Abstract
The de novo biosynthesis of purines is carried out by a highly conserved metabolic pathway that includes several validated targets for anticancer, immunosuppressant, and anti-inflammatory chemotherapeutics. The six enzymes in humans that catalyze the 10 chemical steps from phosphoribosylpyrophosphate to inosine monophosphate were recently shown to associate into a dynamic multiprotein complex called the purinosome. Here, we demonstrate that heat shock protein 90 (Hsp90), heat shock protein 70 (Hsp70), and several cochaperones functionally colocalize with this protein complex. Knockdown of expression levels of the identified cochaperones leads to disruption of purinosomes. In addition, small molecule inhibitors of Hsp90 and Hsp70 reversibly disrupt purinosomes and are shown to have a synergistic effect with methotrexate, an anticancer agent that targets purine biosynthesis. These data implicate the Hsp90/Hsp70 chaperone machinery in the assembly of the purinosome and provide a strategy for the development of improved anticancer therapies that disrupt purine biosynthesis.
Keywords: purine metabolism, J-domain protein, Bcl-2-associated anthogene domain protein
Purines are essential molecules for all life, serving not only as the building blocks of DNA and RNA, but also playing roles in energy storage and in signaling pathways. The de novo biosynthesis of these molecules from 5-phospho-α-D-ribose 1-diphosphate (PRPP) through to inosine monophosphate (IMP) requires 10 highly conserved chemical steps (Fig. 1). Although the reactions of this pathway are catalyzed by 10 separate enzymes in most prokaryotes, humans use only six gene products, including one trifunctional and two bifunctional enzymes. Because of the essential nature of purines, particularly in rapidly dividing cells, the de novo purine biosynthetic pathway has long been considered an ideal target for anticancer, antiviral, and antimicrobial chemotherapeutics.
Fig. 1.
De novo purine biosynthetic pathway in humans. In eukaryotes, purines are synthesized in 10 chemical steps that are catalyzed by six enzymes (shown in bold). The six enzymes are phosphoribosylpyrophosphate amidotransferase (PPAT), a trifunctional enzyme (TGART) that is composed of glycinamide ribonucleotide synthetase (GARS), GAR formyltransferase (GART) and aminoimidazole ribonucleotide synthetase (AIRS), formylglycinamidine ribonucleotide synthase (FGAMS), a bifunctional enzyme (PAICS) that is composed of carboxyaminoimidazole ribonucleotide synthase (CAIRS) and succinoaminoimidazolecarboxamide ribonucleotide synthetase (SAICARS), adenylosuccinate lyase (ASL), and a bifunctional enzyme (ATIC) that is composed of aminoimidazolecarboxamide ribonucleotide transformylase (AICART) and inosine monophosphate cyclohydrolase (IMPCH).
Although it has been postulated that enzymes of biosynthetic pathways may complex in response to cellular signals (1, 2) and recent reports have provided strong indications that physiologically significant transient complexes occur (3, 4), little direct evidence for such metabolic machines has been reported. For purine biosynthesis, several indications that a macromolecular protein complex forms have been observed (5), including the copurification of proteins from native source tissues (6), the instability of a key chemical intermediate, and the kinetic arguments against free diffusion of this intermediate (7). Despite repeated attempts at in vitro and in vivo reconstitution of such a complex, however, it was not until recently that live-cell fluorescence microscopy revealed that all six of the enzymes in the pathway colocalize into protein clusters, especially under conditions of purine starvation (8). These structures, called purinosomes, were demonstrated to be dynamically regulated by changing purine levels in the media or by adding exogenous small molecules, such as kinase inhibitors or G-protein coupled receptor (GPCR) ligands (9, 10).
Although the conditions that promote purinosome formation are well characterized and we are beginning to understand the organization of purinosomes (11), little is known about the assembly of this complex or the involvement of any accessory factors. In this work, we provide evidence that the heat shock protein 90 (Hsp90)/heat shock protein 70 (Hsp70) chaperone machinery is involved in stabilization or assembly of the purinosome. We demonstrate that these chaperones and several cochaperones associate with purinosome proteins under conditions that promote complex assembly. In addition, we provide evidence that inhibitors of Hsp90 or Hsp70 disrupt purinosomes and are synergistic with methotrexate, a molecule known to block de novo purine biosynthesis. These results implicate the chaperone machinery in purinosome assembly and provide an alternative mechanism to disrupt purine biosynthesis to generate improved cancer chemotherapeutics.
Results and Discussion
To investigate the composition of the purinosome and the factors that drive its assembly and disassembly, we carried out an immunoprecipitation (IP) of myc-tagged formylglycinamidine synthetase (FGAMS) from transiently transfected cells that had been treated with a chemical cross-linking reagent. FGAMS, the enzyme that catalyzes the fourth step in purine biosynthesis, is an established marker for purinosomes. This protein is an ideal marker for purinosomes because it has been shown to colocalize with all other members of the pathway and has been speculated to act as a scaffold for the formation of purinosomes (8, 11). The purified samples, confirmed by Western blot to be enriched in FGAMS (Fig. S1) were analyzed by MS to identify coprecipitated proteins (Table S1). Among the proteins pulled down with FGAMS, of particular interest was a group of cochaperone proteins including Bcl-2–associated anthogene (BAG) domain proteins, J-domain proteins (DnaJ or Hsp40), and a heat shock organizing protein (HOP) (Stip1) that led us to investigate the possible role of the Hsp70/Hsp90 chaperone machinery in purinosome formation.
Hsp70 and Hsp90 belong to a family of ubiquitously expressed proteins that are up-regulated in response to stress and serve many functions, including assisting in protein folding and the transport, degradation, and prevention of unspecific aggregation of proteins (12–15). In addition, these chaperones have been demonstrated to participate in the formation and stabilization of protein complexes (16, 17) and have recently emerged as important targets for cancer chemotherapeutics (18, 19). To investigate whether the Hsp70/Hsp90 machinery plays a role in the assembly of the purinosome, we first determined whether these proteins colocalize with FGAMS under conditions of purine starvation. Live cell imaging of fluorescently labeled constructs of Hsp70 or Hsp90 confirmed that these proteins colocalize with this protein in HeLa cells (Fig. 2 A–F), as well as in skin and liver cancer cell lines (Fig. S2). In addition, both Hsp70 and Hsp90 were shown to colocalize with other members of the pathway (Fig. S3 A–L). Cells transfected with Hsp90 or Hsp70 alone yielded a diffuse staining pattern (Fig. S3 M–P). This colocalization was not observed for control proteins C1-tetrahydrofolate synthase (C1-THF) or serinehydroxymethyl transferase 1 (SHMT1) (Fig. 2 J–M). In addition, both Hsp70 and Hsp90 were shown to be dynamically associated with the purinosomes as purine levels changed in the media (Fig. 2 G–I). As a further validation that Hsp70 and Hsp90 associate with purinosome proteins, we used an in vivo protein proximity reporter assay to measure the interaction of Hsp70 or Hsp90 with FGAMS (11). These results indicate that both Hsp70 and Hsp90 strongly interact with FGAMS in vivo and that the level of interaction is increased in conditions that favor purinosome formation (Fig. S4). Note that a lower level of interaction was also observed in purine rich media, in agreement with the hypothesis that the purinosome is present under normal conditions and up-regulated in response to purine starvation (11).
Fig. 2.
Hsp90 and Hsp70 colocalize with purinosomes. Hsp90-GFP (A) and Hsp70-GFP (D) colocalize with the purinosome marker FGAMS-OFP (B and E, Pearson’s coefficient of 0.92 and overlap coefficient of 0.93 for Hsp90, Pearson’s coefficient of 0.90 and overlap coefficient of 0.91 for Hsp70) in HeLa cells. C and F show the merged images. The merged images of Hsp90-GFP and FGAMS-OFP serve to demonstrate that purinosomes appear under purine depleted conditions (G), disappear when conditions are changed to purine rich (H, 120 min), and reappear when purine-depleted conditions are restored (I, additional 90 min after media change). Hsp90-OFP (K and M) does not cocluster with control proteins C1-THF-GFP (J) or SHMT1-GFP (L). (Scale bar, 10 µm.)
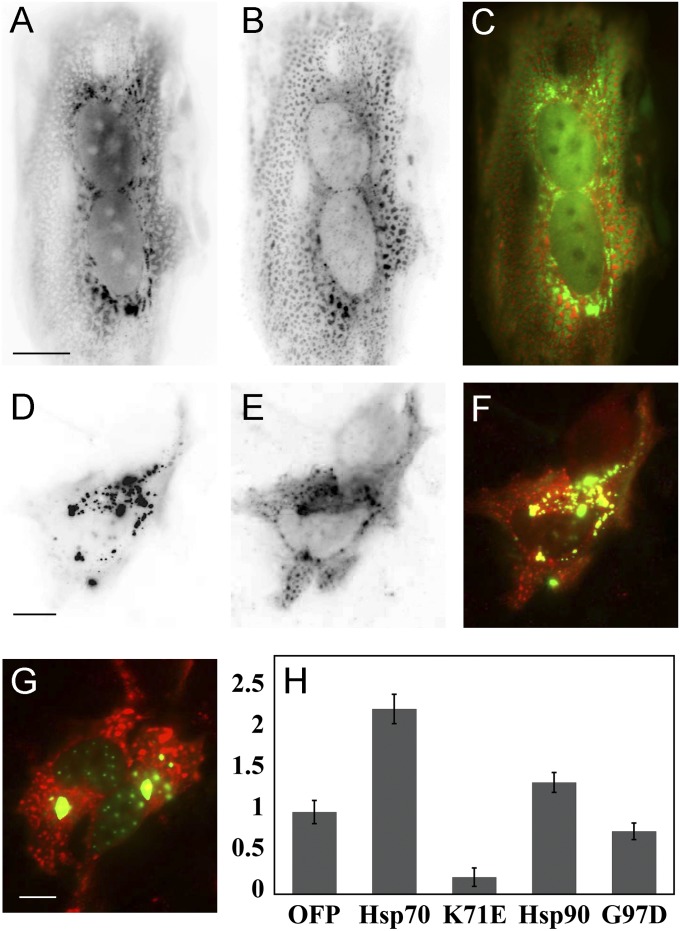
To further ensure that the association of Hsp70 and Hsp90 with purinosomes was not simply a result of cellular stress, we examined the colocalization of these proteins with stress granules (20) or aggresomes (21). We first examined the colocalization of orange fluorescent protein tagged FGAMS (FGAMS-OFP) with a known marker of stress granules, the RasGAP-associated endoribonuclease G3BP (22, 23). After cotransfection with GFP-G3BP, some cells do show stress granules, presumably due to stress caused by the transfection protocol. However, the percentage of cells with granules and the number of granules per cell did not vary between purine-rich and purine-depleted conditions. In addition, the stress granule morphology was distinctly different from that of purinosomes, and dual color imaging revealed that the two structures do not colocalize (Fig. 3 A–C). Similarly, known markers for aggresomes, gp170* (24) and gp250 (25), did not colocalize with purinosomes (Fig. 3 D–G). To provide further evidence that the purinosomes do not represent misfolded or aggregated protein, we quantified the effect of Hsp70, Hsp90, and inactive mutant forms of these proteins on the purinosome content in cells. The percentage of cells with purinosomes increased by 120% or 40% in the presence of Hsp70 or Hsp90, respectively, whereas cotransfection with the inactive Hsp70-K71E (26) or Hsp90-G97D (27) mutants led to lower numbers of cells with purinosomes (Fig. 3H). Western blots were used to confirm that the Hsp70 and Hsp90 mutants were expressed at detectable levels in the cells (Fig. S5). If the purinosomes were clusters of misfolded or aggregated protein, we would expect the numbers of these clusters to decrease in the presence of higher concentrations of Hsp70 and Hsp90. In addition to these observations, our IP results did not show a difference in the level of ubiquitin or polyubiquitin between samples with overexpressed FGAMS or untransfected controls. We conclude from these data that purinosomes are not stress granules or aggresomes and that the function of Hsp70 and Hsp90 in these structures is the trafficking of protein members to, or the assembly or stabilization of, this protein complex.
Fig. 3.
Purinosomes are not stress bodies or aggresomes and are dependent on functional Hsp70 and Hsp90. The stress body marker GFP-G3BP (A) does not colocalize with FGAMS-OFP (B). C shows the merge of A in green and B in red (Pearson’s coefficient of 0.08 and overlap coefficient of 0.12). Similarly, FGAMS-OFP (E) does not colocalize with markers for aggresomes, gp250 (D). F shows the merge of D in green and E in red (Pearson’s coefficient of 0.4 and overlap coefficient of 0.41) or gp170* (G, merge of gp170* and FGAMS-OFP, Pearson’s coefficient of 0.08 and overlap coefficient of 0.09). (H) The overexpression of both Hsp70-OFP and Hsp90-OFP increased the level of purinosome formation over that of the empty OFP vector control, whereas the nonactive mutant forms of these proteins decreased the purinosome level. The data represent the amount of purinosome formation when cells coexpressed the proteins listed with FGAMS-GFP. The y axis reports the number of cells with purinosomes normalized to the result of FGAMS-GFP cotransfected with the OFP empty vector. (Scale bar, 10 µm.)
The functions of Hsp90 and Hsp70 are facilitated and regulated in vivo by a large number of cochaperones. Accessory proteins such as J-domain proteins, BAG domain proteins, HOP, and p23 assist in client binding and nucleotide exchange (28–32). Investigation of our IP results revealed that several of these cochaperone proteins were precipitated with FGAMS. We identified two J-domain proteins (DnaJ-A1 and DnaJ-C7), two BAG proteins (BAG2 and BAG5), and a HOP (Stip1) in our IP data (Table S1). Colocalization experiments revealed that these proteins associate with FGAMS in purinosomes (Fig. 4 A–L; Fig. S6 A–F). As controls, two additional J-domain proteins (DnaJ-B1 and -C14, not identified in our IP experiment) were examined and did not colocalize with FGAMS (Fig. S6 G–J). To confirm the requirement of these cochaperones for purinosome assembly, we examined the effect of siRNA knockdown on the identified J-domain proteins. Depletion of DnaJ-A1 or DnaJ-C7 levels significantly reduced the number of cells harboring purinosomes compared with the control reaction or knockdown of a J-domain protein not identified in our pulldown (Fig. 4 M and N). The presence of these proteins establishes that a complete complement of chaperones and cochaperones are associated with purinosomes and implicates the Hsp90/Hsp70-based chaperone machinery in the assembly of the purinosome.
Fig. 4.
Several cochaperones are associated with purinosomes and promote their formation. BAG5-GFP (A), Stip1-GFP (D), DnaJ-C7-GFP (G), and p23-GFP (J) colocalize with FGAMS-OFP (B, E, H, and K). Merged images are shown in C, F, I, and L, respectively. Overlap coefficients with FGAMS-OFP for BAG5, Stip1, DnaJ-C7, and p23 are 0.89, 0.86, 0.90, and 0.84, respectively. (Scale bar, 10 µm.) (M) Knockdown of the expression of cochaperones identified in the pulldown (DnaJ-C7 and DnaJ-A1) leads to a decrease in purinosome formation, whereas knockdown of an unrelated cochaperone (DnaJ-C14) has no effect on purinosome formation (these data represent the mean of four replicates, P = 0.0018). DnaJ-C7, DnaJ-A1, and DnaJ-C14 are knocked down by 51%, 67%, and 98%, respectively (N).
To further examine the role of Hsp70 and Hsp90 in the assembly of purinosomes, we tested known inhibitors of the chaperones for their ability to disrupt these protein complexes. Cells that were treated with the Hsp90 inhibitors 17-AAG (33) or NVP-AUY922 (34) lost the punctate staining pattern that is characteristic of purinosomes (Fig. 5; Fig. S7 G and H). The effect was observed to be concentration and time dependent. In addition, a media change to remove the presence of the inhibitors reversed the effect and resulted in a reappearance of purinosomes (Fig. 5 A–C). Similarly, treatment of cells with the Hsp70 inhibitors 2-phenylethynesulfonamide (Pifithrin-µ) (35), MKT-077 (36), or methylene blue (37) produced a similar reversible disruption of the purinosomes (Fig. S7 A–F). Note that, although the Hsp70 inhibitors tested are expected to be less specific than those used against Hsp90, the data are suggestive of a role for both proteins. These results are consistent with the hypothesis that the chaperone machinery is necessary to assemble or stabilize the purinosome and indicate that these proteins may represent a novel target for the inhibition of purinosome formation and function.
Fig. 5.
Hsp90 inhibitors disrupt purinosomes and are synergistic with methotrexate. A shows HeLa cells transfected with the purinosome marker FGAMS-GFP. Treatment of cells with 1.5 nM of the Hsp90 inhibitor NVP-AUY922 for 35 min disrupts purinosomes (B). After washing the cells three times with BSS and a return to purine-depleted media, purinosomes begin to reform after 45 min (C). Treatment of HeLa cells for 72 h with the Hsp90 inhibitor 17-AAG alone (D, filled squares and dotted line) kills cells with an EC50 of 0.14 ± 0.4 µM, whereas treatment with methotrexate alone (D, filled diamonds and dashed line) gives an EC50 of 10.1 ± 0.4 nM. The combination of 17-AAG and methotrexate (D, filled circles and solid line) leads to a decrease in the EC50 to 4.7 ± 0.3 nM. A Chou plot of the results (F) verifies that the combination is synergistic (CI < 1). Similarly, the combination of the Hsp90 inhibitor NVP-AUY922 and methotrexate (E, filled circles and solid line) gives a decrease in EC50 (4.4 ± 0.07 nM) over either NVP-AUY922 alone (E, filled diamonds and solid line, EC50 of 9.1 ± 0.8 µM) or methotrexate alone (E, filled squares and dotted line, EC50 of 10.1 ± 0.4 nM) alone. A Chou plot of the results (G) verifies that the combination is synergistic (CI < 1). (Scale bar, 10 µm.)
Considering the importance of the purine biosynthetic pathway as a target for cancer chemotherapeutics and the role that purinosomes play in increasing flux through the pathway (38), we speculated that drugs that target Hsp70 or Hsp90 may work cooperatively with current cancer treatments that disrupt purine biosynthesis. To test this hypothesis, we investigated the effect of combining Hsp70 or Hsp90 inhibitors with the known cancer drug methotrexate. This compound, which is currently used to treat several forms of cancer, inhibits purine biosynthesis by preventing the production of a cofactor that is essential at two separate steps of the pathway (39). The combination of methotrexate and either of the Hsp90 inhibitors NVP-AUY922 or 17-AAG led to an increased cytotoxic effect on cervical cancer cells with a greater than twofold decrease in the EC50 (Fig. 5 D and E). Analysis of the results using the Chou-Talalay method (40) verified that the drug combinations were synergistic (Fig. 5 F and G). A similar synergistic effect was observed for the Hsp70 inhibitor Pifithrin-µ (Fig. S8 D and E). Conversely, when the Hsp70 activator geranylgeranylacetone (41) was combined with methotrexate, it had an antagonistic effect, increasing the EC50 and stimulating the production of purinosomes (Fig. S8 A–C). These data confirm a role for the chaperone machinery in the assembly of the purine biosynthetic protein complex and validate the purinosome as a viable target for improved cancer chemotherapeutics.
The recent discovery of the purinosome has led to a paradigm shift regarding how metabolic enzymes associate intracellularly and has raised questions about the advantage of such multiprotein structures to the cell. Our findings reveal that together Hsp90, Hsp70, and several cochaperones play a role in the assembly or stabilization of the purinosome. The involvement of this chaperone machinery begins to explain why the in vitro reconstitution of a purine biosynthetic complex has eluded researchers for so long. Although these findings add another level of complexity, they help to elucidate the dynamic nature of the purinosome and may be the key to understanding the signals that drive the assembly and disassembly of this protein complex. The recent enthusiasm over the development of inhibitors to chaperones as anticancer drugs provides a rich resource that can be used to further our understanding of purinosome structure and function. In addition, the role of these proteins in assembling the purinosome can be exploited in the development of novel pharmaceuticals that target the purinosome alone or that combine disruption of the purinosome with some other complementary effect such as inhibition of purine biosynthesis.
Methods
Construction of Plasmids.
Plasmids were constructed by standard molecular biology techniques as previously described (8) or were generously provided as gifts in the case of G3BP-GFP (Jamal Tazi, Institut de Genetique Moleculaire de Montpellier, Montpellier, France), GFP170* and GFP250 (Elizabeth Sztul, University of Alabama at Birmingham, Birmingham, AL), and SHMT1 (Patrick Stover, Cornell University, Ithaca, New York). Template DNA was obtained from the Arizona State University Biodesign Institute Plasmid Repository (DNASU) or ATCC. Details of the cloning are provided in SI Methods.
Coimmunoprecipitation and MS.
HeLa cells were transiently transfected with FGAMS-9xcMyc and treated with the chemical cross-linking reagent dithiobis(succinimidyl propionate) (DSP) before harvesting the cells. The FGAMS-9xcMyc and associated proteins were purified from the lysates using immobilized anti-myc monoclonal antibodies (Clontech). The presence of FGAMS-9xcMyc (∼162 kDa) in the final IP eluate samples was confirmed by Western blot using anti-cMyc antibody (Abcam ab9132) (Fig. S1). Protein was precipitated with trichloroacetic acid (TCA) and resuspended in 8 M urea in 50 mM Tris, pH 8.0. Each sample was reduced with 10 mM tris(2-carboxyethyl)phosphine (TCEP) and alkylated with 12.5 mM iodoacetamide. Samples were diluted to 2 M urea with 50 mM Tris, pH 8.0, and digested overnight in the presence of 1 mM CaCl2 and trypsin (1 µL of 0.5 µg/µL). Digested samples were acidified to 5% (vol/vol) final formic acid and centrifuged. Peptides were loaded onto a biphasic column with strong cation exchange and C18 for analysis on a LTQ XL ion trap mass spectrometer (Thermo Scientific) using a five-step standard procedure (42). The mass spectrometer was set in a data-dependent acquisition mode, with dynamic exclusion enabled with a repeat count of 1, a repeat duration of 20 s, exclusion duration of 60 s, and an exclusion list size of 300. All tandem mass spectra were collected using normalized collision energy of 35% and an isolation window of 2 Da. One microscan was applied for all experiments in the LTQ. Spray voltage was set to 2.50 kV. Each full MS survey scan was followed by seven MS/MS scans.
Transient Transfection and Live-Cell Imaging.
Three human cancer cell lines, HeLa, C3A, and A431, were obtained from the American Type Culture Collection. Cell lines were maintained in purine-rich or purine-depleted media as previously described (8). Cells were transiently transfected with plasmid DNA using either Lipofectamine 2000 (Invitrogen) (for HeLa) or X-tremeGENE HP (Roche) (for A431 and C3A) following the manufacturer’s protocol. For siRNA (Santa Cruz Biotechnology and Abcam), transfections were carried out using Lipofectamine 2000 following the manufacturer’s protocol. Note that in all cases, transfection efficiency was between 25% and 35%. Quantification of protein expression levels from Western blots was carried out using a Bio-Rad ChemiDoc XRS+ system. Before imaging, all samples were washed three times for 5 min with BSS [20 mM Hepes (pH 7.4), 135 mM NaCl, 5 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, and 5.6 mM glucose]. Cells were imaged at ambient temperature (∼25 °C) using an S484/15× excitation filter and an S517/30m emission filter for GFP and an S555/25× excitation filter and an S605/40m emission filter for OFP. All images were created using the ImageJ program and were in some cases cropped, inverted, or shown in color for clarity, but were otherwise unmodified.
Luciferase Reporter Assay.
The assay was carried out as previously described (11) to measure interactions between FGAMS and either Hsp70 or Hsp90. For these experiments, the tTa reporter was fused to the C terminus of FGAMS (FGAMS-tTa), whereas the modified tobacco etch virus (TEV) protease was fused to the C terminus Hsp70 or Hsp90 (Hsp70-TEV and Hsp90-TEV).
Cell Death Assay.
A standard 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used to monitor cytotoxic effect of the various inhibitors. HeLa cells were plated in 96-well plates and grown at 37 °C in 5% CO2 for 16 h. Varying concentrations of the drug or vehicle control were added, and the cells were grown for an additional 72 h. The media were removed, and 5 mg/mL MTT was added to the wells and incubated for 3 h. The media were removed, and DMSO was added to solubilize the formazan product. After a 1-h incubation, the plate was read at 550 nm. The data were plotted as factor affected vs. dose and fit with the median-effect equation fa = 1/[1 + (Dm/D)m], where fa is the factor affected, Dm is the median-effect dose (EC50), m is the slope of the curve, and D is the dose (40). For combinations of drugs with methotrexate, the drugs were combined at a 1:1 ratio, and the assay was carried out as detailed above. To determine whether the drug combination had a synergistic effect, a Chou plot of factor affected vs. combination index (CI) was constructed, where CI = (D)1/(Dx)1 + (D)2/(Dx)2. The curves shown on the Chou plots are smooth curves connecting 100 simulated data points for values of 0–1 fraction affected using the equation Dx = Dm[fa/(1 − fa)]1/m. All samples were carried out in triplicate and are reported as mean ± SE.
Supplementary Material
Acknowledgments
We thank Y. Fang (Corning Inc.), E. Sztul (University of Alabama at Birmingham), R. Y. Tsien (University of California, San Diego), and P. Stover (Cornell University) for providing compounds and plasmids, and Patricia Tu for help with MS analysis. J.B.F. thanks the Canadian Institutes of Health Research for fellowship support. This work was supported in part by National Institutes of Health Grants GM024129 (to S.J.B.) and CA087660 (to B.F.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300173110/-/DCSupplemental.
References
- 1.Alberts B. The cell as a collection of protein machines: Preparing the next generation of molecular biologists. Cell. 1998;92(3):291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 2.Srere PA. Complexes of sequential metabolic enzymes. Annu Rev Biochem. 1987;56:89–124. doi: 10.1146/annurev.bi.56.070187.000513. [DOI] [PubMed] [Google Scholar]
- 3.James JR, Vale RD. Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature. 2012;487(7405):64–69. doi: 10.1038/nature11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li P, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483(7389):336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schendel FJ, Cheng YS, Otvos JD, Wehrli S, Stubbe J. Characterization and chemical properties of phosphoribosylamine, an unstable intermediate in the de novo purine biosynthetic pathway. Biochemistry. 1988;27(7):2614–2623. doi: 10.1021/bi00407a052. [DOI] [PubMed] [Google Scholar]
- 6.Smith GK, Mueller WT, Wasserman GF, Taylor WD, Benkovic SJ. Characterization of the enzyme complex involving the folate-requiring enzymes of de novo purine biosynthesis. Biochemistry. 1980;19(18):4313–4321. doi: 10.1021/bi00559a026. [DOI] [PubMed] [Google Scholar]
- 7.Rudolph J, Stubbe J. Investigation of the mechanism of phosphoribosylamine transfer from glutamine phosphoribosylpyrophosphate amidotransferase to glycinamide ribonucleotide synthetase. Biochemistry. 1995;34(7):2241–2250. doi: 10.1021/bi00007a019. [DOI] [PubMed] [Google Scholar]
- 8.An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320(5872):103–106. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- 9.An S, Kyoung M, Allen JJ, Shokat KM, Benkovic SJ. Dynamic regulation of a metabolic multi-enzyme complex by protein kinase CK2. J Biol Chem. 2010;285(15):11093–11099. doi: 10.1074/jbc.M110.101139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verrier F, et al. GPCRs regulate the assembly of a multienzyme complex for purine biosynthesis. Nat Chem Biol. 2011;7(12):909–915. doi: 10.1038/nchembio.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng Y, et al. Mapping protein-protein proximity in the purinosome. J Biol Chem. 2012;287(43):36201–36207. doi: 10.1074/jbc.M112.407056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246(4928):382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 13.Frydman J. Folding of newly translated proteins in vivo: The role of molecular chaperones. Annu Rev Biochem. 2001;70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 14.Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 2003;228(2):111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 15.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11(7):515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 16.Young JC, Barral JM, Ulrich Hartl F. More than folding: Localized functions of cytosolic chaperones. Trends Biochem Sci. 2003;28(10):541–547. doi: 10.1016/j.tibs.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Makhnevych T, Houry WA. The role of Hsp90 in protein complex assembly. Biochim Biophys Acta. 2012;1823(3):674–682. doi: 10.1016/j.bbamcr.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Goloudina AR, Demidov ON, Garrido C. Inhibition of HSP70: A challenging anti-cancer strategy. Cancer Lett. 2012;325(2):117–124. doi: 10.1016/j.canlet.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 19. Whitesell L, Santagata S, Lin NU (2012) Inhibiting HSP90 to treat cancer: A strategy in evolution. Curr Mol Med 12(9):1108–1124. [DOI] [PubMed]
- 20.Buchan JR, Parker R. Eukaryotic stress granules: The ins and outs of translation. Mol Cell. 2009;36(6):932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10(12):524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 22.Tourrière H, et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160(6):823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Tourrière H, et al. RasGAP-associated endoribonuclease G3Bp: Selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol. 2001;21(22):7747–7760. doi: 10.1128/MCB.21.22.7747-7760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu L, et al. Nuclear aggresomes form by fusion of PML-associated aggregates. Mol Biol Cell. 2005;16(10):4905–4917. doi: 10.1091/mbc.E05-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García-Mata R, Bebök Z, Sorscher EJ, Sztul ES. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J Cell Biol. 1999;146(6):1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Brien MC, Flaherty KM, McKay DB. Lysine 71 of the chaperone protein Hsc70 Is essential for ATP hydrolysis. J Biol Chem. 1996;271(27):15874–15878. doi: 10.1074/jbc.271.27.15874. [DOI] [PubMed] [Google Scholar]
- 27.Hawkins TA, et al. The ATPase-dependent chaperoning activity of Hsp90a regulates thick filament formation and integration during skeletal muscle myofibrillogenesis. Development. 2008;135(6):1147–1156. doi: 10.1242/dev.018150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James P, Pfund C, Craig EA. Functional specificity among Hsp70 molecular chaperones. Science. 1997;275(5298):387–389. doi: 10.1126/science.275.5298.387. [DOI] [PubMed] [Google Scholar]
- 29.Johnson JL, Beito TG, Krco CJ, Toft DO. Characterization of a novel 23-kilodalton protein of unactive progesterone receptor complexes. Mol Cell Biol. 1994;14(3):1956–1963. doi: 10.1128/mcb.14.3.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabbage M, Dickman MB. The BAG proteins: A ubiquitous family of chaperone regulators. Cell Mol Life Sci. 2008;65(9):1390–1402. doi: 10.1007/s00018-008-7535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morishima Y, et al. The hsp90 cochaperone p23 is the limiting component of the multiprotein hsp90/hsp70-based chaperone system in vivo where it acts to stabilize the client protein: hsp90 complex. J Biol Chem. 2003;278(49):48754–48763. doi: 10.1074/jbc.M309814200. [DOI] [PubMed] [Google Scholar]
- 32.Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63(22):2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schulte TW, Neckers LM. The benzoquinone ansamycin 17-allylamino-17-demethoxygeldanamycin binds to HSP90 and shares important biologic activities with geldanamycin. Cancer Chemother Pharmacol. 1998;42(4):273–279. doi: 10.1007/s002800050817. [DOI] [PubMed] [Google Scholar]
- 34.Eccles SA, et al. NVP-AUY922: A novel heat shock protein 90 inhibitor active against xenograft tumor growth, angiogenesis, and metastasis. Cancer Res. 2008;68(8):2850–2860. doi: 10.1158/0008-5472.CAN-07-5256. [DOI] [PubMed] [Google Scholar]
- 35.Leu JI, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell. 2009;36(1):15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rousaki A, et al. Allosteric drugs: The interaction of antitumor compound MKT-077 with human Hsp70 chaperones. J Mol Biol. 2011;411(3):614–632. doi: 10.1016/j.jmb.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang AM, et al. Inhibition of hsp70 by methylene blue affects signaling protein function and ubiquitination and modulates polyglutamine protein degradation. J Biol Chem. 2010;285(21):15714–15723. doi: 10.1074/jbc.M109.098806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An S, Deng Y, Tomsho JW, Kyoung M, Benkovic SJ. Microtubule-assisted mechanism for functional metabolic macromolecular complex formation. Proc Natl Acad Sci USA. 2010;107(29):12872–12876. doi: 10.1073/pnas.1008451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allegra CJ, Hoang K, Yeh GC, Drake JC, Baram J. Evidence for direct inhibition of de novo purine synthesis in human MCF-7 breast cells as a principal mode of metabolic inhibition by methotrexate. J Biol Chem. 1987;262(28):13520–13526. [PubMed] [Google Scholar]
- 40.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 41.Hirakawa T, Rokutan K, Nikawa T, Kishi K. Geranylgeranylacetone induces heat shock proteins in cultured guinea pig gastric mucosal cells and rat gastric mucosa. Gastroenterology. 1996;111(2):345–357. doi: 10.1053/gast.1996.v111.pm8690199. [DOI] [PubMed] [Google Scholar]
- 42.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.