Abstract
SNPs in the first intron of FTO (fat mass and obesity associated) are strongly associated with human obesity. While it is not yet formally established that this effect is mediated through the actions of the FTO protein itself, loss of function mutations in FTO or its murine homologue Fto result in severe growth retardation, and mice globally overexpressing FTO are obese. The mechanisms through which FTO influences growth and body composition are unknown. We describe a role for FTO in the coupling of amino acid levels to mammalian target of rapamycin complex 1 signaling. These findings suggest that FTO may influence body composition through playing a role in cellular nutrient sensing.
Keywords: genetics, translation, tRNA synthetase, demethylase
In 2007, single nucleotide polymorphisms (SNPs) in the first intron of fat mass and obesity related (FTO) were found to be powerfully associated with body mass index and predisposing to childhood and adult obesity (1). Many subsequent studies, covering multiple populations of European, African, and Asian ancestries, across different age ranges, have confirmed the association of FTO with body mass index (reviewed in ref. 2). It is clear from the weight of evidence that the major effect of SNPs in FTO is on increased energy intake, with reduction in satiety (3–8). To date, no conclusive link has been made between the risk alleles and expression or function of FTO. However, transgenic manipulation in mice supports the notion that FTO itself regulates body weight, with overexpression resulting in obesity (9), whereas Fto-null mice (10) and humans homozygous for a loss-of-function allele (11) display postnatal growth retardation and have high rates of early mortality.
FTO is widely expressed across multiple tissues, although it is most highly expressed in the brain, especially in the hypothalamus, a region that plays a key role in the control of energy homeostasis (12). We have found that expression of FTO, specifically in the arcuate nucleus of the hypothalamus, is bidirectionally regulated as a function of nutritional status—decreasing following a 48-h fast and increasing after 10 wk of exposure to a high-fat diet—and that modulating FTO levels specifically in the arcuate nucleus can influence food intake (13). Carriers of the obesity predisposing allele are hyperphagic and show altered macronutrient preference.
FTO shares sequence motifs with Fe(II)- and 2-oxoglutarate–dependent oxygenases (12). In vitro, recombinant FTO is able to catalyze the Fe(II)- and 2-oxoglutarate–dependent demethylation of nucleic acids. It is more active at RNA than DNA (12, 14) and is capable of demethylating N6-methyladenosine (15) (more prevalent on mRNAs) or 3-methyluracil (15) (more prevalent in tRNAs and ribosomal RNAs). To date, however, there has been no understanding of how FTO might functionally link to energy homeostasis, growth, or nutrient sensing.
Results
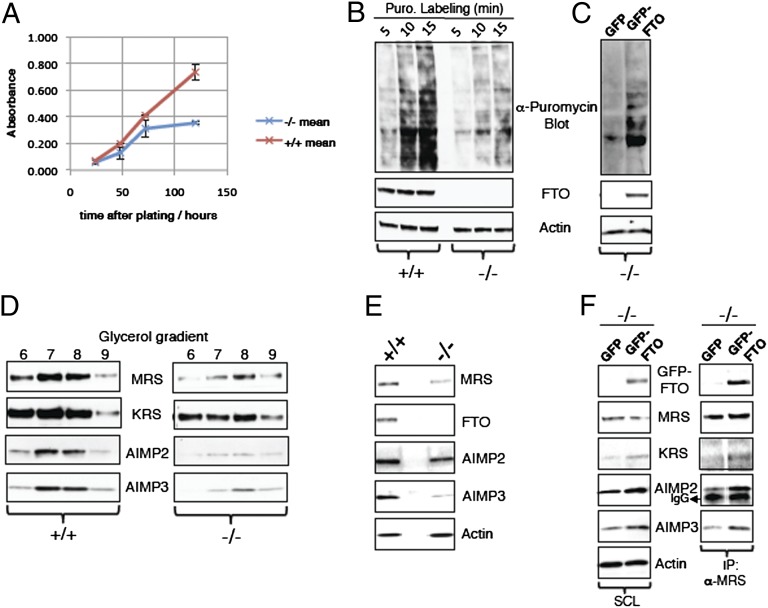
One of the most striking features of mice lacking FTO (10), or humans homozygous for an enzymatically inactive mutated FTO (11), is severe growth retardation. We hypothesized that exploring the mechanisms underlying this phenotype might provide clues to the physiological functions of FTO. We examined cellular proliferation rates in embryonic fibroblasts from Fto−/− mice and Fto+/+ littermates. The growth rate of Fto−/− cells, measured over 5 d, was ∼50% of that seen in Fto+/+ mouse embryonic fibroblasts (MEFs; Fig. 1A). To examine whether this reduction in growth rate was associated with a global decrease in protein synthesis, we examined the rate of translation of mRNA by using the technique of puromycin labeling of MEFs followed by antipuromycin Western blotting (16). The rates of puromycin incorporation into nascent proteins were measured at three time points. Fto−/− MEFs showed markedly decreased puromycin incorporation at all time points, indicating reduced rates of mRNA translation (Fig. 1B). mRNA translational rates in Fto−/− MEFs could be normalized by the expression of exogenous GFP-WT FTO but not by GFP alone (Fig. 1C).
Fig. 1.
Fto−/− MEFs have reduced rates of cell growth, mRNA translation, and low levels of MSC. (A) For measurement of growth rate, Fto+/+ and Fto−/− MEFs were allowed to grow for different time periods. After each time point, 250 µL of phenol red-free DMEM with 10 µM phenazine methosulfate and 50 µM WST-1 was added for 2 h. Absorbance of media samples was measured as described in Materials and Methods. All data are expressed as mean ± SEM. (B) Fto+/+ and Fto−/− MEFs were treated with 2 µM puromycin for different time periods. MEFs were then harvested, and the resulting cell extracts analyzed by Western blotting utilizing the indicated antibodies. (C) Fto−/− MEFs were transfected with GFP alone or GFP-WT FTO using a Neon transfection system. At 48 h after transfection, cells were labeled with 2 µM puromycin for 15 min and the mRNA translation rate was measured as described in B. (D) Equal amounts of total protein containing cell extracts from Fto+/+ and Fto−/− MEFs were subjected to 10% to 45% glycerol gradients fractionation as described in Materials and Methods. Different fractions obtained were analyzed by Western blotting using the indicated antibodies. (E) Tissue extracts of mouse hypothalamus, obtained from different Fto+/+ and Fto−/− mice, were analyzed for components of the MSC by Western blotting using the indicated antibodies. (F) Fto−/− MEFs were transfected with GFP or GFP-WT FTO as in C. At 48 h after transfection, cells were harvested, and MRS protein was immunoprecipitated as described in Materials and Methods. Cell extracts and MRS immunoprecipitates were analyzed for components of the MSC by Western blotting using the indicated antibodies.
mRNA translational rates are dependent on the activity and integrity of the multi-tRNA synthetase complex (MSC), a multiprotein complex of aminoacyl-tRNA synthetases (AARSs) and accessory proteins including AIMP1, AIMP2, and AIMP3 (AARS complex-interacting multifunctional proteins 1, 2, and 3, respectively) (17). AARS tether free amino acids to their cognate tRNAs in a process known as tRNA charging. Each of the 20 aa has its own AARS. During tRNA charging, at least nine different AARS and three AIMPs (18) cooperate to form the MSC. We compared the level of proteins and mRNAs for several members of this complex in Fto−/− and Fto+/+ MEFs as well as in the hypothalamic tissue from adult Fto−/− and Fto+/+ mice. To ensure we were comparing the MSC components levels, samples were subject to glycerol gradient fractionation and multiple fractions were subsequently immunoblotted (Fig. 1D). Protein levels of AIMP3 and AIMP2 [which are solely present in the MSC and maintain integrity of the MSC (18)] were markedly reduced in Fto−/− compared with Fto+/+ MEFs, and levels of methionyl-tRNA synthetase (MRS) and lysyl-tRNA synthetase (KRS) were markedly reduced, especially in the glycerol gradient fractions in which they comigrated with AIMPs (Fig. 1D). In a complementary analysis, we found reduced levels of MSC components in the MRS/KRS pull-downs obtained from Fto−/− MEFs cell lysates compared with pull-down performed in parallel from Fto+/+ MEFs cell lysates (Fig. S1A). A similar reduction in several components of the MSC was seen in hypothalamic tissue from adult Fto−/− mice (Fig. 1E).
The expression of GFP-WT FTO in Fto−/− MEFs was able to rescue protein levels of many of the MSC components, including AIMP2, AIMP3, and KRS, but was less effective at restoring MRS protein levels (Fig. 1F). In contrast to the protein levels, mRNA levels for MRS, KRS, AIMP2, and AIMP3 were indistinguishable between Fto−/− and Fto+/+ MEFs (Fig. S1B). Thus, lack of FTO markedly impairs the formation and/or stability of the MSC.
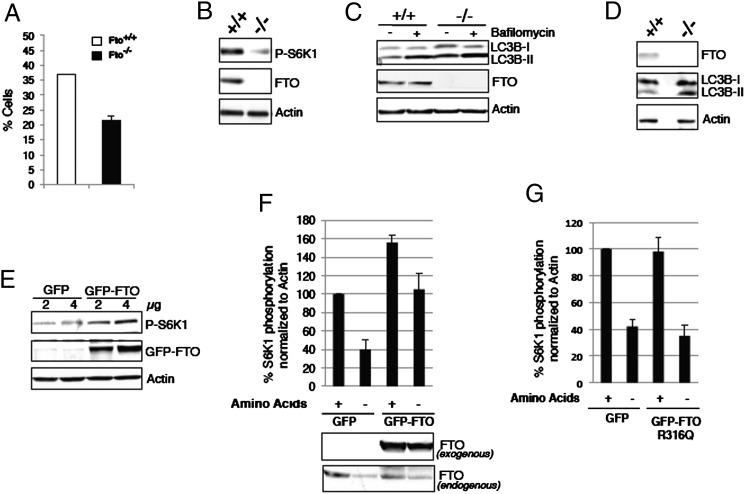
Members of the AARS family have recently been identified as having a role in linking amino acid levels to the mammalian target of rapamycin complex 1 (mTORC1) signaling (19, 20). Thus, we wondered if FTO-deficient cells might exhibit disturbed amino acid sensing. Consistent with this notion, Fto−/− MEFs showed a 50% decrease in survival rate after 24 h of amino acid deprivation compared with Fto+/+ MEFs (Fig. 2A).
Fig. 2.
FTO is necessary for the normal responsiveness of mTORC1 signaling to amino acid status. (A) Fto+/+ and Fto−/− MEFs grown for 24 h were subjected to total amino acid deprivation for a further 24 h in the presence of dialyzed serum. After amino acid deprivation, cells were stained with trypan blue and the number of live cells was counted. (B) Cell extracts from Fto+/+ and Fto−/− MEFs grown for 24 h were analyzed for mTORC1 signaling by Western blotting using indicated antibodies. (C) Fto+/+ and Fto−/− MEFs grown for 24 h were amino acid-deprived and/or treated with 400 µM bafilomycin for 4 h in the presence of dialyzed serum. After 4 h treatment, cells were harvested, and the resulting cell extracts were analyzed by Western blotting using indicated antibodies. (D) Tissue extracts of mouse liver obtained from different Fto+/+ and Fto−/− mice were analyzed by Western blotting using indicated antibodies. (E) HEK cells were transfected with 2 µg or 4 µg of vector alone or GFP-WT FTO and cultured for 48 h as described in Materials and Methods. At 48 h after transfection, cells were harvested and cell extracts were analyzed for mTORC1 signaling by Western blotting using indicated antibodies. (F) HEK cells were transfected with 4 µg of vector alone or GFP-WT FTO and cultured for 48 h as described in Materials and Methods. After 48 h of transfection, cells were deprived of all amino acids for 4 h in the presence of dialyzed serum. Cells were then harvested, and cell extracts analyzed by Western blotting. Western blots were quantitated by densitometry and data normalized to actin levels. Data are shown as mean ± SEM. (G) HEK cells were transfected with 4 µg of vector alone or GFP-FTO R316Q mutant construct and were cultured for 48 h. Afterward, cells were treated and harvested and lysates were analyzed as in F. Quantification of Western blotting results obtained from separate experiments was performed and plotted as in F.
As the mTORC1 pathway is a key regulator of cell growth and mRNA translation (21), and is sensitive to amino acid levels, we measured the status of mTORC1 signaling in Fto−/− MEFs. Consistent with the reduced mRNA translation, phosphorylation of S6 kinase-1 (S6K1; an effector of mTORC1) was reduced in the Fto−/− MEFs, indicating decreased basal levels of mTORC1 signaling (Fig. 2B). To confirm that mTORC1 signaling is compromised in the absence of FTO, we examined the rate of autophagy, another key pathway regulated by mTORC1. To compare autophagy in Fto+/+ and Fto−/− MEFs, we measured levels of the autophagosome component LC3B-II (22) (Fig. 2C). Bafilomycin was used to inhibit the degradation of LC3B-II and thus amplify the signal (22). Fto−/− MEFs expressed more LC3B-II than Fto+/+ MEFs, indicating more autophagic flux in Fto−/− MEFs (Fig. 2C). To test the in vivo relevance of these observations, we analyzed LC3B-II amounts in mouse liver: this revealed increased amounts of LC3B-II in liver of Fto−/− compared with Fto+/+ mice (Fig. 2D). To examine whether an increase in FTO expression in the cell would influence activation of mTORC1 pathways, HEK293 cells were transiently transfected with vectors expressing GFP- WT FTO or GFP alone. Transfection of GFP-WT FTO, but not GFP alone, resulted in a dose-dependent increase in the phosphorylation of S6K1 (Fig. 2E).
We have previously reported that amino acid deprivation results in a rapid reduction of cellular FTO protein, likely involving active degradation (23). We wondered if this active suppression of FTO levels was necessary for the cellular response to amino acid deprivation. To answer this question, we subjected HEK293 cells transfected with GFP vector or GFP-WT FTO construct to amino acid deprivation (Fig. 2F). As observed earlier, amino acid deprivation reduces FTO levels and S6K1 phosphorylation in the empty vector-transfected HEK293 cells (Fig. 2F). However, even though endogenous levels of FTO decrease during amino acid deprivation, overexpression of WT FTO in these cells prevents the expected reduction in S6K1 phosphorylation, in essence rendering them “insensitive” to amino acid deprivation (Fig. 2F). A mutant (R316Q) form of FTO lacking demethylase activity (11) was, in contrast, ineffective in this regard (Fig. 2G).
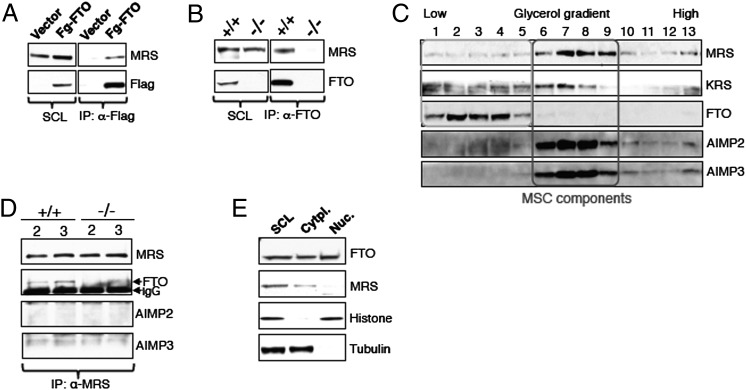
To begin to identify a molecular mechanism whereby FTO might impact on the pathways controlling mRNA translation, we undertook proteomic studies to identify proteins that physically interact with FTO. We immunoprecipitated Flag-WT FTO from lysates of transiently transfected HEK293 cells and subjected eluates to MS analysis. Among the proteins identified, four different AARSs—methionyl-, tyrosyl-, arginyl-, and phenylalanyl-tRNA synthetases (MRS, YRS, RRS, and FRS, respectively)—were detected in the Flag-WT FTO immunoprecipitates, but were absent in the Flag-control immunoprecipitates.
The interaction of MRS and FTO was validated by Western blotting of Flag-WT FTO immunoprecipitates from lysates of HEK293 cells transfected with Flag-WT FTO construct. We demonstrated that endogenous MRS coimmunoprecipitated with Flag-FTO (Fig. 3A). We next investigated if endogenous FTO could interact with MRS by immunoprecipitating endogenous FTO from Fto+/+ MEFs. We performed the same experiments on Fto−/− MEFs as a negative control. An antibody against native FTO coimmunoprecipitated endogenous MRS (Fig. 3B), and an antibody against endogenous MRS was able to coimmunoprecipitate native FTO (Fig. S2A). These results confirm that the MRS is an interacting protein partner of FTO. We also tested the interaction of FTO with other AARSs, and observed that FTO coimmunoprecipitated with endogenous KRS (Fig. S2B) and RRS (Fig. S2C), indicating that FTO physically interacts with multiple members of the AARS family.
Fig. 3.
FTO physically interacts with multiple members of MSC. (A) HEK cells overexpressing vector alone or Flag-WT FTO were cultured for 48 h. Cells were then harvested and cell extracts subjected to Flag immunoprecipitation using an antibody against Flag protein. Flag immunoprecipitates were analyzed by Western blotting using the indicated antibodies. (B) FTO was immunoprecipitated from cell extracts of Fto+/+ and Fto−/− MEFs (grown for 24 h) using an anti-FTO antibody, as described in Materials and Methods. FTO immunoprecipitates were analyzed by Western blotting using the indicated antibodies. (C) Fto+/+ MEF cell extracts were loaded onto 10% to 45% glycerol gradients prepared as described for Fig. 1D. Different fractions were subjected to Western blotting using the indicated antibodies. (D) Part of the glycerol gradient fractions 2 and 3 prepared from the extracts of Fto+/+ and Fto−/− MEFs in C were used for the MRS IP as in B. (E) Extracts derived from Fto+/+ MEFs were subjected to subcellular fractionation using the compartmental protein extraction kit from Millipore per manufacturer protocol.
Considering our observed interaction of FTO with multiple members of AARS, all of which are also the part of MSC, we asked whether FTO was itself present in the MSC. To answer this question, we performed glycerol gradient fractionation by using Fto+/+ MEFs cell lysates and loaded 13 fractions from low to high densities onto an SDS/PAGE gel (Fig. 3C). We found that the bulk of AIMP3, AIMP2, MRS (Fig. 3C), and KRS, and thus components of the MSC, are present in fractions 6 to 9 of glycerol gradients (Fig. 3C). In contrast, FTO migrates with the lower-density fractions (i.e., fractions 1–5) of glycerol gradients and is clearly not a part of the MSC (Fig. 3C). However, MRS (and KRS) is also present in these lower-density fractions with FTO (Fig. 3C), and, crucially, when we perform anti-MRS immunoprecipitations from fractions 2 and 3 of the glycerol gradient obtained from lysates of Fto+/+ and Fto−/− MEFs, we continue to see coimmunoprecipitation of FTO with MRS (Fig. 3D). Thus, FTO appears to interact with the pool of AARS that is free of the MSC.
Previous studies, by using immunofluorescence, showed that FTO is present in the nucleus (12). AARSs, however, are predominantly, but not exclusively, cytoplasmic proteins. To establish whether FTO colocalizes in the same cellular compartment as MRS, we performed subcellular fractionation. We found that, in addition to the previously reported nuclear localization, FTO, along with MRS, is also present in the cytoplasmic fraction derived from Fto+/+ MEFs (Fig. 3E). This cytoplasmic localization of FTO is also found in mouse pancreatic β-cells (Fig. S3A) and hypothalamic N46 cells (Fig. S3B). Although it is formally possible that some nuclear-localized FTO that is weakly bound leaks into the cytosolic fraction during subcellular fractionation, it is also worth noting that a proportion of AARSs have been localized to the nucleus (24–26). Thus, the precise subcellular localization of the interaction between FTO and AARS remains an open question.
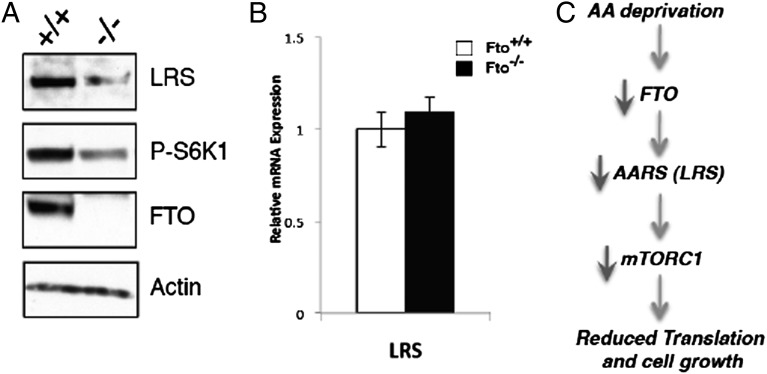
Considering recent reports implicating leucyl-tRNA synthetase (LRS) as a critical link between amino acid availability and activation of mTORC1 signaling (19, 20) and our findings that FTO deficiency leads to altered AARS levels, we measured LRS levels in Fto−/− MEFs. We found that, as with the other AARSs described earlier, lack of FTO also results in reduced amounts of LRS at the protein level (Fig. 4A) but not the mRNA level (Fig. 4B). Our data as a whole, taken in context with the recent data implicating LRS as being a key amino acid sensor upstream of RAG and mTORC1, suggest a model whereby FTO is upstream of AARS (i.e., LRS) in the link between amino acid levels and mTORC1 signaling (Fig. 4C).
Fig. 4.
Levels of LRS, known to link amino acid signaling to mTORC1, are reduced in Fto−/− MEFs. (A) Cell extracts from Fto+/+ and Fto−/− MEFs were analyzed for the mTORC1 signaling pathway as in Fig. 2B using the indicated antibodies. (B) Fto+/+ and Fto−/− MEFs were grown for 24 h, after which mRNAs were extracted and used for quantitative RT-PCR analysis of LRS mRNA levels. (C) Working model for the cellular role of FTO based on our current data.
Discussion
This study shows that cells lacking FTO display decreased activation of the mTORC1 pathway, decreased rates of mRNA translation, and increased autophagy, all of which are likely to contribute to the phenotype of stunted growth seen in humans and mice homozygous for loss-of-function mutations in FTO. The rapid degradation of FTO seen when cells are deprived of amino acids appears to a necessary component of the physiological response to amino acid deprivation, as artificial maintenance of FTO levels results in inappropriately maintained levels of mTORC1 activation in the setting of amino acid deprivation.
It is as yet unclear how the nucleic acid demethylase activity of FTO is related to its role in amino acid signaling. Clearly, a mutant form of FTO lacking demethylase activity is unable to mimic WT FTO in its ability to sustain mTORC1 activity in the setting of amino acid deprivation, so such enzymatic activity appears to be crucial to its signaling function. FTO has been shown to demethylate N6-methyladenine (15), which is predominantly found in mRNA, but can also demethylate 3-methyluridine (12, 14), which is largely found in ribosomal RNA. Two independent studies recently published the human and mouse 6meA modification landscape in a transcriptome-wide manner (27, 28). They report that 6meAs are common, are highly conserved between mice and man, and crucially, are dynamically, developmentally, and tissue-specifically regulated. 6meA at specific mRNA sites appears to perform a fundamental role in regulating gene expression; in particular, CAP structures with 6meA are most efficiently translated, as well as exerting varying effects on mRNA splicing and transport (27, 28). Changing levels of FTO would presumably alter the 6meA landscape, possibly influencing rates of translation. The elucidation of the precise demethylation activities responsible for the physiological effects and the determination of the site of such actions within the cell are priorities for future research.
How do these findings relate to the association between FTO SNPs and obesity? In humans, this association appears to be largely driven by effects on appetite and food intake rather than energy expenditure or nutrient partitioning (3–8). The sensing of amino acid levels in the brain has critical impacts on the activity of orexigenic and anorexigenic pathways controlling energy balance (29), and FTO is most highly expressed in the brain (12). Subtle and tissue-specific effects of intronic SNPs on expression of FTO in key brain centers concerned with the control of energy balance are likely to influence the way in which these cells sense amino acid levels. It is notable that human carriers of the obesity predisposing SNPs in FTO not only consume more calories at test meals but also show an alteration in nutrient preference, suggesting that FTO status can influence the central sensing of dietary macronutrient composition. The role in amino acid sensing we describe here may provide some clues toward understanding the cellular basis for this physiological phenomenon.
Methods
Reagents.
Anti-rabbit FTO antibody was raised against recombinant full-length FTO. Mouse antipuromycin was a gift from the laboratory of Philippe Pierre (Université de la Méditerranée, Marseille, France). Mouse monoclonal anti-FTO antibody was from PhosphoSolutions; rabbit anti-actin, mouse monoclonal anti-MRS, rabbit anti-KRS, rabbit anti-RRS, rabbit anti-LRS, rabbit anti-AIMP3, and rabbit anti-LC3B antibodies were from Abcam; rabbit anti-Flag, rabbit anti-GFP antibodies, and anti-Flag M2 beads magnetic beads were from Sigma; rabbit anti–phospho T-389 S6K-1 antibody was from Cell Signaling; and rabbit anti-AIMP2 antibody was from Proteintech. Bafilomycin, puromycin, and WST-1 (Dojindo's highly water-soluble tetrazolium salt or (2-(4-Iodophenyl)- 3-(4-nitrophenyl)-5-(2,4-disulfophenyl)- 2H-tetrazolium, monosodium salt)) were from Sigma. Protein sepharose A and G and HRP-conjugated secondary antibodies were from Amersham. All reagents not mentioned here were from Sigma.
Cell Culture and Transfections.
MEFs and HEK293 cells were maintained in DMEM supplemented with 10% (wt/vol) FCS. For ectopic expression studies, transfections were performed using CalPhos kit (for HEK293) from Clontech and Neon System (for MEFs) from Invitrogen according to manufacturer protocol.
WST Assay.
For measurement of the metabolic rate, cells were plated at 3,000 cells per well on a 24-well plate and grown. After different time periods as indicated, 250 µL of phenol red-free DMEM was added with 10 µM phenazine methosulfate and 50 µM WST-1, and cells were further kept at 37 °C for 2 h. After 2 h incubation, 100 μL of the media was used for absorbance measurements. Readings were taken on the Oasys Expert plus plate reader in a dual mode with a recording wavelength of 450 nm and a reference wavelength of 620 nm.
Puromycin Labeling.
Protein translation rates were determined by using puromycin labeling as described previously (16).
Immunoprecipitation.
Cells were rinsed twice with ice-cold PBS solution and lysed in 500 µL of ice-cold lysis buffer [50 mM Tris⋅HCl, pH 8.0, 120 mM NaCl, 5 mM EDTA, 10 mM pyrophosphate, 5 mM NaF, 0.1% Nonidet P-40, 0.1% SDS, and one tablet of EDTA-free protease inhibitors (Roche) per 25 mL]. Immunoprecipitation of Flag-hFTO was performed by adding 15 µL of anti-Flag M2 beads magnetic beads per 0.6 to 0.8 mg of protein lysate from cells transfected with Flag-WT FTO construct. Cell extracts with anti-Flag M2 beads magnetic beads were incubated overnight at 4 °C on a rocking platform. Afterward, the immunocomplex was washed three times with the lysis buffer. Endogenous FTO were immunoprecipitated by using 2.0 µg of rabbit anti-FTO antibody and 35 µL of 50% slurry of protein A–Sepharose beads. Endogenous MRS, RRS, and KRS were immunoprecipitated as described in the manufacturer protocol. Immune complexes were washed three times with the lysis buffer and were then resuspended in 1× sample buffer for Western blot analysis.
Glycerol Gradient Fractionation.
Cells were disrupted in ice-cold lysis buffer (50 mM Tris⋅HCl, pH 8.0, 120 mM NaCl, 5 mM EDTA, 10 mM pyrophosphate, 5 mM NaF, 0.1% Nonidet P-40, 0.1% SDS, and one tablet of EDTA-free protease inhibitors (Roche) per 25 mL], cell extracts were clarified at 55,000 × g for 25 min, and 900 μL was layered on a 10% to 45% glycerol gradient and centrifuged in an SW40 rotor for 16 h at 36,000 rpm at 4 °C.
MS Analysis.
Gel bands from SDS/PAGE were excised and subjected to in-gel tryptic digestion, and eluted peptides analyzed by LC/tandem MS by using an OrbiTrap XL coupled to a nanoACQUITY System. Raw files were processed using MaxQuant version 1.0.13.13 Quant module, and the resulting .msm files were searched against SwissProt version 57.1 by using Mascot Daemon 2.3. Species was restricted to human with precursor and fragment ion mass tolerances 2 ppm and 0.5 Da, respectively. Carbamidomethyl was defined as a fixed modification, with deamidation, oxidation, and acetylation (protein N terminus) allowed as potential variable modifications. Mascot search results were exported to Scaffold version 3.0 for validation. Peptide and protein probability were set at 90% and 99%, respectively, and reported proteins required a minimum of two peptides.
Preparation of Tissue Extracts.
Approximately 100 mg of each sample from a panel of tissues were homogenized in 500 μL of RIPA buffer (with protease inhibitors and phosphatase inhibitors) by using the FastPrep-24 System (MP Biomedicals) according to the manufacturer’s instruction. Lysates were collected after sedimenting the debris at 11,000 × g for 30 min at 4 °C and stored at −80 °C until analyses.
RT-PCR.
Total RNA was isolated from Fto+/+ and Fto−/− MEFs by using the RNeasy Minikit (Qiagen) and was treated with DNase-1 (Qiagen) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 1 μg of total RNA by using M-MLV Reverse Transcriptase (Promega) according to the manufacturer’s protocol. Quantitative real-time PCR analysis was performed on a TaqMan ABI Prism 7900 Sequence Detector System (Applied Biosystems). Expression results were analyzed relative to GAPDH and β-actin mRNA content in the same sample.
Supplementary Material
Acknowledgments
We thank P. Pierre for the provision of surface sensing of translation (SUnSET) reagent. This study was supported by UK Medical Research Council (MRC) Programme Grant G9824984, European Union FP7-HEALTH-2009-241592 EurOCHIP and FP7-FOOD-266408 Full4Health, Wellcome Trust Grant 093136, and the MRC Centre for Obesity and Related Metabolic Disorders.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222796110/-/DCSupplemental.
References
- 1.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fawcett KA, Barroso I. The genetics of obesity: FTO leads the way. Trends Genet. 2010;26(6):266–274. doi: 10.1016/j.tig.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CN. An obesity-associated FTO gene variant and increased energy intake in children. N Engl J Med. 2008;359(24):2558–2566. doi: 10.1056/NEJMoa0803839. [DOI] [PubMed] [Google Scholar]
- 4.Haupt A, et al. Variation in the FTO gene influences food intake but not energy expenditure. Exp Clin Endocrinol Diabetes. 2009;117(4):194–197. doi: 10.1055/s-0028-1087176. [DOI] [PubMed] [Google Scholar]
- 5.Jonsson A, Franks PW. Obesity, FTO gene variant, and energy intake in children. N Engl J Med. 2009;360(15):1571–1572. doi: 10.1056/NEJMc090017. [DOI] [PubMed] [Google Scholar]
- 6.Speakman JR, Rance KA, Johnstone AM. Polymorphisms of the FTO gene are associated with variation in energy intake, but not energy expenditure. Obesity (Silver Spring) 2008;16(8):1961–1965. doi: 10.1038/oby.2008.318. [DOI] [PubMed] [Google Scholar]
- 7.Wardle J, et al. Obesity associated genetic variation in FTO is associated with diminished satiety. J Clin Endocrinol Metab. 2008;93(9):3640–3643. doi: 10.1210/jc.2008-0472. [DOI] [PubMed] [Google Scholar]
- 8.Wardle J, Llewellyn C, Sanderson S, Plomin R. The FTO gene and measured food intake in children. Int J Obes (Lond) 2009;33(1):42–45. doi: 10.1038/ijo.2008.174. [DOI] [PubMed] [Google Scholar]
- 9.Church C, et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet. 2010;42(12):1086–1092. doi: 10.1038/ng.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer J, et al. Inactivation of the Fto gene protects from obesity. Nature. 2009;458(7240):894–898. doi: 10.1038/nature07848. [DOI] [PubMed] [Google Scholar]
- 11.Boissel S, et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am J Hum Genet. 2009;85(1):106–111. doi: 10.1016/j.ajhg.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerken T, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tung YC, et al. Hypothalamic-specific manipulation of Fto, the ortholog of the human obesity gene FTO, affects food intake in rats. PLoS ONE. 2010;5(1):e8771. doi: 10.1371/journal.pone.0008771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia G, et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582(23-24):3313–3319. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6(4):275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 17.Quevillon S, Robinson JC, Berthonneau E, Siatecka M, Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: Identification of protein-protein interactions and characterization of a core protein. J Mol Biol. 1999;285(1):183–195. doi: 10.1006/jmbi.1998.2316. [DOI] [PubMed] [Google Scholar]
- 18.Park SG, Ewalt KL, Kim S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem Sci. 2005;30(10):569–574. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Bonfils G, et al. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell. 2012;46(1):105–110. doi: 10.1016/j.molcel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Han JM, et al. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149(2):410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 21.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10(5):307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 22.Rubinsztein DC, et al. In search of an “autophagomometer”. Autophagy. 2009;5(5):585–589. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 23.Cheung MK, Gulati P, O’Rahilly S, Yeo GS. FTO expression is regulated by availability of essential amino acids. Int J Obes (Lond) 2012 doi: 10.1038/ijo.2012.77. [DOI] [PubMed] [Google Scholar]
- 24.Nathanson L, Deutscher MP. Active aminoacyl-tRNA synthetases are present in nuclei as a high molecular weight multienzyme complex. J Biol Chem. 2000;275(41):31559–31562. doi: 10.1074/jbc.C000385200. [DOI] [PubMed] [Google Scholar]
- 25.Popenko VI, et al. Compartmentalization of certain components of the protein synthesis apparatus in mammalian cells. Eur J Cell Biol. 1994;65(1):60–69. [PubMed] [Google Scholar]
- 26.Ko YG, Kang YS, Kim EK, Park SG, Kim S. Nucleolar localization of human methionyl-tRNA synthetase and its role in ribosomal RNA synthesis. J Cell Biol. 2000;149(3):567–574. doi: 10.1083/jcb.149.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 28.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149(7):1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cota D, et al. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312(5775):927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.