Abstract
Energy-coupling factor (ECF) transporters are a recently discovered family of primary active transporters for micronutrients and vitamins, such as biotin, thiamine, and riboflavin. Found exclusively in archaea and bacteria, including the human pathogens Listeria, Streptococcus, and Staphylococcus, ECF transporters may be the only means of vitamin acquisition in these organisms. The subunit composition of ECF transporters is similar to that of ATP binding cassette (ABC) importers, whereby both systems share two homologous ATPase subunits (A and A′), a high affinity substrate-binding subunit (S), and a transmembrane coupling subunit (T). However, the S subunit of ECF transporters is an integral membrane protein, and the transmembrane coupling subunits do not share an obvious sequence homology between the two transporter families. Moreover, the subunit stoichiometry of ECF transporters is controversial, and the detailed molecular interactions between subunits and the conformational changes during substrate translocation are unknown. We have characterized the ECF transporters from Thermotoga maritima and Streptococcus thermophilus. Our data suggests a subunit stoichiometry of 2S:2T:1A:1A′ and that S subunits for different substrates can be incorporated into the same transporter complex simultaneously. In the first crystal structure of the A–A′ heterodimer, each subunit contains a novel motif called the Q-helix that plays a key role in subunit coupling with the T subunits. Taken together, these findings suggest a mechanism for coupling ATP binding and hydrolysis to transmembrane transport by ECF transporters.
Keywords: membrane transport, structural biology, vitamin uptake
Vitamins are small organic molecules that function as essential enzymatic cofactors in all organisms. For example, riboflavin (Vitamin B2) is the precursor to the indispensible redox cofactors flavin mononucleotide and flavin adenine dinucleotide. Notably, the genomes of the human pathogens Listeria, Streptococcus, and Enterococcus lack the enzymes for de novo riboflavin synthesis. Instead, these bacteria possess energy-coupling factor (ECF) transporters with the S subunit for riboflavin, RibU (1). Consequently, ECF-RibU–mediated uptake may be the sole source of riboflavin in these pathogenic bacteria (1).
Active membrane transporters share common mechanistic features: a binding site for the transport substrate; a means for coupling to cellular energy, such as transmembrane ion gradients or ATP; and a pathway for substrate translocation across the membrane. In ECF transporters, these roles are divided between three types of subunits: two homologous ATPase subunits (A and A′), a transmembrane coupling subunit (T), and a high-affinity substrate-binding subunit (S) (2). The subunit composition of ECF transporters is similar to that of ATP binding cassette (ABC) importers, which possess a symmetrical architecture of two ATPase domains (also called ABCs or nucleotide binding domains), two homologous transmembrane domains, and one bilobed substrate binding protein (Fig. S1). However, the S subunit of ECF transporters is an integral membrane protein, and the transmembrane coupling subunits do not share an obvious sequence homology between ECF and ABC transporters. In addition, the subunit stoichiometry of ECF transporters is controversial (3–6), and it is unclear whether the two transporter families share a common mechanism.
ECF transporters can be divided into two groups. Both groups use an energizing module composed of the A, A′, and T subunits, also known as the ECF. In group I transporters, the energizing module associates with a single dedicated type of S subunit, as typified by the ECF transporter for biotin from Rhodobacter capsulatus (2). In contrast, in group II transporters multiple S subunits for distinct substrates share a common energizing module (1) (Fig. S1). Therefore, group II ECF transporters essentially use the same transporter to import a variety of vitamin substrates. In both groups, ATP-driven conformational changes in the A–A′–T energizing module are presumably coupled to the release of the tightly bound substrate from the S subunits. A framework for understanding the transmembrane transport mechanism of ECF transporters requires elucidation of the subunit stoichiometry, the interactions between subunits, and the role of ATP binding and hydrolysis. To address these questions, we have characterized the group II ECF transporters from Thermotoga maritima and Streptococcus thermophilus.
Results
Assembly of the ECF Riboflavin Transporter from T. maritima.
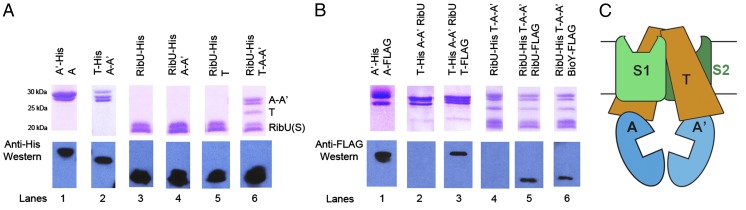
We first determined the composition of the group II ECF riboflavin transporter from T. maritima. Individually, neither the riboflavin-binding S subunit (RibU) nor the A–A′–T energizing module was able to transport riboflavin as judged by a complementation assay (Table S1). In contrast, coexpression of all four genes supported growth of an Escherichia coli riboflavin auxotroph, suggesting that the A, A′, T, and RibU subunits form a functional protein complex for riboflavin uptake. To investigate the assembly of the ECF riboflavin transporter, we conducted a series of pull-down experiments. First, His-tagged EcfA′ and untagged EcfA were coexpressed in E. coli, and a heteromeric A–A′ subcomplex was isolated by metal affinity chromatography. (Fig. 1A, lane 1). Similarly, the A, A′, and T subunits also copurified, indicating that the energizing module assembles into a stable core complex in the absence of an S subunit (Fig. 1A, lane 2). In contrast, no binary or tertiary subcomplexes containing the RibU S subunit were detected (Fig. 1A, lanes 3–5). Instead, coexpression of the three genes of the energizing module and RibU led to isolation of an intact complex between the four proteins (Fig. 1A, lane 6), indicating that assembly of the A–A′–T core is necessary for incorporation of the S subunit.
Fig. 1.
Assembly of the ECF transporter from T. maritima. (A and B) SDS/PAGE and Western blots from elutions of ECF subcomplexes isolated by affinity chromatography from E. coli cells expressing the indicated subunits. (C) Schematic showing the 1A:1A′:2T:2S (3 × 2) subunit model of an ECF transporter.
Subunit Stoichiometry of the ECF Riboflavin Transporter.
The subunit stoichiometry of ECF transporters is controversial. Models with one A, A′, T, and S (1 × 4 subunit model, Fig. S1) have been proposed (1, 5–7), but more recent data suggest the presence of multiple T and S subunits in the functional transporter (3, 4). To investigate the relative subunit stoichiometry of the Thermotoga ECF riboflavin transporter, the four genes were coexpressed with distinct His- and FLAG-tagged copies of the same subunit. As metal affinity chromatography will pull down complexes with the His-tagged subunit, the presence of a second subunit copy could be detected by Western blotting for the FLAG-tagged version. Indeed, His-tagged A′ pulled down a FLAG-tagged copy of A (Fig. 1B, lane 1), which agrees with the EcfA–A′ heterodimer observed by both cross-linking and size exclusion chromatography (SEC) (Fig. S2). Therefore, we concluded that this method could be used to probe the relative contents of the T. maritima ECF transporter.
As our data suggest that the ECF transporter contains a dimer of the ATPase subunits, we next investigated whether more than one T subunit was present in the complex. Isolation of ECF riboflavin transporter complexes via His-tagged EcfT led to copurification of a second, FLAG-tagged T subunit (Fig. 1B, lane 3), suggesting that the transporter contains a homodimeric T subunit. Importantly, no anti-FLAG reactivity was observed in control experiments lacking a FLAG-tagged subunit (Fig. 1B, lanes 2 and 4). A two T model is also supported by cross-linking experiments on the biotin ECF transporter, which suggested a dimeric T subunit in this system (4). Sequences of T subunits lack internal repeats and are not homologous to the transmembrane domains of ABC transporters (8). However, T subunits contain two conserved Ala–Arg motifs that have been identified as possible sites of interaction with A and A′ (2, 9). In an ECF transporter model with two T subunits, these motifs may contribute to a binary interface with each ATPase subunit (4). Alternatively, the Ala–Arg motifs could contact both ATPase subunits (7), as has been observed in the coupling interfaces of ABC exporters (10). In such a 1A:1A′:2T model, the energizing module resembles an ABC transporter with heterodimeric ATPases and homodimeric T subunits (Fig. S1).
Using the same methodology, we showed that at least two copies of the RibU S subunit are present in the transporter (Fig. 1B, lane 5). As Thermotoga also contains an S subunit for biotin, BioY, we tested whether S subunits for two different substrates could assemble into the same complex simultaneously. Indeed, BioY copurified in a complex pulled down with RibU (Fig. 1B, lane 6), indicating the presence of two different S subunits in the same ECF transporter. Given that the group II S subunits are monomeric in isolation (6, 11, 12), these observations immediately suggest that the A–A′–T energizing module from Thermotoga contains two separate binding sites for S subunits. These data are consistent with the presence of a dual S subunit in the related Co2+ transporter (13) as well as studies of the ECF transporter for biotin from R. capsulatus (3). Therefore, we propose that there are two copies of the S subunit in the functional transporter complex. Furthermore, cross-linking of the purified homologous transporter from S. thermophilus are consistent with a transporter complex with 2T, 2S, and 2 ATPase subunits (discussed below). Taken together, our results (Fig. 1 A and B) support a 1A:1A′:2T:2S (3 × 2) subunit model for the ECF transporter from Thermotoga. In this model, the intact ECF transporter is composed of an energizing module with two identical transmembrane coupling subunits (T subunits) and two homolgous ATPase subunits (A and A′) with two substrate-binding proteins (S subunits) bound within the membrane (Fig. 1C). However, we acknowledge the need for further experimentation, particularly in the membrane environment, to conclusively determine the physiologically relevant subunit stoichiometry of ECF transporters.
Mechanistic Implications of the 1A:1A′:2T:2S (3 × 2) Model of ECF Transporters.
The proposed 1A:1A′:2T:2S (3 × 2) architecture (Fig. 1C) predicts several features of the assembly and transport mechanism of ECF transporters that differ from a 1A:1A′:1T:1S (1 × 4) model (1, 5, 6) (Fig. S1). First, the 3 × 2 model does not strictly require an A–S or A′–S interaction, an essential feature of one version of the 1 × 4 model (6). The absence of an A–A′–RibU subcomplex (Fig. 1A) and the lack of a conserved cytoplasmic surface among the available S subunit structures (6, 7, 14) is consistent with this prediction (Fig. 1A). Second, a homodimeric T assembly suggests that each T subunit interacts with the A and A′ subunits via a homologous interface. The isolation of the A–A′–T core (Fig. 1A) and the presence of a second T subunit in the intact transporter (Fig. 1B) point to such a 1A:1A′:2T core assembly. Third, attachment of two S subunits to this energizing core yields the functional transporter. Consequently, the 3 × 2 assembly (Fig. 1C) displays twofold symmetry, which is also a critical aspect of ABC transporter function (15) (Fig. S1). Finally, our data on the Thermotoga ECF transporter indicate that S subunits for two different vitamins can simultaneously assemble into the same complex (Fig. 1B). This result raises the intriguing possibility that two substrate molecules could be taken up during one transport cycle.
Although these experiments shed light on the general assembly of the TmECF transporter, they also raised more specific questions, such as the molecular interactions that stabilize the A–A′ heterodimer, the structural basis for coupling between the ATPase and T subunits, and how ATP binding and hydrolysis drives membrane transport. To address these key mechanistic questions, we determined the first crystal structure of an EcfA–A′ heterodimer.
Structure of the EcfA–A′ Heterodimer from T. maritima.
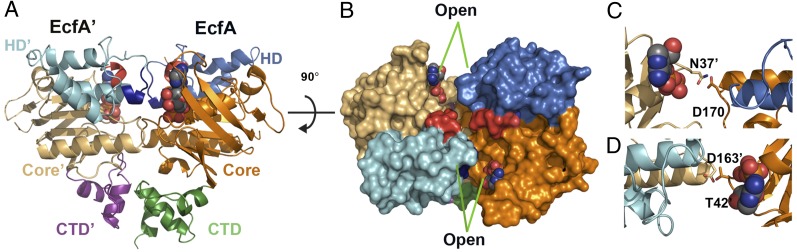
The EcfA and A′ subunits form a stable heterodimer in solution (Fig. S2) and directly interact with the T subunits (Fig. 1A). To define the structural basis of these interactions, we determined the crystal structure of the adenosine diphosphate (ADP)-bound EcfA–A′ heterodimer from T. maritima to 2.7 Å resolution (Table 1). Each subunit of the heterodimer contains three subdomains (Fig. 2A): an ATP binding core (Core subdomain) that contains the phosphate binding P-loop and Walker B motif, a three helix bundle (Helical subdomain, HD) that contains the critical LSGGQ signature motif, and a novel ∼40 residue subdomain at the carboxy-terminus of each subunit that forms the subunit interface.
Table 1.
Data collection and refinement statistics
| Data collection | |
| Space group | P32 |
| Cell dimensions (Å) | 67.9, 67.9, 251.2 |
| Resolution (Å) | 48.1–2.7 (2.77–2.70) |
| Rmerge | 0.092 (0.86) |
| I/σI | 37.2 (3.3) |
| Completeness (%) | 99.9 (100.0) |
| Redundancy | 11.5 (11.5) |
| Refinement | |
| No. reflections | 35,599 |
| Rwork/Rfree | 0.212/0.256 |
| No. atoms | |
| Protein | 8101 |
| Ligand | 112 |
| Solvent | 107 |
| Average B-factor (Å2) | |
| Protein | 60.6 |
| Ligands | 47.1 |
| Solvent | 42.2 |
| rmsd | |
| Bond lengths (Å) | 0.007Å |
| Bond angles (°) | 1.2 |
Values in parenthesis are for the highest resolution shell.
Fig. 2.
The crystal structure of ADP bound EcfA–A′ heterodimer from T. maritima. (A) Ribbon diagram of the EcfA–A′ heterodimer structure colored by subdomain, with the ATP-bindng Core in tan and orange, the Helical subdomain (HD) in cyan and blue, and the CTDs in purple and green for EcfA′ and A, respectively. The Q-helices are colored red, and ADP is shown in sphere representation. (B) Top view of the heterodimer colored as in A and shown in surface representation. The opening between the P-loop and LSGGQ is indicated. (C and D) Interaction between the P-loop and D-loop of each subunit, with relevant side chains shown as sticks.
EcfA–A′ Dimerizes Via Novel Subdomains at the Carboxy-Termini.
The carboxyl-terminal subdomain (CTD) of the EcfA subunit (residues 221–266) consists of a three-helix bundle in which the first and third helices are perpendicular to the second (Fig. S3A). In the A′ subunit, this domain is formed by residues 213–259 and is composed of two short orthogonal helices linked by an extended structure (Fig. S3B). A search for similar protein structures (16) failed to reveal any structural homologs for either of the CTDs (Fig. S3C). At the A–A′ dimer interface, the CTDs interact primarily along helix-3 of the A subunit and helix-2 of A′ with a salt bridge between Asp257 and Lys232′ and hydrophobic contacts between Leu260 and Leu233′ (Fig. S3D). This interface buries ∼1,700 Å2 of solvent-accessible surface area and displays a surface complementarity (17) of 0.62. Notably, deletion of the EcfA CTD completely abolished the association of the heterodimer, indicating that the CTDs form the primary subunit interface in the absence of ATP (Fig. S3E). Although the CTDs are a general feature of the ATPase subunits from group II ECF transporters, many group I transporters lack these subdomains.
ADP-Bound Structure Represents the Open State of the ATPase.
In ABC ATPases, ATP binding drives a transition from an open to a closed state in which two molecules of ATP are sandwiched between the P-loop of one subunit and the LSGGQ signature sequence of the other (18–21). In the ADP-bound EcfA–A′ structure, the distance between the P-loop and the LSGGQ motif is ∼15 Å in each subunit (Fig. 2B), which agrees with the open state observed in several ABC importer structures (22–26). Indeed, the only intersubunit contacts between the ATPase domains involve the P-loop of one monomer and the D-loop of the opposing subunit. Specifically, Asn37 of the EcfA′ P-loop forms a hydrogen-bond across the subunit interface with the D-loop Asp170 of EcfA (Fig. 2C), whereas the equivalent Thr42 of EcfA contacts Asp163′ (Fig. 2D). As a result, the ADP-bound EcfA–A′ structure likely represents the physiologically relevant open state of the ATPase dimer. Notably, the homologous intersubunit contacts were observed in the nucleotide-free, inward facing structure of the E. coli maltose transporter in the pretranslocation state (22). In this system, ATP binding remodels this “loose” interface leading to closure of the ATPase dimer and conversion of the attached transmembrane domains to the outward-facing conformation (22, 27). Similarly, in ECF transporters, the ATP driven open to closed transition of the A and A′ subunits could push the cytoplasmic regions of the attached T subunits toward one another potentially leading to an outward-facing conformation of the transporter.
Interaction of EcfA–A′ with the EcfT Subunits.
Each subunit of the EcfA–A′ heterodimer contains an acidic, highly conserved groove (Fig. S4 A–C) that may interact with the conserved Ala–Arg motifs of the T subunits (4). This groove is formed by the first helix of the helical subdomain (HD) and a highly conserved helical turn that is unique to ECF–ATPases. The consensus sequence of this six-residue helix is xPD/ExQϕ (where ϕ is a hydrophobic residue) and we have termed this structure the “Q-helix” due to the invariant Gln residue (Fig. 3A). In each Q-helix, the backbone carbonyl oxygens of the first two residues form H-bonds with the backbone amides of the i+3 residues to form a short 310 helix. In the EcfA Q-helix, Tyr86 and Pro87 interact with Asp89 and Gln90 (Fig. 3B), whereas Asn80 and Pro81 of EcfA′ interact with Ser83 and Gln84 (Fig. 3C). As a result, these interactions place the conserved acidic residue on one side of the Q-helix and the invariant Gln on the opposing face (discussed below).
Fig. 3.
The ECF-specific Q-helix forms the T subunit interface. (A) Sequence alignment of the Q-helix region in ABC and ECF-type ATPases. (B and C) The Q-helix (pink sticks) in the EcfA and EcfA′ subunits, respectively. The H-bonds that form the helix are shown as dashed lines, and the invariant Pro is labeled. (D) Superposition of the EcfA structure onto the maltose transporter (with the MalK ATPase omitted and the MalG transmembrane domain colored gray) reveals that the conserved groove formed by the Q-helix coincides with the coupling interface in ABC transporters. (E) In the T. maritima EcfA monomer structure (2YZ2), Glu88 of the Q-helix forms a salt bridge with the highly conserved Arg145 that follows the LSGGQ signature sequence. (F) The homologous residues in the EcfA′ structure are Ser82 of the Q-helix and Gln138 of the helical subdomain. In most EcfA′ subunits, the equivalent of Ser82 is an Asp residue. (G) In the EcfA crystal structure, the Glu88-Arg145 salt bridge is broken.
As each Q-helix is located ∼14 Å from the ATPase active site and the conserved Gln side chain points toward the interior of the groove (Fig. S4D), we hypothesize that this motif is involved in coupling to the T subunits. Superposition of the A subunit with the MalK ATPase from the maltose transporter structure (27) supports this inference, as the Q-helix resides at the NBD-transmembrane domain interface in ABC transporters (Fig. 3D). Indeed, this groove is lined with conserved residues, including Arg77/71′ of the core domain, Phe102/96′ and Asn106/Ile100′ of the helical subdomain, and Gln90/84′ of the Q-helix (Figs. S4D and S5). Consistently, the homologous region of the BioM ECF ATPase could be cross-linked to the Ala–Arg motifs of the BioN T subunit (Fig. S4E) (4). As a result, these data indicate that the conserved groove formed by the Q-helix represents the coupling interface between the ATPase and T subunits in ECF transporters.
Intrasubunit Coupling Between the Q-Helix and the LSGGQ Signature Sequence.
The Q-helix of ECF transporters is in a prime position to couple conformational changes in the T subunits to the ATPase active site. In each subunit of the A–A′ heterodimer, the conserved acidic residue of the Q-helix is poised to interact with the amino acid following the LSGGQ motif in the same subunit. In a previously determined structure of the monomeric, nucleotide-free EcfA subunit from Thermotoga (28), Glu88 of the Q-helix forms an intrasubunit salt bridge with Arg145 (Fig. 3E), which is two residues after the LSGGQ and is completely conserved in ECF A subunits (Fig. S5). Similarly, Ser82 of the EcfA′ Q-helix, which is more commonly an Asp residue in other A′ subunits (Fig. 3A), is in position to interact with the invariant Gln138 that follows the signature sequence in this subunit (Fig. 3F). In the A–A′ dimer structure, the Q-helix has retracted by ∼6 Å toward the core subdomain, which breaks the Glu88–Arg145 salt bridge (Fig. 3G) leading to a distinct position for the LSSGQ motif compared with the EcfA monomer structure (Fig. S6A). Notably, this conformational transition is consistent with rotation of the helical subdomain into the ATP-bound state (22, 27) (Fig. S6B). Therefore, these intrasubunit interactions between the Q-helix and the helical subdomain could function as a molecular brake to prevent futile nontransporting cycles of ATP hydrolysis. Potentially, high-affinity binding of substrate to the S subunits may promote a conformational change in the attached T subunits that is transduced to the ABC subunits via the Q-helix. In this way, disruption of the interaction between the Q-helix and the signature sequence could release the helical subdomain and prealign the ATPase active sites into a conformation that favors formation of the closed ATP bound dimer (Fig. S6C).
Q-Helices Are Essential for Membrane Transport.
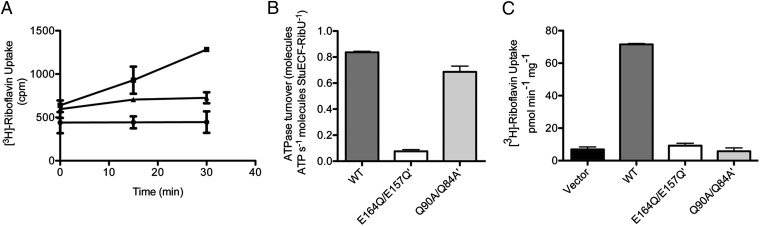
The EcfA–A′ structure suggests that the Q-helices interact with the T subunits but are not directly involved in ATP binding and hydrolysis. To determine the functional importance of the Q-helix, we characterized the ECF riboflavin transporter from S. thermophilus, StuECF–RibU. These proteins display superior biochemical stability and activity at ambient temperatures compared with the T. maritima orthologs. In detergent, the purified StuECF–RibU complex elutes as a single peak on SEC (Fig. S7A), and treatment of the complex with glutaraldehyde yielded a dominant band at ∼160 kDa (Fig. S7B), which is consistent with the inferred 3 × 2 stoichiometry of the T. maritima homologs (Fig. 1C). Upon expression in E. coli, the StuECF–RibU complex catalyzed the uptake of radioactive riboflavin (Fig. 4A).
Fig. 4.
The Q-helix is essential for transmembrane transport. (A) 3H-riboflavin uptake of energized E. coli cells expressing the vector (●) or the ECF riboflavin transporter from S. thermophilus, StuECF-RibU (▲). 3H-riboflavin uptake of de-energized E. coli cells expressing StuECF-RibU is represented by ■. (B) ATPase activity of the wild-type, Walker B mutants, and Q-helix mutants of purified StuECF–RibU in detergent. (C) 3H-riboflavin uptake of energized E. coli cells expressing the wild-type and mutant transporters.
As the Q-helix resides outside of the ATPase active site, mutations of the conserved Gln would be expected to have limited impact on the ATPase activity. Wild-type StuECF–RibU displayed a robust ATPase turnover rate at 37 °C. Mutation of each Q-helix Gln to Ala (Gln90 and Gln84′) had a minimal effect on the ATPase turnover (<20%), indicating that this residue does not play a direct role in ATP hydrolysis (Fig. 4B). In contrast, mutation of the Walker B catalytic Glu to Gln in both of the A (Glu164) and A′ (Glu157′) subunits reduced the rate by ∼11-fold (Fig. 4B). As expected, both types of mutations greatly reduced riboflavin uptake by the transporter (Fig. 4C). Given the sequence conservation and position of this motif, these functional data indicate that the Q-helix plays an essential role in coupling to the T subunits in ECF transporters.
Discussion
3 × 2 Model for ECF Transporters.
Herein we present data that the functional assembly of the group II ECF transporters from T. maritima and S. thermophilus consist of two substrate-binding S subunits, two identical T subunits, and two homolgous ATPase subunits. In this 3 × 2 model for ECF transporters (Fig. 1C), each T subunit interacts with a conserved groove in the ATPase subunits, A and A′ (4) (Fig. 3 and Fig. S4). Consequently, this 1A:1A′:2T assembly can present two membrane-embedded surfaces for interaction with the integral membrane S subunits. This arrangement uses the modularity of this transporter family (1) whereby two different S subunits can assemble into the same transporter complex (Fig. 1B). Moreover, by positioning the S subunits at the T subunit interface via the surface formed by transmembrane helices 1–2–3 (7, 14), ATP-driven, symmetrical conformational changes in the T subunits could be coupled to displacement of the substrate from the binding site (7).
Proposed Transport Mechanism of ECF Transporters.
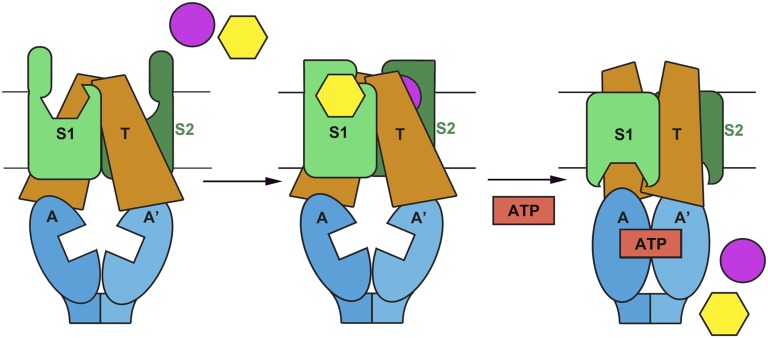
The structural, functional, and assembly data presented herein suggest a potential model for the transport mechanism of ECF transporters (Fig. 5). Substrate binding to the S subunits results in a structural transition from an open apo to a closed, substrate-bound, occluded conformation (6, 7, 14). The free energy of high-affinity substrate binding (11, 12) could be coupled to a conformational change in the T subunits that prealigns the two ATPase active sites into a conformation that favors ATP binding (Fig. S6C). This conformational change could be transduced from motifs on the conserved surface formed by TM1–2–3 of the S subunits to the coupling helices and the Q-helices in A and A′. Subsequently, ATP binding drives the transition to a closed dimer of A and A′, and this closure leads to conversion of the T subunits to an outward facing conformation. In the maltose transporter, this conformational change “splits open” the substrate binding protein resulting in release of maltose into a low-affinity extracellular binding site between the transmembrane subunits (21, 27). In ABC exporters, the analogous domain rearrangement exports compounds from the cell (10). Accordingly, ATP binding to the A and A′ subunits could drive a similar conformational change in the T subunits that releases the tightly bound substrate from the attached S subunits (Fig. 5). Whether the substrate is then bound to a closed, low-affinity binding site or translocated directly into the cytoplasm will need to be explored by further investigation.
Fig. 5.
Transport mechanism of ECF transporters. Schematic illustrating how ATP binding to the A–A′ heterodimer may drive substrate translocation across the membrane in ECF transporters.
Materials and Methods
Protein Expression and Purification of TmECF–RibU Complexes.
The EcfA and A′ genes were cloned into pACYC-Duet (EMD4Biosciences) for co-expression. TmRibU was cloned into a modified pET15 vector (EMD4Biosciences) fused to an Nt decahistidine tag. For coexpression of the TmRibU and EcfT subunits, these genes were linked by a ribosome binding site with the following sequence: 5′ATAATTTTGTTTAACTTTAAGAAGGAGATATACC3′. The ECF FLAG-tagged subunits (EcfA, EcfT, RibU, and BioY) were cloned into pCDF-Duet (EMD4Biosciences). Protein expression was performed in E. coli BL21(DE3) at 37 °C in LB media. Cells were resuspended in Buffer A (50 mM Tris, pH 7.5, 500 mM NaCl, 20% (vol/vol) glycerol, and 10 mM imidazole). After cell disruption, unlysed cells were removed by centrifugation and the supernatant was treated with 1% (wt/vol) dodecyl-maltoside (DDM) for membrane protein solublization. Protein complexes were purified on Talon resin (Clontech) in 0.05% DDM and eluted with 300 mM imidazole. Copurification was analyzed by SDS/PAGE of elution fractions and Western blotting for the FLAG tag with the mouse anti-FLAG M2 antibody (Sigma-Aldrich).
Expression and Purification of the EcfA–A′ Heterodimer from T. maritima.
The gene for the EcfA′ subunit (TM1663) was amplified from T. maritima genomic DNA and cloned into a modified pET28 vector (EMD4Biosciences) with an amino-terminal decahistidine tag linked by a thrombin recognition site. The gene for EcfA subunit (TM0222) was amplified from genomic DNA and cloned into MCS1 of pACYC-Duet. These constructs were cotransformed into E. coli BL21(DE3), and protein expression was performed in LB media at 37 °C. Cells were resuspended in Buffer A. After cell disruption, unlysed cells and the membrane fraction were removed by centifugation and the supernatant was bound to Talon resin. After washing, the A and A′ heterodimer was eluted with a buffer containing 85 and 120 mM imidazole. The Talon fractions were diluted into 50 mM Tris, pH 8.5, and 2 mM EDTA; loaded onto a Q-Sepharose column; and eluted with a linear gradient from 5 to 500 mM NaCl. Peak fractions were pooled and treated with thrombin to remove the purification tag from the A′ subunit. After thrombin digestion, the sample was further purified by gel filtration on a Superdex 200 column equilibrated in 20 mM Tris, pH 7.5, 100 mM NaCl, and 1 mM EDTA. The best crystals required the mutation of two nonconserved surface patches with high side chain entropy (29) in the A′ subunit to Ala: Glu53, Glu55, Glu125, Lys126, and Glu127. For crystallization, SEC peak fractions were pooled, concentrated to ∼7 mg/mL, and incubated with 10 mM ADP before setup.
Crystallization and Structure Determination of EcfA–A′.
Crystals of ADP-bound EcfA–A′ were grown by vapor diffusion from drops consisting of 1.2 μL protein and 0.6 μL 100 mM NaOAc, pH 5.5, 27.5% 2-methylpentane-2,4-diol (MPD), and 200 mM NaCl at 4 °C. Crystals were flash frozen in liquid nitrogen before data collection in oscillation mode at beamline X29 of the National Synchrotron Light Source (NSLS). Diffraction data were processed with HKL2000 (30). The EcfA–A′ crystal structure was determined by molecular replacement in Phaser (31) with PDB entry 2YZ2 (28) as the search model. Structures were built with Coot (32) and refined with Phenix (33). Electrostatic surface potential was calculated with Adaptive Poisson-Boltzman Solver (APBS) (34). The final model contains residues 1–265 of the A subunit and 1–247 of A′.
ATPase Activity Determination of the StuECF–RibU Complex.
The ATPase activity of the wild-type and mutant StuECF–RibU complexes was measured in 100 mM NaCl, 0.2 mM EDTA, 50 mM Tris, pH 8.0, 0.1% DDM, and 5 mM MgATP with a protein concentration of ∼1 μM. All reactions were performed for 300 s at 37 °C. Hydrolyzed phosphate was detected by the Malachite Green assay (35). Samples lacking enzyme were performed in parallel for background subtraction.
3H-Riboflavin Uptake Assays.
Plasmids for expression of wild-type and mutant StuECF–RibU and the empty vector were transformed into E. coli BL21(DE3). Cells were grown in LB medium containing ampicillin at 37 °C and induced with 0.002% arabinose for 2 h at an OD595 of ∼0.6. Cells were harvested and washed with cold wash buffer (50 mM potassium phosphate, pH 7.0) and resupended to an OD595 of 10. Fresh cells were energized by the addition of 10 mM glucose for 10 min at 30 °C. Transport assays were initiated by the addition of 50 nM [3H]-riboflavin (American Radiolabeled Chemicals), and at selected time points, 200 μL samples were mixed with 2 mL cold wash buffer and rapidly filtered through 0.45 μm cellulose nitrate filters. Filters were washed by an additional 2 mL wash buffer, dried, and added to 1.6 mL scintillation fluid. The levels of incorporated radioactive riboflavin were determined with a Wallac scintillation counter.
Supplementary Material
Acknowledgments
We thank Romina Mancusso and Jin-Mei Song for technical assistance, and Bryan Czyzewski and Jennifer Marden for comments on the manuscript. We also thank the staff at beamlines X25 and X29 of the NSLS at the Brookhaven National Laboratory for assistance in X-ray diffraction experiments. N.K.K. thanks the American Heart Association and the National Institutes of Health (F32-HL091618) for postdoctoral fellowships. This work was financially supported by National Institutes of Health Grants R01-DK073973, R01-GM093825, R01-DA019676, and R01-MH083840.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4HLU).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217361110/-/DCSupplemental.
References
- 1.Rodionov DA, et al. A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol. 2009;191(1):42–51. doi: 10.1128/JB.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hebbeln P, Rodionov DA, Alfandega A, Eitinger T. Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc Natl Acad Sci USA. 2007;104(8):2909–2914. doi: 10.1073/pnas.0609905104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finkenwirth F, et al. Subunit composition of an energy-coupling-factor-type biotin transporter analysed in living bacteria. Biochem J. 2010;431(3):373–380. doi: 10.1042/BJ20100813. [DOI] [PubMed] [Google Scholar]
- 4.Neubauer O, Reiffler C, Behrendt L, Eitinger T. Interactions among the A and T units of an ECF-type biotin transporter analyzed by site-specific crosslinking. PLoS ONE. 2011;6(12):e29087. doi: 10.1371/journal.pone.0029087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ter Beek J, Duurkens RH, Erkens GB, Slotboom DJ. Quaternary structure and functional unit of energy coupling factor (ECF)-type transporters. J Biol Chem. 2011;286(7):5471–5475. doi: 10.1074/jbc.M110.199224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang P, Wang J, Shi Y. Structure and mechanism of the S component of a bacterial ECF transporter. Nature. 2010;468(7324):717–720. doi: 10.1038/nature09488. [DOI] [PubMed] [Google Scholar]
- 7.Erkens GB, et al. The structural basis of modularity in ECF-type ABC transporters. Nat Struct Mol Biol. 2011;18(7):755–760. doi: 10.1038/nsmb.2073. [DOI] [PubMed] [Google Scholar]
- 8.Eitinger T, Rodionov DA, Grote M, Schneider E. Canonical and ECF-type ATP-binding cassette importers in prokaryotes: Diversity in modular organization and cellular functions. FEMS Microbiol Rev. 2011;35(1):3–67. doi: 10.1111/j.1574-6976.2010.00230.x. [DOI] [PubMed] [Google Scholar]
- 9.Neubauer O, et al. Two essential arginine residues in the T components of energy-coupling factor transporters. J Bacteriol. 2009;191(21):6482–6488. doi: 10.1128/JB.00965-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443(7108):180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 11.Duurkens RH, Tol MB, Geertsma ER, Permentier HP, Slotboom DJ. Flavin binding to the high affinity riboflavin transporter RibU. J Biol Chem. 2007;282(14):10380–10386. doi: 10.1074/jbc.M608583200. [DOI] [PubMed] [Google Scholar]
- 12.Erkens GB, Slotboom DJ. Biochemical characterization of ThiT from Lactococcus lactis: A thiamin transporter with picomolar substrate binding affinity. Biochemistry. 2010;49(14):3203–3212. doi: 10.1021/bi100154r. [DOI] [PubMed] [Google Scholar]
- 13.Siche S, Neubauer O, Hebbeln P, Eitinger T. A bipartite S unit of an ECF-type cobalt transporter. Res Microbiol. 2010;161(10):824–829. doi: 10.1016/j.resmic.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Berntsson RP, et al. Structural divergence of paralogous S components from ECF-type ABC transporters. Proc Natl Acad Sci USA. 2012;109(35):13990–13995. doi: 10.1073/pnas.1203219109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins CF, Linton KJ. The ATP switch model for ABC transporters. Nat Struct Mol Biol. 2004;11(10):918–926. doi: 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- 16.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234(4):946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 18.Hopfner KP, et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101(7):789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 19.Smith PC, et al. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol Cell. 2002;10(1):139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fetsch EE, Davidson AL. Vanadate-catalyzed photocleavage of the signature motif of an ATP-binding cassette (ABC) transporter. Proc Natl Acad Sci USA. 2002;99(15):9685–9690. doi: 10.1073/pnas.152204499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Sharma S, Quiocho FA, Davidson AL. Trapping the transition state of an ATP-binding cassette transporter: Evidence for a concerted mechanism of maltose transport. Proc Natl Acad Sci USA. 2001;98(4):1525–1530. doi: 10.1073/pnas.041542498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oldham ML, Chen J. Crystal structure of the maltose transporter in a pretranslocation intermediate state. Science. 2011;332(6034):1202–1205. doi: 10.1126/science.1200767. [DOI] [PubMed] [Google Scholar]
- 23.Pinkett HW, Lee AT, Lum P, Locher KP, Rees DC. An inward-facing conformation of a putative metal-chelate-type ABC transporter. Science. 2007;315(5810):373–377. doi: 10.1126/science.1133488. [DOI] [PubMed] [Google Scholar]
- 24.Locher KP, Lee AT, Rees DC. The E. coli BtuCD structure: A framework for ABC transporter architecture and mechanism. Science. 2002;296(5570):1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 25.Khare D, Oldham ML, Orelle C, Davidson AL, Chen J. Alternating access in maltose transporter mediated by rigid-body rotations. Mol Cell. 2009;33(4):528–536. doi: 10.1016/j.molcel.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rees DC, Johnson E, Lewinson O. ABC transporters: The power to change. Nat Rev Mol Cell Biol. 2009;10(3):218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J. Crystal structure of a catalytic intermediate of the maltose transporter. Nature. 2007;450(7169):515–521. doi: 10.1038/nature06264. [DOI] [PubMed] [Google Scholar]
- 28.Ethayathulla AS, et al. Purification, crystallization and preliminary X-ray diffraction analysis of the putative ABC transporter ATP-binding protein from Thermotoga maritima. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64(Pt 6):498–500. doi: 10.1107/S1744309108013778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Derewenda ZS. Rational protein crystallization by mutational surface engineering. Structure. 2004;12(4):529–535. doi: 10.1016/j.str.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Otwinoski ZM, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kodama T, Fukui K, Kometani K. The initial phosphate burst in ATP hydrolysis by myosin and subfragment-1 as studied by a modified malachite green method for determination of inorganic phosphate. J Biochem. 1986;99(5):1465–1472. doi: 10.1093/oxfordjournals.jbchem.a135616. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.