and G2 immunostained for α-tubulin (Upper panel, green) and GM130 (upper panel, red; lower panel, grayscale). Maximal projection of confocal z stacks is shown. Gamma adjustment was used to highlight low-light details. Scale bar, 10 µm.
To complement the FACS analysis, the synchronized cells were processed for immunofluorescence and checked for centrosome number and Golgi organization. Synchronized RPE1 and LLC-PK1 cells were fixed and immunostained for α-tubulin and the Golgi marker GM130.
In G1 cells, the single well-organized Golgi apparatus positioned close at one side of the nucleus presumably to the MTOC (Figure 1C-G1, 1D-G1). In G2 cells, there isa less continuous Golgi complex distributed around the nucleus in a non-polarized manner (Figure 1C-G2,1D-G2). Thus, during G1 both cell lines present a compact Golgi ribbon organization, and in G2 Golgi fragmentation starts to prepare cells for division.
Microtubule Nucleation at the Golgi in G1 and G2
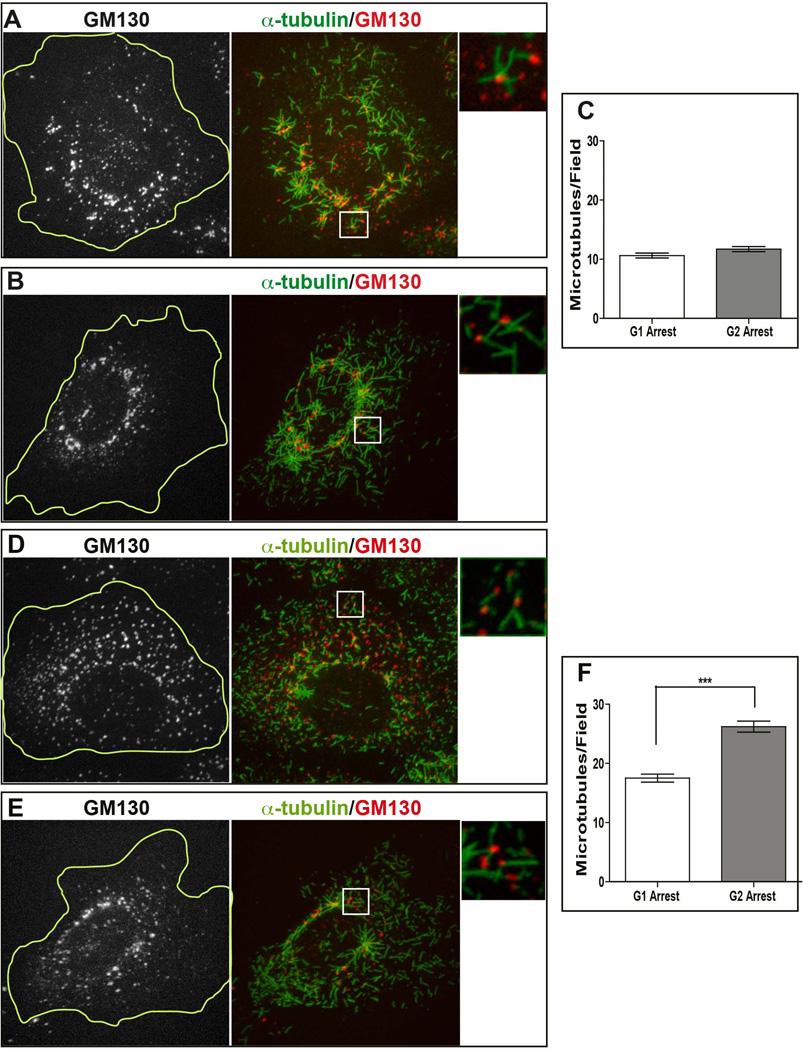
To address potential differences in the ability of G1 and G2 cells to nucleate MTs from the Golgi apparatus, we quantified Golgi-associated MTs in synchronized RPE1 cells. To differentiate Golgi and centrosomal MT nucleation, we treated cells with 2.5 µg/mL of nocodazole that completely depolymerizes MTs and also disrupts Golgi ribbon organization into small stacks (Cole et al., 1996). After nocodazole removal, MT polymerization was allowed to occur for 45 seconds. Cells were subsequently fixed and stained for α-tubulin and GM130 to visualize newly formed MTs and Golgi mini-stacks, respectively, and visualized by confocal microscopy (Figure 2). After nocodazole washout, newly formed MTs were observed at the centrosomes and at dispersed Golgi mini stacks, as described previously (Efimov et al., 2007). Interestingly, both G1 and G2 arrested RPE1 cells were able to nucleate MTs from the Golgi stacks (Figure 2A and B, respectively). To quantify Golgi MT nucleation capacity, the number of MTs closely associated with the Golgi was counted in randomly chosen square fields (five regions per cell) (Figure 2C). Number of Golgi-associated MT number per field was determined, in order to normalize MT numbers to the cell size. This normalization is required because the Golgi network elongates throughout the interphase, so that G2 cells contain higher amounts of Golgi membranes (Sengupta and Linstedt, 2011). MTs closely associated with the Golgi in this assay were assumed to be Golgi-derived MTs, as we showed previously by live-cell imaging (Efimov et al., 2007). The quantifications from G1 and G2 RPE1 cells showed no major differences in the number of Golgi nucleated MTs (the median values were 10 MTs/field in G1 and 11 MTs/field in G2).
Figure 2. MT nucleation at the Golgi during G1 and G2.
(A,B,D,E) Representative immunofluorescence images of RPE (A, B) and LLC-PK1 (D, E) cells synchronized in G1 (A, D) or G2 (B, E) after nocodazole washout. After 45-second period of MT re-growth, cells were fixed and stained for α-tubulin (central and right panels, green) and GM130 (left panel, grayscale; central and right panels, red). Cell border is outlined at the greyscale images (yellow line). Maximal projection of confocal z stacks is shown. Right panel represents enlarged insets (boxed in the central panel). Gamma adjustment was used to highlight low-light details. Scale bar, 10 µm. (C, F) Quantification of MT number per field in G1 (n=57) and G2 (n=58) in synchronized RPE (C) and LLC-PK1 (F) cells was used to analyze the data points. The two populations of RPE cells do not differ statistically (Mann Whitney t test, p>0.05). The two populations of LLC-PK1 cells differ statistically (Mann Whitney t test, p<0.0001, marked by ***).
We also investigated Golgi MT nucleation ability in LLC-PK1 cell line. Since LLC-PK1 were not readily responsive to synchronization, we analyzed drug treated cells with one centrosome as G1 population, and those with two centrosomes as G2 population.
After nocodazole washouts and staining for α-tubulin and GM130, we observed MTs associated with the fragmented Golgi stacks in G1 and G2 (Figure 2D and E, insets). According to these data, Golgi MT nucleation was very robust, in both stages. Quantification showed that 18 Golgi-associated MTs/field in G1 and 26 MTs/field in G2, indicating that number of Golgi-derived MTs in G2 cells is significantly higher than in G1 (Figure 2F).
Thus, both RPE1 and LLC-PK1 cells nucleate Golgi MTs during G1 and G2. Unlike RPE1 cells, LLC-PK1 shows slight but significant regulation of Golgi-associated nucleation depending on the interphase stage.
Detection of Microtubule Nucleation at the Golgi during Mitosis in Steady State
It is well described that the Golgi apparatus fragment into single vesicles upon mitotic entry (Robbins and Gonatas, 1964; Wei and Seemann, 2009). We addressed a long-standing question of whether Golgi membranes retain part of their functionality during mitosis, in particular their ability to nucleate MTs.
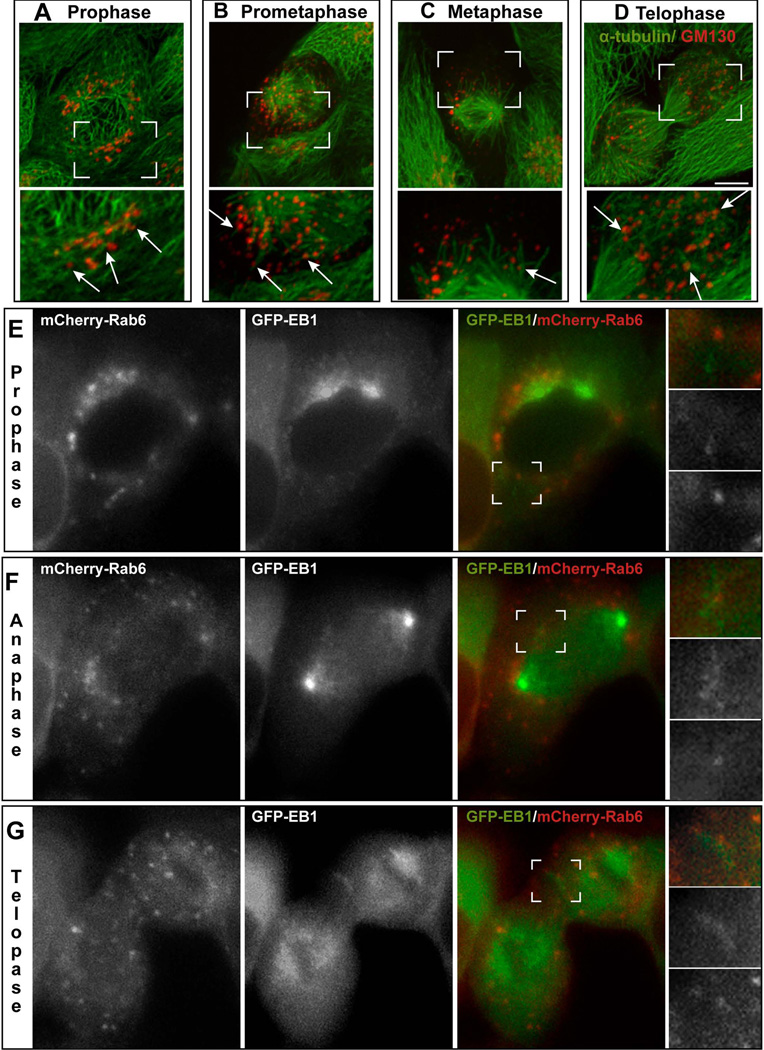
In order to visualize Golgi fragmentation and associated MTs in mitotic cells, asynchronous LLC-PK1 were immunostained for α-tubulin and the cis-Golgi protein GM130, and cells at specific phases of mitosis were located. In prophase, the Golgi is no longer a ribbon network but is dispersed as stacks around the nucleus. At this stage, non-centrosomal MTs associated with the Golgi stacks can be easily detected at the side of the nucleus opposite to the centrosomes (Figure 3A). Once nuclear envelope breaks down, the Golgi undergoes a higher degree of fragmentation and is dispersed throughout the prometaphase and metaphase cytoplasm (Figure 3B) (Figure 3C). Non-centrosomal MTs radiating from the Golgi fragments can be detected in prometaphase (Figure 3B). In metaphase, MTs associated with the Golgi can also be located though less frequently (Figure 3C). In telophase and cytokinesis, emerging clusters of Golgi stacks accumulate throughout the cytoplasm. Non-centrosomal MTs associated with the Golgi are readily recognizable at this stage (Figure 3D).
Figure 3. Mitotic Golgi Membranes are able to Nucleate Microtubules.
(A–D) Representative images of LLC-PK1 cells in prophase (A), prometaphase (B), metaphase (C), and telophase (D) immuno-stained for α-tubulin (green) and GM130 (red). Scale bar, 10µm. Lower panel represents enlarged insets (boxed in the upper panel). Arrows, non-centrosomal MTs at the Golgi stacks. (E–G) A representative dividing LLC-PK1 cell expressing GFP-EB1 (right panel, green; central pane, grayscale) and transiently transfected with mCherry-Rab6 (right panel, red; left panel, greyscale), visualized by epifluorescence live cell microscopy. Images show the cell in prophase (E), anaphase (D), and telophase (G). Single plane confocal slices are shown. Insets represent high magnifications of the boxed area. Gamma adjustment was used to highlight low-light details. Scale bar, 10 µm.
To directly test whether Golgi membranes retain the ability to nucleate MTs during mitosis, we applied confocal live cell imaging to LLC-PK1 cells expressing GFP-EB1 as a marker for growing MT plus ends and mCherry-Rab6 as a marker for the Golgi. In order to clearly distinguish Golgi-derived MTs from the centrosomal and kinetochoreMTs, we closely observed Golgi fragments distant from the centrosomes and chromosomes. We directly detected EB1 comets coming out of the Golgi stacks in prophase (Figure 3E and Movie S1), prometaphase (not shown), anaphase (Figure 3F and Movie S2), and telophase (Figure 3G and Movies S3 and S4). MTs were quantified throughout movies corresponding to each mitotic stage; however, no statistical analysis was performed in this experiment due to the low number of data points (1–3 movies per stage were analyzed). Interestingly, MT formation during prophase, anaphase and telophase was significant and close to each other (on average, 19, 18 and 21, respectively). During prometaphase only nine MTs were detected, and no MTs were observed in metaphase cells, where detection was especially challenging due to size and dynamics of the mitotic spindle.
These data indicate that Golgi-derived MT nucleation actively occurs in prophase, anaphase and telophase. Significantly fewer MTs are formed in prometaphase, and especially metaphase.
Detection of Microtubule Nucleation at the Golgi during Mitosis in Cold Recovery
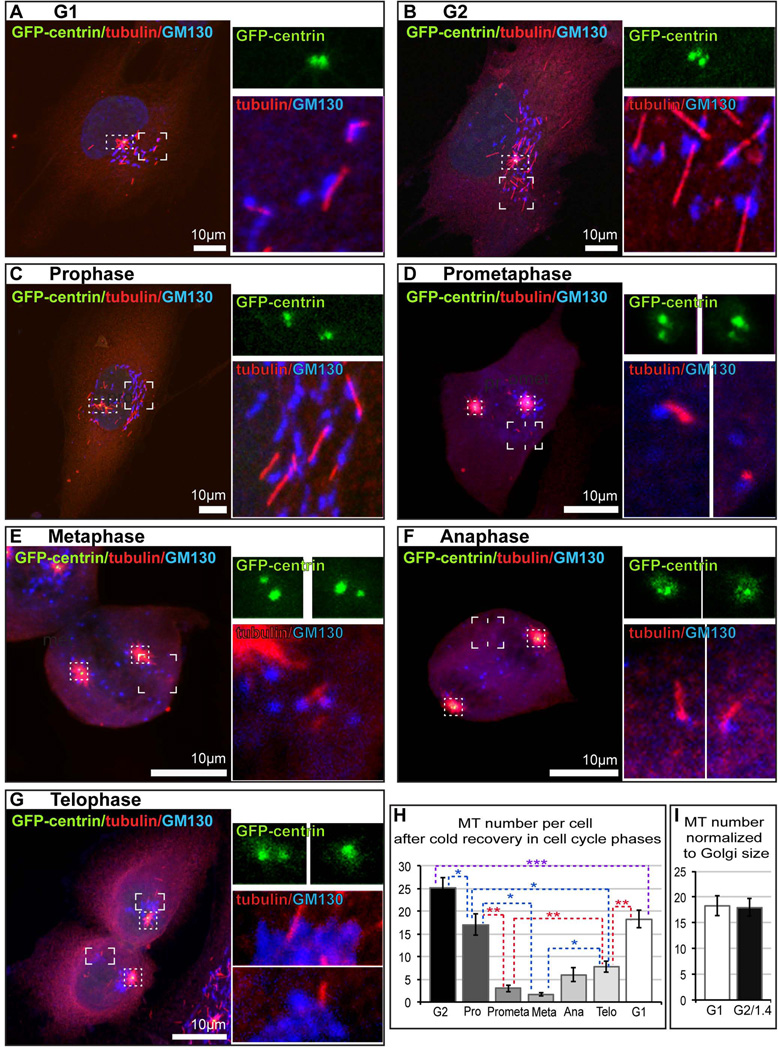
In order to quantitatively approach MT formation at the Golgi in mitosis, we applied MT re-growth assay after cold treatment (Fig. 4). Non-synchronized RPE1 cells stably expressing GFP-centrin were located on ice for 40 minutes for complete depolymerization of all MTs (Rivero et al., 2009; Vinogradova et al., 2012) and then allowed to recover at room temperature for 5 minutes. Under these conditions, MT regrowth is slow and inefficient, resulting in low number of Golgi-derived and centrosomal MTs and significantly decreasing non-specific MT formation in the cytoplasm, as compared to nocodazole washout. Cell were fixed, immunostaining for GM130 and tubulin and subjected to confocal microscopy. Golgi-associated MTs located away from the centrosome and chromosomes were detected. Cell cycle stage was determined by the centrosome number (Fig. 4) considered together with condensation and location of chromosomes detected by DIC microscopy (not shown). We detected significant MT formation at the Golgi in G1 and G2 (Fig. 4 A, B, H), with slight difference which was eliminated when results were normalized to Golgi size (Fig. 4I), consistently with the Figure 2C. MT formed at the Golgi in LLCPK cells in G1 versus G2 in similar settings differed with ratio 1:1.7 (not shown), also confirming the result of nocodazole washout (Fig. 2F). Upon mitotic entry, MT formation at the Golgi was detected throughout cell division, though with clear distinction between stages. MT number was slightly decreased in prophase (Fig. 4C, H), and noticeably reduced in prometaphase (Fig. 4D, H), and metaphase (Fig. 4E, H). In anaphase (Fig. 4F, H) and telophase (Fig. 4G, H), MT formation was gradually enhanced again.
Figure 4. Ability of Golgi Membranes to Nucleate Microtubules is Regulated Throughout the Cell Cycle.
(A–G). Representative images of RPE1 cells at indicated cell cycle stages after short-term cold recovery. Confocal Z-stack maximal projections. For all stages, overview cell images (left panels) show maximal projections of the whole cells expressing GFP-centrin (green) immuno-stained for α-tubulin (red) and GM130 (blue, false color for far red). Scale bar, 10 µm. At the top right, insets visualize centrosomes (GFP-centrin channel enlarged form dotted boxes at the overview images). At the bottom right, insets visualize Golgi-associated MTs (GM130 and tubulin channels enlarged from cornered boxes at the overview images). Insets represent maximal projections of confocal planes only through the depicted structure. (H). Number of MTs associated with the Golgi per cell, based on data as in (A–G). (I). Number of MTs per cell in G1 compared to G2 corrected for the Golgi size difference.
These data confirm dynamic changes in MT formation at the Golgi detected by live cell imaging, and indicate that Golgi membrane is capable of MT nucleation throughout mitosis, however this ability is diminished as compared to interphase, with distinct down-regulation at metaphase.
Golgi Re-assembly during Telophase is Attenuated in the Absence of Golgi-Derived MTs
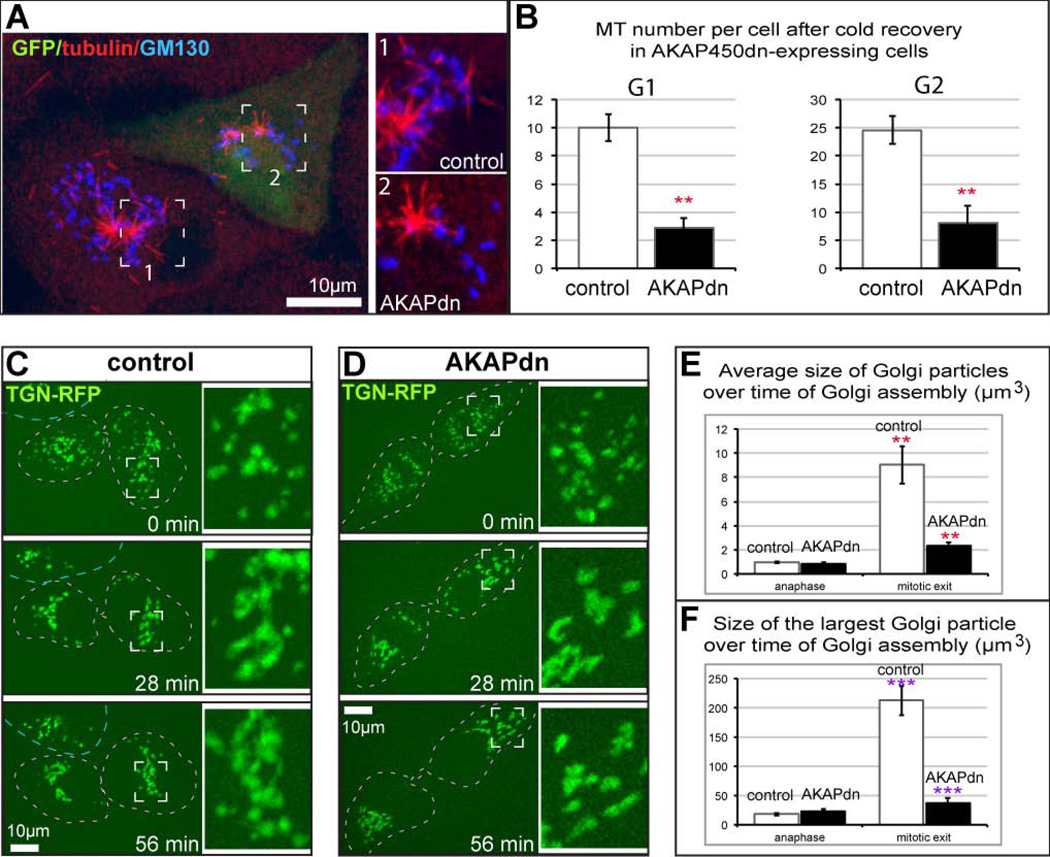
We have previously determined that both Golgi-derived and centrosomal MTs are necessary for the Golgi assembly after nocodazole washout (Miller et al., 2009; Vinogradova et al., 2012). Golgi-derived MTs drive Golgi stack clustering in the cytoplasm away from the centrosome. Similar clustering was observed during post-mitotic Golgi assembly in RPE1 cells (Miller et al., 2009). Here, we directly addressed whether Golgi assembly after mitosis is influenced by Golgi-derived MTs. Because depletion of molecular factors required for MT formation at the Golgi (CLASPs or AKAP450) (Efimov et al., 2007; Rivero et al., 2009) may influence mitotic spindle assembly, we took advantage of a dominant negative approach previously developed to eliminate AKAP450 function at the Golgi but not the centrosome by ectopic expression of truncated fragment of AA159-463 of AKAP450 (AKAPdn) (Hurtado et al., 2011). Expression of this construct was shown to block Golgi-derived MT formation and function in Golgi stack clustering during nocodazole washout (Hurtado et al., 2011). Consistently with these data, we found that MT formation at the Golgi was significantly decreased in LLCPK cells expressing AKAPdn and identified by concurrent GFP expression (Fig. 5A,B). Then, we followed live Golgi re-assembly during mitotic exit in LLC-PK1 cells after either control or AKAPdn transfection. TGN-RFP was used as a Golgi marker. In control cells, we observed that the Golgi assembly started in the transition from anaphase to telophase and included both continuous enlargement of Golgi fragments (stack clustering) and concentration of these fragments into a single Golgi complex, presumably close to the centrosome (Fig. 5C). Accordingly, the size of Golgi fragments (Fig. 5E), and especially the largest entity in the cell center (Fig. 5F), enlarged in the course ofmitotic exit. In AKAPdn-expressing cells, in contrast, stacks were collected in the cell center but remained fragmented and small, suggestive of impaired stack clustering (Fig. 5 D–F). Based on these and our previous data (Miller et al., 2009), we propose that Golgi-derived MTs formed at late stages of mitosis are responsible for Golgi stack clustering and post-mitotic efficient Golgi assembly.
Figure 5. Golgi Reassembly during Mitotic Exit is attenuated in the absence of Golgi-derived MTs.
(A) Two G2 LLCPK cells after cold recovery (as in Figure 4) are shown. A non-transfected cell is on the left. Boxed region featuring MTs (red) at the Golgi (blue) in enlarged (inset 1). A cell expressing AKAPdn (marked by GFP, green) contains very few Golgi-associated MTs (right, inset 2). (B) Expression of AKAPdn significantly decreases Golgi-associated MT number in LLCPK in both G1 and G2. Student’s t test, p<0.01, marked by **. (C, D) Representative frames from time-lapse sequences dividing LLC-PK1 cells expressing RFP-TGN (false-colored green), visualized by spinning disk confocal microscopy. Maximal projection of confocal z stacks. Time, minutes. Scale bars, 10 µm. Boxed insets are enlarged on the left. (C) In control, Golgi mini-stacks are collected in a tight complex within 30 minutes. The average Golgi fragment volume increases from 1.3µm3 to 11.3µm3; the volume of the largest particle increases from 7.9µm3 to 77.1µm3. A representative example out of four movies. (D) A cell expressing AKAPdn (identified by GFP expression, not shown). Golgi stack clustering is incomplete after one hour of assembly. The average Golgi fragment volume increases from 1.1µm3 to 4.4µm3; the volume of the largest particle increases from 5.1µm3 to 39.6µm3. A representative example out of three movies. (E) Increase of the average volume of Golgi stacks in control and AKAP-dn-expression cells from anaphase to mitotic exit, based on LLCPK cells immunostained for GM130. N=5. Student’s t test, p<0.01, marked by **.(F) Increase of the largest Golgi stack (Golgi complex) in control and AKAP-dn-expression cells from anaphase to mitotic exit, based on LLCPK cells immunostained for GM130. N=5. Student’s t test, p<0.001, marked by ***.
Discussion
Golgi-derived MTs emerge at diverse stages of the cell cycle, which are characterized by diverse Golgi organization. High MT nucleating capacity of the Golgi persists throughout interphase when the Golgi is assembled in highly coordinated membrane system (reviewed in (Wei and Seemann, 2009)) that is consistent with previous findings (Efimov et al., 2007; Miller et al., 2009; Rivero et al., 2009; Vinogradova et al., 2012). We observed subtle variations in MT nucleating activity of the Golgi membrane between G1 and G2, but only in one of the two analyzed cell lines. It is not clear how this difference is regulated though it might reflect high protein processing activity of the Golgi as cells prepare for division.
More surprisingly, we have identified MTs emerging from the Golgi fragments at almost all mitotic stages, including prometaphase and anaphase when the Golgi membranes are unstacked and split into small vesicles (reviewed in (Wei and Seemann, 2009)). This is the first description that the Golgi fragments retain the capacity to nucleate MTs during mitosis, supporting the idea that the Golgi membranes retain its functionality during mitosis (Wei and Seemann, 2009). Mitotic Golgi vesicles are thought to maintain their compartmental identity, so that TGN membrane vesicles are clearly distinguished from those arising from the cis cisternae (Shima et al., 1997; Tang et al., 2010). Both TGN and cis-Golgi proteins were found necessary for MT formation at the Golgi in interphase (Efimov et al., 2007; Rivero et al., 2009), and a potential interplay between these molecular factors has been suggested (Sutterlin and Colanzi, 2010; Vinogradova et al., 2012). In mitosis, we can detect Golgi-derived MTs emerging from sites containing a TGN marker (Rab6), as well as cis-Golgi marker GM130, which was detected as a potential component of MT-organizing activity at the Golgi (Rivero et al., 2009). Though exact contributions of TGN-localized and cis-localized machineries in Golgi-derived MT formation are still a matter of investigation, our data suggest that mitotic Golgi-derived MTs arise from sites of mitotic Golgi clusters which were shown to contain closely associated vesicles of differential origin (Cabrera-Poch et al., 1998; Shima et al., 1997). Most importantly, the interaction between CLASPs and GCC185 described to be required for Golgi MT stabilization and anchoring during interphase (Efimov et al., 2007) is preserved during mitosis (Maffini et al., 2009). That said, in our experiments the number of Golgi-derived MTs in mitosis was significanltly lower than in interphase, hitting its minimum in metaphase when these MTs were hardly detectable. It is plausible to suggest thatMT formation at the Golgi is gradually attenuated concurrent with Golgi fragmentation, and is almost obliterated in metaphase due to maximal Golgi dispersal and loss of connection between TGN and cis- located machineries.
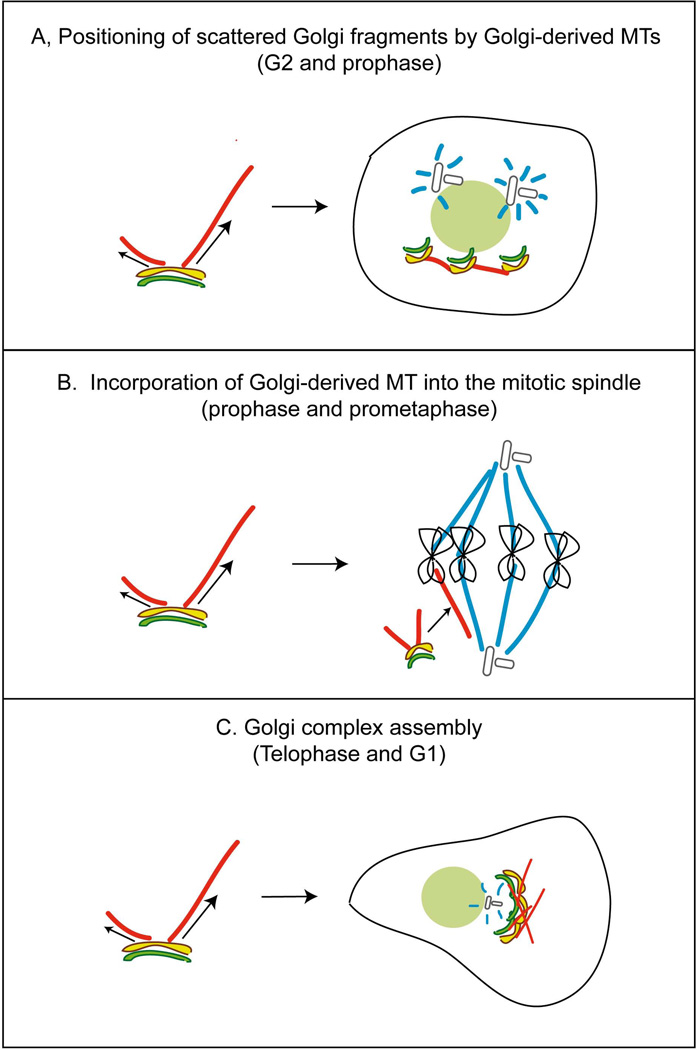
Existence of Golgi-derived MTs at diverse cell cycle stages suggests that they may participate in a number of cellular tasks (Fig. 6). Our previous and current findings show that this MT subset is essential for bringing together emerging Golgi stacks in the course of post-mitotic Golgi assembly to enable further Golgi stack fusion and efficient Golgi complex assembly (Miller et al., 2009) (Fig. 5). This function has to be carried out by MTs formed in late mitosis and G1 (Fig. 6C). Functions of Golgi derived MTs in G2 and early mitosis are yet to be identified. We speculate that MTs formed in G2 may be involved in positioning of Golgi fragments around the nucleus in the course of initial fragmentation (Fig. 6A). Similarly, Golgi fragments may be positioned by these MTs at any stage of mitosis. Another intriguing possibility is that Golgi-derived MTs contribute to the mitotic spindle itself (Fig. 6B) as was hypothesized previously (Liu et al., 2007). In this scenario, MTs formed in prophase and prometaphase could represent a non-centrosomal MT subset recruited to the mitotic spindle from the cytoplasm (Tulu et al., 2003). In metaphase, when the spindle is already assembled and mature, MT recruitment is unnecessary, which could justify less frequent MT nucleation at the Golgi at this phase.
Figure 6. Proposed functions of Golgi-derived MTs at specific cell cycle phases.
Model. A, In G2 and prophase, Golgi-derived MTs may position of Golgi fragments around the nucleus. B, In prophase and prometaphase, Golgi-derived MTs may be recruited to the mitotic spindle from the cytoplasm. C, In telophase and G1, Golgi-derived MTs bring together emerging Golgi stacks in the course of post-mitotic Golgi assembly.
Overall, while exact functions of Golgi-derived MTs at distinct stages of the cell cycle require further insights, our study is the first to indicate cell cycle regulation of this MT sub-population and to identify it in mitosis. We hypothesize that this is an indication that the mitotic Golgi membranes can bear an essential cellular function. Moreover, here we directly show that participation of Golgi-derived MT in Golgi stack clustering, previously identified in experimental settings, is indeed essential for efficient post-mitotic Golgi assembly.
Materials and Methods
Cell Culture and Synchronization
Immortalized human retinal pigment epithelial cells hTert-RPE1 (Clontech) were maintained in DMEM/F12 with 10% fetal bovine serum (FBS). GFP-centrin-expressing RPE1 stable line was kindly provided by A. Khodjakov (Wadsworth Center, Albany, NY). GFP-tubulin expressing LLC-PK1 stable line (renal epithelial cell line derived from porcine kidney, a gift from P. Wadsworth, UMass, Amherst, MA) were cultured in a mixture of Opti-MEM and F10 (1:1) supplemented with 10% of FBS. Cells were grown in 5% CO2 at 37°C and were plated on fibronectin-coated glass coverslips 24 hours before experiments were performed. Cell cycle synchronization was achieved through different treatments in RPE1 and LLC-PK1 cells. Exponentially growing cells were treated with 5 µg/µL Aphidicolin for 24 hours. Aphidicolin is an inhibitor of the DNA polymerase, and allows an arrest in G1 (Ikegami et al., 1978). The G2 arrest was done through the inhibition of Cdk1 kinase activity by the specific small molecule inhibitor RO-3306 (Vassilev et al., 2006), by adding 10 µM of the inhibitor (in DMSO) for 20 hours. To enrich for mitotic cells, 100 ng/mL of nocodazole was added to the medium 14 hours before harvesting cells.
DNA constructs and transfection
TGN-RFP was kindly provided by E. Rodriguez-Boulan (Weil Cornell Medical College, New York, NY). mCherry-Rab6 was kindly provided by A. Akhmanova (Utrecht University, Utrecht, The Netherlands). To generate AKAPdn construct, sequence according to AA 159–463 was amplified by PCR. To provide non-tagged AKAPdn in GFP-marked cells, it was cloned after GFP followed by T2A cleavage site, and then inserted in PCIG vector by SalI/XhoI+ECORV. Cells were transfected by Amaxa nucleofection.
Fluorescence Activated Cell Sorting
After cell harvesting by centrifugation, cells were washed once with cold PBS and fixed dropwise with cold ethanol, while vortexing. Thereafter, cells were washed by 70% ethanol twice with PBS. Cell pellet was resuspended with 100 µg/mL RNAse in PBS, and incubated for 30 minutes to 3 hours at 37°C. To this solution was added 5 µg/mL of Propidium Iodide in PBS and cells were analysed by FACS. Cells were analyzed in a 3-laser BD LSRII fluorescence cytometer.
Nocodazole washout
For microtubule depolymerization and Golgi dispersal, 2.5 µg/mL of nocodazole was added to the culture medium for 2 hours. LLC-PK1 cells were previously incubated in ice for 30 minutes followed by 5 µg/µL of nocodazole for full MT depolymerization. For Golgi washout experiments, cells were rinsed five times with ice-cold medium to remove nocodazole and then moved to a dish with warm (37°C) medium. Soluble tubulin was extracted with the extraction buffer (PIPES, 60 mM ; EGTA, 10 mM; HEPES, 25 mM; MgCl2, 2 mM; Saponin, 0,1%; pH 6,9) supplemented with 2,5µM of taxol and nocodazole (at 37°C), and the plate swirled for 40 seconds. Cells were fixed with MetOH at −20°C. MT absence before recovery was controlled by immunostaining for tubulin (not shown).
Cold recovery
RPE1 cells were plated on coverslips in a petri dish, and then were located on ice for 40 minutes. Then the cells were allowed to recover at room temperature in the same medium for 5 minutes, fixed and stained. LLCPK1 cells were plated on coverslips in a petri dish, located on ice for 40 minutes, then nocodazole (5µg/ml) added for 2 hour, and then washed 8 times with ice-cold medium. Then the cells were allowed to recover at room temperature in the same medium for 10 minutes, fixed in 4% PFA plus 0.2% glutaraldehyde, and immunostained for tubulin and GM130. MT absence before recovery was controlled by immunostaining for tubulin (not shown).
Immunofluorescence Analysis
Rabbit polyclonal antibodies against CLASP2 VU-83 have been described previously (Efimov et al., 2007). Rabbit polyclonal antibodies against CLASP1 were provided by F. Severin (MPI, Dresden, Germany). For Golgi compartment identification, a mouse monoclonal antibody against GM130 (1:300; Transduction Laboratories) was used. Microtubules were stained with a rat monoclonal YL1/2 antibody (1:800; Abcam). Cells were fixed in cold methanol (10 min at –20°C) for CLASP stainings. For VU-83 stainings cells were additionally treated for 1 minute in methanol/acetone 1:1 mixture. Antibodies for CLASPs were used at a 1:60 dilution for immunofluorescence. For MT and Golgi staining, cells were fixed (15 min at room temperature) in 2% paraformaldehyde, 0.1% glutaraldehyde, 0.5% saponin in cytoskeleton buffer (10 mM MES, 150 mM NaCL, 5 mM EGTA, 5 mM glucose, and 5 mM MgCl2, pH 6.1). Alexa488 and Alexa568-conjugated highly crossabsorbed goat anti-mouse IgG antibodies, Alexa568-conjugated goat anti-rat IgG antibodies, and Alexa568 donkey anti-sheep IgG (Molecular Probes) were used as secondary antibodies. All secondary antibodies were used at a 1:500 dilution.
Image acquisition and processing
Fluorescence imaging was performed using confocal stacks taken by a Yokogawa cQLC-100/CSU-10 spinning disk head (Visitec assembled by Vashaw) attached to a Nikon TE2000E microscope using a CFI PLAN APO VC 1003 oil lens, NA 1.4, with or without 1.5 intermediate magnification, and a backilluminated EM-CCD camera Cascade 512B (Photometrics) driven by IPLab software (Scanalytics). A krypton-argon laser (75 mW 488/568; Melles Griot) with AOTF was used for two-color excitation. Custom double dichroic mirror and filters (Chroma) in a filter wheel (Ludl) were used in the emission light path. Z steps (0.8 µm) were driven by a Nikon built-in Z motor. Live cells plated on MatTech glass bottom dishes were maintained at 37°C by heated stage (Warner Instruments) on a Nikon TE2000E inverted microscope equipped with a PerfectFocus automated focusing device. Single-plane or 3D confocal video sequences were taken as described for single confocal stacks. A similar setup with a Pinkel triple-filter set (Semrock) was used for nearly simultaneous two-color wide-field imaging.
Microtubule Quantifications and statistical analysis
Golgi MT nucleation in nocodazole washout was quantified by counting the number of MTs overlapping with Golgi fragments in different fields of immunofluorescence microscopy images of different cells. The ratio of Golgi-associated MT number per field was determined, in order to normalize the number of Golgi stacks counted in each cell. Golgi MT nucleation in cold recovery was quantified by counting the number of MTs associated with the Golgi per cell. In all experiments, only MTs directly associated with the Golgi as detected at fluorescent microscopy level were considered. MTs, which were closely associated with both Golgi and chromosomes, were excluded from the analysis to assure exclusion of kinetochore-derived MTs. Volume of the Golgi fragments was calculated applying ImageJ 3D object counter to confocal stacks. Mann Whitney t test or Student t test were used to analyze the data points, and values were considered statistically different whenever p<0.05.
Supplementary Material
Acknowledgments
We thank Ms. Nadia Efimova and Huancheng Dong for technical assistance and advice. This work was supported by National Institutes of Health grants R01-GM078373, and by American Heart Association grant-in-aid 10GRNT4230026 (to I.K.). A.R.R.M. was funded by the grant SFRH/BD/32976/2006 from the Portuguese Fundação para a Ciência e Tecnologia (FCT), through the GABBA Program (10th edition). P.M.M. funded by an American Heart Association predoctoral fellowship 09PRE2260729. Flow Cytometry experiments were performed in the VMC Flow Cytometry Shared Resource. The VMC Flow Cytometry Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). Work in the laboratory of H.M. is funded by grants PTDC/SAU-GMG/099704/2008 and PTDC/SAU-ONC/112917/2009 from FCT (COMPETE-FEDER), the Human Frontier Research Program and the 7th framework program grant PRECISE from the European Research Council. Work in the laboratory of G.G. is funded by National Institutes of Health grant RO1 DK065949.
References
- Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, Galjart N. Clasps are CLIP-115 and-170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci. 2006;119:4155–4163. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- Cabrera-Poch N, Pepperkok R, Shima DT. Inheritance of the mammalian Golgi apparatus during the cell cycle. Biochim Biophys Acta. 1998;1404:139–151. doi: 10.1016/s0167-4889(98)00051-2. [DOI] [PubMed] [Google Scholar]
- Chabin-Brion K, Marceiller J, Perez F, Settegrana C, Drechou A, Durand G, Pous C. The Golgi complex is a microtubule-organizing organelle. Mol Biol Cell. 2001;12:2047–2060. doi: 10.1091/mbc.12.7.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NB, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol Biol Cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Brabander MJ, Van de Veire RM, Aerts FE, Borgers M, Janssen PA. The effects of methyl (5-(2-thienylcarbonyl)-1H-benzimidazol-2-yl) carbamate, (R 17934; NSC 238159), a new synthetic antitumoral drug interfering with microtubules, on mammalian cells cultured in vitro. Cancer Res. 1976;36:905–916. [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, Yates JR, 3rd, Maiato H, Khodjakov A, Akhmanova A, Kaverina I. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado L, Caballero C, Gavilan MP, Cardenas J, Bornens M, Rios RM. Disconnecting the Golgi ribbon from the centrosome prevents directional cell migration and ciliogenesis. J Cell Biol. 2011;193:917–933. doi: 10.1083/jcb.201011014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami S, Taguchi T, Ohashi M, Oguro M, Nagano H, Mano Y. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-alpha. Nature. 1978;275:458–460. doi: 10.1038/275458a0. [DOI] [PubMed] [Google Scholar]
- Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- Liu Z, Vong QP, Zheng Y. CLASPing microtubules at the trans-Golgi network. Dev Cell. 2007;12:839–840. doi: 10.1016/j.devcel.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Lucocq JM, Berger EG, Warren G. Mitotic Golgi fragments in HeLa cells and their role in the reassembly pathway. J Cell Biol. 1989;109:463–474. doi: 10.1083/jcb.109.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucocq JM, Warren G. Fragmentation and partitioning of the Golgi apparatus during mitosis in HeLa cells. EMBO J. 1987;6:3239–3246. doi: 10.1002/j.1460-2075.1987.tb02641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- Maffini S, Maia AR, Manning AL, Maliga Z, Pereira AL, Junqueira M, Shevchenko A, Hyman A, Yates JR, 3rd, Galjart N, Compton DA, Maiato H. Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr Biol. 2009;19:1566–1572. doi: 10.1016/j.cub.2009.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Fairley EA, Rieder CL, Swedlow JR, Sunkel CE, Earnshaw WC. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 2003;113:891–904. doi: 10.1016/s0092-8674(03)00465-3. [DOI] [PubMed] [Google Scholar]
- Maiato H, Rieder CL, Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol. 2004;167:831–840. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazia D. The chromosome cycle and the centrosome cycle in the mitotic cycle. Int Rev Cytol. 1987;100:49–92. doi: 10.1016/s0074-7696(08)61698-8. [DOI] [PubMed] [Google Scholar]
- Miller PM, Folkmann AW, Maia AR, Efimova N, Efimov A, Kaverina I. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat Cell Biol. 2009;11:1069–1080. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho-Pereira S, Debec A, Maiato H. Microtubule cytoskeleton remodeling by acentriolar microtubule-organizing centers at the entry and exit from mitosis in Drosophila somatic cells. Mol Biol Cell. 2009;20:2796–2808. doi: 10.1091/mbc.E09-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell CB, Khodjakov AL. Cooperative mechanisms of mitotic spindle formation. J Cell Sci. 2007;120:1717–1722. doi: 10.1242/jcs.03442. [DOI] [PubMed] [Google Scholar]
- Rivero S, Cardenas J, Bornens M, Rios RM. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 2009;28:1016–1028. doi: 10.1038/emboj.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins E, Gonatas NK. The Ultrastructure of a Mammalian Cell during the Mitotic Cycle. J Cell Biol. 1964;21:429–463. doi: 10.1083/jcb.21.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta D, Linstedt AD. Control of organelle size: the Golgi complex. Annu Rev Cell Dev Biol. 2011;27:57–77. doi: 10.1146/annurev-cellbio-100109-104003. [DOI] [PubMed] [Google Scholar]
- Shima DT, Cabrera-Poch N, Pepperkok R, Warren G. An ordered inheritance strategy for the Golgi apparatus: visualization of mitotic disassembly reveals a role for the mitotic spindle. J Cell Biol. 1998;141:955–966. doi: 10.1083/jcb.141.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima DT, Haldar K, Pepperkok R, Watson R, Warren G. Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J Cell Biol. 1997;137:1211–1228. doi: 10.1083/jcb.137.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin C, Colanzi A. The Golgi and the centrosome: building a functional partnership. J Cell Biol. 2010;188:621–628. doi: 10.1083/jcb.200910001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin C, Hsu P, Mallabiabarrena A, Malhotra V. Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell. 2002;109:359–369. doi: 10.1016/s0092-8674(02)00720-1. [DOI] [PubMed] [Google Scholar]
- Tang D, Yuan H, Wang Y. The role of GRASP65 in Golgi cisternal stacking and cell cycle progression. Traffic. 2010;11:827–842. doi: 10.1111/j.1600-0854.2010.01055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulu US, Rusan NM, Wadsworth P. Peripheral, non-centrosome-associated microtubules contribute to spindle formation in centrosome-containing cells. Curr Biol. 2003;13:1894–1899. doi: 10.1016/j.cub.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Vasquez RJ, Howell B, Yvon AM, Wadsworth P, Cassimeris L. Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol Biol Cell. 1997;8:973–985. doi: 10.1091/mbc.8.6.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci U S A. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradova T, Paul R, Grimaldi AD, Loncarek J, Miller PM, Yampolsky D, Magidson V, Khodjakov A, Mogilner A, Kaverina I. Concerted effort of centrosomal and Golgi-derived microtubules is required for proper Golgi complex assembly but not maintenance. Mol Biol Cell. 2012 doi: 10.1091/mbc.E11-06-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei JH, Seemann J. Mitotic division of the mammalian Golgi apparatus. Semin Cell Dev Biol. 2009;20:810–816. doi: 10.1016/j.semcdb.2009.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.