Abstract
Dimerization of single-pass membrane receptors is essential for activation. In the human thrombopoietin receptor (TpoR), a unique amphipathic RWQFP motif separates the transmembrane (TM) and intracellular domains. Using a combination of mutagenesis, spectroscopy, and biochemical assays, we show that W515 of this motif impairs dimerization of the upstream TpoR TM helix. TpoR is unusual in that a specific residue is required for this inhibitory function, which prevents receptor self-activation. Mutations as diverse as W515K and W515L cause oncogenic activation of TpoR and lead to human myeloproliferative neoplasms. Two lines of evidence support a general mechanism in which W515 at the intracellular juxtamembrane boundary inhibits dimerization of the TpoR TM helix by increasing the helix tilt angle relative to the membrane bilayer normal, which prevents the formation of stabilizing TM dimer contacts. First, measurements using polarized infrared spectroscopy show that the isolated TM domain of the active W515K mutant has a helix tilt angle closer to the bilayer normal than that of the wild-type receptor. Second, we identify second-site R514W and Q516W mutations that reverse dimerization and tilt angle changes induced by the W515K and W515L mutations. The second-site mutations prevent constitutive activation of TpoR W515K/L, while preserving ligand-induced signaling. The ability of tryptophan to influence the angle and dimerization of the TM helix in wild-type TpoR and in the second-site revertants is likely associated with its strong preference to be buried in the headgroup region of membrane bilayers.
Keywords: myeloproliferation, transmembrane domain, JAK2, NMR spectroscopy, FTIR spectroscopy
The thrombopoietin receptor (TpoR, c-MPL) is a typical single-pass membrane receptor with a large extracellular ligand-binding domain, a single transmembrane (TM) helix, and a cytoplasmic domain that is involved in intracellular signaling (1, 2). In these TM receptors, activation is mediated by ligand-induced dimerization of receptor monomers or structural rearrangement within preformed dimers, which induce changes in the rotational orientation and/or association of the TM and juxtamembrane (JM) regions of the receptor. In many single-pass membrane receptors, receptor activity is regulated through these structural elements (3).
TpoR is unique in that it is expressed in hematopoietic stem cells, where it is essential for maintaining stem cell reservoirs (4–6), and in early myeloid progenitors and megakaryocytes, where it regulates the differentiation and production of platelets (7, 8). In contrast to the TpoR, the erythropoietin receptor (EpoR), a related cytokine receptor, is essentially expressed only in erythroid progenitors, where it regulates red blood cell production.
Tight control of receptor activity is particularly important for the TpoR, because mutations within its TM and JM domains both lead to constitutive activation and myeloproliferative disorders. Within the TM domain, the S505N mutation is associated with familial forms of thrombocytosis (9) and induces stable, active receptor dimers (10, 11). At the intracellular TM–JM boundary, we have described a conserved amphipathic motif (RWQFP) that is specific to the TpoR (12) in which mutations at W515 (W515L/K/A) lead to constitutive receptor activation and are associated with JAK2 V617F-negative myeloproliferative disorders, namely essential thrombocythemia and primary myelofibrosis (13–17).
Biochemical studies and assays for membrane-spanning domain dimerization indicated that the TM domain of TpoR is a dimer and that TpoR can signal from several different helix orientations (18). To establish how the relative rotational orientations of the TM helices influence receptor activation, we have engineered dimers of both the TpoR and EpoR. In the TpoR, we have shown that different in vivo phenotypes are imparted by distinct TM helix orientations (19). We identified one rotational orientation corresponding to an inactive dimeric state, and another orientation corresponding to the activated receptor. Several other orientations induce partial and pathologic signaling. The helix interface identified as inactive using the engineered dimeric coiled coils (19) is identical to the inactive interface identified previously (18). Moreover, the ability of the TpoR to signal through several different dimeric orientations contrasts with the EpoR, where only a single active interface was identified (20).
Here we focus on the role of the JM domain of the TpoR in the regulation of its activity. We address the ability of wild-type TpoR to dimerize in membrane bilayers and discuss how dimerization is controlled by the unique RWQFP motif at the intracellular TM–JM boundary. We describe a unique function for the tryptophan residue within this motif in preventing dimerization of the upstream TM α-helix and productive signaling. We propose a mechanism that explains TpoR activation by mutations at W515, as well as the reversal of constitutive activation by second-site mutations.
Results
Tryptophan Is Absolutely Required at JM Position 515 to Maintain the Unliganded TpoR Inactive.
Several specific mutations have been described in the RWQFP insert that lead to myeloproliferative disorders—namely W515 mutations to leucine, lysine, arginine, alanine, and serine (12–17, 21). The diversity of amino acids of both hydrophobic and hydrophilic character that are able to activate the TpoR suggests that Trp515 is unusual and is regulatory as a result of its size and/or aromatic character. We compared the influence of mutations at position 515 that have been identified in patients with myeloproliferative neoplasms (MPNs) and then the influence of additional mutations at adjacent positions to test whether Trp515 interacts in concert with Arg514 and Gln516 to stabilize the TpoR in the inactive state.
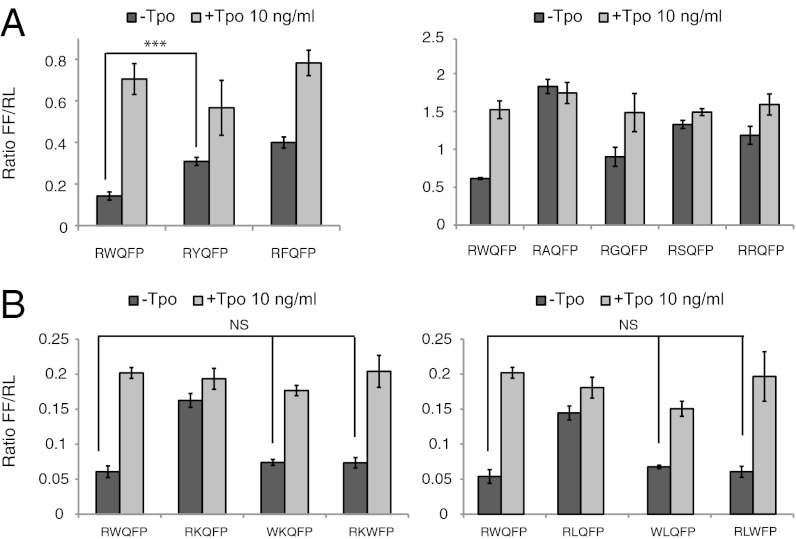
In the murine (KWQFP) and human (RWQFP) sequences, the first and second residues are conserved as a positively charged amino acid and tryptophan, respectively, suggesting a cation–π interaction (Fig. S1). To address whether an aromatic residue is required at the second position for function, W515 was mutated to phenylalanine or tyrosine. As shown in Fig. 1A (Right), both tyrosine and phenylalanine lead to constitutive TpoR activation, indicating that aromatic character alone is not sufficient to prevent self-activation. The activity of W515Y and W515F TpoR is significantly higher than wild-type TpoR, although lower than mutations identified in MPN patients (K, L, A, R, or S; Fig. 1 A and B). The results roughly correlate with the interfacial hydrophobicity scale (22) in which tryptophan has the highest free energy among the amino acids to partition into the headgroup region of the membrane bilayer.
Fig. 1.
Mutation of the TpoR W515 residue induces constitutive signaling via JAK2 and STAT5. (A) The indicated TpoR W515 mutants, where the W515 residue was replaced by aromatic residues (Left) or other residues (Right) were expressed in γ-2A cells cotransfected with JAK2 and tested for ligand-dependent and -independent induction of STAT5-dependent transcriptional activity. Two reporters are used in the dual luciferase assay: the experimental firefly (FF) luciferase reporter, which responds to JAK/STAT pathway, and a cotransfected renilla (RL) luciferase control reporter driven by a constitutive promoter that provides a baseline signal proportional to transfection levels. The firefly/renilla (FF/RL) ratio normalizes the FF response for experimental variability due to cell viability and transfection efficiency. (B) Constitutive activation of TpoR W515 mutants is reverted by substituting the preceding (R514) or subsequent (Q516) residue by tryptophan. Replacing residue 514 or 516 with a tryptophan residue (R514W or Q516W) reverts TpoR W515K (Left) and TpoR W515L (Right) to a wild-type TpoR phenotype. Shown in A and B are averages of three replicates ± SD in one representative experiment out of at least three. NS, nonsignificant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001, Student t test.
We asked whether residues in the vicinity of W515 impact its inhibitory function. Substitution to lysine of residues 516, 517, and 518 did not induce activation (Fig. S2). Similarly, mutating R514 to A, W, or F did not induce activation. However, all these mutants responded to Tpo addition like the wild-type TpoR.
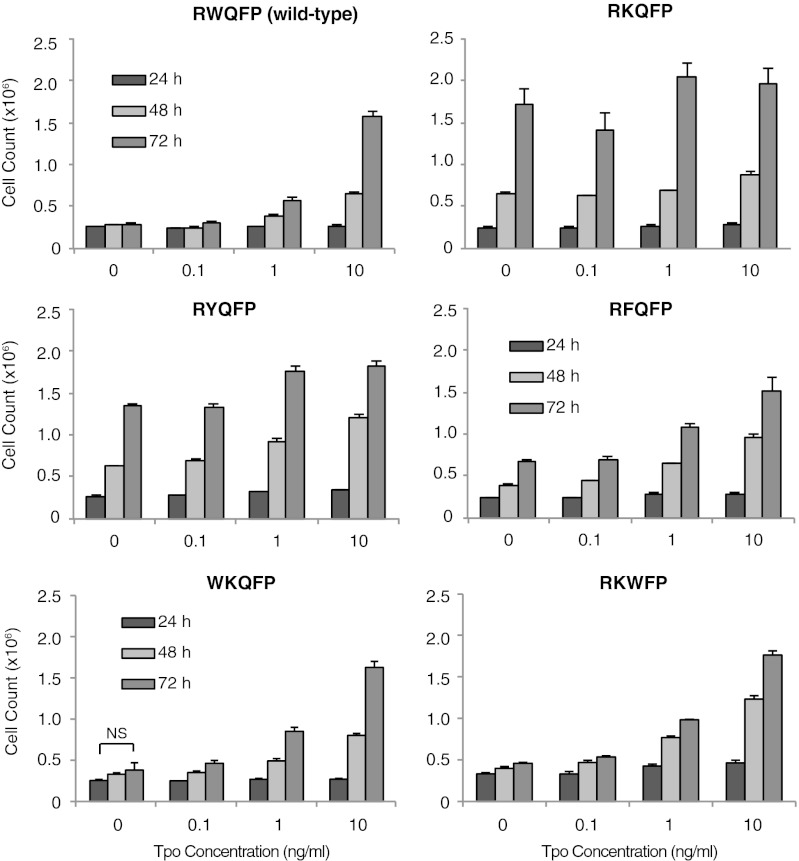
The data obtained using transient expression in γ-2A cells were confirmed in stably transduced IL-3–dependent Ba/F3 cells (Fig. 2). All mutants of TpoR that exhibited constitutive activation in luciferase assays in γ-2A cells were able to induce IL-3–independent proliferation of Ba/F3 cells after expression at levels similar to that of the wild-type TpoR, which was only activated in the presence of Tpo. Equal levels of expression were confirmed on the basis of equivalent GFP levels by flow cytometry cell sorting (23) and HA-TpoR expression by Western blotting.
Fig. 2.
Dose-dependent effects of Tpo on proliferation of Ba/F3 cells stably expressing TpoR mutants. Constitutive activation of TpoR W515 mutants is reverted by substituting the precedent (514) or subsequent (516) residue by tryptophan. The R514W and Q516W mutations revert the transforming phenotype of TpoR W515K to a wild-type phenotype in Ba/F3 cells stably expressing TpoR mutants. Shown are average of triplicate cell counts at the indicated times ± SD from a representative experiment out of three. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001, Student t test.
In addition, we tested STAT-dependent transcriptional signaling of W515 mutants using a luciferase reporter in the JAK2-deficient γ-2A cells (24), in which TYK2 and not JAK2 was coexpressed with the TpoR and luciferase reporters. TpoR W515 mutants did not exhibit constitutive activity in the absence of JAK2 but responded to Tpo like wild-type TpoR. These data indicate that W515 mutants constitutively activate signaling of TpoR via ligand-independent activation of JAK2 (Figs. S3 and S4).
Reversal of Constitutive Activity of TpoR W515 Mutants by Substituting Surrounding Residues to Tryptophan.
To further test the hypothesis that tryptophan plays a unique role at the intracellular TM–JM boundary of the TpoR, we engineered R514W or Q516W mutations into the active TpoR W515K and W515L mutants. The idea was that in the context of the active TpoR W515 mutants, a tryptophan residue before or after residue 515 might reverse activation. As shown in Fig. 1B, R514W suppressed the constitutive STAT5 activation of TpoR W515K and W515L mutants in γ-2A cells. The doubly mutated receptors behaved as wild-type receptors, in that they responded normally to Tpo. Similarly, the Q516W reversed activation of TpoR W515K and W515L mutants, conferring a wild-type phenotype, with no activation in the absence of Tpo and normal response to Tpo. These data were reproduced upon stable retroviral expression in Ba/F3 cells. Ligand-independent proliferation of Ba/F3 cells induced by the TpoR W515K mutant was prevented by the R514W and Q516W mutations, whereas Tpo-induced proliferation remained intact (Fig. 2). As a control, individual mutations at the rescuing positions, or the WWQFP mutant did not induce constitutive activation and retained response to the Tpo ligand. Overall these data indicate that placing tryptophan in the vicinity of the W515 mutations prevents receptor self-activation and renders a wild-type phenotype to the TpoR W515 mutant.
Impact of W515 on Dimerization of Full-Length TpoR.
To assess the oligomerization status of the TpoR and selected mutants in cell membranes, we used the Gaussia princeps luciferase complementation assay (25). This protein fragment complementation assay can be used at levels of expression that are close or similar to those of normal cells (25). Two fragments of Gaussia princeps luciferase, Gluc1 and Gluc2, were fused in-frame to the carboxyl terminus of the TpoR after introduction of a short flexible linker and subcloned in pcDNA3.
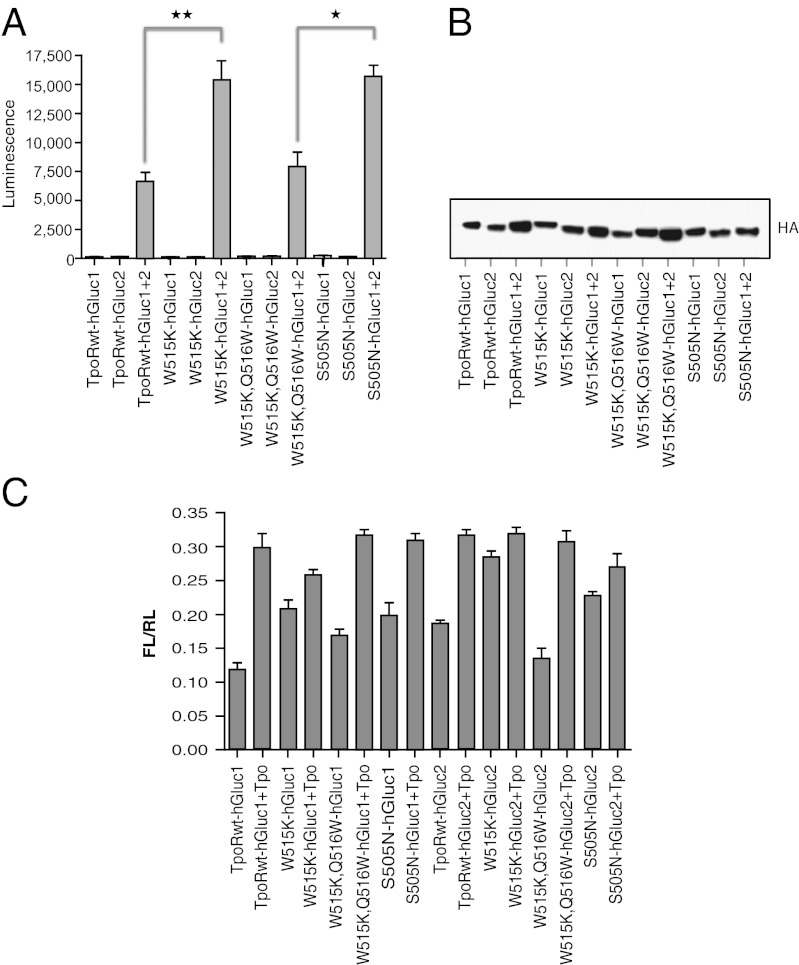
Fig. 3A shows that when the wild-type TpoR-Gluc1 or -Gluc2 constructs were individually transfected in HEK293 cells, no reconstituted luciferase signal could be detected. When the wild-type TpoR-Gluc1 and -Gluc2 constructs were cotransfected, a signal was obtained that reflects a basal level of receptor dimerization. This level is significantly weaker than that of EpoR fused to the same Gluc1 and Gluc2 fragments (∼2.5- to 3-fold weaker; Fig. S5), consistent with weaker levels of dimerization previously reported for the TM domain of TpoR (18), compared with EpoR (26–29). We then tested the activating mutations found in the TM (S505N) and JM (W515K) regions of the TpoR. As shown in Fig. 3A, both mutations increased the luciferase signal to statistically significant levels (P < 0.01 and P < 0.05 for the S505N and W515K mutants, respectively). Furthermore, when the double mutant W515K, Q516W was tested, the luciferase signal was not statistically different from that of wild-type TpoR. Taken together these data indicate that the W515K mutation, and as expected the S505N mutation, impact the dimerization status of the full-length receptor. Levels of expression of TpoR mutants are shown in Fig. 3B. Fusion of TpoR with the Gluc1 or Gluc2 did not change signaling in the presence of Tpo or activation by the W515K or S505N mutations (Fig. 3C).
Fig. 3.
Detection of TpoR dimers by split-luciferase assay. (A) Luciferase assay of HEK293 cell lysates expressing hGluc-tagged TpoR and its mutants (W515K, W515KQ516W, S505N). The average luminescence of triplicate values ± SD is shown for one representative experiment out of at least three. (B) Immunoblotting of transfected HEK293 cell lysates by anti-HA antibodies. (C) STAT5 transcriptional assays in γ-2A cells transfected with cDNAs coding for hGluc-tagged TpoR, JAK2, and its mutants. *P < 0.01; **P < 0.001, one-way ANOVA.
Constitutive TpoR Activation Is Associated with TM Helix Dimerization and Modulated by the RWQFP Motif.
To address whether dimerization is directly modulated by the RWQFP motif, measurements of dimerization were performed in lipid membrane environments using deuterium magic angle spinning (MAS) NMR spectroscopy and in detergent environments using analytical ultracentrifugation. Deuterium MAS NMR spectroscopy provides a simple approach to compare membrane dimerization of TM peptides (30). The intensities of the side bands in a deuterium MAS spectrum map out the deuterium line shape and are sensitive to molecular motion. Methyl-deuterated leucines facing the lipid have greater rotational freedom and exhibit weaker side band intensities than leucine side chains facing the dimer interface.
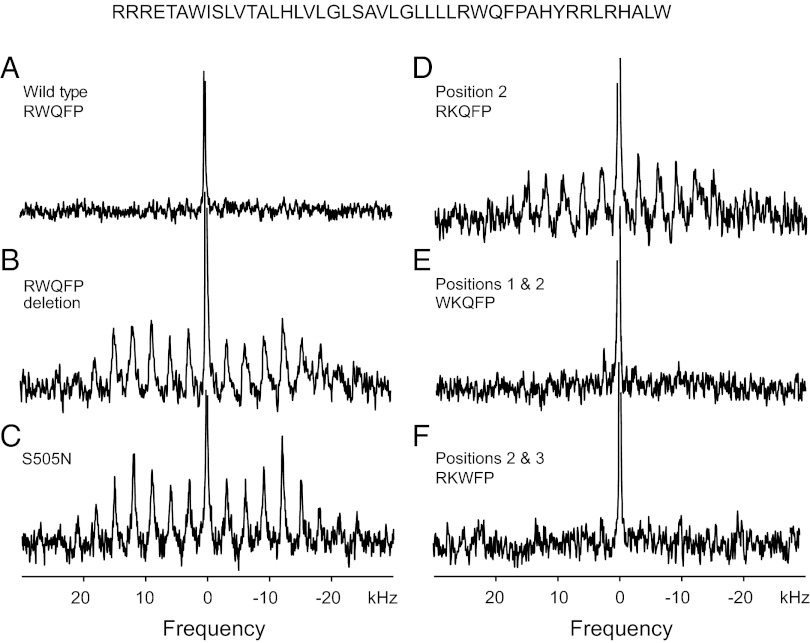
Fig. 4A shows the deuterium MAS spectrum of the wild-type TpoR TM peptide reconstituted into 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1,2-dimyristoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (DMPG) bilayers. The spectrum lacks the side band pattern characteristic of constrained deuterated methyl groups. The sharp resonance at zero frequency is from deuterated water in the sample. We interpret the lack of MAS sidebands as resulting from rotational motion of the peptide on the time scale of MAS. Lowering the temperature restricts rotational diffusion of the peptide and results in a spectrum with a distinctive stronger pattern of rotational side bands (Fig. S6).
Fig. 4.
Deuterium MAS NMR spectra of the TpoR TM domain. Deuterium MAS NMR spectra were obtained of the wild-type Tpo receptor TM peptide (residues 488–529) and of peptides with mutations at S505 and in RWQFP insert: (A) wild-type, (B) RWQFP deletion mutant, (C) S505N, (D) W515K, (E) R514W, W515K, and (F) W515K, Q516W double mutation. The peptides were deuterium labeled at Leu512 and reconstituted into DMPC:DMPG bilayers at a peptide/lipid molar ratio of 1:50. Each experiment was repeated using separate reconstitutions.
Fig. 4B presents the deuterium NMR spectrum of the TpoR TM domain lacking the five-residue RWQFP motif. The deletion was previously shown to be activating (12) and is similar to the sequence tested by Engelman and coworkers (18) in which the RWQFP was partially deleted. The deletion of the insert restores the pattern of rotational side bands spaced at the MAS frequency of 3 kHz. The width of the side band envelope is characteristic of a TM domain dimer. Fig. 4 C and D show deuterium MAS spectra of the TpoR TM peptide with the S505N and W515K substitutions. Both mutations are activating. The spectra of both the S505N and W515K mutants exhibit the same pattern of rotational side bands as seen in Fig. 4B, indicating that the mutation stabilizes the TM dimer of TpoR.
To test whether inactivation of the TpoR W515K mutant by insertion of tryptophan at positions 514 or 516 influences dimerization, deuterium NMR spectra were obtained of TM peptides containing the R514W, W515K (Fig. 4E), and W515K, Q516W (Fig. 4F) double mutations. In both of the double mutants, the intensity in the rotational side bands is lost, consistent with greater mobility of L512 due to a shift to monomeric TpoR.
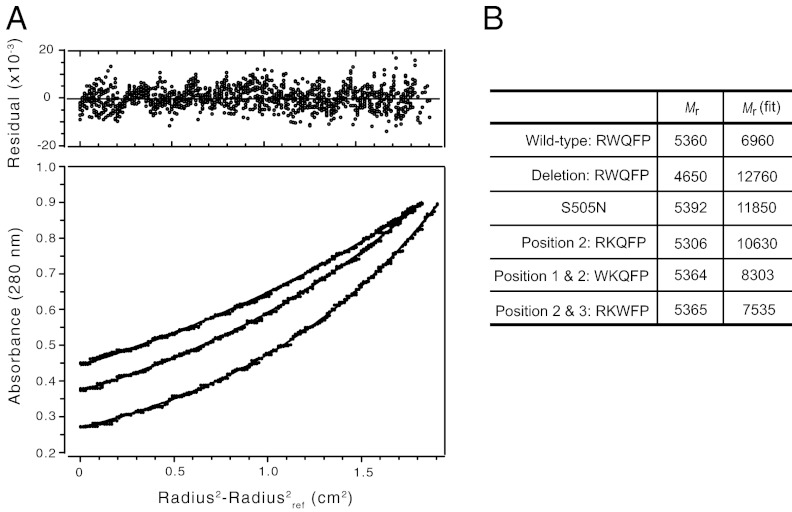
Sedimentation equilibrium analytical ultracentrifugation was also performed to compare the ability of the TM peptides to dimerize in detergent micelles. The wild-type and mutant TpoR TM peptides were reconstituted into DPC micelles at three concentrations (∼20 μM, 35 μM, and 50 μM), and data were acquired at three centrifugation speeds (98,784, 129,024 and 185,785 × g). Fig. 5A presents the sedimentation equilibrium measurements of the wild-type TpoR TM peptide at peptide concentration of 35 μM. The data were analyzed using a single component analysis. Fig. 5B presents the Mr of the monomeric peptide and the single component fit to data from all concentrations and rotor speeds. The results roughly match those obtained by deuterium MAS NMR of the TM peptides reconstituted into membrane bilayers. The wild-type TpoR peptide exhibits a Mr close to that of the monomer, whereas the S505N and W515K mutants sediment at an Mr close to that of a dimer. The apparent dimerization of the W515K mutant is reversed in the W515K, Q516W and R514W, W515K double mutants.
Fig. 5.
Analytical ultracentrifugation of the TpoR TM domain. (A) Sedimentation equilibrium experiments of the wild-type TpoR TM peptide in DPC micelles were analyzed using nonlinear least-squares global curve-fitting. Representative data are shown at rotor speeds of 98,784, 129,024, and 185,785 × g using a peptide concentration of 35 μM. Data were collected at 25 °C after 20 h of centrifugation. (Upper) Distribution of residuals. (Lower) Open circles for experimental data and solid lines for a fit to a single component model. (B) Monomer Mrs and Mrs derived from single component fits are shown for wild-type TpoR and its mutants. The experimental Mrs represent the global fit to three concentrations and three rotor speeds.
Dimerization of the TpoR TM Domain Is Regulated by the TM Helix Tilt Angle.
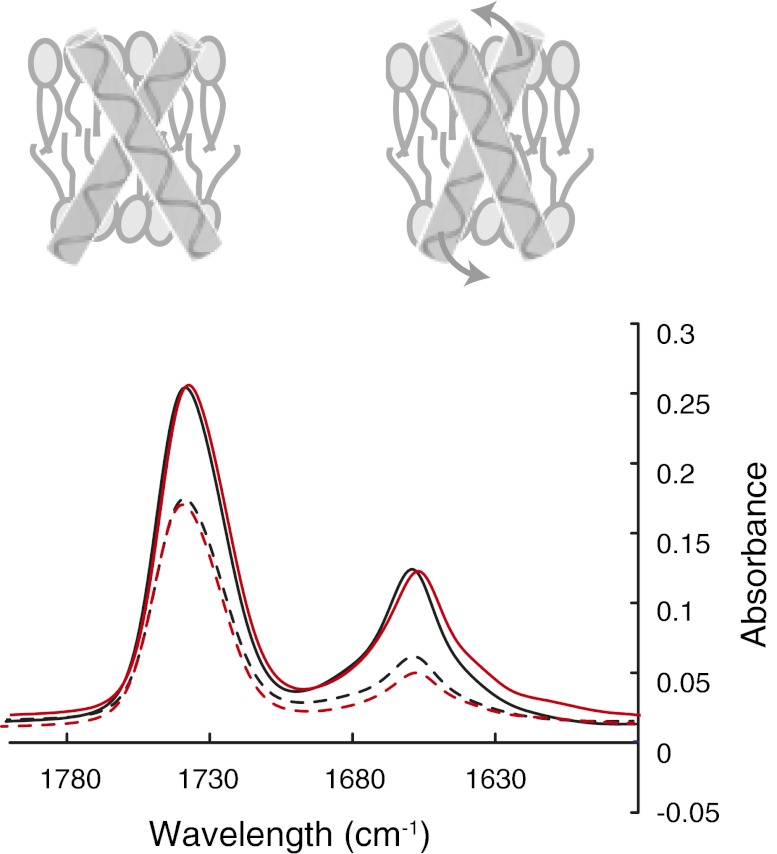
Polarized FTIR spectroscopy of TM peptides was used to assess the extent of helical secondary structure and the angle of the TM helix relative to the bilayer normal. For the TM domain of the TpoR reconstituted in DMPC bilayers, we typically observe an amide I vibration at 1,657 cm−1 (Fig. 6A). Fourier deconvolution of the amide I band reveals no significant nonhelical structure. The dichroic ratio of the amide I band is sensitive to the angle of the TM helix relative to the plane of membrane. For the wild-type TpoR TM domain, we observe a dichroic ratio of 2.7 ± 1, which corresponds to a helix angle of ∼30° relative to the membrane normal. The W515K mutation results in an increase in the dichroic ratio to 3.0 ± 1 and a helix angle (∼23°) closer to the bilayer normal (Fig. 6B). In contrast, both the WKQFP and RKWFP sequences lead to lower dichroic ratios (∼2.5), consistent with an increase in the helix tilt angle (∼38°). We interpret the results from analytical ultracentrifugation, deuterium MAS NMR and IR spectroscopy in terms of a model in which the RWQFP sequence also controls the tilt of the helix in the bilayer, which in turn controls the ability of the helix to dimerize.
Fig. 6.
Polarized FTIR spectra of wild-type and W515K TpoR TM peptides. The wild-type (black) and W515K (red) TpoR spectra were obtained with light polarized parallel (solid line) or perpendicular (dashed line) to the membrane normal. The cartoon illustrates the proposed change in helix tilt angle upon W515K mutation. The IR band at 1,745 cm−1 corresponds to the lipid carbonyl vibration, whereas the band at ∼1,657 cm−1 corresponds to the protein amide I vibration. The dichroic ratios of the amide I band correspond to the average of three separate reconstitutions.
Discussion
Our main finding is that W515 in the RWQFP sequence impairs TpoR TM helix dimerization, and by doing so prevents constitutive receptor activation. Mutations (W515K or S505N) that induce TM helix dimerization activate signaling. Second-site mutations (R514W or Q516W), which reverse dimerization of the TM α-helix, prevent signaling. The RWQFP motif seems to regulate a receptor that is primed for activation. NMR and analytical ultracentrifugation measurements of the TpoR TM domain lacking the RWQFP motif demonstrate that the TM sequence alone has an inherent propensity to dimerize and that inclusion of the motif inhibits dimerization.
These observations seem to be associated with several unique functional features of TpoR. First, the receptor is expressed at several distinct stages of hematopoiesis, from stem cells to mature platelets. Second, TpoR functions differently in different cellular contexts, from regulating hematopoietic stem cell quiescence (4–6) to controlling megakaryopoiesis (7, 8), both promoting and inhibiting megakaryocyte proliferation as a function of cellular JAK2 levels (31). Our data indicate that W515 maintains the receptor in the inactive conformation and imparts specific signaling upon ligand activation. In this regard, in our engineered coiled-coil receptor dimers, mutation of W515 to lysine removes the inhibition for the one inactive orientation and abolishes differences in biologic effects induced by several other TpoR dimers (19).
Regulation of dimerization is a critical aspect of all single-pass TM receptors. The isolated TM domains for both the EpoR (26, 27) and TpoR (18) have previously been shown to have a propensity to dimerize, although to different extents; EpoR TM dimerization seems to be comparable to that of the TM domain of glycophorin A (26), whereas TpoR TM dimerization seems to be weaker than glycophorin A (18). Our experiments using the Gaussia princeps luciferase assays show that the dimerization of full-length TpoR is weaker than that of EpoR, which is known to be a preformed dimer (32, 33). Our data indicate that mutations that promote TM α-helix dimerization and that induce constitutive activation also impact full-length receptor dimerization.
Our working model for how the RWQFP motif regulates dimerization is that it extends the hydrophobic length of the TM helix, which tilts as the tryptophan side chain partitions into the bilayer. R514 may partition into the membrane owing to favorable cation–π interactions with W515. Solution NMR studies show that the RW region is helical and the TM helix unravels at Phe517-Pro518 (Fig. S7). The increase in the tilt angle reduces the surface contact area between TM helices and disfavors dimerization.
Tryptophan plays a unique role in this mechanism. Tryptophan exhibits a strong preference for the ends of TM helices in single-pass membrane proteins (34) and has the largest free energy for partitioning into the headgroup region of membrane bilayers (22). Model peptide studies have shown that the position of tryptophan and lysine at the ends of TM helices can influence the mismatch between the hydrophobic length of the peptide and the bilayer thickness, which in turn can alter the helix tilt angle (35). In contrast, tryptophan at central positions in single-pass membrane proteins may actually mediate helix association (36).
Ligand binding to the extracellular domain (or the activating S505N mutation) can overcome the favorable membrane interactions of W515 and drive helix dimerization by changing the tilt angle and orientation of the TM helix. The activating W515 mutants remove the stabilizing cation–π interaction and leave the positively charged R514 side chain, as well as the polar Q516, in an unfavorable hydrophobic environment. Arginine and glutamine have unfavorable transfer free energies (22). As a result, the tilt angle of the helix decreases in the W515 mutants as R514 moves out of the membrane. The second site mutations adjacent to W515K and W515L mutations reintroduce a tryptophan at the JM boundary.
On the basis of sequence database searches, the conserved RWQFP sequence seems to be unique at the intracellular end of single-pass membrane receptors. The balance of hydrophilic and hydrophobic amino acids within the motif likely contributes to its ability to partition between membrane and cytosolic environments upon ligand binding. However, a second possible function of the motif may be to guide dimerization of the ligand-activated receptor. Q516 is predicted to be in the interface of the active receptor dimer (18, 19) and has the potential to mediate helix interactions. Thus, this simple five amino acid insert in the TpoR may both hold the TM helix in an inactive conformation in the unliganded receptor and contribute to the rotational dependence of activation in the ligand-bound receptor.
Methods
cDNA Constructs and Cells.
The wild-type and mutant human TpoR cDNAs cloned in bicistronic pMX-IRES-GFP contained an HA tag at the amino terminus (2). Human JAK2 and TYK2 cDNAs were cloned in pMX-IRES-CD4 (2). Site-directed mutagenesis was performed with the Stratagene Quick mutagenesis kit or overlapping PCR. All constructs were verified by Sanger sequencing. Ba/F3 cells are IL-3–dependent proB mouse cells that were maintained in RPMI supplemented with 10% (vol/vol) FBS and Walter and Eliza Hall Institute (WEHI) cell supernatant (19) as a source of IL-3. Gamma-2A cells are human fibrosarcoma cells deficient in JAK2 (24).
Retroviral Transduction and Generation of Cell Lines Stably Expressing TpoR Mutants.
Ecotropic retroviral supernatants were generated in BOSC cells, and retroviral transduction was performed on Ba/F3 cells (19). Flow cytometry cell sorting was used to isolate pools of cells expressing equivalent levels of GFP (15). Total and cell surface TpoR levels in transduced cells were assessed by Western blotting and flow cytometry with anti-HA antibodies, respectively (15).
Dual Luciferase Transcriptional Assays.
Transcriptional activation of STAT5 and STAT3 was analyzed in γ-2A, Ba/F3, and HEK293 cells expressing TpoR or TpoR mutants, as previously described (19), using the STAT5/STAT3 pGRR5 luciferase reporter and pRL-TK driven renilla luciferase. Gamma-2A cells or Ba/F3 cells were transiently transfected using Lipofectamine and electroporation, respectively (2). A Perkin-Elmer Victor X Light analyzer was used for detecting luminescence in cell lysates.
Synthesis and Reconstitution of TpoR TM Helix Peptides.
2H-Labeled amino acids were purchased from Cambridge Isotope Laboratories. Lipids were obtained from Avanti Polar Lipids, as a lyophilized powder and used without further purification. Peptides (45 residues; Fig. 4) corresponding to the TM domain of TpoR were synthesized using solid-phase methods (Keck Facility, Yale University, New Haven, CT). TpoR peptides were purified as previously described for EpoR (37) and reconstituted by detergent dialysis using a 10:3 DMPC/DMPG molar ratio and a ∼1:60 peptide/lipid molar ratio. FTIR measurements were made after dialysis. For NMR measurements, the membrane reconstituted peptides were pelleted, lyophilized, rehydrated with deuterium-depleted water (50% ± 5% weight), and incubated at 37 °C for 24 h.
Polarized IR Spectroscopy.
Polarized attenuated total reflection FTIR spectra were obtained on a Bruker IFS 66V/S spectrometer. Membranes containing TpoR peptides (∼150 μg) were layered on a germanium internal reflection element using a slow flow of nitrogen gas directed at an oblique angle to the IR plate to form an oriented multilamellar lipid–peptide film. One thousand scans were acquired and averaged for each sample at a resolution of 4 cm−1.
Analytical Ultracentrifugation.
Sedimentation equilibrium experiments were performed using a Beckman XL-I analytical ultracentrifuge at 25 °C. The solution [15 mM dodecylphosphocholine (DPC), 50 mM Tris·HCl, and 0.1 M NaCl (pH 7.5)] was density matched to account for the buoyancy of DPC micelles by adding 52.5% D2O (38). Absorbance was measured radially across the cell. Data points were collected in radial increments of 0.001 cm, with each data point representing the average of 10 replicates. Three different peptide concentrations (∼20 μM, 35 μM, and 50 μM) and three different speeds (98,784, 129,024, and 185,785 × g) were used to ensure the quality of the data. All of the data were analyzed by nonlinear least-squares global curve fitting using the UltraScan II version 9.9 data analysis software. The analysis uses a solvent density ρ of 1.0580 and the partial specific volume for the TpoR peptide of 0.7645 cm3/g.
Deuterium MAS NMR Spectroscopy.
Deuterium NMR spectra were obtained at a 2H frequency of 55.2 MHz on a Bruker Avance NMR spectrometer using a 4-mm MAS probe and a MAS frequency of 3 kHz. Single pulse excitation was used using a 4- to 7.5-μs 90° pulse, followed by a 4.5-μs delay before data acquisition. The repetition delay was 0.25 s. A total of 600,000–1,500,000 transients were averaged for each spectrum and processed using a 100-Hz exponential line broadening function. Spectra were obtained at 25 °C.
Protein-Fragment Complementation Assay.
The cDNA of human wild-type TpoR was inserted between Notl and Clal restriction sites of pcDNA3.1/Zeo vector upstream of either the hGluc fragment coding amino acids 1–93 (hGluc1) or the fragment coding amino acids 94–169 (hGluc2) of Gaussia princeps luciferase (25). Mutants were obtained by QuikChange Site-Directed Mutagenesis (Stratagene). Statistics were calculated using one-way ANOVA using GraphPad. Plasmids for the split luciferase assay coding TpoR and mutants were cotransfected in a ratio of 1:1 (2 μg total DNA per well) in HEK293 cells using Lipofectamine (Invitrogen). Forty-eight hours after transfection, cells were trypsinized, resuspended in 500 μL of cold PBS 1×, pelleted by centrifugation at 4 °C for 1 min, and each pellet was resuspended in 400 μL of DMEM/F-12 (Gibco) containing protease inhibitors. Cell samples were flash-frozen in dry ice for 10 min and thawed in a water bath at 37 °C for 10 min (cycle was repeated twice). After centrifugation (10,000 × g, 5 min) the supernatants were collected and aliquoted (100 μL per well) in 96-well white plates (Nunc) to measure the luciferase activity. Native coelenterazine (NanoLight Technology) was reconstituted as a stock solution of 1 mg/mL in methanol and diluted to a final concentration 20 μM in DMEM F12 at room temperature for injection (100 μL). Signal intensities (integrated over 10 s, with an injection delay of 2 s) were read on a Perkin-Elmer Victor X Light analyzer. Aliquots of the supernatants were used also for Western blot determination of TpoR expression levels.
Supplementary Material
Acknowledgments
We thank Stephen Michnick (University of Montreal) for the Gaussia princeps system and Céline Mouton and Julie Klein for technical and graphics support. Financial support was received from National Institutes of Health Grant GM-46732 (to S.O.S.); a Fonds pour la formation à la Recherche dans l'Industrie et dans l'Agriculture (FRIA) doctoral fellowship (to J.-P.D.); a Delori postdoctoral fellowship (to V.G.); the Fonds de la Recherche Scientifique-Fonds National de la Recherche Scientifique (FRS-FNRS) and Fonds de la Recherche Scientifique Médicale (FRSM) (S.N.C.), Belgium; the Salus Sanguinis Foundation; the Action de Recherche Concertée (ARC) MEXP31C1 and ARC10/15-027 program of the Université catholique de Louvain; the Fondation contre le Cancer; the Belgium Interuniversity Attraction Pole (IAP) Program (S.N.C.); and de Duve Institute and FNRS fellowships (to C.P.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211560110/-/DCSupplemental.
References
- 1.Drachman JG, Griffin JD, Kaushansky K. The c-Mpl ligand (thrombopoietin) stimulates tyrosine phosphorylation of Jak2, Shc, and c-Mpl. J Biol Chem. 1995;270(10):4979–4982. doi: 10.1074/jbc.270.10.4979. [DOI] [PubMed] [Google Scholar]
- 2.Royer Y, Staerk J, Costuleanu M, Courtoy PJ, Constantinescu SN. Janus kinases affect thrombopoietin receptor cell surface localization and stability. J Biol Chem. 2005;280(29):27251–27261. doi: 10.1074/jbc.M501376200. [DOI] [PubMed] [Google Scholar]
- 3.Hubbard SR. Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat Rev Mol Cell Biol. 2004;5(6):464–471. doi: 10.1038/nrm1399. [DOI] [PubMed] [Google Scholar]
- 4.Solar GP, et al. Role of c-mpl in early hematopoiesis. Blood. 1998;92(1):4–10. [PubMed] [Google Scholar]
- 5.Yoshihara H, et al. Thrombopoietin/MPL signaling regulates hematopoietic stem cell quiescence and interaction with the osteoblastic niche. Cell Stem Cell. 2007;1(6):685–697. doi: 10.1016/j.stem.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 6.Qian H, et al. Critical role of thrombopoietin in maintaining adult quiescent hematopoietic stem cells. Cell Stem Cell. 2007;1(6):671–684. doi: 10.1016/j.stem.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 7.de Sauvage FJ, et al. Physiological regulation of early and late stages of megakaryocytopoiesis by thrombopoietin. J Exp Med. 1996;183(2):651–656. doi: 10.1084/jem.183.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaushansky K. Thrombopoietin: Understanding and manipulating platelet production. Annu Rev Med. 1997;48:1–11. doi: 10.1146/annurev.med.48.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Ding J, et al. Familial essential thrombocythemia associated with a dominant-positive activating mutation of the c-MPL gene, which encodes for the receptor for thrombopoietin. Blood. 2004;103(11):4198–4200. doi: 10.1182/blood-2003-10-3471. [DOI] [PubMed] [Google Scholar]
- 10.Onishi M, et al. Identification of an oncogenic form of the thrombopoietin receptor MPL using retrovirus-mediated gene transfer. Blood. 1996;88(4):1399–1406. [PubMed] [Google Scholar]
- 11.Ding J, et al. The Asn505 mutation of the c-MPL gene, which causes familial essential thrombocythemia, induces autonomous homodimerization of the c-Mpl protein due to strong amino acid polarity. Blood. 2009;114(15):3325–3328. doi: 10.1182/blood-2008-04-149047. [DOI] [PubMed] [Google Scholar]
- 12.Staerk J, et al. An amphipathic motif at the transmembrane-cytoplasmic junction prevents autonomous activation of the thrombopoietin receptor. Blood. 2006;107(5):1864–1871. doi: 10.1182/blood-2005-06-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pikman Y, et al. MPLW515L is anovel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006;3(7):1140–1151. doi: 10.1371/journal.pmed.0030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pardanani AD, et al. MPL515 mutations in myeloproliferative and other myeloid disorders: A study of 1182 patients. Blood. 2006;108(10):3472–3476. doi: 10.1182/blood-2006-04-018879. [DOI] [PubMed] [Google Scholar]
- 15.Pecquet C, et al. Induction of myeloproliferative disorder and myelofibrosis by thrombopoietin receptor W515 mutants is mediated by cytosolic tyrosine 112 of the receptor. Blood. 2010;115(5):1037–1048. doi: 10.1182/blood-2008-10-183558. [DOI] [PubMed] [Google Scholar]
- 16.Boyd EM, et al. Clinical utility of routine MPL exon 10 analysis in the diagnosis of essential thrombocythaemia and primary myelofibrosis. Br J Haematol. 2010;149(2):250–257. doi: 10.1111/j.1365-2141.2010.08083.x. [DOI] [PubMed] [Google Scholar]
- 17.Beer PA, et al. MPL mutations in myeloproliferative disorders: Analysis of the PT-1 cohort. Blood. 2008;112(1):141–149. doi: 10.1182/blood-2008-01-131664. [DOI] [PubMed] [Google Scholar]
- 18.Matthews EE, et al. Thrombopoietin receptor activation: Transmembrane helix dimerization, rotation, and allosteric modulation. FASEB J. 2011;25(7):2234–2244. doi: 10.1096/fj.10-178673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staerk J, et al. Orientation-specific signalling by thrombopoietin receptor dimers. EMBO J. 2011;30(21):4398–4413. doi: 10.1038/emboj.2011.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seubert N, et al. Active and inactive orientations of the transmembrane and cytosolic domains of the erythropoietin receptor dimer. Mol Cell. 2003;12(5):1239–1250. doi: 10.1016/s1097-2765(03)00389-7. [DOI] [PubMed] [Google Scholar]
- 21.Abe M, Suzuki K, Inagaki O, Sassa S, Shikama H. A novel MPL point mutation resulting in thrombopoietin-independent activation. Leukemia. 2002;16(8):1500–1506. doi: 10.1038/sj.leu.2402554. [DOI] [PubMed] [Google Scholar]
- 22.Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;3(10):842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 23.Koffie RM, et al. Oligomeric amyloid beta associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc Natl Acad Sci USA. 2009;106(10):4012–4017. doi: 10.1073/pnas.0811698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohlhuber F, et al. A JAK1/JAK2 chimera can sustain alpha and gamma interferon responses. Mol Cell Biol. 1997;17(2):695–706. doi: 10.1128/mcb.17.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Remy I, Michnick SW. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat Methods. 2006;3(12):977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- 26.Gurezka R, Laage R, Brosig B, Langosch D. A heptad motif of leucine residues found in membrane proteins can drive self-assembly of artificial transmembrane segments. J Biol Chem. 1999;274(14):9265–9270. doi: 10.1074/jbc.274.14.9265. [DOI] [PubMed] [Google Scholar]
- 27.Ebie AZ, Fleming KG. Dimerization of the erythropoietin receptor transmembrane domain in micelles. J Mol Biol. 2007;366(2):517–524. doi: 10.1016/j.jmb.2006.11.035. [DOI] [PubMed] [Google Scholar]
- 28.Ruan WM, Becker V, Klingmüller U, Langosch D. The interface between self-assembling erythropoietin receptor transmembrane segments corresponds to a membrane-spanning leucine zipper. J Biol Chem. 2004;279(5):3273–3279. doi: 10.1074/jbc.M309311200. [DOI] [PubMed] [Google Scholar]
- 29.Kubatzky KF, et al. Self assembly of the transmembrane domain promotes signal transduction through the erythropoietin receptor. Curr Biol. 2001;11(2):110–115. doi: 10.1016/s0960-9822(01)00018-5. [DOI] [PubMed] [Google Scholar]
- 30.Liu W, Crocker E, Constantinescu SN, Smith SO. Helix packing and orientation in the transmembrane dimer of gp55-P of the spleen focus forming virus. Biophys J. 2005;89(2):1194–1202. doi: 10.1529/biophysj.104.057844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Besancenot R, et al. A senescence-like cell-cycle arrest occurs during megakaryocytic maturation: Implications for physiological and pathological megakaryocytic proliferation. PLoS Biol. 2010;8(9):e1000476. doi: 10.1371/journal.pbio.1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Constantinescu SN, et al. Ligand-independent oligomerization of cell-surface erythropoietin receptor is mediated by the transmembrane domain. Proc Natl Acad Sci USA. 2001;98(8):4379–4384. doi: 10.1073/pnas.081069198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown RJ, et al. Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat Struct Mol Biol. 2005;12(9):814–821. doi: 10.1038/nsmb977. [DOI] [PubMed] [Google Scholar]
- 34.Arkin IT, Brünger AT. Statistical analysis of predicted transmembrane α-helices. Biochim Biophys Acta. 1998;1429(1):113–128. doi: 10.1016/s0167-4838(98)00225-8. [DOI] [PubMed] [Google Scholar]
- 35.de Planque MRR, Killian JA. Protein-lipid interactions studied with designed transmembrane peptides: Role of hydrophobic matching and interfacial anchoring. Mol Membr Biol. 2003;20(4):271–284. doi: 10.1080/09687680310001605352. [DOI] [PubMed] [Google Scholar]
- 36.Ried CL, Kube S, Kirrbach J, Langosch D. Homotypic interaction and amino acid distribution of unilaterally conserved transmembrane helices. J Mol Biol. 2012;420(3):251–257. doi: 10.1016/j.jmb.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 37.Itaya M, Brett IC, Smith SO. Synthesis, purification, and characterization of single helix membrane peptides and proteins for NMR spectroscopy. Methods Mol Biol. 2012;831:333–357. doi: 10.1007/978-1-61779-480-3_18. [DOI] [PubMed] [Google Scholar]
- 38.Kochendoerfer GG, et al. Total chemical synthesis of the integral membrane protein influenza A virus M2: Role of its C-terminal domain in tetramer assembly. Biochemistry. 1999;38(37):11905–11913. doi: 10.1021/bi990720m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.