Abstract
Bacterial biofilm formation is a complex developmental process involving cellular differentiation and the formation of intricate 3D structures. Here we demonstrate that exposure to ferric chloride triggers rugose biofilm formation by the uropathogenic Escherichia coli strain UTI89 and by enteric bacteria Citrobacter koseri and Salmonella enterica serovar typhimurium. Two unique and separable cellular populations emerge in iron-triggered, rugose biofilms. Bacteria at the air–biofilm interface express high levels of the biofilm regulator csgD, the cellulose activator adrA, and the curli subunit operon csgBAC. Bacteria in the interior of rugose biofilms express low levels of csgD and undetectable levels of matrix components curli and cellulose. Iron activation of rugose biofilms is linked to oxidative stress. Superoxide generation, either through addition of phenazine methosulfate or by deletion of sodA and sodB, stimulates rugose biofilm formation in the absence of high iron. Additionally, overexpression of Mn-superoxide dismutase, which can mitigate iron-derived reactive oxygen stress, decreases biofilm formation in a WT strain upon iron exposure. Not only does reactive oxygen stress promote rugose biofilm formation, but bacteria in the rugose biofilms display increased resistance to H2O2 toxicity. Altogether, we demonstrate that iron and superoxide stress trigger rugose biofilm formation in UTI89. Rugose biofilm development involves the elaboration of two distinct bacterial populations and increased resistance to oxidative stress.
Keywords: functional amyloid, biofilm matrix, wrinkled colony
By adopting a biofilm lifestyle, bacteria gain resistance to antibiotics, the host immune system, and environmental stresses (1, 2). Gene expression and metabolic processes can vary drastically from cell to cell in a biofilm community (3, 4), and cellular differentiation within a biofilm contributes to antibiotic resistance (1). Therefore, an in-depth understanding of biofilm formation is critical for the development of antibacterial agents.
Iron is an essential nutrient for almost all bacterial species, but it is efficiently sequestered by the host during an infection (5) and it is largely insoluble in aerobic environments (6). Additionally, ferrous iron can react with H2O2 in the Fenton reaction to form reactive hydroxyl radicals, which can damage proteins, DNA, and lipids (7, 8). To counter iron scarcity and toxicity, most bacteria express efficient iron acquisition systems and tightly regulate intracellular iron homeostasis (6, 8).
Iron affects biofilm formation in a variety of bacteria (9–13). In Escherichia coli, the master biofilm regulator csgD appears to have several indirect ties to iron sensing, transport, and availability. Overexpression of csgD inhibits transcription of the iron regulator fecR and the outer-membrane ferric-coprogen receptor fhuE (14). Furthermore, CsgD binding sites are present upstream of the fepDGC and entS operons, which encode components of a ferric enterobactin transporter and an enterobactin exporter, respectively (15). Finally, iron chelators promote csgD expression in Salmonella enterica serovar typhimurium (16, 17).
In this study we investigated the relationship between curli expression, biofilm formation, and environmental iron levels in an E. coli cystitis isolate, UTI89. Uropathogenic E. coli (UPEC) is the predominant agent of urinary tract infections, and biofilm formation and the production of curli fibers contribute to UPEC pathogenesis (18, 19). Curli fibers are functional amyloids that form an integral component of the extracellular matrix (20). CsgD controls E. coli biofilm formation largely through induction of the curli subunit operon csgBAC (21) and the cellulose activator adrA (22, 23). Along with curli, cellulose aids cell-to-cell attachment and adherence to inorganic surfaces and host cells (24–26). UTI89 and a variety of other bacterial species form rugose biofilms (also known as rdar) on agar plates (4, 23, 27-29). In both S. typhimurium and E. coli, the CsgD-regulated matrix components curli and cellulose are necessary for development of rugose biofilms (23, 27, 30).
This work revealed several features of E. coli biofilms in response to iron. First, we observed that iron induced UTI89 rugose biofilms without increasing total matrix production. Within a rugose biofilm, distinct and separable bacterial populations emerged. Curli production was limited to bacteria at the air–biofilm interface, and non–curli-producing bacteria filled the interior of rugose biofilm wrinkles. Furthermore, superoxide could activate the rugose biofilm developmental pathway in place of high iron, and rugose biofilm formation coincided with increased survival after H2O2 treatment. In summary, we describe an iron-induced biofilm pathway involving formation of a bimodal bacterial population and oxidative stress resistance in enteric bacteria.
Results
Iron Triggers the Formation of csgD-Dependent Rugose Biofilms.
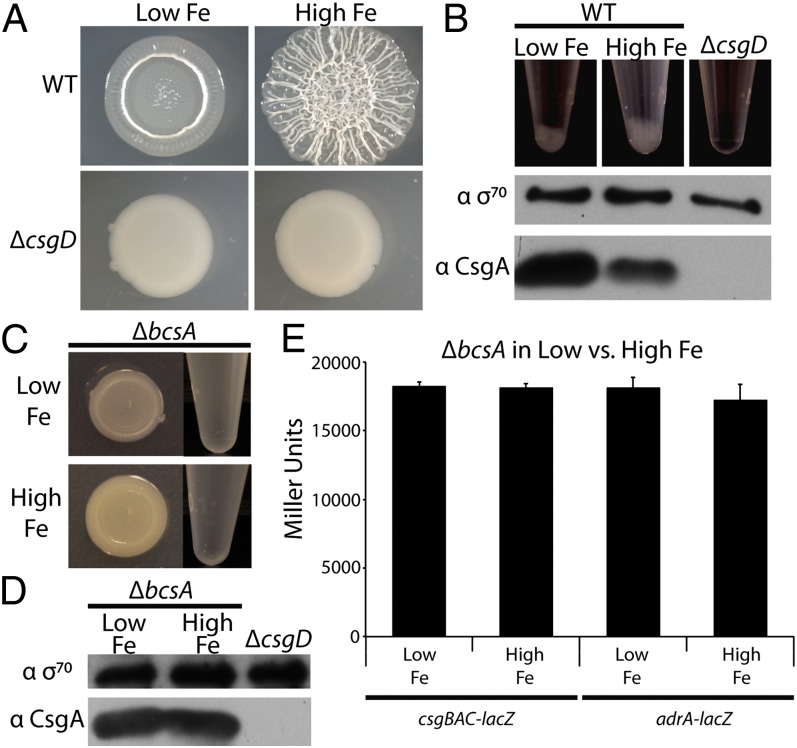
UTI89 forms csgD-dependent rugose biofilms on YESCA agar plates (Fig. S1). To test the effect of iron levels on rugose biofilm development, we treated YESCA with Chelex-100, a chelating resin. WT UTI89 grown on Chelex-treated YESCA agar plates (referred to as low-iron conditions) did not form rugose biofilms. Addition of 2 mM FeCl3 to the cell mixture before plating (referred to as high-iron conditions) restored biofilm formation (Fig. 1A). As expected, a csgD mutant was unable to form rugose biofilms in either low- or high-iron conditions (Fig. 1A). To determine whether iron-induced rugose biofilm formation coincided with increased curli fiber production, whole-cell Western blot analysis probing for the major curli subunit CsgA was performed on UTI89 grown in low- or high-iron conditions. Bacteria grown in low-iron conditions unexpectedly expressed more CsgA than rugose-forming UTI89 grown in high-iron conditions (Fig. 1B). However, rugose biofilms coincided with an increase in cell aggregation after treatment with a tissue homogenizer (Fig. 1B). Because aggregation was dependent on csgD (Fig. 1B), we hypothesized that curli and cellulose were components of the aggregate; this could bias Western blot analysis, because noncurliated, suspended bacteria would more likely be sampled. To prevent aggregation, we produced a UTI89 mutant that cannot synthesize cellulose. In most, but not all, E. coli strains, CsgD activates cellulose production by inducing transcription of adrA (23, 31). In turn, the diguanylate cyclase AdrA stimulates the cellulose synthase BcsA, presumably through production of cyclic di-GMP, because BcsA contains a cyclic di-GMP binding domain (31–33). We verified that UTI89 rugose biofilm formation was dependent on csgD, adrA, and bcsA (22, 23, 31) (Fig. 1C; Fig. S1). No aggregates collected when the bcsA mutant was tissue homogenized (Fig. 1C). We therefore repeated the CsgA Western blot described above in the bcsA mutant background and found that overall levels of CsgA remained unchanged in low- vs. high-iron conditions (Fig. 1D). Furthermore, β-galactosidase assays of a bcsA mutant transformed with plasmids encoding csgBAC-lacZ or adrA-lacZ transcriptional fusions revealed that expression of neither operon changed in response to iron (Fig. 1E). Our results demonstrate that iron drives rugose biofilm development and cell aggregation in UTI89. However, because overall curli production does not change as a result of varying iron concentrations, we inferred that iron exposure may trigger clustering of curli-producing cells within a rugose biofilm.
Fig. 1.
UTI89 forms a csgD-dependent rugose biofilm in high-iron conditions. (A) WT UTI89 or a csgD mutant was grown on Chelex-treated YESCA plates with or without 2 mM FeCl3 added to the cell mixture before plating. (B) After tissue homogenization, aggregates immediately began falling out of suspension and were fully settled within 10 min. Aggregation increased with rugose biofilm formation and was dependent on csgD (Top). Suspended cells from low iron colonies demonstrate an increase in CsgA protein levels by whole-cell Western blot compared with suspended cells from high iron conditions, whereas σ70 levels remain constant (Middle and Bottom). (C) A bcsA mutant does not form rugose biofilms (Left) and does not form aggregates after tissue homogenization (Right) when grown in either low- or high-iron conditions. (D) A Western blot of the bcsA mutant shows that total CsgA levels do not change between low- and high-iron conditions. (E) A UTI89 bcsA mutant strain carrying csgBAC (pBA14) or adrA (pRJ800-adrA) transcriptional lacZ fusion plasmids or an empty vector (pRJ800) were plated on Chelex-treated YESCA plates with or without the addition of FeCl3. β-Galactosidase assays were performed, and error bars represent the SD of biological triplicates. Average Miller units obtained from bcsA carrying an empty vector (pRJ800) were subtracted from each respective sample.
Rugose Biofilms Contain Two Distinct, Separable Bacterial Populations.
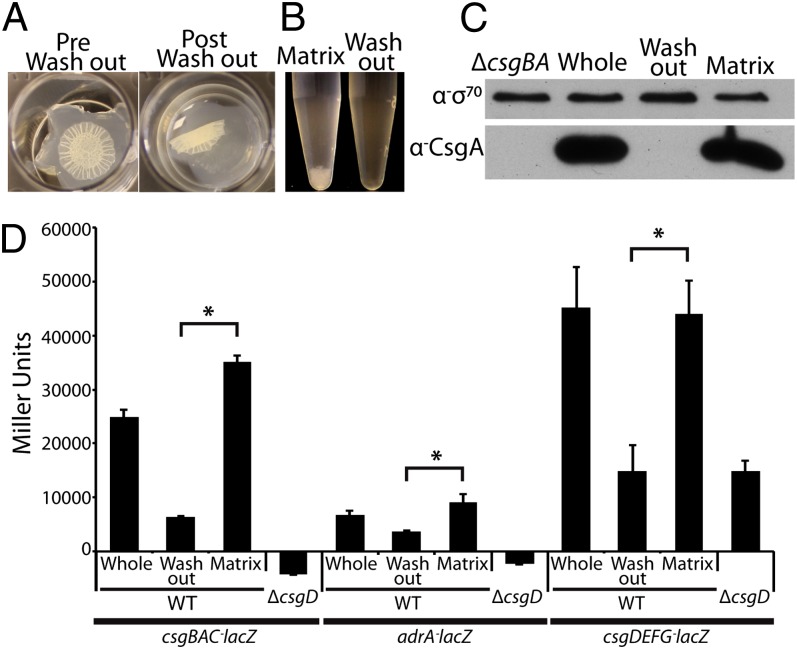
Our data lead us to hypothesize that two bacterial populations are present in a rugose biofilm—(i) matrix associated and (ii) non-matrix associated. To mechanically separate these populations, we flooded a rugose biofilm in potassium phosphate buffer pH 7.2 (KPi) with light shaking, reasoning that a non–matrix-encased population would be washed into suspension. Upon shaking, the biofilm floated off the agar surface (Fig. 2A). Bacteria that washed freely into suspension (washout fraction) were separated from the aggregated bacteria (matrix fraction), which maintained its general shape after loss of the washout cells (Fig. 2A). Aggregates only settled out of suspension after tissue homogenization of the matrix fraction (Fig. 2B), and Western blot analysis demonstrated that CsgA was solely associated with matrix fraction bacteria (Fig. 2C).
Fig. 2.
Two separable populations, matrix producing and non-matrix producing, are present in a rugose biofilm. (A) Washing a UTI89 rugose biofilm in 50 mM KPi buffer after growth on a YESCA agar plate separates suspended washout from aggregative matrix bacteria. (B) Cell aggregates only collect after tissue homogenization of the matrix fraction. (C) Whole-cell Western blot analysis demonstrates that CsgA is localized solely to the matrix fraction bacteria. (D) WT UTI89 or a csgD mutant carrying csgBAC (pBA14), adrA (pRJ800-adrA), or csgDEFG (pD1) transcriptional lacZ fusion plasmids were grown on YESCA plates. β-Galactosidase assays were performed on each strain before (whole) or after the washout assay, and error bars represent the SD of biological triplicates. WT and a csgD mutant carrying an empty vector (pRJ800) were also assayed, and average Miller units from these strains were subtracted from the respective samples. Asterisks represent P values (*P < 0.05) evaluated using a Student t test.
To test whether cellulose, the other major rugose biofilm component, was also absent in the washout fraction, we quantified binding of Congo red (CR)—a diazo dye that binds to both curli and cellulose (31, 34). Washout fraction bacteria bound a similar amount of CR as a csgBA bcsA double mutant (Fig. S2), indicating that they produce neither curli nor cellulose (34). Consistent with the CR-binding results, β-galactosidase assays showed that transcription of csgDEFG, csgBAC, and adrA was significantly reduced in the washout fraction compared with the matrix fraction (Fig. 2D). Therefore, we were able to identify and mechanically separate matrix-encased and non–matrix-encased bacteria in a rugose biofilm.
Curliated Bacteria Are Localized to the Air–Biofilm Interface in Rugose Biofilms.
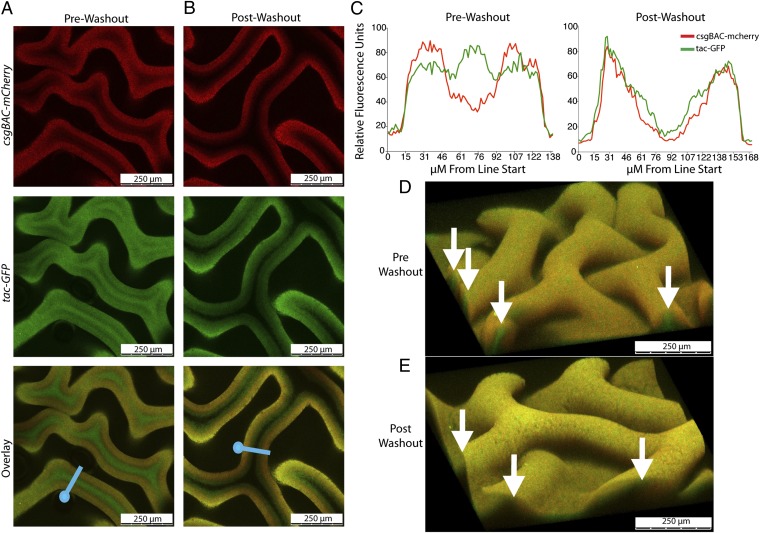
Confocal microscopy was used to locate curli-producing cells within a rugose biofilm. A UTI89 strain harboring a csgBAC-mCherry transcriptional fusion at the attB site was transformed with the IPTG-inducible GFP-expressing plasmid pCKR101-eGFP. We reasoned that all metabolically active cells would express GFP, whereas curli-expressing cells would produce both mCherry and GFP. Z-stack videos taken from a rugose biofilm grown on a YESCA agar plate revealed that bacteria expressing mCherry were localized to the biofilm surface, whereas a population of non–curli-producing bacteria was localized to the interior of the each wrinkle (Movie S1). Cross-section images of the biofilm midpoint, as well as 3D reconstructions of biofilm sections, further demonstrated that each wrinkle is filled with a population of non–curli-producing cells (Fig. 3 A, C, and D). After the washout assay, cells were absent from the wrinkle interiors, indicating that interior, non–curli-producing cells were removed as the washout fraction (Fig. 3 B, C, and E; Movie S2).
Fig. 3.
Confocal microscopy reveals bimodal rugose biofilm architecture. (A) UTI89 attB::csgBAC-mcherry/pCKR101-eGFP was grown on 0.05-μM cellulose filters on YESCA agar plates with 1 mM IPTG added to the cell mixture before plating. Filter sections containing a biofilm were cut out with a razor blade, placed on a slide, and treated with mounting solution. A 20× confocal microscopy cross-section of a UTI89 attB::csgBAC-mcherry/pCKR101-eGFP rugose biofilm before and (B) after the washout assay was imaged. (C) Average relative fluorescent intensity of five separate traces across the blue lines in A and B for each fluorophore was calculated and is graphed against distance along the blue line from each blue circle. 3D reconstructions before (D) and after (E) the washout assay. White arrows indicate wrinkle openings where non–curli-producing bacteria are present before the washout assay but not after.
To specifically probe iron-responsive architectural changes, the csgBAC-mCherry/pCKR101-eGFP reporter strain was grown on Chelex-treated YESCA plates with or without FeCl3 added to the cell mixture. In the low-iron colony, GFP and mCherry were evenly distributed throughout, indicating no large-scale spatial separation between curli-expressing cells and non–curli-expressing cells except for a gradual increase in curli/mCherry-producing cells near the biofilm surface (Fig. S3A). When iron was added to the cell mixture before plating on Chelex-treated YESCA plates, the distribution of cells throughout the rugose biofilms matched that seen on YESCA plates (Fig. 3D; Fig. S3B).
Oxygen levels can affect folding and activity of fluorophores (35). Because oxygen penetration into biofilms is generally limited (3), we also imaged expression patterns of a UTI89 attB::csgBAC-eGFP/pCKR101-mCherry strain. In this strain, mCherry-producing cells were localized to the interior of the wrinkles. Bacteria expressing both mCherry and GFP lined the air–biofilm interface (Fig. S3C). Because this pattern was essentially the inverse of that seen in Fig. 3D, we concluded that both fluorophores can fluoresce in the biofilm interior and that the csgBAC promoter is repressed in the interior washout cells.
Superoxide Stress Drives Rugose Biofilm Formation.
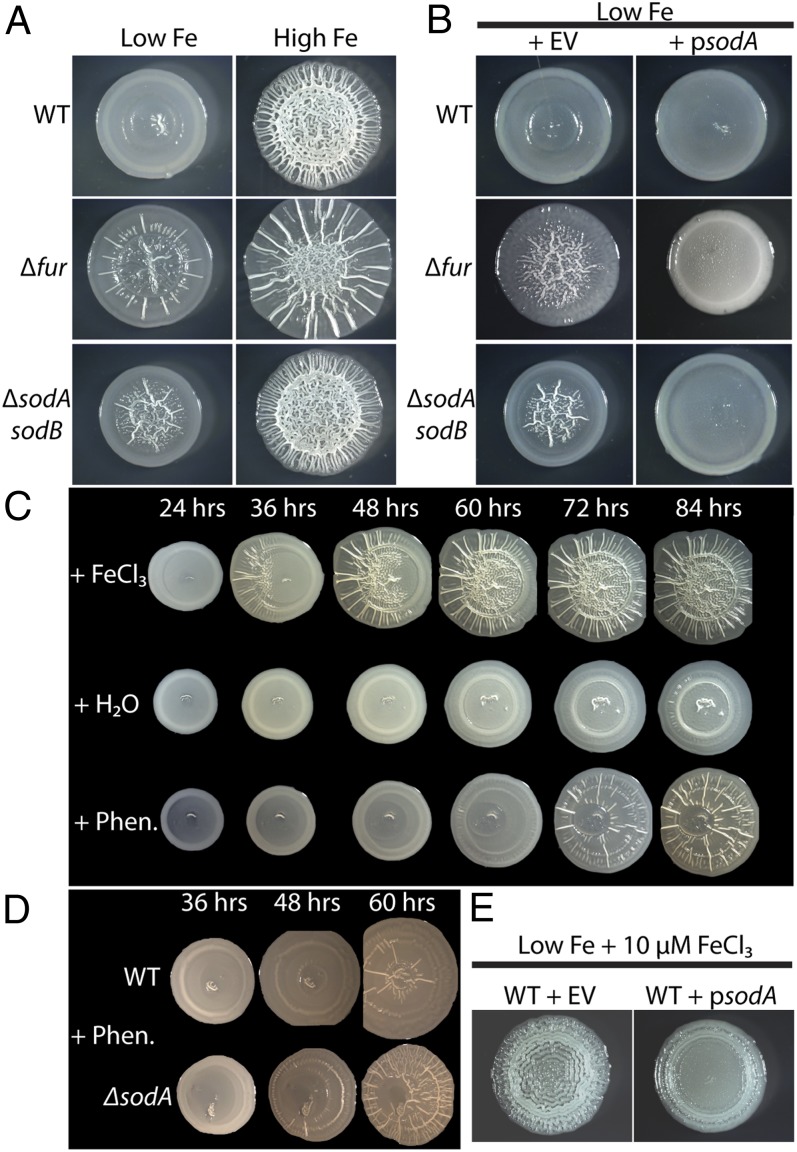
Because iron triggers rugose biofilm formation, we hypothesized that an iron-responsive regulatory protein was involved in the rugose biofilm developmental process. To test this, we knocked out known iron-responsive transcriptional factors that have also been shown to affect biofilm formation. Our candidates included the global iron regulator, Fur (11, 12), the iron-sulfur cluster regulator, IscR (9), the ferric iron-sensing two-component system, BasSR (36), and the small RNA, RyhB (10). All of the mutants still formed rugose biofilms in response to iron (Fig. S4). However, the fur mutant wrinkled more than WT in the low-iron conditions (Fig. 4A; Fig. S4).
Fig. 4.
Iron and superoxide stress drives rugose biofilm formation. (A) WT UTI89, a fur mutant, or a sodA sodB double mutant were grown on Chelex-treated YESCA plates with or without the addition of FeCl3 to the cell mixture before plating. (B) Overexpression of sodA repressed rugose biofilm formation in the fur and the sodA sodB mutant compared with the empty vector (EV) controls. Strains were grown on Chelex-treated YESCA plates supplemented with 100 μM MnCl2 and either 10 μM IPTG (WT and sodA sodB) or 50 μM IPTG (fur). (C) WT UTI89 was exposed to 5 μL H2O, 100 mM FeCl3, or 10 mM phenazine methosulfate on a sterile paper disk (1 cm to the left of the pictured biofilms). (D) Rugose biofilm formation in response to phenazine is more rapid in a sodA mutant compared with WT. (E) Rugose biofilm formation in WT UTI89 grown on Chelex-treated YESCA plates supplemented with 100 μM MnCl2, 50 μM IPTG, and 10 μM FeCl3 was repressed by overexpression of sodA.
A fur mutant constitutively expresses various iron acquisition systems and accumulates toxic amounts of cytoplasmic free ferrous iron (37). To investigate the possibility that the increase in rugose biofilm formation in the fur mutant is due to iron-induced toxicity, we constructed a UTI89 sodA sodB double mutant that cannot produce cytoplasmic superoxide dismutase. In a sodA sodB mutant, cytoplasmic superoxide accumulates and breaks down solvent-exposed [4Fe-4s] clusters in vulnerable proteins, freeing ferrous iron (38–40). The sodA sodB mutant formed a rugose biofilm under low-iron conditions in a similar manner as the fur mutant (Fig. 4A). We also overexpressed sodA in a fur mutant to see if rugose biofilm formation was dampened, because reactive oxygen stress in a fur mutant can be partly negated by sodA overexpression (37). Rugose biofilm formation was decreased under low-iron conditions in both the fur mutant and sodA sodB mutant when sodA was overexpressed (Fig. 4B).
Because accumulation of endogenous superoxide induced rugose biofilm development, we next tested whether superoxide-producing antibiotics can induce rugose biofilm formation in place of high iron levels. Pseudomonas aeruginosa, along with a variety of other bacteria, produce redox-cycling antibiotics called phenazines (29, 41). In susceptible bacteria such as E. coli, phenazines enter the cell and produce superoxide by oxidizing cytoplasmic targets and reducing molecular oxygen (42). Intriguingly, exogenous phenazine methosulfate exposure resulted in rugose biofilm formation in low-iron conditions in UTI89, albeit at a slower rate than iron induction (Fig. 4C). A sodA mutant developed a rugose biofilm more rapidly than WT in response to phenazine exposure (Fig. 4D). Because both iron and superoxide can independently induce rugose biofilms, our next question was whether reactive oxygen stress was involved in iron-driven rugose biofilm formation. To this end, we overexpressed sodA in WT and observed rugose biofilm formation on Chelex-treated YESCA plates supplemented with 10 μM FeCl3 (Fig. 4E). Overexpression of sodA decreased rugose biofilm formation after 48 h. Altogether, these results indicate that both iron and superoxide stress trigger rugose biofilm formation. Furthermore, iron-induced rugose biofilm formation is at least partly dependent on reactive oxygen stress.
Rugose Biofilm Formation Coincides with H2O2 Resistance.
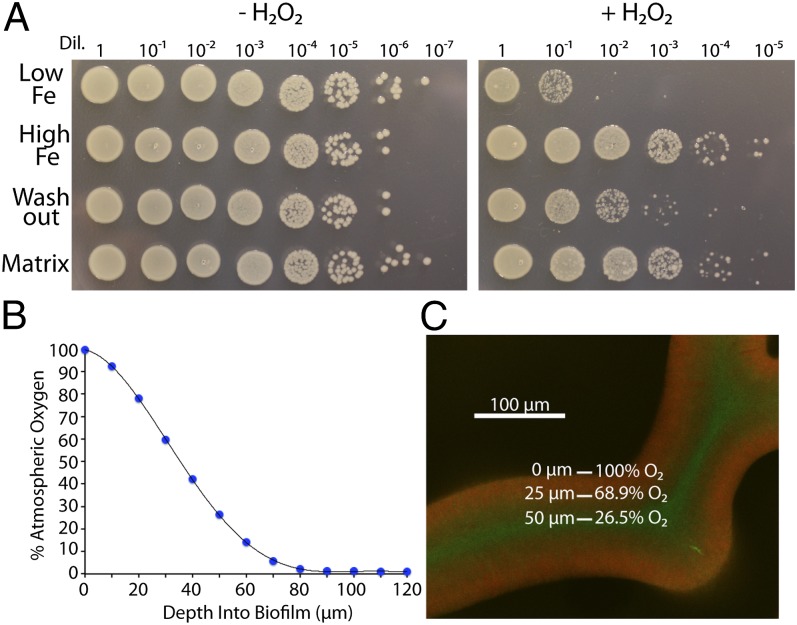
Because rugose biofilm development can be triggered by reactive oxygen stress, we hypothesized that a rugose biofilm would demonstrate increased resistance to H2O2. Therefore, UTI89 grown in low- or high-iron conditions was tissue-homogenized, normalized by OD600, and treated with H2O2. Strikingly, iron-induced rugose biofilm development corresponded with H2O2 resistance (Fig. 5A). We next tested H2O2 resistance of the washout and the matrix fractions of rugose biofilms grown on a Chelex-treated YESCA plate with addition of iron. Intriguingly, washout fraction bacteria were more sensitive to H2O2 than matrix fraction cells, whereas both fractions were more resistant than bacteria grown under low-iron conditions (Fig. 5A).
Fig. 5.
Rugose biofilm formation coincides with H2O2 resistance. (A) WT UTI89 was grown on Chelex-treated YESCA plates with or without the addition of FeCl3 to the cell mixture before plating. Bacteria from each iron condition, or from washout and matrix fractions from high-iron rugose biofilms, were normalized to 1 OD600 and exposed to 1% (vol/vol) H2O2 in KPi for 20 min. Cells were serially diluted and plated on LB plates. (B) A 10-μm oxygen microsensor was used to measure oxygen penetration into WT UTI89 rugose biofilms. Each data point is an average of three technical and three biological replicates. A sixth-order polynomial curve was fit to the data points. (C) Oxygen penetration depth was mapped onto a 40× confocal cross-section image of the midpoint of a UTI89 attB::csgBAC-mcherry/pCKR101-eGFP rugose biofilm induced with IPTG.
Reactive oxygen species are generated as a byproduct of aerobic metabolism (43, 44), and oxygen diffusion into biofilms is generally limited due to respiration by periphery cells (3). We hypothesized that within a rugose biofilm, interior, nonmatrix-encased bacteria would be exposed to less atmospheric oxygen than matrix-encased, exterior cells. Oxygen microsensor measurements of a rugose biofilm demonstrated that oxygen levels decrease with biofilm depth (Fig. 5B). Assuming equal oxygen penetration from both the top and the sides of a single wrinkle, we can then map oxygen penetration depth onto a confocal image of curli-expressing cells in a rugose biofilm (Fig. 5C). The interior, non–matrix-associated cells of a rugose biofilm are therefore exposed to ∼25–70% atmospheric oxygen, depending on their proximity to the surface (Fig. 5C). These data indicate that rugose biofilm formation parallels H2O2 resistance. Within a rugose biofilm, interior, washout cells are partly shielded from oxygen and are more susceptible to reactive oxygen stress than their matrix-encased neighbors.
Iron Triggers Bimodal Rugose Biofilms in Other Enteric Bacteria.
Last, we wanted to determine if iron-induced rugose biofilm formation was widespread among enterics. To this end, S. typhimurium and a clinical isolate of Citrobacter koseri, which form rugose biofilms on LB–salt and YESCA agar plates, respectively (Fig. S5A), were plated on Chelex-treated agar plates with or without addition of FeCl3. Both strains formed rugose biofilms only when exposed to iron (Fig. S5B). Furthermore, CsgA was largely localized to the matrix fractions after performing the washout assay (Fig. S5C). As in UTI89, rugose biofilm formation paralleled H2O2 resistance in each strain (Fig. S5D). We conclude that iron-induced bimodal rugose biofilm development and the increase in H2O2 resistance as a function of rugose biofilm formation are common features of enteric bacteria.
Discussion
Our results demonstrate that iron induces macroscopic architectural restructuring and H2O2 resistance in UTI89 biofilms (Figs. 1 and 5). However, total curli levels did not change in response to varying iron concentrations (Fig. 1), indicating that matrix production is not sufficient for rugose biofilm formation. Previous work has found that low iron levels can affect curli production under certain conditions, because iron chelators added to S. typhimurium results in lower curli expression (16, 17). However, comparing the Salmonella results with ours is difficult because of disparities in strains used and growth conditions. Iron-dependent rugose biofilm formation has likely not been reported before because common laboratory media has sufficient iron to induce rugose biofilm formation, as is evidenced by the fact that WT UTI89, C. koseri, and S. typhimurium form rugose biofilms on YESCA or LB–salt agar plates without the addition of iron (Fig. S5A).
Both iron availability and biofilm formation affect UPEC pathogenesis. During infection, UPEC ascends the urethra into the bladder. Once in the bladder, UPEC is able to attach to and invade urothelial cells (45). After cell invasion, UPEC forms biofilm-like pods, termed intracellular bacterial communities (IBCs) (18), and IBC formation helps UPEC avoid the host immune system (18, 45, 46). Growth in urine induces expression of a variety of iron acquisition systems in UPEC, demonstrating that urine is an iron-limiting environment (47). Additionally, IBC formation correlates with induction of genes involved in acquiring iron from heme and siderophores (48). Because iron and biofilm formation both play a significant role in UPEC pathogenesis, it is tempting to speculate that the iron-induced biofilm formation we describe here may contribute to UPEC virulence.
Experiments with UTI89 grown in low- and high-iron conditions revealed that iron induces aggregation and rugose biofilm development. There are at least two spatially distinct cellular populations within rugose biofilms: matrix encased and non-matrix encased (Figs. 2 and 3). Studies in S. typhimurium have previously demonstrated that levels of CsgD and curli can vary from cell to cell in bacterial communities (17, 49). Here we have demonstrated a distinct bimodal restructuring in which matrix-producing bacteria line the air–biofilm interface, and non–matrix-producing bacteria fill the wrinkle interiors. Additionally, the two populations are easily separated, because expression of curli and cellulose imbues a rigidity to the matrix-encased cells that prevents them from being suspended during buffer washes. Separation via the washout assay allows for examination of the two distinct populations by biochemical and genetic techniques. The dramatic disappearance of curli and cellulose and the elevated H2O2 susceptibility in the washout fraction imply significant physiological differences between washout and matrix fractions.
We have determined that the iron-induced rugose biofilm developmental pathway is triggered, at least in part, by reactive oxygen stress. However, the mechanism by which high extracellular iron would cause intracellular reactive oxygen stress is unclear. E. coli is able to maintain a steady intracellular iron pool, regardless of extracellular iron fluctuations, due to the efficacy of iron homeostasis regulators such as Fur and RyhB (6). Fur is a transcriptional regulator that binds to ferrous iron and represses expression of iron acquisition systems. RyhB is a small RNA that limits production of iron-containing proteins in low-iron conditions (6, 50). Therefore, high extracellular iron should not necessarily lead to cytoplasmic-free iron stress. High iron levels could allow for rapid cell growth and aerobic respiration, resulting in superoxide production from the electron transport chain (43, 44). Additionally, it has been proposed that normal fluxes in intracellular free iron through Fe-Fur cycling can lead to brief periods of iron stress (37); if this is the case, the summation of oxidative stress from multiple free-iron cycles may lead to enough reactive oxygen stress to induce rugose biofilm formation. sodA overexpression could temper the amount of free-iron stress in each cycle. Future work will elucidate the genetic pathways leading from oxidative stress to the development of rugose biofilms.
A wide variety of bacterial species form rugose biofilms (4, 23, 28, 29). Though the triggers of rugose biofilm development no doubt vary between systems, the conserved wrinkled architecture implies a measure of commonality. Rugose biofilm formation in P. aeruginosa is inhibited by self-produced phenazines (29). Intriguingly, we found that E. coli forms rugose biofilms when exposed to phenazines or when it is unable to neutralize endogenous superoxide (Fig. 4 A and C). Furthermore, a recent study in Bacillus subtilis determined that localized cell death contributes to the mechanical buckling necessary for rugose biofilm wrinkle formation (28). It is tempting to speculate that superoxide stress could accelerate cell death, although we found that both the washout and matrix fraction cells are viable under conditions that promote rugose biofilm formation (Fig. 5A). Additionally, we would expect superoxide toxicity to increase with proximity to molecular oxygen, and we have demonstrated that interior bacteria in a rugose biofilm are exposed to less atmospheric oxygen than their surface-exposed neighbors.
Biofilm formation and, in particular, cellular differentiation within biofilms, contributes to bacterial antibiotic resistance (1). The sophisticated genetic tools available in E. coli, the large body of literature dedicated to E. coli metabolism, and the prevalence of E. coli-mediated uropathogenic infections make UTI89 an attractive model for biofilm development. Within iron-induced rugose biofilms, two populations (matrix encased and non-matrix encased) emerge. These populations are easily separable due to the loose association of interior cells with the biofilm matrix. Rugose biofilm development also parallels tiered H2O2 resistance, with surface-exposed bacteria being more resistant than interior cells. Analysis of the mechanisms of dual-population formation and the genetic pathways involved in H2O2 resistance will provide further insight into a biofilm developmental pathway.
Materials and Methods
Strains, Growth Conditions, and Biofilm Formation.
Reagents, strains, growth conditions, primers, and cloning methods are detailed in SI Text. Strains and plasmids are listed in Table S1, and primers are listed in Table S2. Bacteria were routinely grown in LB broth at 37 °C under aeration. Rugose biofilms were grown for 48 h on agar plates at 26 °C unless otherwise noted. Chelex-100 resin was used to remove metals from media, as detailed in SI Text.
Washout Assay.
After 48 h of growth on agar plates, a sterile metal spatula was used to cut out and transfer a slab of agar containing an intact colony to a well of a Falcon tissue-treated polystyrene 24-well plate. One milliliter of sterile 50 mM KPi (pH 7.2) was added to the well, and the plate was shaken gently until the matrix fraction had completely dissociated from the agar surface (typically 5 min). The KPi buffer was removed and the washout cells were spun down. Two more 5-min washes were performed on the colony, and buffer from each rinse was added to the washout tube and spun down. A pipette tip was used to remove the matrix fraction from the third rinse, and the entire matrix was placed in a separate Eppendorf tube with 1 mL KPi. The matrix fraction was then tissue homogenized. Briefly, a Fisher Scientific Tissuemiser Homogenizer was inserted into the Eppendorf tube, set to medium speed, and run for 20 s. Aggregates were allowed to settle, and suspended cells from each fraction were then assayed.
Detailed protocols for Western blot analysis, β-galactosidase assays, confocal microscopy, H2O2 viability, and oxygen microsensor measurements are described in SI Text.
Supplementary Material
Acknowledgments
We thank Scott Hultgren, Blaise Boles, and James Imlay for materials and strains; Gregg Sobocinski for microscopy sample preparation and microscopy assistance; and members of the M.R.C., Blaise Boles, and Robert Bender laboratories for helpful discussion. This work was supported by National Institutes of Health Grants R01 A1073847-01 (to M.R.C.) and T32 GM007544-32 (to W.H.D.), and the University of Michigan Rackham Merit Fellowship (W.H.D.)
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218703110/-/DCSupplemental.
References
- 1.Lewis K. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol. 2008;322:107–131. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- 2.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 3.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6(3):199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 4.López D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harb Perspect Biol. 2010;2(7):a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skaar EP. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010;6(8):e1000949. doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cornelis P, Wei Q, Andrews SC, Vinckx T (2011) Iron homeostasis and management of oxidative stress response in bacteria. Metallomics 3(6):540–549. [DOI] [PubMed]
- 7.Imlay JA, Chin SM, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240(4852):640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 8. Cabiscol E, Tamarit J, Ros J (2000) Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol 3(1):3–8. [PubMed]
- 9.Wu Y, Outten FW. IscR controls iron-dependent biofilm formation in Escherichia coli by regulating type I fimbria expression. J Bacteriol. 2009;191(4):1248–1257. doi: 10.1128/JB.01086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mey AR, Craig SA, Payne SM. Characterization of Vibrio cholerae RyhB: The RyhB regulon and role of ryhB in biofilm formation. Infect Immun. 2005;73(9):5706–5719. doi: 10.1128/IAI.73.9.5706-5719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banin E, Vasil ML, Greenberg EP. Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci USA. 2005;102(31):11076–11081. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson M, Cockayne A, Williams PH, Morrissey JA. Iron-responsive regulation of biofilm formation in Staphylococcus aureus involves fur-dependent and fur-independent mechanisms. J Bacteriol. 2005;187(23):8211–8215. doi: 10.1128/JB.187.23.8211-8215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banin E, Brady KM, Greenberg EP. Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl Environ Microbiol. 2006;72(3):2064–2069. doi: 10.1128/AEM.72.3.2064-2069.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brombacher E, Baratto A, Dorel C, Landini P. Gene expression regulation by the Curli activator CsgD protein: Modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J Bacteriol. 2006;188(6):2027–2037. doi: 10.1128/JB.188.6.2027-2037.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogasawara H, Yamamoto K, Ishihama A. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J Bacteriol. 2011;193(10):2587–2597. doi: 10.1128/JB.01468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Römling U, Sierralta WD, Eriksson K, Normark S. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol Microbiol. 1998;28(2):249–264. doi: 10.1046/j.1365-2958.1998.00791.x. [DOI] [PubMed] [Google Scholar]
- 17.White AP, et al. Aggregation via the red, dry, and rough morphotype is not a virulence adaptation in Salmonella enterica serovar Typhimurium. Infect Immun. 2008;76(3):1048–1058. doi: 10.1128/IAI.01383-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson GG, et al. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301(5629):105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 19.Cegelski L, et al. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat Chem Biol. 2009;5(12):913–919. doi: 10.1038/nchembio.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapman MR, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295(5556):851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammar M, Arnqvist A, Bian Z, Olsén A, Normark S. Expression of two Csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18(4):661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 22.Römling U, Rohde M, Olsén A, Normark S, Reinköster J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol Microbiol. 2000;36(1):10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- 23.Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39(6):1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 24.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saldaña Z, et al. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ Microbiol. 2009;11(4):992–1006. doi: 10.1111/j.1462-2920.2008.01824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou YZ, et al. Promiscuous cross-seeding between bacterial amyloids promotes interspecies biofilms. J Biol Chem. 2012;287(42):35092–35103. doi: 10.1074/jbc.M112.383737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim JY, May JM, Cegelski L. Dimethyl sulfoxide and ethanol elicit increased amyloid biogenesis and amyloid-integrated biofilm formation in Escherichia coli. Appl Environ Microbiol. 2012;78(9):3369–3378. doi: 10.1128/AEM.07743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Asally M, et al. (2012) Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc Natl Acad Sci USA 109(46):18891–18896. [DOI] [PMC free article] [PubMed]
- 29.Dietrich LE, Teal TK, Price-Whelan A, Newman DK. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science. 2008;321(5893):1203–1206. doi: 10.1126/science.1160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Römling U, Bian Z, Hammar M, Sierralta WD, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180(3):722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Da Re S, Ghigo JM. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J Bacteriol. 2006;188(8):3073–3087. doi: 10.1128/JB.188.8.3073-3087.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22(1):3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 33.Ryjenkov DA, Simm R, Römling U, Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: The PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem. 2006;281(41):30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 34.Ma Q, Wood TK. OmpA influences Escherichia coli biofilm formation by repressing cellulose production through the CpxRA two-component system. Environ Microbiol. 2009;11(10):2735–2746. doi: 10.1111/j.1462-2920.2009.02000.x. [DOI] [PubMed] [Google Scholar]
- 35.Hansen MC, Palmer RJ, Jr, Udsen C, White DC, Molin S. Assessment of GFP fluorescence in cells of Streptococcus gordonii under conditions of low pH and low oxygen concentration. Microbiology. 2001;147(Pt 5):1383–1391. doi: 10.1099/00221287-147-5-1383. [DOI] [PubMed] [Google Scholar]
- 36.Ogasawara H, Shinohara S, Yamamoto K, Ishihama A. Novel regulation targets of the metal-response BasS-BasR two-component system of Escherichia coli. Microbiology. 2012;158(Pt 6):1482–1492. doi: 10.1099/mic.0.057745-0. [DOI] [PubMed] [Google Scholar]
- 37.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: Protective role of superoxide dismutase. J Bacteriol. 1995;177(9):2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keyer K, Imlay JA. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci USA. 1996;93(24):13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liochev SI, Fridovich I. The role of O2.- in the production of HO: In vitro and in vivo. Free Radic Biol Med. 1994;16(1):29–33. doi: 10.1016/0891-5849(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 40.Liochev SL. The role of iron-sulfur clusters in in vivo hydroxyl radical production. Free Radic Res. 1996;25(5):369–384. doi: 10.3109/10715769609149059. [DOI] [PubMed] [Google Scholar]
- 41.Mavrodi DV, et al. Diversity and evolution of the phenazine biosynthesis pathway. Appl Environ Microbiol. 2010;76(3):866–879. doi: 10.1128/AEM.02009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hassan HM, Fridovich I. Intracellular production of superoxide radical and of hydrogen peroxide by redox active compounds. Arch Biochem Biophys. 1979;196(2):385–395. doi: 10.1016/0003-9861(79)90289-3. [DOI] [PubMed] [Google Scholar]
- 43.Messner KR, Imlay JA. The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J Biol Chem. 1999;274(15):10119–10128. doi: 10.1074/jbc.274.15.10119. [DOI] [PubMed] [Google Scholar]
- 44.Imlay JA, Fridovich I. Assay of metabolic superoxide production in Escherichia coli. J Biol Chem. 1991;266(11):6957–6965. [PubMed] [Google Scholar]
- 45.Mulvey MA, et al. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282(5393):1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 46.Justice SS, et al. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc Natl Acad Sci USA. 2004;101(5):1333–1338. doi: 10.1073/pnas.0308125100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snyder JA, et al. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect Immun. 2004;72(11):6373–6381. doi: 10.1128/IAI.72.11.6373-6381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reigstad CS, Hultgren SJ, Gordon JI. Functional genomic studies of uropathogenic Escherichia coli and host urothelial cells when intracellular bacterial communities are assembled. J Biol Chem. 2007;282(29):21259–21267. doi: 10.1074/jbc.M611502200. [DOI] [PubMed] [Google Scholar]
- 49.Grantcharova N, Peters V, Monteiro C, Zakikhany K, Römling U. Bistable expression of CsgD in biofilm development of Salmonella enterica serovar typhimurium. J Bacteriol. 2010;192(2):456–466. doi: 10.1128/JB.01826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Massé E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2002;99(7):4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.