Abstract
TRIM5 is a host antiviral gene with an evolutionary history of genetic conflict with retroviruses. The TRIMCyp gene encodes a protein fusion of TRIM5 effector domains with the capsid-binding ability of a retrotransposed CyclophilinA (CypA), resulting in novel antiviral specificity against lentiviruses. Previous studies have identified two independent primate TRIMCyp fusions that evolved within the past 6 My. Here, we describe an ancient primate TRIMCyp gene (that we call TRIMCypA3), which evolved in the common ancestor of simian primates 43 Mya. Gene reconstruction shows that CypA3 encoded an intact, likely active, TRIMCyp antiviral gene, which was subject to selective constraints for at least 10 My, followed by pseudogenization or loss in all extant primates. Despite its decayed status, we found TRIMCypA3 gene fusion transcripts in several primates. We found that the reconstructed “newly born” TrimCypA3 encoded robust and broad retroviral restriction activity but that this broad activity was lost via eight amino acid changes over the course of the next 10 My. We propose that TRIMCypA3 arose in response to a viral pathogen encountered by ancestral primates but was subsequently pseudogenized or lost due to a lack of selective pressure. Much like imprints of ancient viruses, fossils of decayed genes, such as TRIMCypA3, provide unique and specific insight into paleoviral infections that plagued primates deep in their evolutionary history.
Keywords: paleovirology, restriction factor, retroviral capsids
Ancient viruses have selected for changes in host antiviral genes throughout primate evolution (1, 2). Understanding when these adaptive changes occurred, together with how they altered the antiviral specificities of these genes, can lead to strong inferences about the existence of ancient viruses and their consequences on the modern function and specificity of the primate innate immune system. For example, although the TRIM5α protein encodes a retroviral restriction factor that blocks the viral life cycle of several retroviruses (3–8), retroviral specificity varies among primates as a result of ancient selection for changes in antiviral specificity (9–12). These species-specific differences in TRIM5α are due to dramatic variation in both the coiled-coil and B30.2 domains, which are responsible for the interaction with the viral capsid protein of a variety of retroviruses (13, 14). Thus, innovation for capsid-binding specificity has directly resulted in rapid changes in TRIM5α.
An additional form of genetic innovation in the TRIM5 locus involves novel gene fusions. Such a gene fusion was first identified in owl monkeys (Aotus trivirgatus), which encode a fusion protein between the TRIM5 gene and a retrotransposed CyclophilinA (CypA1) gene, called the TRIMCyp gene fusion (15). The retrotransposition of CypA between exons 7 and 8 of owl monkey TRIM5 (15) occurred 4.5–6 Mya (16, 17). Like TRIM5α, the resulting TRIMCyp protein contains RING, B-box 2, and coiled-coil domains. However, a CypA domain has structurally and functionally replaced the B30.2 domain as the capsid-binding determinant (18, 19). The resulting fusion of TRIM5 effector domains with the capsid-binding ability of CypA in owl monkeys generated a protein with novel antiviral defense activity against HIV-1 (15, 20). This restriction occurs at the same early, postentry stage before RT as TRIM5α (3, 13, 21–23).
Several macaque species also encode TRIMCyp, which is the consequence of another, independent CypA retrotransposition (CypA2 retrogene) downstream of the TRIM5 gene (24–27). This event is also estimated to have occurred 5–6 Mya (28). Unlike CypA1 in owl monkeys, the CypA2-encoding TRIM5 allele is found at varying frequencies across macaque species (24, 25, 27, 28). These two TRIMCyp gene fusions thus represent a remarkable case of convergent evolution in the generation of novel antiviral specificity in the TRIM5 locus.
Here, by reconstructing a detailed evolutionary history of CypA retrogenes proximal to TRIM5 across primates, we find two additional currently pseudogenized CypA retrogenes that inserted downstream of the TRIM5 gene 18–43 Mya in primate evolution. One of these (which we refer to as CypA3 in keeping with prior nomenclature) is still expressed as a novel TRIMCyp gene fusion transcript in several Old World monkeys. Our phylogenetic analyses date the origin of CypA3 to 43 Mya and find that TRIMCypA3 was maintained as an intact gene for at least 10 My, making it the most ancient TRIMCyp yet identified in primates. Although CypA3 is decayed in all extant primates and the resulting TRIMCyp gene fusion is defective, our evolutionary reconstruction and virological assays suggest that TRIMCypA3 encoded broad and potent restriction activity following its birth. Our findings reveal that convergent evolution has led to at least four independent CypA retrogene insertions proximal to TRIM5 and, consequently, the formation of TRIMCyp at least three independent times in primate history. This further reflects the intense, recurrent pressure imposed by ancient viruses. We posit that the currently inactive TRIMCypA3 gene fusion represents the fossil remnants of an ancient antiviral innovation that points to a retroviral challenge before the common ancestor of all simian primates. Our study highlights the utility of antiviral gene evolution for the study of paleovirology (1, 2).
Results
CypA Retrogenes Proximal to TRIM5.
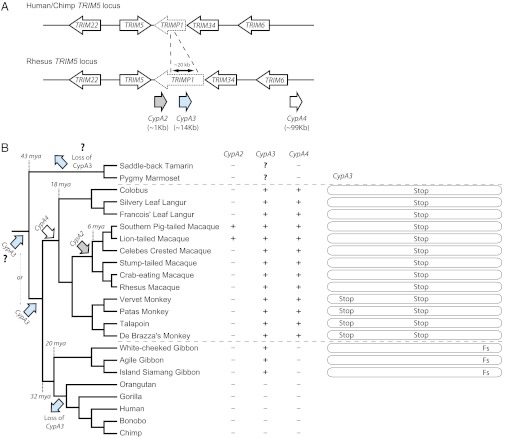
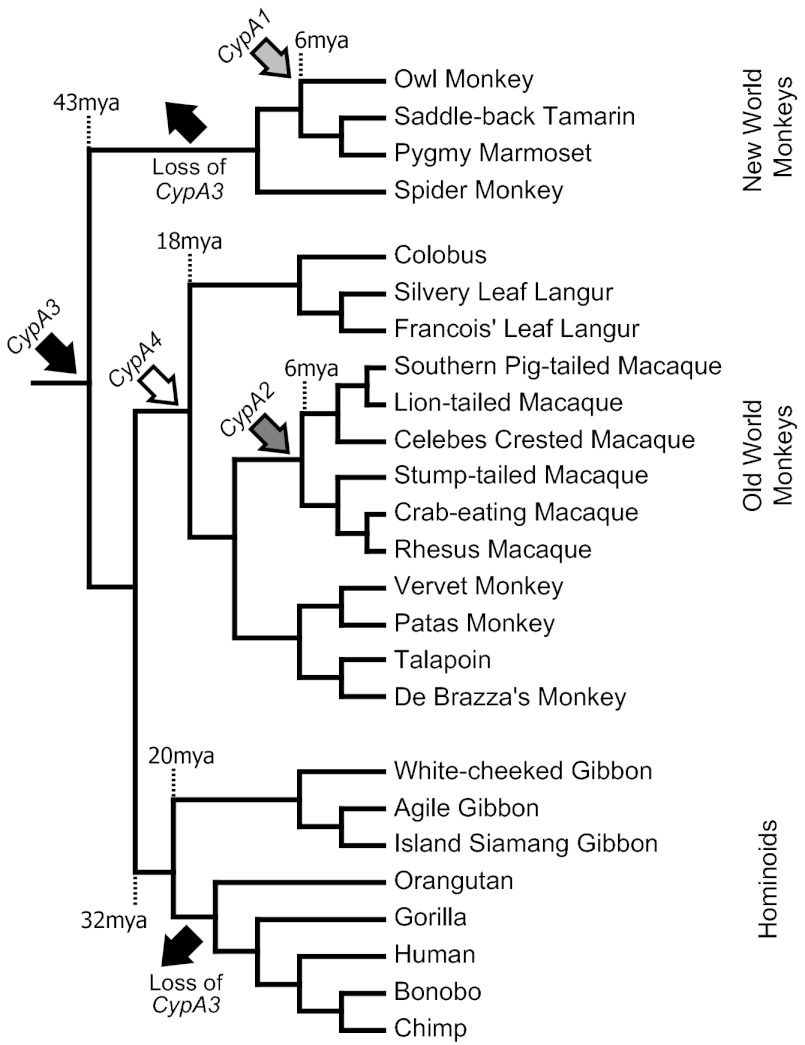
The TRIM5 locus in primates consists of four intact TRIM genes: TRIM22, TRIM5, TRIM34, and TRIM6 (Fig. 1A), as well as a TRIM pseudogene, TRIMP1 (Fig. 1A, dotted outline). Our analyses revealed the presence of three CypA retrogenes proximal to and downstream of TRIM5 (Fig. 1A). The most proximal of these, located ∼1 kb downstream of TRIM5 (Fig. 1A and Fig. S1), is the polymorphic CypA2 identified in previous studies (24–27, 29) but missing from the reference rhesus macaque genome. We discovered another CypA, located ∼14 kb downstream of TRIM5, which we labeled as CypA3 (Fig. 1A and Fig. S1). CypA3 lies within TRIMP1, which is ∼40 kb long in the rhesus macaque genome but ∼20 kb shorter in the human and chimpanzee genomes. Finally, we discovered CypA4, located ∼99 kb downstream of TRIM5 (Fig. 1A and Fig. S1) in the rhesus macaque genome. Neither CypA3 nor CypA4 was found in the human and chimpanzee genomes (Fig. 1B and Fig. S1).
Fig. 1.
Summary of CypA retrogenes proximal to TRIM5. (A) Relative locations of the CypA retrogenes are displayed below the representations of the TRIM5 locus. CypA2 (light gray arrow), CypA3 (light blue arrow), and CypA4 (white arrow) retrogenes are ∼1 kb, ∼14 kb, and ∼99 kb downstream of TRIM5, respectively. The rhesus macaque TRIMP1 region contains an additional ∼20 kb not present in the human and chimpanzee TRIMP1. (B) Panel of primates investigated by PCR for CypA2, CypA3, and CypA4 is shown in the phylogeny with the notation of retrogene presence or absence indicated to the right. The plus (+) symbol indicates the presence of the CypA retrogene. The minus (−) symbol indicates the absence of the CypA retrogene. Arrows with labels (CypA2, CypA3, Loss of CypA3, and CypA4) indicate the point at which the retrogene was acquired or lost in primate evolution. In our analysis, we recovered CypA2 from southern pig-tailed and lion-tailed macaques. However, given previous reports of the origin and spread of CypA2 (28), we could place the date of its acquisition at the root of the macaque lineage. CypA3 sequences, along with pseudogenizing mutations, are represented for those primates found to encode the retrogene. Stop and Fs denote a stop codon and a frameshift mutation in the CypA3 sequence, respectively.
To determine if the recurrence of CypA retrotranspositions into the TRIM5 locus was greater than what we would expect from random insertions into the genome, we calculated the probability of finding three independent CypA retrogenes within 100 kb of rhesus macaque TRIM5. We queried available primate genomes for all CypA retrogenes and found over 100 CypA retrogenes distributed in the human, chimpanzee, and rhesus macaque genomes, consistent with previous analyses of the human genome (30). Based on the number of CypA retrogenes and their distribution, we found the probability of the three retrogenes in such close proximity to be highly nonrandom (P < 0.0233; Methods). We therefore conclude that some recurrently acting selective pressure must have preserved CypA retrogenes within the TRIM5 locus.
Estimating the Age of CypA2, CypA3, and CypA4.
We sought to understand the temporal distribution of the CypA retrogenes proximal to the TRIM5 locus among primate species. Using PCR and primers to flanking regions of each retrogene, we genotyped the panel of primate genomes for the presence or absence of CypA2, CypA3, and CypA4 (Fig. S1). All CypA retrogenes recovered in this analysis were subsequently sequenced to determine their potential to encode a full-length ORF and for phylogenetic analysis. Consistent with previous reports (25, 26), we found CypA2 to be present in lion-tailed macaque (Macaca silenus) and pig-tailed macaque (Macaca nemestrina) genomes, each generating an ∼2.3-kb band (Fig. S1). We found no evidence of CypA2 outside of macaques. Our results are consistent with a previous study that showed this retrogene is present only within the macaque lineage that arose 5–6 Mya (25).
In contrast to CypA2, we found CypA3 to be present throughout Old World monkeys as well as in gibbons (Fig. 1B and Fig. S1). CypA3 primers were not expected to generate a PCR product from human and chimpanzee genomes due to an ∼20-kb region deletion in TRIMP1 that corresponds to the genomic region containing CypA3 (Fig. 1A). Results from other hominoids (gorilla and orangutan) suggest that this ∼20-kb deletion occurred before the branching of humans and orangutans. We did not observe the presence of CypA3 in any New World monkey genomes. Investigations of the assembled marmoset genome (WUGSC3.2/calJac3 and GenBank accession no. AC148555) revealed no evidence of CypA3 within the ∼20-kb stretch between TRIM5 and TRIM34 (Fig. S2). Additional searches of another New World monkey, Nancy Ma’s night monkey (Aotus nancymaae; Genbank accession no. AC183999), also did not reveal the presence of CypA3 in TRIMP1. Therefore, based on orthologous CypA3 retrogenes in gibbons and Old World monkeys (see below), we can estimate that the ancestral CypA3 retrogene was acquired in primates at least before the Old World monkey/hominoid split (Fig. 1B), which occurred 32 Mya (17).
Similar assays revealed that CypA4 is present in all Old World monkeys assayed but not outside this clade (Fig. 1B and Fig. S1). This suggests that CypA4 retrotransposed before the common ancestor of Old World monkeys, at least 18 Mya (17). Thus, both CypA3 and CypA4 considerably predate the macaque-specific CypA2 retrogene.
Extant Transcriptional Expression of a Pseudogenized TRIMCypA3 Gene Fusion.
To determine whether the retrotransposition of CypA3 or CypA4 into the TRIM5 locus led to the formation of novel TRIMCyp gene fusions, we probed total mRNA from fibroblasts from 16 primate species by RT-PCR, with the forward primer located in the RING domain of TRIM5 and the reverse primer designed to either CypA3 or CypA4. We identified four Old World monkeys (vervet monkey, De Brazza’s monkey, patas monkey, and talapoin) that expressed TRIMCyp transcripts, which included the CypA3 retrogene on their 3′-end (Fig. 2). We found three distinct isoforms of TRIM5-CypA3 (TRIMCypA3) transcripts. Only one of these, isoform-1, has its CypA3 in-frame with TRIM5 exons, where it would be translated as a TRIMCyp gene fusion. Isoform-1 encodes TRIM5 exons 2–7, a short stretch of the upstream region of CypA3, and the CypA3 coding region. The other two isoforms would not result in an in-frame TRIMCyp gene fusion (Fig. 2).
Fig. 2.
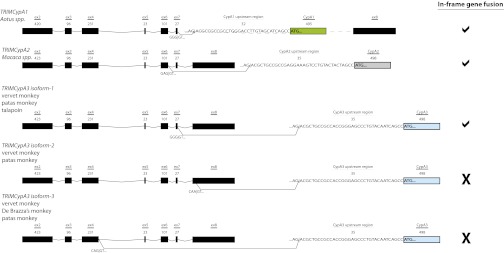
Structure of TRIMCyp transcripts. From a subset of Old World monkeys, we found three TRIMCypA3 isoforms transcribed. TRIM5 exons (black blocks) are joined to a stretch of the upstream region of CypA and the subsequent CypA coding sequence (CypA1, green; CypA2, light gray; CypA3, light blue). We included the sequence of the intron boundaries and the approximate size of each exon. The structures of owl monkey TRIMCypA1 and macaque TRIMCypA2 are also presented for comparison (15, 24, 27). The splice acceptor sequence “AG|AC,” also present in the CypA upstream region, is shown for each retrogene. Isoform-1 encodes TRIM5 exons 2–7 fused in-frame to the CypA3 upstream region and CypA. In contrast, isoform-2 encodes TRIM5 exons 2–8 (62 nt of exon 8) to the CypA3 upstream region and CypA3 coding region, whereas isoform-3 encodes TRIM5 exons 2–4 to the CypA3 upstream region and CypA3 coding region. To the right, we indicated whether the gene fusion produces a product, where TRIM5 effector domains are in-frame with CypA, with a “check mark,” indicating an in-frame product or an “X,” indicating that the product would not be in-frame. Both isoform-2 and isoform-3 would result in an “out-of frame” gene fusion with CypA.
Intriguingly, a shared feature of the gene fusions, including those found in the owl monkey and macaque, is the inclusion of a short segment corresponding to the region immediately upstream of the CypA retrogene coding region (Fig. 2, labeled CypA upstream region, and Fig. S3A). This short DNA segment, which appears to originate from the 5′-untranslated region of the parental CypA gene, encodes a cryptic splice acceptor site that appears conserved throughout mammals (Fig. S3B). At least among primates, this region provides the splice acceptor site and sequences necessary for an in-frame fusion of the CypA retrogene with the TRIM5 effector domains, thereby facilitating formation of the TRIMCyp fusion transcripts.
Sequencing the CypA3 retrogenes from our PCR survey (Fig. S1) revealed signs of pseudogenization in each case (Fig. 1B and Fig. S3), with either a nonsense mutation or a frameshift in their ORF (Fig. 1B). A premature stop codon was identified at the 19th codon of CypA3 in a subset of Old World monkeys (talapoin, patas monkey, and De Brazza’s monkey) of the family Cercopithecidae. In addition, all Old World monkey CypA3 sequences shared a premature stop codon at the site corresponding to the 90th codon of the CypA coding sequence. However, neither stop codon is found within the three orthologous, syntenic gibbon CypA3 sequences from the agile gibbon, island siamang, and white-cheeked gibbon. Instead, all three gibbons encode a frameshift mutation that is predicted to truncate the 3′-end of the CypA3 coding sequence by 69 nt. Thus, gibbons maintain a large portion of their CypA3 coding sequence and do not encode the pseudogenizing mutations found in Old World monkeys. Despite encoding a longer intact ORF than Old World monkeys, we did not detect any evidence of a TRIMCypA3 transcript in gibbons. The CypA3 found in Old World monkeys was likely pseudogenized in their common ancestor, suggesting that the retrogene has existed as a pseudogene in that lineage of primates for 18 My (17). Likewise, we estimate that gibbon CypA3 sequences acquired their frame shift mutation 9–20 Mya in either the gibbon or the hominoid common ancestor (17). Thus, although modern CypA3 sequences are expressed as a TRIMCyp gene fusion, the product is likely defective in all extant primates. However, because the pseudogenizing nonsense mutations found in Old World monkey CypA3 sequences are completely distinct from the frameshift mutation found in gibbon sequences, our analysis strongly implies that CypA3 in the Old World monkey/hominoid ancestor (32myoCypA3) encoded an active ORF (17).
Several attempts to identify a TRIMCyp transcript that includes CypA4 yielded no such product from fibroblast mRNA. It is possible that such a product could be expressed in different tissues. However, it is also likely that CypA4 never contributed to the formation of a TRIMCyp gene fusion, because sequencing of CypA4 retrogenes revealed a shared indel (2-bp deletion) at the position corresponding to the seventh codon, resulting in a pseudogenizing frameshift. Because the CypA4 from all Old World monkeys shares this common pseudogenizing mutation, CypA4 may have become pseudogenized at or shortly after birth.
Evolutionary Analysis of CypA3.
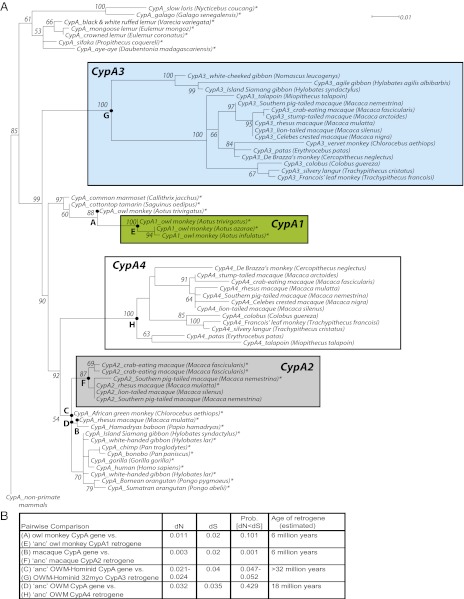
Despite being decayed in all extant primates, the disparate pattern of pseudogenizing mutations in gibbons vs. Old World monkeys suggests that TRIMCypA3 might have encoded an active antiviral gene at one time. To test this, we built a phylogeny (Fig. 3A) composed of intact functional owl monkey CypA1 and macaque CypA2 retrogenes, pseudogenized retrogenes CypA3 and CypA4, as well as primate CypA (parental) genes to calibrate the ages of the retrogenes. The phylogeny shows that all four CypA retrogenes split into four distinct monophyletic groups and that the tree topology and branch lengths are consistent with the estimated evolutionary origins of the retrogenes. For instance, the closest outgroup to the CypA retrogenes that gave rise to Aotus TRIMCyp (CypA1) is the Aotus CypA gene, confirming that Aotus CypA1 retrogenes are derived from a CypA gene within the Aotus genus (16). We are unable to gain high resolution within the hominoid and Old World monkey CypA (parental) genes due to the very high identity of these sequences, likely the consequence of extremely strong purifying selection (31). However, our phylogenetic analysis also places CypA2 retrogenes close to the macaque genus, albeit with poor resolution due to the phylogenetic proximity of the Old World monkey and hominoid CypA genes.
Fig. 3.
Phylogeny of CypA retrogenes. (A) We built a phylogeny of parental CypA genes and retrogenes using maximum likelihood methodologies (57). CypA gene sequences were collected from rodents (outgroup), prosimians, New World monkeys, Old World monkeys, and hominoids. The CypA retrogenes that we included were CypA1 (green-filled box) from owl monkeys, CypA2 (light gray-filled box) from macaques, CypA3 (light blue-filled box), and CypA4 (white-filled box). Bootstrap support values are shown at the nodes. The phylogeny has been rooted to the rodent parental CypA genes: mouse (Mus musculus), rat (Rattus norvegicus), and squirrel (Ictidomys tridecemlineatus). (B) We used the K-Estimator program (58) to evaluate the rates of dN and dS for CypA retrogenes and computed the probability that dN is significantly different from dS by confidence interval tests.
Our PCR genotyping for the presence or absence of CypA retrogenes in primate genomes allowed a tentative dating of their age on a primate phylogenetic tree (Fig. 1). Consistent with our genotyping results, we find that a phylogenetic analysis of the CypA sequences themselves shows that CypA3 and CypA4 retrogenes are older than CypA2 retrogenes, with CypA4 appearing to branch slightly before the common ancestor of the Old World monkeys and hominoids (Fig. 3A). Surprisingly, based on the phylogeny, with 99% bootstrap support, we find that CypA3 was acquired in the simian common ancestor (Fig. 3A) at least 43 Mya (17), which is even earlier than the 32 Mya that we had inferred from PCR genotyping (Fig. S1). However, our attempts to detect CypA3 in New World monkeys were unsuccessful (Fig. S2). This discordance between our genotyping and phylogenetic analysis could be a result of discordance in mutation rates between retrogenes compared with the parental CypA genes. However, because we assume that all these retrogenes were the product of a single cycle of retrotransposition (i.e., retrogenes did not give rise to other retrogenes), we do not believe this difference in mutation rates is sufficient to skew our phylogenetic analysis. We also note the consistency between the genotyped and phylogenetic “age” inferences for CypA1 (16), despite CypA1 having also gone through a retrotransposition event. Instead, we conclude that the CypA3 retrogene was independently lost in the lineage of New World monkeys, similar to its loss in some hominoids. Thus, our phylogenetic results clearly reveal CypA3 as being at least 43 My old, which means that it is, by far, the oldest of the four primate CypA retrogenes.
Using both parsimony and likelihood criteria, we were able to reconstruct the sequence of this “intact” Old World monkey/hominoid ancestral CypA3 (hereafter referred to as 32myoCypA3) based on extant CypA3 sequences. Only a single site, residue 144 of CypA3, could not be resolved (Fig. S3), and it could encode a proline (P), arginine (R), or histidine (H). To determine whether selective constraints (either diversifying or purifying selection) acted on CypA3, we compared the dN/dS ratio of 32myoCypA3 with that of the ancestral parental CypA gene from which it likely derived (Fig. 3B). Finding a high dN/dS ratio would indicate CypA3 was under pressure to evolve adaptively, a low dN/dS ratio would be evidence of selective pressure for protein constraint, and a dN/dS ratio ∼1 would indicate an absence of selective pressure. CypA3 showed evidence suggestive of purifying selection (probability of [dN < dS] = 0.0470–0.0520; Fig. 3B). Applying the same analysis to evaluate the modern, functional owl monkey and macaque TRIMCyps (Fig. 3B), we also find evidence for purifying selection in owl monkey CypA1 (probability of [dN < dS] = 0.1010) and in macaque CypA2 (probability of [dN < dS] = 0.0010). In contrast, an analysis of the ancestral version of CypA4 shows no evidence of selective constraint (probability of [dN < dS] = 0.429). Thus, during the period that it encoded an intact ORF, CypA3 seems to have evolved under similar selective pressures as both owl monkey and macaque CypA retrogenes (Fig. 3B).
The dS values of these retrogene-parental gene comparisons are also informative as a rough proxy for their age of divergence (assuming roughly equal rates of evolution at silent sites). Both the owl monkey and macaque TRIMCyp gene comparisons have a dS of 0.02 (Fig. 3B), consistent with their birth ∼4.5–6 Mya (16, 28). In contrast, the CypA3 comparison between the parental gene and 32myoCypA3 reveals a dS of 0.04 (Fig. 3B), which is twice that of the value estimated for the owl monkey or macaque, suggesting that CypA3 was preserved as an ORF for at least twice as long as the currently intact TRIMCyp gene fusions. This suggests that CypA3 was preserved as an intact retrogene from the time it was acquired 43 to 32 Mya, when we begin to observe evidence of independent pseudogenization (or loss) events across the primate lineages. The signature of purifying selection (Fig. 3B) further suggests the TRIMCypA3 gene fusion was functional during this period of ∼10 My. Because no extant CypA3 sequences could be isolated in New World monkeys, the “oldest” version of ancestral CypA3 that we could faithfully reconstruct represents the version that existed in the last common ancestor of hominoids and Old World monkeys (32myoCypA3) 10 My following the birth of CypA3.
Testing Ancient and de Novo TRIMCyp Proteins for Restriction of Modern or Ancient Lentiviruses.
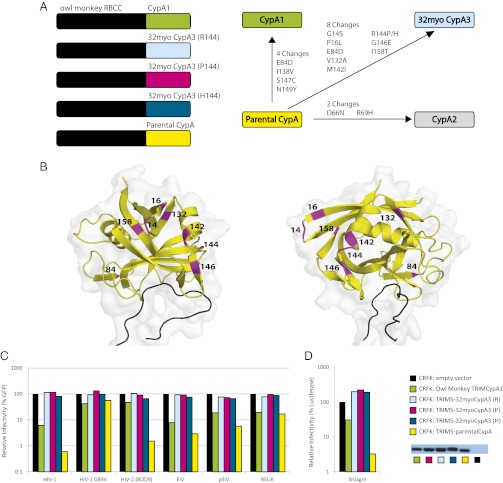
Previous studies have explored the interactions between CypA genes and retrogenes with lentiviral capsids (15, 24–27, 32–34). We were therefore interested in assessing whether ancient, potentially active versions of TRIMCyA3 might have interacted with lentiviral capsids. Both naturally occurring and artificial TRIMCyp genes have highlighted the modularity of the TRIM5-CypA gene arrangement (35). We therefore designed two synthetic TRIMCypA3 versions. We elected to use the TRIM5 effector domains from the owl monkey because the exon structure of TRIMCypA1 closely resembles TRIMCypA3. Thus, the first version (TRIM5-32myoCypA3) consisted of owl monkey TRIM5 effector domains fused to 32myoCypA3 (Fig. 4A). Because we were not able to resolve the identity of residue 144 unambiguously from our evolutionary reconstruction, we constructed three separate TRIM5-32myoCypA3 versions, which encoded a P, R, or H at this site. We also designed a chimera composed of owl monkey TRIM5 effector domains and the inferred parental CypA gene (TRIM5-parentalCypA), representing the CypA gene from which CypA3 derived at the time of its birth. Because CypA genes have been evolving under strong purifying selection throughout primate history, this approach allows us to evaluate the lentiviral specificity of a de novo CypA retrogene unambiguously, representing TRIMCypA3 immediately following its birth (36).
Fig. 4.
Reconstructed ancestral CypA3 vs. modern and extinct (reconstructed) lentiviruses. (A) Cartoon representations of the owl monkey TRIMCypA1 (CypA1, green), owl monkey TRIM5-32myoCypA3 [32myoCypA3 (R144), light blue; 32myoCypA3 (P144), magenta; 32myoCypA3 (H144), teal], and owl monkey TRIM5-parental CypA (yellow) gene fusions, with the owl monkey TRIM5 effector domains (RING, B-box, coiled-coil) represented as a black block. Differences in residues encoded by CypA1, parental CypA, CypA2 (light gray), and 32myoCypA3 have been identified and listed according to the direction of the arrow. (B) Eight residues unique to 32myoCypA3 (P144) (magenta) can be mapped onto a structure of parental CypA (yellow) interacting with capsid (black) using PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC). (C) Stable CRFK cell lines encoding an empty vector (black box), owl monkey TRIMCypA1 (green box), TRIM5-32myoCypA3 (R/P/H144) (light blue, magenta, and teal boxes, respectively), and owl monkey TRIM5-parentalCypA (yellow box) were assayed against chimeric EIAV encoding the ancient (resurrected) capsid of paleolentiviruses RELIK and pSIV (38) and the modern lentiviruses HIV-1 (LAI strain), HIV-1 G89V0, HIV-2 (ROD9 strain), and FIV (9, 56). (D) SIVagm. Viruses are listed along the x axis. The y axis reflects virus infectivity, determined by the percentage of cells infected with GFP-expressing virus, normalized to 100% for infections against CRFK cells encoding an empty vector. The virus inoculums were standardized to give the absolute percentage of GFP between 15% and 30%. In the case of SIVagm, this system used a luciferase reporter. Shown is a representative experiment that was repeated three times. We confirmed the stable expression of TRIMCyp proteins by Western blot analysis, using 30 μg of protein extract for each sample (lane 1, owl monkey TRIMCypA1; lane 2, TRIM5-32myoCypA3 (R144); lane 3, P144; lane 4, H144; lane 5, TRIM5-parental CypA; lane 6, CRFK with an empty LPCX vector).
Consistent with previous results, we observed that owl monkey TRIMCypA1 was not able to restrict HIV-2 (4) but was able to restrict HIV-1 (15), simian immunodeficiency virus from african green monkeys (SIVagm) (18), and feline immunodeficiency virus (FIV) (37), as well as chimeric viruses encoding the reconstructed capsid of the “paleoviruses” RELIK and pSIV (38) (Fig. 4 C and D). Remarkably, we found that the TRIM5-parentalCypA encodes broad and potent antiviral activity, because it restricts all these lentiviruses tested except a mutant that disrupts the CypA binding site on capsid (HIV-1 G89V) (Fig. 4 C and D). Thus, TRIM5-parentalCypA could restrict all representatives tested from the modern-day lentiviruses and the paleolentiviruses (Fig. 4 C and D). The variation between the slightly narrowed binding specificity of CypA1 and the broad specificity of parentalCypA is attributed to four amino acid differences that occurred during CypA1 evolution (Fig. 4A). On the other hand, we also found that TRIM5-32myoCypA could not restrict any of the paleolentiviruses or modern-day lentiviruses (Fig. 4 B and C). Depending on the ambiguous residue 144, parentalCypA and 32myoCypA3 differ at seven (or eight) residues (Fig. 4 A and B), one of which has also independently occurred during CypA1 evolution. We attempted to explore the loss of restrictive ability by evaluating residues unique to 32myoCypA3 within the parentalCypA backbone using both 32myoCypA3 (P144)/parentalCypA and parentalCypA/32myoCypA3 (P144) chimeras (Fig. S4A). We found that the loss of restriction activity in 32myoCypA3 could not be reversed by single residue changes (Fig. S4B), suggesting that this observed loss of restrictive ability in 32myoCypA3 is attributable to a combination of multiple residues among the seven (or eight) residues specific to the 32myoCypA3-encoded protein (Fig. 4 A and B). These results indicate that the most ancient version of the Trim5-CypA fusion gene had the broadest specificity for restriction of retroviruses but that subsequent evolution either narrowed its specificity to (ancient) retroviral capsids that we were not able to test in our assays or destroyed this activity.
Discussion
Recurrent TRIM5-CypA Gene Fusions Across the Primate Phylogeny.
Retrotransposition of CypA retrogenes proximal to the TRIM5 locus has the instantaneous effect of creating a new restriction factor, potentially expanding the restrictive range of primate genomes (24–27). Including the present study, at least three such instances of TRIMCyp gene fusion are now documented in primate genomes. In addition to the still active CypA1 and CypA2 retrogenes that were born in the owl monkey and macaque species 4.5–6 Mya, we have identified a third, much more ancient retrogene that is still present as a fusion transcript and likely encoded a putative restriction factor 43 Mya in primate history. This remarkably convergent retrotransposition proximal to TRIM5, in contrast to the frequent but otherwise random pattern of CypA retrogene insertions elsewhere in primate genomes, strongly suggests that the CypA retrogene-bearing haplotype must have had a strong enough selective advantage to sweep through populations and species. Based on the potent antiviral activity of TRIMCyp fusion proteins, we posit that it is most likely that this selective advantage was conferred by protection against an ancient viral infection.
Previous studies have suggested that CypA fusions function in concert with a variety of TRIM genes (35, 39); however, only TRIM5 has recurrently been revealed to accommodate a functional gene fusion with CypA naturally. It may be that the expression patterns of other TRIM genes could not accommodate a functional antiviral gene fusion without compromising endogenous function. Alternatively, given that TRIM genes homomultimerize via their B-box and coiled-coil domains, homomultimerization of a TRIM gene with CypA might have had deleterious consequences that would only be tolerated when this involved a canonical restriction factor like TRIM5 but perhaps not a TRIM gene that plays an essential housekeeping function in the cell. Consistent with this hypothesis, our survey of rhesus macaque, chimpanzee, and human genomes has not revealed any other TRIMCyp candidates in which a CypA retrogene was found within 20 kb of a TRIM gene.
The retention pattern of TRIMCyp genes could pose a cost to antiviral defense and the cell. In the case of CypA1 and CypA2, it precludes the production of a B30.2-containing TRIM5α from that allele, providing a tradeoff in terms of restrictive potential. This would explain why CypA2 has variably swept through to fixation in macaque species (40), likely as a result of balancing selection, as seen previously in the TRIM5 locus of macaque populations (41). An additional explanation could be that the fusion of the TRIM5 E3 ubiquitin ligase domain to a CypA protein that may bind numerous client proteins in the primate proteome increases the toxic burden of such gene fusions, or leads to aberrant cell signaling (42). In light of this “cost,” if the restriction activity of the evolved TRIMCyp is obviated, either because the restricted viral capsid is eliminated or evolves away from TRIMCyp recognition, the advantage to retain the TRIMCyp gene fusion diminishes greatly. It is also possible that rapid evolution of the B30.2 domain of TRIM5α to recognize the target retroviral capsid might obviate the need to maintain TRIMCyp. Thus, it is not surprising that the evolution of these gene fusions is recurrent and dynamic, both remarkably convergent but also relatively short-lived in evolutionary time. This would also help explain why TRIMCypA3, which may have encoded an active antiviral protein in primate history, is now an extinct gene. Such “extinct” TRIMCyp gene fusions have also been identified outside primates. Indeed, such a pseudogenized TRIMCyp gene fusion (ftr52) was recently identified in fish genomes (43). Finally, even though the CypA domain of TRIMCypA3 has decayed, it is formally possible that the TRIMCypA3 fusion transcript in some primates still serves the same function as some of the alternate TRIM5 isoforms, to attenuate TRIM5α function (44).
CypA3 as a Potential Paleoviral Marker in Primate Evolution.
Based on a relatively abundant record of endogenization revealed by sequencing and bioinformatic efforts, retroviral lineages have been shown to date back many millions of years (45, 46). Indeed, lentiviruses have been estimated to be at least 4 My old in primates (47) and ∼12 My old in other mammals based on the presence of endogenous copies within the host genome, although these dates are likely vast underestimations of the true age of lentiviruses (48–51). In response to these retroviral challenges faced throughout their evolution, primates encode a number of intrinsic mechanisms with the capability of inhibiting viral replication. Positive selection of such restriction factors is a potent mechanism for primate genomes to respond to novel or adapted viral pathogens (9), but it is not the only mode of adaptation. Primate genomes also use other mechanisms, such as gene duplications (52), and in the case of TRIMCyp, recurrent gene fusions, to respond to new viral challenges.
TRIMCyp evolution not only serves to belie the traditional view that retrotransposed genes are evolutionary dead ends but suggests that CypA retrogenes are highly labile modules that can be gained and lost throughout primate history. Although whole-gene dN/dS analyses strongly suggest that CypA3 evolved under purifying selection for 10 My following its birth, we identified seven (or eight) residues within 32myoCypA3 that differentiate a loss of capsid-binding from broad-range capsid-binding (as exhibited by parentalCypA). Similarly, only four residues separate the broad binding of parentalCypA to the narrowed binding specificity documented from CypA1. Macaque CypA2 further demonstrates this trajectory of narrowed binding specificity (40). In the cases of CypA1 and CypA2, deviation from broad capsid-binding evolved within 6 My. Therefore, although broad capsid binding appears as a predisposed feature of CypA, the specificity that each CypA retrogene evolves is determined by minor changes that have a great impact on the capsid-binding trajectory (28, 33, 40). Based on our results with TRIM-parentalCypA, we predict that TRIMCypA3 was also capable of interacting with a broad range of lentiviral capsids on birth. Similar to the specificity-narrowing changes that occurred during TRIMCypA1 and TRIMCypA2 evolution, we posit that in the 10 My after its birth, TRIMCypA3 narrowed its specificity to restrict only ancient retroviruses rather than any of the retroviruses we tested. Finally, after the utility of TRIMCypA3 as a retroviral restriction factor was exhausted ∼32 Mya, the TRIMCypA3 gene decayed in all extant primates.
From a paleovirology perspective, even currently inactive or pseudogenized CypA retrogenes may represent remnants of antiviral genes that were active at an earlier time in primate evolution. We propose that ancient TRIMCypA3 arose in response to a pathogen encountered by evolutionarily successful ancestors. Although it is formally possible that the true target of TRIMCypA3 was a nonlentiviral or even a nonretroviral pathogen, there is little precedent for this conjecture. It is also unlikely that TRIMCypA3 was a genomic innovation due to some other “housekeeping” adaptation, based both on the intrinsic costs of TRIM5-CypA gene fusions and the recurrent pseudogenization/loss of TRIMCypA3 in extant primates. Instead, we propose that the birth and demise of TRIMCypA3 are more consistent with the model wherein it helped protect host genomes against viral invasions for at least 10 My of primate history. Thus, “fossil” antiviral genes like CypA3 provide unique paleoviral insight into viral challenges encountered by primate ancestors 43 Mya and complement the incomplete fossil record of retroviral imprints in animal genomes.
Methods
Identifying CypA Retrogenes Proximal to TRIM5.
The human CypA gene (NC_000007) and mRNA (NM_021130) sequences were used as query sequences in a BLAST-like alignment tool (BLAT) analysis to identify CypA homologs (53). BLAT searches were performed from the University of California, Santa Cruz Genome Browser on the human (Homo sapiens), chimpanzee (Pan troglodytes), and rhesus macaque (Macaca mulatta) genomes (54). For each of the primate genomes, the BLAT search results from the two query sequences were combined to assemble a comprehensive list of CypA homologs that was evaluated and compiled into a catalog of CypA retrogenes. CypA retrogenes were mapped back to their respective primate genome, and CypA retrogenes proximal to TRIM5 were identified (Fig. 1). CypA retrogenes were named according to their distance from TRIM5 and based on previously established nomenclature. These CypA retrogenes were then evaluated for an ORF, indels, and premature stop codons. We also catalogued the distribution of CypA retrogenes and organized these based on the number of CypA retrogenes found in a random stretch of 100 kb of the evaluated primate genomes. To calculate the probability of multiple CypA insertions within a given distance, 100 kb in this case, we counted the number of 100-kb stretches that contained 0–10 CypA retrogenes. We focused on rhesus macaques because the largest number of events in which multiple CypA retrogenes could be found in any 100-kb stretch of its genome was reported in this species. We identified 116 cases of only finding 1 CypA retrogene within 100 kb. In addition, we identified four cases of finding 2 CypA retrogenes and one case of finding 3 CypA retrogenes within 100 kb of each other in the rhesus macaque genome. Thus, of 129 total CypA retrogenes in the rhesus macaque genome, only 3 CypA retrogenes could be found within 100 kb of each other (proximal to the TRIM5 locus), which we can calculate as a probability (P = 0.02325).
Determining the Presence or Absence of Proximal CypA Retrogenes.
Genomic DNA (gDNA) was isolated from primate fibroblast cells purchased from Coriell Cell Repositories. The primate panel was composed of human, chimpanzee (ID no. 3448), bonobo (Pan paniscus, ID no. 5253), gorilla (Gorilla gorilla, ID no. 5251), orangutan (Pongo pygmaeus, ID no. 5252), island siamang gibbon (Hylobates syndactylus, PR00722), agile gibbon (Hylobates agilis albibarbis, PR00773), rhesus macaque (ID no. 7098), crab-eating macaque (Macaca fascicularis, ID no. 3446), celebes-crested macaque (Macaca nigra, ID no. 7101), pig-tailed macaque (M. nemestrina, ID no. 8452), stump-tail macaque (Macaca arctoides, ID no. 3443), lion-tailed macaque (M. silenus, OR1890), silvery leaf langur (Trachypithecus cristatus, bl.4381), Francois’ leaf langur (Trachypithecus francoisi, PR01099), colobus (Colobus guereza, PR00980), talapoin (Miopithecus talapoin, PR00716), patas monkey (Erythrocebus patas, ID no. 6254), De Brazza’s monkey (Cercopithecus neglectus, PR01144), pygmy marmoset (Callithrix pygmaea, OR690), and saddle-back tamarin (Saguinus fuscicollis nigrifrons, OR621) species. gDNA from this diverse primate panel, representing New World monkeys, Old World monkeys, and hominoids, was used to determine the presence or absence of CypA2, CypA3, and CypA4 throughout primates in a PCR survey. All PCR reactions were performed using 25-μL reaction volumes and the PCR SuperMix High Fidelity (Invitrogen) reagent. The thermocycler parameters were 94 °C for 3 min; 39 cycles at 94 °C for 15 s, 60 ° for 15 s, and 72 °C for 2 min; and a final extension step at 72 °C for 10 min. All products were directly sequenced using BigDye sequencing (Applied Biosystems).
CypA2 reactions were performed using primers 105 (forward: 5′-CTGTGCTCACCAAGCTCTTGAAC-3′) and 103 (reverse: 5′-TCCCACATAATTCAGTTTGTTTGATAAA-3′), and CypA4 reactions were performed using primers 108 (forward: 5′-AATCTGCTGGCACCTTGTTTTGTAC-3′) and 110 (reverse: 5′-TAGCTTTTGGGCAGCTAGGAGG-3′). We used nested PCR analysis to amplify CypA3 from primates, with the first-round primers being 87 (forward: 5′-GAACTACTTGAATCCAGGAGGCAGA-3′) and 101 (reverse: 5′-TATCCTCTTTTTGAATCAATTCCTTTGTCA-3′) and the second round primers being 100 (forward: 5′-GCAGGAGTAAGTCCTCACCTATC-3′) and 84 (reverse: 5′-TTATTCGAGTTGTCCACAGTCAGCAG-3′).
Detecting TRIM5-CypA3 and TRIM5-CypA4 Transcripts.
A two-step RT-PCR/seminested PCR-based method was used to amplify TRIMCyp from primate RNA. The primates used were human, chimpanzee, island siamang gibbon, agile gibbon, talapoin, patas monkey, De Brazza’s monkey, vervet monkey (Cercopithecus aethiops, PR01190), Francois’ leaf monkey, colobus, rhesus macaque, woolly monkey (Lagothrix lagotricha, ID no. 5356), spider monkey (Ateles belzebuth, KB6701), titi monkey (Callicebus donacophilus, OR1522), and owl monkey (Aotus trivirgatus, CRL-1556). Total RNA was isolated from fibroblast cells purchased from Coriell Cell Repositories.
The initial RT-PCR step was performed using a primer designed to the start of the coding region of the TRIM5 gene (primer 80) and an oligo-dT reverse primer. This primer combination was used to amplify all products encoded by the TRIM5 gene. Next, we used either a CypA3- or CypA4-specific reverse primer in combination with primer 80 to confirm the transcription of a TRIMCyp gene fusion. RT-PCR reactions were performed using SuperScript III Reverse Transcriptase with Platinum Taq (Invitrogen) in 12.5-μL volume reactions. The RT-PCR parameters were an initial RT step at 50 °C for 30 min; followed by 34 cycles at 94 °C for 15 s, 60 °C for 15 s, and 68 °C for 3 min; and a final extension at 72 °C for 10 min. A 1:300 dilution of the RT-PCR product was then prepared for the subsequent seminested PCR step. This was performed using primers 80 and 73 (reverse: 5′-TTATTMGAGTTGTCCACAGTCAGCARTRGTGA-3′) to amplify TRIMCyp without targeting a specific CypA retrogene. The PCR parameters were kept unchanged. All products were TOPO TA (Invitrogen) cloned and BigDye sequenced using M13 universal primers.
Construction of CypA Phylogeny, Alignment, and 32myoCypA3.
The nucleotide sequences of modern CypA genes and CypA1–4 retrogenes were used to build an alignment of all CypA sequences using Clustal W2 (55). This was done for CypA sequences at the nucleotide and protein levels. The nucleotide alignment was used in reconstructing the 32-My-old form of CypA3 (32myoCypA3). We were able to use a parsimony-based approach to reconstruct the sequence of 32myoCypA, which was in agreement with a maximum likelihood reconstruction.
Phylogenetic trees were generated using Mr. Bayes (version 3.1) in the construction of the CypA phylogeny. We performed 1 million Markov chain Monte Carlo generations with a sampling every 1,000th generation and discarded the first 250 samplings as run-in.
Assessing Ancient and de Novo TRIMCyp Proteins for Antiviral Activity.
To test 32myoCypA3 for the ability to interact with viral capsid protein, we used “stitch-PCR” to join the TRIM5 effector domains (RING, B-box, and coiled-coil) from owl monkey TRIMCypA1 to 32myoCypA. All PCR parameters were as previously mentioned. The first-round set of stitch-PCR used primers 144 (forward: 5′-GCGCTTCTCGAGGCCACCAT-3′) and 134 (reverse: 5′-GGGGTTGACCATGGCTGATGCTAC-3′) to amplify the TRIM5 region of owl monkey TRIMCypA1 and primers 133 (forward: 5′-GTAGCATCAGCCATGGTCAACCCC-3′) and 177 (reverse: 5′-GCGCGCTTATCGATGAATTCTTATTC-3′) to amplify 32myoCypA. Dilutions of the first-round products were combined and stitched together by PCR using primers 144 and 177. This PCR product was sequenced to verify the successful construction of the TRIM5-32myoCypA3 gene fusion. Owl monkey TRIMCypA1 and TRIM5-32myoCypA3 were cloned into the expression vector pLPCX and then transduced into Crandell-Rees feline kidney (CRFK) cell lines to establish the following stable cell lines: CRFK (owl monkey TRIMCypA1) and CRFK (TRIM5-32myoCypA). TRIM5-32myoCypA3 P144R and P144H were generated using a QuikChange II Site-Directed Mutagenesis Kit (Agilent). A CRFK cell line containing pLPCX without an insert was also established to serve as a negative control. The gene fusion of owl monkey TRIM5 effector domains and parentalCypA (TRIM5-parentalCypA) was built by first amplifying the mRNA from the rhesus macaque CypA gene with primers: 262 (forward: 5′-CTGGGACCTTGTAGCATCAGCCATGGTCAACCCCACCGTGTTCTTC-3′) and 264 (reverse: 5′-GCGCGCTTATCGATGAATTATTCGAGTTGTCCACAGTCAGCAATG-3′). Next, the owl monkey TRIM5 amplicon was combined with the rhesus macaque CypA gene using primers 177 and 264. We confirmed TRIMCyp protein expression in the appropriate cell lines by Western blot analysis.
We used the following viruses in assessing our cell lines: HIV-1 (LAI strain), HIV-2 (ROD9), FIV, RELIK, and pSIV, and we used no virus as a control. RELIK and pSIV were prepared by cotransfection of 293T cells with pL-vesicular stomatitis virus-G, pCMV-tat, equine infectious anemia virus (EIAV) GFP 6.1 (encoding the genome of EIAV with a GFP expression cassette), and either pEIAV-RELIK or pEIAV-pSIV (38) (a kind gift from Melvyn Yap, MRC National Institute for Medical Research, London). Viruses were harvested by collecting supernatant and titered on CRFK cells to determine the dose of virus that would infect between 15% and 30% of the cells in a 12-well plate based on flow cytometry for GFP expression. Other viruses were similarly constructed and assayed (9, 56). For infection assays, cell lines were seeded onto 12-well plates and subsequently infected with the aforementioned viruses using the predetermined viral titers. Three days postinfection, cells were collected from the 12-well plates and suspended in fixing agent for immediate analysis by flow cytometry. SIVagm infections were performed using 96-well plates. We did not need to fix RELIK- or pSIV-infected samples before flow cytometry. In all experiments, mock-infected cells were used to set the GFP gate.
Supplementary Material
Acknowledgments
We thank Matt Daugherty, Oliver Fregoso, Patrick Mitchell, and Maulik Patel for comments on the manuscript and members of the Malik and Emerman laboratories, as well as the Thursday Morning Retrovirus Club, for useful discussions. This work was supported by Fred Hutchinson Cancer Research Center Interdisciplinary Training Grant T32 CA 80416 and Ruth L. Kirschstein National Research Service Awards for Individual Predoctoral Fellowship F31 A1084620 (to R.M.-B.), National Institutes of Health Grant R01 AI3097 (to M.E.), and a National Science Foundation CAREER award (to H.S.M.). H.S.M. is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KC146412–KC146414 and JX896147–JX896164).
See Author Summary on page 2447 (volume 110, number 7).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1216542110/-/DCSupplemental.
References
- 1.Patel MR, Emerman M, Malik HS. Paleovirology—Ghosts and gifts of viruses past. Curr Opin Virol. 2011;1(4):304–309. doi: 10.1016/j.coviro.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emerman M, Malik HS. Paleovirology—Modern consequences of ancient viruses. PLoS Biol. 2010;8(2):e1000301. doi: 10.1371/journal.pbio.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 4.Zhang F, Hatziioannou T, Perez-Caballero D, Derse D, Bieniasz PD. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology. 2006;353(2):396–409. doi: 10.1016/j.virol.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 5.Song B, et al. Retrovirus restriction by TRIM5alpha variants from Old World and New World primates. J Virol. 2005;79(7):3930–3937. doi: 10.1128/JVI.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yap MW, et al. Restriction of foamy viruses by primate Trim5alpha. J Virol. 2008;82(11):5429–5439. doi: 10.1128/JVI.02462-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yap MW, Nisole S, Lynch C, Stoye JP. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci USA. 2004;101(29):10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahm N, et al. Unique spectrum of activity of prosimian TRIM5alpha against exogenous and endogenous retroviruses. J Virol. 2011;85(9):4173–4183. doi: 10.1128/JVI.00075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102(8):2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaiser SM, Malik HS, Emerman M. Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science. 2007;316(5832):1756–1758. doi: 10.1126/science.1140579. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Li X, Stremlau M, Lee M, Sodroski J. Removal of arginine 332 allows human TRIM5alpha to bind human immunodeficiency virus capsids and to restrict infection. J Virol. 2006;80(14):6738–6744. doi: 10.1128/JVI.00270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirmaier A, et al. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 2010;8(8):e10000462. doi: 10.1371/journal.pbio.1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebastian S, Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maillard PV, Ecco G, Ortiz M, Trono D. The specificity of TRIM5 alpha-mediated restriction is influenced by its coiled-coil domain. J Virol. 2010;84(11):5790–5801. doi: 10.1128/JVI.02413-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430(6999):569–573. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 16.Ribeiro IP, et al. Evolution of cyclophilin A and TRIMCyp retrotransposition in New World primates. J Virol. 2005;79(23):14998–15003. doi: 10.1128/JVI.79.23.14998-15003.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perelman P, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7(3):e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin TY, Emerman M. Cyclophilin A interacts with diverse lentiviral capsids. Retrovirology. 2006;3:70. doi: 10.1186/1742-4690-3-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thali M, et al. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372(6504):363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- 20.Nisole S, Lynch C, Stoye JP, Yap MW. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc Natl Acad Sci USA. 2004;101(36):13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatziioannou T, Perez-Caballero D, Yang A, Cowan S, Bieniasz PD. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc Natl Acad Sci USA. 2004;101(29):10774–10779. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perron MJ, et al. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc Natl Acad Sci USA. 2004;101(32):11827–11832. doi: 10.1073/pnas.0403364101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103(14):5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brennan G, Kozyrev Y, Hu SL. TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc Natl Acad Sci USA. 2008;105(9):3569–3574. doi: 10.1073/pnas.0709511105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman RM, et al. Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 2008;4(2):e1000003. doi: 10.1371/journal.ppat.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Virgen CA, Kratovac Z, Bieniasz PD, Hatziioannou T. Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc Natl Acad Sci USA. 2008;105(9):3563–3568. doi: 10.1073/pnas.0709258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson SJ, et al. Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc Natl Acad Sci USA. 2008;105(9):3557–3562. doi: 10.1073/pnas.0709003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dietrich EA, Jones-Engel L, Hu SL. Evolution of the antiretroviral restriction factor TRIMCyp in Old World primates. PLoS ONE. 2010;5(11):e14019. doi: 10.1371/journal.pone.0014019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoye JP, Yap MY. Chance favors a prepared genome. Proc Natl Acad Sci USA. 2008;105(9):3177–3178. doi: 10.1073/pnas.0800667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Harrison PM, Liu Y, Gerstein M. Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13(12):2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ortiz M, Bleiber G, Martinez R, Kaessmann H, Telenti A. Patterns of evolution of host proteins involved in retroviral pathogenesis. Retrovirology. 2006;3:11. doi: 10.1186/1742-4690-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Towers GJ, et al. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat Med. 2003;9(9):1138–1143. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- 33.Price AJ, et al. Active site remodeling switches HIV specificity of antiretroviral TRIMCyp. Nat Struct Mol Biol. 2009;16(10):1036–1042. doi: 10.1038/nsmb.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dietrich EA, et al. Variable prevalence and functional diversity of the antiretroviral restriction factor TRIMCyp in Macaca fascicularis. J Virol. 2011;85(19):9956–9963. doi: 10.1128/JVI.00097-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yap MW, Mortuza GB, Taylor IA, Stoye JP. The design of artificial retroviral restriction factors. Virology. 2007;365(2):302–314. doi: 10.1016/j.virol.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Neagu MR, et al. Potent inhibition of HIV-1 by TRIM5-cyclophilin fusion proteins engineered from human components. J Clin Invest. 2009;119(10):3035–3047. doi: 10.1172/JCI39354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diaz-Griffero F, et al. Comparative requirements for the restriction of retrovirus infection by TRIM5alpha and TRIMCyp. Virology. 2007;369(2):400–410. doi: 10.1016/j.virol.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstone DC, et al. Structural and functional analysis of prehistoric lentiviruses uncovers an ancient molecular interface. Cell Host Microbe. 2010;8(3):248–259. doi: 10.1016/j.chom.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Javanbakht H, et al. The ability of multimerized cyclophilin A to restrict retrovirus infection. Virology. 2007;367(1):19–29. doi: 10.1016/j.virol.2007.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ylinen LM, et al. Conformational adaptation of Asian macaque TRIMCyp directs lineage specific antiviral activity. PLoS Pathog. 2010;6(8):e1001062. doi: 10.1371/journal.ppat.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newman RM, et al. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5alpha. Proc Natl Acad Sci USA. 2006;103(50):19134–19139. doi: 10.1073/pnas.0605838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pertel T, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472(7343):361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boudinot P, et al. Origin and evolution of TRIM proteins: New insights from the complete TRIM repertoire of zebrafish and pufferfish. PLoS ONE. 2011;6(7):e22022. doi: 10.1371/journal.pone.0022022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berthoux L, Sebastian S, Sayah DM, Luban J. Disruption of human TRIM5alpha antiviral activity by nonhuman primate orthologues. J Virol. 2005;79(12):7883–7888. doi: 10.1128/JVI.79.12.7883-7888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katzourakis A, Gifford RJ, Tristem M, Gilbert MT, Pybus OG. Macroevolution of complex retroviruses. Science. 2009;325(5947):1512. doi: 10.1126/science.1174149. [DOI] [PubMed] [Google Scholar]
- 46.Han GZ, Worobey M. An endogenous foamy-like viral element in the coelacanth genome. PLoS Pathog. 2012;8(6):e1002790. doi: 10.1371/journal.ppat.1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilbert C, Maxfield DG, Goodman SM, Feschotte C. Parallel germline infiltration of a lentivirus in two Malagasy lemurs. PLoS Genet. 2009;5(3):e1000425. doi: 10.1371/journal.pgen.1000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gifford RJ. Viral evolution in deep time: Lentiviruses and mammals. Trends Genet. 2012;28(2):89–100. doi: 10.1016/j.tig.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 49.Keckesova Z, Ylinen LM, Towers GJ, Gifford RJ, Katzourakis A. Identification of a RELIK orthologue in the European hare (Lepus europaeus) reveals a minimum age of 12 million years for the lagomorph lentiviruses. Virology. 2009;384(1):7–11. doi: 10.1016/j.virol.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han GZ, Worobey M. Endogenous lentiviral elements in the weasel family (mustelidae) Mol Biol Evol. 2012;29(10):2905–2908. doi: 10.1093/molbev/mss126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui J, Holmes EC. Endogenous lentiviruses in the ferret genome. J Virol. 2012;86(6):3383–3385. doi: 10.1128/JVI.06652-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han K, Lou DI, Sawyer SL. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet. 2011;7(12):e1002388. doi: 10.1371/journal.pgen.1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kent WJ. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 56.Yamashita M, Emerman M. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol. 2004;78(11):5670–5678. doi: 10.1128/JVI.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 58.Comeron JM. K-Estimator: Calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics. 1999;15(9):763–764. doi: 10.1093/bioinformatics/15.9.763. [DOI] [PubMed] [Google Scholar]