Abstract
The epigenetic regulator Bmi1 controls proliferation in many organs. Reexpression of cell cycle proteins such as cyclin-dependent kinases (CDKs) is a hallmark of neuronal apoptosis in neurodegenerative diseases. Here we address the potential role of Bmi1 as a key regulator of cell cycle proteins during neuronal apoptosis. We show that several cell cycle proteins are expressed in different models of retinal degeneration and required in the Rd1 photoreceptor death process. Deleting E2f1, a downstream target of CDKs, provided temporary protection in Rd1 mice. Most importantly, genetic ablation of Bmi1 provided extensive photoreceptor survival and improvement of retinal function in Rd1 mice, mediated by a decrease in cell cycle markers and regulators independent of p16Ink4a and p19Arf. These data reveal that Bmi1 controls the cell cycle-related death process, highlighting this pathway as a promising therapeutic target for neuroprotection in retinal dystrophies.
Keywords: blindness, neurodegeneration, polycomb
Retinitis pigmentosa (RP) is a group of genetic disorders known as a major cause of inherited blindness that is currently untreatable and affects millions of patients worldwide. Around 200 different mutated loci or genes cause this disease (1) (also https://sph.uth.edu/RetNet/sum-dis.htm). Rd1 mice, a widely used model of RP, carry a mutation in the rod-specific Phosphodiesterase-6β (Pde6b) gene, which is also mutated in around 4–5% of human RP patients in the United States (1). Rd1 mice exhibit a rapid loss of rod photoreceptors, which are responsible for night vision, followed by a more gradual loss of cones, which are necessary for day vision and visual acuity. Several studies have attempted to rescue or delay retinal degeneration in Rd1 mice with neurotrophic factors (2), calcium blockers (3), or antiapoptotic gene transfer (4), but these neuroprotective approaches never exceeded modest or temporary effects. Interestingly, deleting cyclic nucleotide channel-b1 (CNGB1), which is constantly stimulated by the accumulation of cyclic GMP (cGMP) in Rd1 photoreceptors, leads to a marked rescue of the sensory cells (5). Because an elevated level of cGMP is believed to be an early event in the induction of cell death in this model, these results can be considered as a milestone reference to conduct the identification of other candidates involved in the photoreceptor death process.
Various models of neurodegenerative diseases have shown that neurons committed to death reexpress cell cycle-related proteins. This phenomenon has been observed in mouse models of Alzheimer’s disease (6, 7), Parkinson disease (8), and amyotrophic lateral sclerosis (9), where postmitotic neurons start to express nuclear cyclin-dependent kinase 4 (CDK4), implicated in the reentry into the cell cycle and in the transition from G1 to S phase. However, these cells fail to complete S phase and undergo apoptosis (reviewed in ref. 10). Using CDK inhibitors, interference with CDKs in vitro enhances the survival of motoneurons (11) and sympathetic neurons (12) after trophic factor withdrawal, but these effects remain modest. CDK inhibition before induction of stroke in vivo protects around 80% of neurons (13). However, such success has not been achieved in other types of neuronal degeneration.
At early stages of retinal degeneration in Rd1 mice, the synthetic thymidine analog Bromodeoxyuridine (BrdU) is incorporated in the outer nuclear layer (ONL) where photoreceptors reside, but this BrdU incorporation has been attributed to DNA repair (14). Nevertheless, DNA replication does occur in neurons of an Alzheimer’s disease model (7) as well as in dopaminergic neurons of a patient with Parkinson disease (8), showing that neurons synthesize de novo DNA in an S-like phase. In view of the interesting observations made in animal models and in patient samples of neurodegenerative disorders, we asked whether retinal degeneration is also linked to cell cycle proteins and machinery and tested the potential role of cell cycle regulators in photoreceptor cell death.
During retinogenesis, cell cycle arrest is controlled by a panel of factors, among which the Rb/E2F pathway plays a central role. For example, in the developing retina, retinoblastoma protein (Rb) is required to couple terminal differentiation to cell cycle exit, and thus its absence leads to E2F1-dependent ectopic division (15). In dividing cells, Rb is inactivated by CDK4- or CDK6-mediated phosphorylation, which inhibits its binding to E2F1 (reviewed in ref. 16). Upstream of this pathway, the polycomb group protein Bmi1 promotes CDK4 and CDK6 activity by repressing the Ink4a/Arf locus, encoding the CDK4/6 inhibitor p16Ink4a and p19Arf that activates p53 (17). Because the Rb/E2F1 pathway partially contributes to neuronal death in Parkinson disease (8), we investigated whether Bmi1 and E2F1 are required to promote photoreceptor death during retinal degeneration.
Results
Expression of Cell Cycle Proteins in Degenerating Rd1 Retinas.
We analyzed the expression of cell cycle markers in Rd1 mice at early stages of photoreceptor loss. At postnatal day 9 (P9), when the first apoptotic cells were detected, CDK4 was present in few cells of the ONL, and then the number of CDK4-positive photoreceptors markedly increased until P12 (Fig. 1 A–C and G), during the peak of cell death in this model. Quantification revealed that the amount of CDK4-expressing cells doubled between P10 and P11, as well as between P11 and P12 (Fig. 1G) in the Rd1 ONL, whereas almost no CDK4-positive cells were detected in the wild-type (WT) retina (Fig. 1B). Costaining with Rhodopsin antibodies and subsequent confocal analysis demonstrated that CDK4 was expressed in rod photoreceptors of degenerating Rd1 retinas (Fig. 1C and Fig. S1). Although not expressed in the WT ONL, CDK2 and CDK6 were also present in the mutant ONL, however, to a lesser extent than CDK4 (Fig.1 D–F). No variation of CDK4 and CDK6 expression was detected between the Rd1 and the WT retina by RT-PCR analysis of total mRNA extracts (Fig. S1), suggesting that the positive signal observed in the cell body and processes of other retinal cells masks the variations observed in the ONL. In addition, phosphorylation of Rb, which is the substrate of CDK4 and CDK6, on serine 807 and 811, was increased in the Rd1 ONL, showing a link with CDK expression (Fig. S1). The specificity of the antibody was checked by Western blot. We used ethynyl deoxyuridine (EdU) incorporation to test the possibility that dying photoreceptors enter S phase. Very few cells in the ONL integrated EdU (Fig. 1H), and their number was much lower than that of the CDK4- and TUNEL-positive cells.
Fig. 1.
Cell cycle proteins are expressed in the Rd1 retina. (A and B) The Rd1 retina contains CDK4-expressing cells in the ONL (brown nuclei highlighted by arrows) (A), whereas CDK4 is not expressed in WT ONL (B). (C) At P12, double labeling for CDK4 (red) and Rhodopsin (green) clearly shows that photoreceptor cells express CDK4 in Rd1 retina (confocal image and enlargement). Arrows point to some representative cells positive for CDK4. (D) CDK2 is expressed in sparse Rd1 photoreceptor nuclei (red, arrows) at P12, evidenced by localization of CDK2+ nuclei rhodopsin-positive cells (green). (E) Similarly, CDK6-positive nuclei are detected in the P11 Rd1 retina (arrows). (F) The quantification of CDK-positive nuclei at P11 shows that Rd1 photoreceptors express CDK4 to a greater extent (more than fivefold) than CDK2 and CDK6. The graph represents the number of CDK-expressing cells in the ONL of central retina ± SEM (n = 4 for CDK4, n = 3 for CDK2 and CDK6, **P < 0.01). (G) From their appearance at P9, the number of CDK4-positive nuclei increases with age (P10–P12). The graph represents the mean value of positive cell number in the ONL for each age ± SEM (n = 5 at P9, 6 at P11, and 2 at P12, *P < 0.05, **P < 0.01). (H) Rare EdU (red)-positive cells are present in Rd1 ONL nuclei after a 24-h pulse in Rd1 mice at P12. A representative labeling is marked by an arrow. (I) Most CDK4-expressing photoreceptor cells in the Rd1 ONL at P11 are dying (TUNEL, red) and CDK4 (green) double-positive cells (arrows). Nevertheless, some single-positive cells can also be detected (open arrows). (J) Quantification of TUNEL-positive cells in the Rd1 central retina shows a gradual increase from P9 to P12 (for TUNEL, n = 2 at P9, 6 at P11, and 3 at P12, *P < 0.05, ***P < 0.001) correlating with CDK4 expression (G). (K) Decreased cell death, as revealed by TUNEL (red), in roscovitine-treated (Right) vs. control (Left) Rd1 retinal explants at P6 + 6 DIV (n = 6 control and 9 treated Rd1 explants spread over three series of experiments). (L) Quantification of TUNEL+ cells in roscovitine- vs. control-treated Rd1 explants. Data are expressed as mean ± SEM (**P < 0.01). INL, inner nuclear layer; ONL, outer nuclear layer. (Scale bars: 50 μm.)
We performed a time-course analysis of CDK4 expression and TUNEL staining in the Rd1 retina to determine how cell cycle protein expression is related to DNA fragmentation. In the Rd1 ONL at P9, few cells were TUNEL single positive, and we detected only rare TUNEL/CDK4 double-positive cells and no CDK4 single-positive cells (Fig.1 G and J). In P11 Rd1 retinas, a large majority of cells in the death process were double positive for TUNEL and CDK4, around 25% were positive only for TUNEL, and we also detected some CDK4 single-positive cells. Colocalization analyses suggest that CDK4 expression is tightly related to late processes of photoreceptor cell death (Fig. 1I).
These results are consistent with a previous study reporting BrdU incorporation in degenerating photoreceptors in Rd1 mice at P15, where it was attributed to DNA repair because of Ligase IV expression in photoreceptors (14). We did not observe any cells positive for the M-phase marker phospho-H3 in the Rd1 ONL (Fig. S1), suggesting that CDKs and related proteins of the G1 phase may have a role during photoreceptor death, similar to that in other neurodegenerative diseases.
Reexpression of Cell Cycle Proteins Also Occurs in Other Genetic or Acute Models of Retinal Degeneration.
To assess whether cell cycle protein reactivation in photoreceptors during retinal degeneration was specific to Rd1 mice or common to other models, we analyzed the expression of CDKs in four additional genetic or acute models of retinal degeneration. The Rd10 mouse is another recessive animal model of PDE6β deficiency (missense mutation) exhibiting a delayed rod degeneration, starting around P18 and complete at P30 in our hands. At the time of photoreceptor loss, CDK4 was expressed in photoreceptor nuclei, and the number of CDK4-positve cells increased considerably from P19 to P23 (Fig. S2). We investigated whether mutations inducing a dominant form of RP are also associated with the reactivation of cell cycle proteins during photoreceptor loss. P23H and S334ter rats bear mutations in the Rhodopsin gene. In both cases, the ONL contained many CDK4-positive nuclei (Fig. S2). Moreover, degenerating retinas of Rd10 mice, as well as P23H, and S334ter rats showed higher levels of phosphorylated Rb than WT (Fig. S2). Finally, an acute model of retinal degeneration induced by light damage was investigated. We observed intense and widespread nuclear expression of CDK4 and CDK2 in the ONL 36 h after light damage (Fig. S3), when the cell death process reaches its peak (18). Similar to the P12 WT retina (Fig. 1), control adult retinas did not contain any CDK4- or CDK2-positive photoreceptor nuclei (Fig. S3). Thus, we conclude that reactivation of cell cycle proteins is associated with photoreceptor death in five distinct models of retinal degeneration.
Interference with CDK Function Reduces Apoptosis in Rd1 Retinal Explants.
We next aimed to interfere with cell cycle protein reactivation to reveal their roles during retinal degeneration. We focused on the Rd1 mouse, which is one of the most severe and therapeutically challenging models of retinal degeneration. We isolated P6 Rd1 retinas and cultured them as retinal explants ex vivo with or without addition of the general CDK inhibitor roscovitine. Retinal explants were exposed to roscovitine (50 μM) at P6 + 1 d in vitro (DIV) (corresponding to P7 in vivo), fixed, and analyzed at P6 + 6 DIV (corresponding to P12 in vivo). We performed a TUNEL staining on treated and control retinal explant sections to analyze the degree of neuroprotection provided by the CDK inhibitor. Roscovitine treatment caused a 42% reduction in the number of apoptotic photoreceptors (Fig. 1 K and L), thus implying that roscovitine treatment is neuroprotective and that CDK reactivation plays a key role in the photoreceptor death process.
Activating E2fs Contribute to Photoreceptor Cell Death in Rd1 Mice.
Consistent with the above data implicating CDK activation in photoreceptor death, phosphorylated Rb was detected in Rd1 but not in WT ONL (Fig. S1). E2F1 is released and activated upon Rb phosphorylation and is known to contribute to apoptosis in Parkinson disease (8) and stroke (13). Moreover, Rb null rod photoreceptors undergo E2F1-dependent apoptosis (15), and either directed expression of E2f1 in these cells (19) or inhibition of pRb through T-antigen expression also leads to cell death. Thus, we next asked whether E2Fs contribute to Rd1 photoreceptor degeneration. E2F1 is one of three activating E2Fs that interchangeably drive the cell cycle in fibroblasts (20) and promote division redundantly with N-Myc in retinal progenitors together (21). We therefore analyzed the effect of deleting combinations of all three factors, using germ-line E2f1 and E2f2 null alleles and a floxed E2f3 allele, which was conditionally deleted using a Cre transgene expressed in the peripheral retina only (15). In the Rd1;E2f1+/+;E2f2−/−;E2f3f/f;α-Cre P18 retina, the absence of E2F2 in the central retina or E2F2 and E2F3 in the periphery had no effect (Fig. 2 A–C). In Rd1;E2f1+/−;E2f2−/−;E2f3f/f;α-Cre mice, loss of one E2f1 allele and both E2f2 alleles did not prevent photoreceptor loss in the central retina (Fig. 2C). However, in Rd1;E2f1−/−;E2f2+/−;E2f3f/f;α-Cre mice, the absence of two E2f1 alleles and one E2f2 allele increased the number of rescued photoreceptor rows 4.7-fold in the central ONL (39% of WT, Fig. 2C). These data imply that E2F1 has a greater role in rod photoreceptor death than E2F2. To assess E2F3 function we studied the periphery, where degeneration is delayed relative to the central retina and where the E2f3 deletion occurs in this model (Fig. 2). In the Rd1;E2f1+/−;E2f2−/−;E2f3f/f;α-Cre retina periphery, which lacks one allele of E2f1 and both alleles of E2f2 and E2f3, ONL rows were increased 1.9-fold, corresponding to 35% of WT retina (Fig. 2). This effect in the E2f1+/−;E2f2−/−;E2f3−/− periphery was greater than in the central E2f1+/−;E2f2−/− retina, which could reflect a role for E2F3 in rod death or central vs. peripheral differences. Finally, the peripheral retina of Rd1;E2f1−/−;E2f2+/−;E2f3f/f;α-Cre showed improved rescue over that seen in the Rd1;E2f1+/−;E2f2−/−;E2f3f/f;α-Cre periphery (Fig. 2C), confirming that E2F1 is more important than E2F2. E2f removal slowed but did not halt photoreceptor death, because at P26, there was no difference in the central or peripheral retina of Rd1 mice and any of the above E2f-deficient genotypes. These data encouraged us to ask whether upstream factors involved in the cell cycle regulation cascade may have even greater protective potential.
Fig. 2.
E2f1 deletion provides a transient neuroprotection in Rd1 mice. A and B depict the rescue of photoreceptors in Rd1 E2f1 mutants, respectively, in the central and peripheral retina at P18. For the genotypes, refer to figures, main text, and Materials and Methods. For quantification, refer to C, which presents the photoreceptor row number in percentage of WT values ± SEM (n = 3 animals for each genotype, ***P < 0.001 compared with Rd1 and Rd1;E2f1+/+;E2f2−/−;E2f3f/f;α-Cre). Note the rescue obtained with Rd1;E2f1+/−;E2f2−/−;E2f3f/f;α-Cre and Rd1;E2f1−/−;E2f2+/−;E2f3f/f;α-Cre mice. INL, inner nuclear layer; ONL, outer nuclear layer. (Scale bars: 50 μm.)
Extended Protection of Rod Photoreceptors in Rd1;Bmi1−/− Mice.
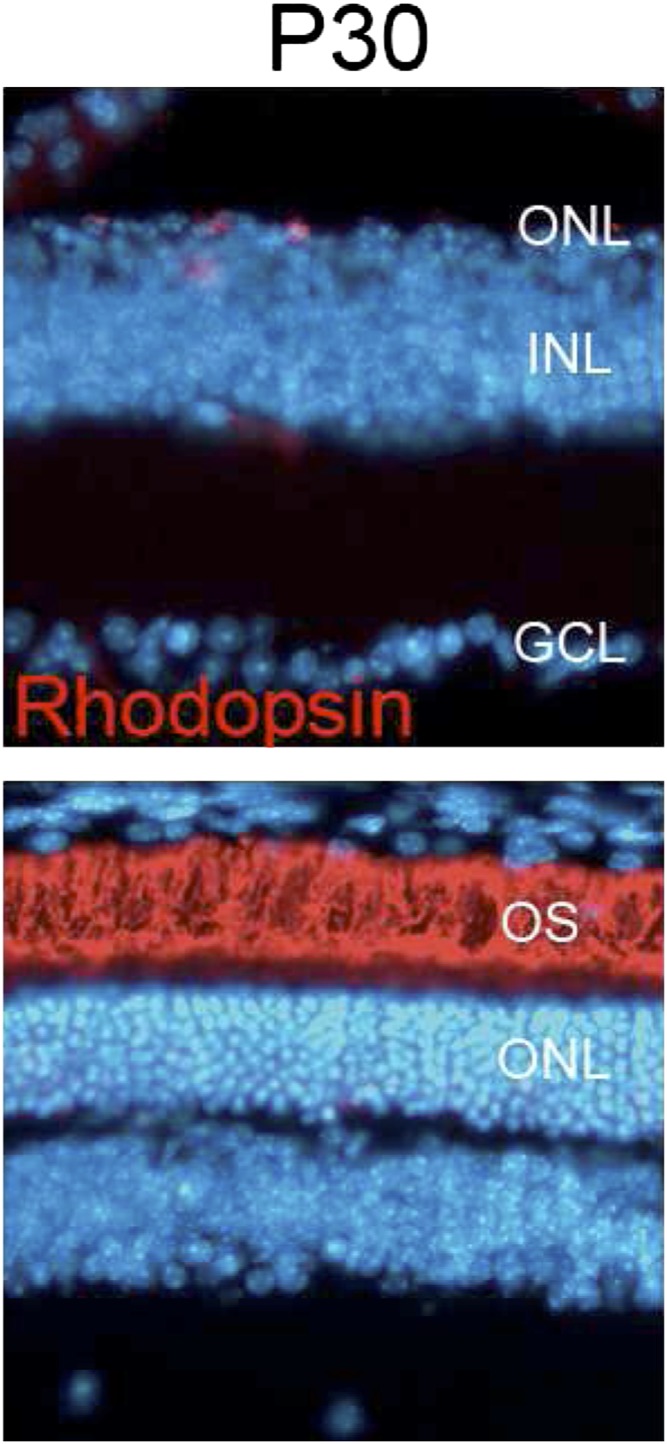
Bmi1 promotes cell cycle progression by repressing several tumor suppressor genes, including the CDK4/6 inhibitor p16Ink4a (17, 22). We hypothesized that deleting Bmi1 would reach a broader spectrum of targets than E2f1 deletion and may thus have a stronger neuroprotective effect. First, we characterized Bmi1 expression at the end of retinal development in WT mice and at early stages of retinal degeneration in Rd1 mice. Bmi1 was expressed in the nuclei of all cell types of the inner nuclear layer (INL) and ONL, including rods and cones in both WT and Rd1 retinas at P12 (Fig. 3A and Fig. S1). RT-PCR analysis did not reveal any substantial change in the level of Bmi1 expression between WT and Rd1 retinas, either at P11 or at P12 (Fig. 3A, Right). Nevertheless, the presence of Bmi1, which may have several targets (23–26), in the Rd1 ONL encouraged us to study its potential action. To test the hypothesis that Bmi1 deletion may limit retinal degeneration, we analyzed the histology of WT, Rd1, and Rd1;Bmi1−/− retinas at early (P12), middle (P15 and P18), and late (P30 and P34) stages of the disease (Fig. 3B). Removing only one Bmi1 allele in Rd1;Bmi1+/− retinas did not induce any change in the progression of retinal degeneration (Fig. 3C). Rhodopsin-stained rod outer segments were similar in Rd1 and Rd1;Bmi1−/− retinas at P12 (Fig. 3B). At P15, rhodopsin staining and the thickness of the ONL were already strongly reduced in Rd1 mice whereas Rd1;Bmi1−/− animals displayed an almost normal ONL (Fig. 3B). At P18, Rd1 and Rd1;Bmi1−/− retinas harbored 10% and 60% of photoreceptor rows, respectively, compared with WT controls (Fig. 3 B–D). At P30, Rd1;Bmi1−/− mice still displayed well-preserved rhodopsin-positive outer segments (Fig. 3B) and even at P34, they harbored 58.5% of the photoreceptor rows normally present in WT retinas, whereas only a single row of scattered photoreceptors (<10% of a WT retina and consisting mostly of cones) was present in Rd1 mice (Fig. 3 B–E). Analysis of older mice was not possible due to the ataxia and epilepsy characteristic of Bmi1 null animals. Because loss of rods is followed by slower degeneration of cones, we quantified GNAT2- and S-opsin–positive cone segments at P34. Whereas both markers were already strongly diminished in Rd1 mice, GNAT2- and S-opsin–positive cells were reduced only by 25% in Rd1;Bmi1−/− littermates compared with those in age-matched WT retinas (Fig. 3 D and E).
Fig. 3.
Bmi1 deletion delays photoreceptor loss in Rd1 mice. (A) Bmi1 expression (brown) at P12 in WT, Rd1, and Rd1;Bmi1−/− (negative control) retinas. (Right) Confirmation of this expression by RT-PCR of total mRNA extracts of P11 and P12 retinas. (B) The thickness of the ONL and rhodopsin staining (green) decrease rapidly in Rd1 mice. P30 Rd1 retinas harbor only a single scattered row of photoreceptors. By contrast, numerous rhodopsin-expressing photoreceptors are present between P12 and P34 in the Rd1;Bmi1−/− retinas. Note the large band of photoreceptor outer segment (OS). (C) Quantification of the number of rows of photoreceptor nuclei in Rd1 and Rd1;Bmi1−/− mice at P18 and P34 vs. those in age-matched WT retinas. (D) Quantification of cones expressing S-opsin and GNAT2 in P34 Rd1 retina compared with those in age-matched WT controls. (C and D) All data are expressed as percentages compared with those in age-matched WT retinas ± SEM (n = 3 animals for each genotype). ***P < 0.001 between Rd1 and Rd1;Bmi1−/−. (E) S-opsin and GNAT2 expression in cone outer segments of Rd1, Rd1;Bmi1−/−, and WT retinas at P34. INL, inner nuclear layer; ONL, outer nuclear layer; OS, outer segments. (Scale bars: 50 μm.)
These data suggest that removing Bmi1 may have a beneficial effect on the loss of vision. Thus, we next investigated retinal function, by electroretinogram recordings under scotopic conditions (low-intensity light stimuli after dark adaptation, followed by light stimuli at increasing intensities) to determine the minimal intensity of light that is required to elicit a photoreceptor response. Rd1;Bmi1−/− mice ranging from age P27 to P30 responded in a dose-dependent manner to increasing light stimuli (Fig. 4A shows explanation and Fig. 4 B–D and Fig. S4 show results). The response threshold in Rd1;Bmi1−/− was situated between a light stimulus of 0.03 and 0.1 cd⋅s/m2, which corresponds to the intensity at which cones start to be activated (27). This pattern of response is consistent with the fact that rods are inactive due to the absence of the PDE6β protein essential for the phototransduction, but that their rescue promotes cone survival. Moreover, this response was remarkable in comparison with the almost complete absence of any response in the Rd1 mouse and is particularly significant compared with previous reports showing only limited rescue of photoreceptors using calcium blockers (3), neurotrophic factors (2), or cell transplantation (28) and gene replacement (29). Insulin treatment of the Rd1 mouse also increased cone survival, but to a lesser extent (30). Their function has not been investigated in this study. Although we cannot exclude the possibility that Bmi1 deletion directly promotes cone survival in Rd1;Bmi−/− mice, this protection is probably due to an indirect effect of rod survival (31). Together, these data show that deletion of Bmi1 results in a robust neuroprotection to an extent never reached by other neuroprotective approaches (2–4, 31) or by gene therapy (29, 32).
Fig. 4.
Rescue of retinal function in Rd1;Bmi1−/− mice. (A) Schematic representation of a WT ERG response. The a-wave is due to photoreceptor activity, whereas the b-wave is caused by subsequent activation of retinal interneurons. (Right) Scale bars for amplitude (μV) and implicit time (ms) are shown. (B) Representative ERG recordings of Rd1, Rd1;Bmi1−/−, and WT mice at P27–P30 in scotopic conditions at high light intensities. Refer to Fig. S4 for complete ERG curves. Quantification is shown for the b-wave amplitude (C) and the implicit time (delay of the response) of the b-wave (D). *P < 0.05, **P < 0.01 between Rd1 and Rd1;Bmi1−/−.
Bmi1 Regulates Cell Cycle Proteins in the Degenerating Retina.
One of the best known roles of Bmi1 is to support cell cycle progression by promoting CDK activity during the G1 phase and at the G1/S checkpoint. Therefore, we analyzed cell cycle markers in Rd1;Bmi1−/− mice at P12 and compared them to those in the WT and Rd1 littermates mentioned above. Deletion of Bmi1 decreased the amount of CDK4-positive photoreceptors by 46% compared with that in Rd1 mice (Fig. 5 A and B). Fewer CDK2-positive photoreceptor nuclei were found in Rd1;Bmi1−/− retinas in comparison with those of Rd1. Very few CDK2-positive cells were observed in Rd1 and Rd1;Bmi1−/− retinas (2 ± 0.9 and 1.1 ± 1 CDK2-positive cells per 100 μm for P12 Rd1 and Rd1;Bmi1−/− retinas, respectively). Consistent with a Bmi1-dependent regulation of cell cycle proteins in the Rd1 ONL, phosphorylation of Rb on serine 807 and 811 was not detected in Rd1;Bmi1−/− photoreceptors at P12.
Fig. 5.
Bmi1 loss acts on the Rd1 photoreceptor rescue independently on the Ink4a/Arf locus and by decreasing Cdk4 expression. (A) CDK4 (brown spots and arrows) stainings in the P12 Rd1 and Rd1;Bmi1−/− retina. (B) Quantification of CDK4-positive photoreceptors in Rd1 and Rd1;Bmi1−/− retinas, values ± SEM (n = 3 animals for each genotype). *P < 0.05. (C and D) Amount of photoreceptors present at P18 and P34, respectively, in different genetic models. (C) Whereas the Rd1 retina shows one to two rows of photoreceptors positive for rhodopsin (green), the deletion of Bmi1 rescues these cells markedly. The ablation of Inka4/Arf does not reverse the rescue induced by Bmi1 deletion (Rd1;TKO, Rd1;triple knockout) nor does it affect the process of retinal degeneration (Rd1;Ink4a/Arf−/−). Bmi1 has no dose effect to prevent retinal degeneration (Rd1;Bmi1+/−). (D) At P34 the rescue is still robust both in Rd1;Bmi1−/− and in Rd1;TKO, which shows similar rescue. The presence of one Bmi1 allele in Rd1;Ink4a/Arf−/− is sufficient to sustain retinal degeneration. For quantification, refer to E, which presents the photoreceptor row number in percentage of the WT values ± SEM (n = 3 animals for each genotype). ***P < 0.001 compared with Rd1, Rd1;Ink4a/Arf−/−, and Rd1;Bmi1+/−. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; OS, outer segments. (Scale bar: 50 μm.)
During the course of retinal degeneration, DNA repair is part of the cascade of events that trigger photoreceptor death (14). In other neurodegenerative diseases, DNA repair is considered to precede the reactivation of CDKs in dying neurons (33). Thus, we analyzed retinas for expression of the DNA repair-specific Ligase IV at P12 and observed similar numbers of sporadic positive photoreceptors in Rd1 and Rd1;Bmi1−/− retinas (Fig. S5). Taken together, these data show that Bmi1 deletion efficiently reduces retinal degeneration in Rd1 mice, that its effect likely occurs downstream of DNA repair events, and that it prevents the induction of CDKs and phosphorylation of Rb.
Ink4a/Arf Locus Is Not Required to Delay Retinal Degeneration.
The Ink4a/Arf locus, encoding the tumor suppressors p16Ink4a and p19Arf, is one of the best known targets of Bmi1 in the regulation of cell cycle and apoptosis. Briefly, p16Ink4a inhibits CDK4/6 activity and its induction in the absence of Bmi1 (17) could, therefore, mediate protection of Rd1 photoreceptors. p19Arf stabilizes p53, and although p53 is not required for retinal degeneration in Rd1 mice (34), p19Arf also has p53-independent effects (35). To determine whether the rescue of retinal degeneration in Rd1;Bmi1−/− mice was dependent on p16Ink4a and/or p19Arf, we analyzed Rd1;Bmi1−/−;Ink4a/Arf−/− triple null mice at ages P18 (Fig. 5C) and P34 (Fig. 5D) and quantified the number of photoreceptor rows (Fig. 5E) in the peripheral and in the central retina. Surprisingly, we did not observe any difference between Rd1;Bmi1−/− and Rd1;Bmi1−/−;Ink4a/Arf−/− mice (Fig. 5), suggesting that Bmi1 loss delays retinal degeneration and down-regulates CDK expression in Rd1 mice in a process that is Ink4a/Arf independent.
Discussion
Previous work has implicated the reactivation of cell cycle components as a key feature of neurodegenerative diseases. The relevance of this mechanism in photoreceptor degeneration was unclear and the role of Bmi1 had never been investigated in any neurodegenerative disease. Here, we show that nuclear expression of CDK4, CDK6, and CDK2 and phosphorylation of Rb precede photoreceptor death in models of retinal degeneration. Roscovitine treatment of Rd1 retinal explants decreased photoreceptor death, supporting the relevance of a relationship between CDK activation and cell death. Deleting E2fs (mainly E2f1) on the Rd1 background resulted in a transient neuroprotection, whereas deletion of Bmi1 (upstream of E2F1 and CDKs) led to an extensive delay in photoreceptor loss. Mortality at 5–6 wk precludes the determination of the full protective extent of Bmi1 deletion. The future availability of conditional knockouts or knockdown approaches may reveal the full extent of photoreceptor survival conferred by Bmi1 depletion and whether Müller cells play a certain function (protection, cell replacement). In other neurodegenerative diseases, severe DNA damage is a major signal to induce cell death and is considered to be upstream of CDK expression (36). The presence of Ligase IV in Rd1 mouse retina (this paper and ref. 14) at early stages of the degeneration process also suggests a similar phenomenon for the retina. The presence of Ligase IV in Rd1;Bmi1−/− retina suggests that the neuroprotective effect of Bmi1 deletion occurs downstream of DNA repair, via CDK inactivation. Nonetheless, we cannot exclude a modest effect of Bmi1 deletion on DNA repair, because Bmi1 has been reported to be involved in DNA repair initiation (37, 38). In contrast, there is recent evidence that Bmi1 loss increases oxidative stress in neurons (39) as well as in other cell types (40). In consequence, our results show that Bmi1 deletion can have a dual effect on cell survival, making investigations on the direct and indirect Bmi1 targets a priority for future studies, to generate appropriate tools for therapy. Other known epigenetic regulators were recently shown to control photoreceptor survival in Rd1 retina explants, such as histone deacetylase 4 (HDAC4) (41), as well as photoreceptor death, such as unidentified members of the HDAC classes I and II, as revealed by the nonspecific inhibitor trichostatin (42). However, no direct evidence on epigenetic modification has been gathered so far. Moreover, HDAC4 survival action occurs in the cytoplasm through hyopxia-inducible factor 1 (HIF1) stabilization (41), revealing the different action modes of HDACs. Binding of the Bmi1-containing polycomb-repressing complex to DNA requires histone deacetylation and Histone-3 methylation. Analyzing these epigenetic regulators in the cytoplasm and nucleus, as well as modifications in Rd1 and Rd1;Bmi1−/− retinas, will help to identify important targets that mediate photoreceptor death.
We have now implicated several cell cycle regulators in photoreceptor death. The broader effect of Bmi1 deletion compared with Rd1;E2f1−/− may be explained by its position upstream of E2F1 and its regulatory action on a large panel of tumor suppressors and CDKs. The wide spectrum of targets for Bmi1 and polycomb complexes (23–26) may also clarify why the photoreceptor rescue observed in Rd1;Bmi1−/− mice does not depend exclusively on the Ink4a/Arf locus. The use of the Cre system to specifically delete E2Fs in photoreceptors, the observation of photoreceptor survival after the inhibition of CDK expression or activity, either by Bmi1 loss or roscovitine treatment, and the variation of Rb phosphorylation paralleling CDK expression imply the cell-intrinsic importance of cell cycle protein reactivation in the control of photoreceptor death. Although we cannot exclude that the survival effect mediated by Bmi1 deletion may also be partially attributed to other retinal cell types, such as Müller glia, our results highlight the role of Bmi1 in the regulation of cell cycle proteins during the cell death process in an animal model of retinal degeneration. In the retina, Müller cells support photoreceptors by regulating their metabolism and through neurotrophic factor delivery (43). However, such neurotrophic factors had only mild neuroprotective effects that are not comparable to the Bmi1 loss-mediated delay of retinal degeneration. In consequence in Rd1;Bmi1−/− retina, a potential role in neurotrophic support by glial cells is probably minor.
A recent report revealed that cGMP is an important mediator during the early phases of photoreceptor death (5). Indeed, deletion of the Cngb1 gene coding for a channel regulated by cGMP strongly delayed photoreceptor death in Rd1 mice. Around 50% of the photoreceptor cells survived at P30 and 39% at P60. The relationship between Bmi1 and cGMP remains to be identified and may reveal important mechanisms controlling sensory neuron death.
Our results with the Rd1 model of RP raise the possibility that the Bmi1-dependent reactivation of cell cycle components may be relevant to a much wider range of retinopathies. Indeed, we also detected CDK4 and CDK2 reactivation coincident with light damage-induced retinal degeneration and other recessive and dominant rodent models of retinal dystrophies. The relevance of Bmi1 in neurodegeneration is unique and should be investigated in other models of retinal degeneration, such as light-induced damage, as well as in other CNS conditions where a cell cycle-related mechanism has already been implicated in apoptosis (10). Indeed, a small neuroprotective effect (E2f1 deletion, CDK inhibition) measured in the rapidly degenerating Rd1 model would likely translate into considerable therapeutic benefit in humans where photoreceptor loss occurs over years rather than days. These results, coupled with the CDK analyses above, provide two promising targets for therapeutic intervention in patients with RP and show clearly that reactivated cell cycle components contribute to the demise of photoreceptors. Interestingly, in the adult cerebral cortex and cerebellum, Bmi1 is expressed at low levels by virtually all neurons (44), supporting a potential broad role for this Polycomb repressor in controlling neuronal cell death. The Bmi1 pathway thus merits being studied in different forms of retinal degeneration and neurodegenerative diseases.
Materials and Methods
Animals and Genotypes.
Rd1, Rd1;Bmi1−/−, and Rd1;Bmi1−/−;Ink4a/Arf−/− (Rd1;TKO) mice on a FVB background were maintained and genotyped by standard PCR as previously described (17, 22, 45). For production of Rd1;Bmi1−/−, Rd1;Bmi+/− breeders were used because homozygotes are viable until around P30 only. Rd1;TKO mice were generated by crossing Bmi1+/− and Ink4a/Arf−/− mice (46), both having an FVB (Rd1) genetic background. To assess the role of E2F factors in Rd1 mice, E2f1 and E2f2 mutants on the FVB background, as well as FVB mice carrying a E2f3 floxed allele (15), were interbred with a-Cre mice (P. Gruss, Göttingen, Germany) to obtain the following genotypes: Rd1;E2f2−/−;E2f3−/−, Rd1;E2f1+/−;E2f2−/−;E2f3−/−, and Rd1;E2f1−/−;E2f2+/−;E2f3−/−. Note that the α-promoter was active only in the peripheral retina, keeping therefore the central retina wild type for E2f3. FVB.129P2;Pde6b+ mice obtained from Charles River Laboratories (stock no. 004828) were used as WT sighted controls. Mice were treated according to institutional and national as well as the Association for Research in Vision and Ophthalmology (ARVO) guidelines, and all experimental procedures were approved by national veterinary authorities.
Histological Analysis and Quantification.
Histological analysis of WT, Rd1, and Rd1;Bmi1−/− mutants was performed at P9, P11, P12, P15, P18, P30, and P34. Rd1;E2f mutants were analyzed at P18 and P26. Upon enucleation, eye globes were fixed for 1 h in 4% (wt/vol) paraformaldehyde (PFA) and then transferred either to 25% (wt/vol) sucrose to obtain 14-μm-thick cryosections, or to PBS before being mounted in paraffin and cut into 5-μm-thick sections. Before being processed at the cryostat, the eyes were oriented to allow the discrimination between dorsal and ventral retina. The sections were collected on six serial slides for each eye, allowing multiple labeling throughout the entire eye for each slide. All measurements were performed on the two most central cryosections including (or surrounding) the optic nerve. Measurements of rows of nuclei were taken at 150 and 250 μm from the periphery of the retina at both dorsal and ventral extremities and data were pooled as a mean for the peripheral retina. The measurements for the central retina are the mean of the data obtained at 300 and 400 μm dorsally and ventrally from the optic nerve head. The measures were taken using an Olympus BX60 fluorescence microscope and the AnalySIS 3.0 software (Soft Imaging) for Bmi1 and Ink4a/Arf mutants, whereas E2f mutant retinas were analyzed with a Zeiss Axioplan-2 microscope and the Axiovision 4.0 software.
Immunohistochemistry and Antibodies.
Immunohistochemical analysis was performed on either cryostat sections or paraffin sections of the whole eyecup. All primary antibodies, their source, and their working dilutions for frozen and/or paraffin sections are listed in Table S1. The standard protocol used for immunochemistry on cryostat sections included 1 h of blocking with 10% (vol/vol) normal goat serum and 0.3% (wt/vol) TritonX; followed by overnight incubation with the primary antibody diluted in blocking solution; and finally revelation with the appropriate Alexa 633, Alexa 488, FITC, or Cy3-coupled secondary antibody (details in Table S2). All exceptions to the standard protocol are noted in Table S1. In certain cases, as specified in Table S1, the TSA Amplification System (PerkinElmer) was used to enhance the fluorescent signal in accordance to manufacturer’s protocol. For stainings on paraffin sections, after the hydration steps, the sections were microwave boiled in a retrieving citrate buffer for 1 min at 900 W and 15 min at 250 W. Endogenous peroxidases were quenched with a 30-min treatment with 3% (vol/vol) H2O2. Primary antibodies were added overnight in 1% (wt/vol) BSA in PBS, and stainings were visualized using the appropriate biotinylated antibody (Table S2) with the Vectastain ABC Kit (Vector) according to the manufacturer’s protocol. On both frozen and cryostat sections, the nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). All measurements were taken using an Olympus BX60 fluorescence microscope and the AnalySIS 3.0 software (Soft Imaging). Quantifications of positive cells were performed on pictures of central retina (450-µm length) and normalized to 100 µm. When specified in the figure legends, confocal analysis was performed using a Zeiss öS; 510 Meta confocal microscope and the Zeiss LSM 510 3.2 software.
EdU Injection and Detection.
To reveal DNA synthesis in Rd1 mice, anti-EdU staining was performed on retinal frozen sections from P12 Rd1 mice that had received i.p. injections of EdU (40 µg/g body weight; Click-iT Edu Imaging Kit, Invitrogen) 24 h earlier.
Retinal Explant Cultures.
Retinal explants were cultured in R-16 medium (47, 48) (Gibco). The complete R-16 medium is prepared by adding 19 supplements (all purchased from Sigma), composed of BSA, hormones, and vitamins, to the stock solution (details in ref. 49). In our experimental paradigm, retinal explants generated from P6 Rd1 mice were cultured in complete R-16 medium for 6 d. One day later (P6 + 1 DIV) the medium was changed, and the explants were cultured either in R-16 medium containing 50 μM Roscovitine (Sigma) resuspended in 50:50 PBS/DMSO or in R-16-medium with 0.125% DMSO. Subsequently, the medium (with or without Roscovitine) was replenished every second day. The explants were fixed at P6 + 6 DIV with 4% PFA for 2 h at RT, before being transferred to 10% and 25% sucrose (in PBS). Each explant was processed into 10 series of 12-μm cryosections. TUNEL-positive cells were counted on the most central sections and normalized to the length of the explant.
Electroretinograms.
The procedure for electroretinogram (ERG) recordings is fully described in a previous study (50). Briefly, the mice were dark adapted overnight and anesthetized by an i.p. injection of a mixture of ketamine (100 mg/kg) and xylazine (15 mg/kg), and pupils were dilated. ERG recordings were obtained using a Ganzfeld stimulator from a Multiliner Vision apparatus (Jaeger/Tonnies) under dark-adapted (scotopic) conditions. The ERG was recorded in response to single flashes of white light of the following increasing intensities: 1 × 10−4, 1 × 10−3, 1 × 10−2, 3 × 10−2, 1 × 10−1, 3 × 10−1, 1, 3, 10, and 25 cd⋅s/m2 to determine at which intensity of light the retina is active. For each intensity, 10–30 responses were averaged. Band-pass filter cutoff frequencies were 0.3 and 300 Hz. The a-wave amplitude corresponds to photoreceptor activity and is the negative shape of the curve (Fig. 4). The b-wave amplitude was defined as the difference between b-wave and a-wave peaks (or the baseline level when the a-wave was not detectable) and corresponds to the interneuron signals stimulated by the photoreceptors. Amplitudes are expressed in microvolts and latencies in milliseconds.
RNA Extraction and RT-PCR.
Four P12 WT or Rd1 retinas were tritured with ophthalmic scissors, pooled, homogenized in 1 mL of Tri Reagent (Sigma), processed according to the manufacturer’s datasheet, and subsequently resuspended in 20 μL of RNase-free water. Samples were treated with a DNase Kit (Ambion; 1906) as described on the datasheet. Reverse transcription was performed as previously described. PCRs for Bmi1 and Gapdh were performed using the following primers at a final concentration of 0.2 μM each: Bmi1-F, 5′CAGCAATGACTGTGATGC3′; Bmi1-R, 5′CTCCAGCATTCGTCAGTC3′; Gapdh-F, 5′ACCACAGTCCATGCCATCAC3′; and Gapdh-R, 5′TCCACCACCCTGTTGCTGTA3′, using a standard Taq Polymerase (Invitrogen) and 2 mM MgCl2. Non–reverse-transcribed samples were used to check for genomic DNA contamination. Amplifications were carried out under the following conditions: denaturation for 5 min at 94 °C; followed by 34 cycles of denaturation at 94 °C for 30 s, annealing for 60 s at 52 °C (for Bmi1) or 62 °C (for Gapdh), and extension at 72 °C for 60 s; with a final extension at 72 °C for 10 min. PCR products were visualized using 1.5% agarose gels with SYBRsafe (Invitrogen; diluted 1/10,000).
Light Damage.
Eight-week-old BALB-C mice (Charles River) were kept for 3 wk in cyclic light at or below 80 lux (lx). After being dark adapted overnight, mice were exposed to 5,000 lx (measured on the cage floor) of white fluorescent light in reflective cages for 1 h. In these series of experiments, the eyes were enucleated for histological analysis after 36 h of recovery time in the dark.
Statistical Analysis.
One-way ANOVA followed by a Bonferroni test was applied to data, which were considered statistically different with a P < 0.05. When only two groups were compared, an impaired two-tailed t test was applied to data, and a P < 0.05 implied statistical significance. A minimum of three animals were analyzed for each histological quantification, and a minimum of six eyes were considered for ERG analysis.
Supplementary Material
Acknowledgments
We thank François Paquet-Durand for providing retina slices of P23H and S334ter rats; Dana Wanner and Meriem Tekaya for their excellent technical support; and Sophia Bruggeman, Corinne Kostic, and Francis Munier for rich discussions. This work was supported by Swiss National Foundation Grant 31000A0-122321 and by the AAVEYE (FP7-223445) consortium.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.-H.S. is a guest editor invited by the Editorial Board.
See Author Summary on page 2448 (volume 110, number 7).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1108297110/-/DCSupplemental.
References
- 1.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368(9549):1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 2.LaVail MM, et al. Protection of mouse photoreceptors by survival factors in retinal degenerations. Invest Ophthalmol Vis Sci. 1998;39(3):592–602. [PubMed] [Google Scholar]
- 3.Frasson M, et al. Retinitis pigmentosa: Rod photoreceptor rescue by a calcium-channel blocker in the rd mouse. Nat Med. 1999;5(10):1183–1187. doi: 10.1038/13508. [DOI] [PubMed] [Google Scholar]
- 4.Bennett J, et al. Adenovirus-mediated delivery of rhodopsin-promoted bcl-2 results in a delay in photoreceptor cell death in the rd/rd mouse. Gene Ther. 1998;5(9):1156–1164. doi: 10.1038/sj.gt.3300733. [DOI] [PubMed] [Google Scholar]
- 5.Paquet-Durand F, et al. A key role for cyclic nucleotide gated (CNG) channels in cGMP-related retinitis pigmentosa. Hum Mol Genet. 2011;20(5):941–947. doi: 10.1093/hmg/ddq539. [DOI] [PubMed] [Google Scholar]
- 6.Vincent I, Rosado M, Davies P. Mitotic mechanisms in Alzheimer’s disease? J Cell Biol. 1996;132(3):413–425. doi: 10.1083/jcb.132.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Geldmacher DS, Herrup K. DNA replication precedes neuronal cell death in Alzheimer’s disease. J Neurosci. 2001;21(8):2661–2668. doi: 10.1523/JNEUROSCI.21-08-02661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Höglinger GU, et al. The pRb/E2F cell-cycle pathway mediates cell death in Parkinson’s disease. Proc Natl Acad Sci USA. 2007;104(9):3585–3590. doi: 10.1073/pnas.0611671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen MD, et al. Cell cycle regulators in the neuronal death pathway of amyotrophic lateral sclerosis caused by mutant superoxide dismutase 1. J Neurosci. 2003;23(6):2131–2140. doi: 10.1523/JNEUROSCI.23-06-02131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: Oxymoron or new biology? Nat Rev Neurosci. 2007;8(5):368–378. doi: 10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
- 11.Appert-Collin A, et al. Cyclin dependent kinase inhibitors prevent apoptosis of postmitotic mouse motoneurons. Life Sci. 2006;79(5):484–490. doi: 10.1016/j.lfs.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 12.Park DS, Levine B, Ferrari G, Greene LA. Cyclin dependent kinase inhibitors and dominant negative cyclin dependent kinase 4 and 6 promote survival of NGF-deprived sympathetic neurons. J Neurosci. 1997;17(23):8975–8983. doi: 10.1523/JNEUROSCI.17-23-08975.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osuga H, et al. Cyclin-dependent kinases as a therapeutic target for stroke. Proc Natl Acad Sci USA. 2000;97(18):10254–10259. doi: 10.1073/pnas.170144197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menu dit Huart L, Lorentz O, Goureau O, Leveillard T, Sahel JA. DNA repair in the degenerating mouse retina. Mol Cell Neurosci. 2004;26(3):441–449. doi: 10.1016/j.mcn.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Chen D, et al. Rb-mediated neuronal differentiation through cell-cycle-independent regulation of E2f3a. PLoS Biol. 2007;5(7):e179. doi: 10.1371/journal.pbio.0050179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharpless NE, DePinho RA. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9(1):22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397(6715):164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 18.Wenzel A, Grimm C, Samardzija M, Remé CE. Molecular mechanisms of light-induced photoreceptor apoptosis and neuroprotection for retinal degeneration. Prog Retin Eye Res. 2005;24(2):275–306. doi: 10.1016/j.preteyeres.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Lin SC, Skapek SX, Papermaster DS, Hankin M, Lee EY. The proliferative and apoptotic activities of E2F1 in the mouse retina. Oncogene. 2001;20(48):7073–7084. doi: 10.1038/sj.onc.1204932. [DOI] [PubMed] [Google Scholar]
- 20.Wu L, et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414(6862):457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- 21.Chen D, et al. Division and apoptosis of E2f-deficient retinal progenitors. Nature. 2009;462(7275):925–929. doi: 10.1038/nature08544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruggeman SW, et al. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005;19(12):1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 24.Kallin EM, et al. Genome-wide uH2A localization analysis highlights Bmi1-dependent deposition of the mark at repressed genes. PLoS Genet. 2009;5(6):e1000506. doi: 10.1371/journal.pgen.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng S, et al. Identification and characterization of Bmi-1-responding element within the human p16 promoter. J Biol Chem. 2010;285(43):33219–33229. doi: 10.1074/jbc.M110.133686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tolhuis B, et al. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet. 2006;38(6):694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- 27.Seeliger MW, et al. New views on RPE65 deficiency: The rod system is the source of vision in a mouse model of Leber congenital amaurosis. Nat Genet. 2001;29(1):70–74. doi: 10.1038/ng712. [DOI] [PubMed] [Google Scholar]
- 28.Otani A, et al. Bone marrow-derived stem cells target retinal astrocytes and can promote or inhibit retinal angiogenesis. Nat Med. 2002;8(9):1004–1010. doi: 10.1038/nm744. [DOI] [PubMed] [Google Scholar]
- 29.Bennett J, et al. Photoreceptor cell rescue in retinal degeneration (rd) mice by in vivo gene therapy. Nat Med. 1996;2(6):649–654. doi: 10.1038/nm0696-649. [DOI] [PubMed] [Google Scholar]
- 30.Punzo C, Kornacker K, Cepko CL. Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nat Neurosci. 2009;12(1):44–52. doi: 10.1038/nn.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Léveillard T, et al. Identification and characterization of rod-derived cone viability factor. Nat Genet. 2004;36(7):755–759. doi: 10.1038/ng1386. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi M, Miyoshi H, Verma IM, Gage FH. Rescue from photoreceptor degeneration in the rd mouse by human immunodeficiency virus vector-mediated gene transfer. J Virol. 1999;73(9):7812–7816. doi: 10.1128/jvi.73.9.7812-7816.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park DS, et al. Involvement of retinoblastoma family members and E2F/DP complexes in the death of neurons evoked by DNA damage. J Neurosci. 2000;20(9):3104–3114. doi: 10.1523/JNEUROSCI.20-09-03104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Trogadis J, Bremner R. Rod and cone degeneration in the rd mouse is p53 independent. Mol Vis. 2001;7:101–106. [PubMed] [Google Scholar]
- 35.Sherr CJ. Divorcing ARF and p53: An unsettled case. Nat Rev Cancer. 2006;6(9):663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 36.Morris EJ, et al. Cyclin-dependent kinases and P53 pathways are activated independently and mediate Bax activation in neurons after DNA damage. J Neurosci. 2001;21(14):5017–5026. doi: 10.1523/JNEUROSCI.21-14-05017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Facchino S, Abdouh M, Chatoo W, Bernier G. BMI1 confers radioresistance to normal and cancerous neural stem cells through recruitment of the DNA damage response machinery. J Neurosci. 2010;30(30):10096–10111. doi: 10.1523/JNEUROSCI.1634-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ismail IH, Andrin C, McDonald D, Hendzel MJ. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J Cell Biol. 2010;191(1):45–60. doi: 10.1083/jcb.201003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chatoo W, et al. The polycomb group gene Bmi1 regulates antioxidant defenses in neurons by repressing p53 pro-oxidant activity. J Neurosci. 2009;29(2):529–542. doi: 10.1523/JNEUROSCI.5303-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, et al. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459(7245):387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen B, Cepko CL. HDAC4 regulates neuronal survival in normal and diseased retinas. Science. 2009;323(5911):256–259. doi: 10.1126/science.1166226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sancho-Pelluz J, et al. Excessive HDAC activation is critical for neurodegeneration in the rd1 mouse. Cell Death Dis. 2010;1:e24. doi: 10.1038/cddis.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bringmann A, et al. Cellular signaling and factors involved in Müller cell gliosis: Neuroprotective and detrimental effects. Prog Retin Eye Res. 2009;28(6):423–451. doi: 10.1016/j.preteyeres.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Bruggeman SW, et al. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell. 2007;12(4):328–341. doi: 10.1016/j.ccr.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 45.van der Lugt NM, et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994;8(7):757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 46.Serrano M, et al. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85(1):27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 47.Romijn HJ. Development and advantages of serum-free, chemically defined nutrient media for culturing of nerve tissue. Biol Cell. 1988;63(3):263–268. doi: 10.1016/0248-4900(88)90116-5. [DOI] [PubMed] [Google Scholar]
- 48.Romijn HJ, de Jong BM, Ruijter JM. A procedure for culturing rat neocortex explants in a serum-free nutrient medium. J Neurosci Methods. 1988;23(1):75–83. doi: 10.1016/0165-0270(88)90025-8. [DOI] [PubMed] [Google Scholar]
- 49.Caffé AR, et al. Mouse retina explants after long-term culture in serum free medium. J Chem Neuroanat. 2001;22(4):263–273. doi: 10.1016/s0891-0618(01)00140-5. [DOI] [PubMed] [Google Scholar]
- 50.Bemelmans AP, et al. Lentiviral gene transfer of RPE65 rescues survival and function of cones in a mouse model of Leber congenital amaurosis. PLoS Med. 2006;3(10):e347. doi: 10.1371/journal.pmed.0030347. [DOI] [PMC free article] [PubMed] [Google Scholar]