Abstract
Background
Mass spectrometry imaging (MSI) is a powerful tool that grants the ability to investigate a broad mass range of molecules from small molecules to large proteins by creating detailed distribution maps of selected compounds. Its usefulness in biomarker discovery towards clinical applications has obtained success by correlating the molecular expression of tissues acquired from MSI with well-established histology.
Results
To date, MSI has demonstrated its versatility in clinical applications, such as biomarker diagnostics of different diseases, prognostics of disease severities and metabolic response to drug treatment, etc. These studies have provided significant insight in clinical studies over the years and current technical advances are further facilitating the improvement of this field. Although the underlying concept is simple, factors such as choice of ionization method, sample preparation, instrumentation and data analysis must be taken into account for successful applications of MSI. Herein, we briefly reviewed these key elements yet focused on the clinical applications of MSI that cannot be addressed by other means.
Conclusions
Challenges and future perspectives in this field are also discussed to conclude that the ever-growing applications with continuous development of this powerful analytical tool will lead to a better understanding of the biology of diseases and improvements in clinical diagnostics.
Keywords: Mass spectrometry imaging, MALDI, DESI, clinical, biomarkers
1. Introduction
The integration of gel electrophoresis and liquid chromatography coupled to MS enables high throughput characterization of complex proteomes to detect disease biomarkers [1-4] or evaluate the response to the exposure of drugs or stress [5-8]. Yet spatial localization of the detected proteins and their corresponding expression changes to specific disease or treatment cannot be attained. Conventional immunohistochemical staining (IHC) allows for obtaining high resolution distribution images of targeted proteins. However, a significant limitation to this standard method is the need for labeling, which means that the target molecules must be known prior to the experiment. Alternatively, mass spectrometric imaging (MSI) has evolved as a powerful tool for the analysis of a wide range of molecules, mainly using matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS), but also using other ionization methods such as desorption electrospray ionization mass spectrometry (DESI-MS) and secondary ion mass spectrometry (SIMS). MSI has enormous advantages over conventional protein imaging techniques in that not only it is label-free, but it also enables simultaneous mapping of numerous molecules in tissue samples with great sensitivity and chemical specificity. MALDI-MSI has proven to be a valuable technology with numerous applications in localizing proteins [9,10], examining lipid distributions [11] and mapping neuropeptides [12,13], at both organ and cellular levels, by varying the experimental conditions. Since no prior knowledge of molecular identities is required for MSI applications, it has become a standard biomarker discovery tool to compare analyte expression pattern changes by analyzing multiplexed data sets. To date, MSI has been applied to identify biomarkers directly from tissue sections as an aid to disease diagnosis or prognosis of different disease types [14]. It has also been used to determine the response of the subjects to drugs or other therapeutic regimes, shedding light in studies of personalized medicine. Herein we review current publications that underscore the critical role MSI, especially MALDI-MSI, plays in the study of molecular dynamics in the context of clinical research. With MSI, a better understanding of the molecular pathology of various diseases such as cancer and neurodegenerative diseases can be obtained, and this information can be used for more efficient diagnoses and improved treatments.
2. Methodology
2.1 Ionization
MALDI has shown its revolutionary power with its capability of analyzing a wide mass range of intact molecules spanning from large proteins and peptides to small metabolites and lipids with a “soft” and efficient ionization source. Other than the wide mass range, MALDI also delivers several other unique features in comparison to other ionization techniques, such as a great tolerance for salts, producing mostly singly charged ions and low femtomole to attomole sensitivity. Moreover, its capability to acquire biomolecules’ mass to charge ratio (m/z) and additional sequence information utilizing post source decay (PSD) or LIFT (a short form of “potential lift”) [15] demonstrated its power for unraveling and understanding molecular complexity. Nitrogen or neodymium-doped yttrium aluminum garnet (Nd:YAG) lasers are usually employed to perform MSI experiments. The size of the laser beam and the matrix crystal combinatorially determine the spatial resolution of MALDI-MS images that can be attained. To date, a commercially available MALDI-TOF/TOF with a regular matrix sprayer is usually able to provide a spatial resolution of 30~75 μm. The resolution could be further improved down to cellular scale with the advances discussed in 2.2.2 single-cell MSI.
SIMS is another technique that has long been established for MSI applications. In SIMS, ionization takes place by first bombarding a solid sample under a high vacuum with high energy “primary” ions [16]. Following the primary ion impact, secondary ions are sputtered from the sample surface and then be drawn into a mass analyzer, typically a TOF analyzer, for surface chemistry analysis. SIMS provides excellent spatial resolution due to its highly focused ion beam. Nevertheless, the primary ion source is fairly limited to small molecules due to its high energy that is prone to fragment large molecules in the ablation/ionization process. Recent advances in SIMS, such as metal cluster and polyatomic ion sources, provide significantly greater ion yields of ions in the m/z 400–3000 range with a reasonable resolution of 200 nm [17]. However, as suggested by Jones et al. , while SIMS can easily achieve spatially resolved images of small molecules for MSI applications, it will not likely be able to match MALDI’s analytical capabilities for proteins [17].
MALDI and SIMS-MSI both require vacuum for specimen analyses, complicating MSI procedure and limiting the applications to live biological samples. DESI, in comparison, is a simple, ambient ionization technique [18-21]. DESI channels charged solvent droplets and ions from an electrospray source onto the surface of interest [22]. The surface is impacted by the charged particles, yielding gaseous ions of the originally present biomolecules. By rastering the electrospray liquid jet across the surface of interest, DESI-MSI can be performed. DESI’s ambient nature and softness allow for the examination of various natural surfaces with no need for matrix [23]. Moreover, its clinical perspectives of miniaturization for field applications strongly poised this technique to be an innovative, portable tool since vacuum is no longer necessitated [24]. The downfall DESI has over vacuum MS methods is its spatial resolution that is generally reported to be approximately 180–200 μm [23]. Solutions have been undertaken to increase this resolution to 12 μm by controlled desorption of analytes present in a restricted region of specimen using a minute amount of solvent between two capillaries comprising the nano-DESI probe [20].
2.2 Sample Preparation for MSI
A typical workflow of MSI generally consists of sample preparation, MSI acquisition and data analysis as simplified in Figure 1. These elements strongly determine the outcome of MSI experiments and are thereby elaborated in the following sections.
Figure 1.
A schematic representation of the clinical applications of MSI. MSI mass spectra arising from the diseased regions are recorded, and the molecular MS images are reconstructed. The molecules like lipids or proteins that differentiate the diseased regions from the normal ones are potential biomarkers of the disease. Moreover, the metabolites that changed corresponding to drug treatment are also investigated based on this workflow.
2.2.1 Sample preparation for tissues
2.2.1.1 Sample preparation for fresh tissues
Sample preparation in imaging experiments aim to generate reproducible and reliable MS images directly from tissue sections or cells. The structural integrity and morphology of tissues must be maintained after sample treatment without delocalization and degradation of analytes. The most common procedure after harvesting tissue is snap-freezing in powdered dry ice or liquid nitrogen, followed by storage at −80°C until use [25]. Another method involves loosely wrapping the tissue in aluminum foil and gently placing it into liquid nitrogen, ice-cold ethanol or isopropanol bath for 30-60 sec [26]. We recommend the latter approach since the gentler, longer freezing process avoids tissue cracking and fragmentation. Prior to long-term storage, tissue stabilization is recommended to minimize the sample aging effect. Tissue stabilization methods, such as microwave irradiation [27] and heat denaturation by Denator Stabilizor T1 (Gothenburg, Sweden), have been reported to effectively deactivate proteolytic enzymes, preventing post mortem degradation of proteins or peptides of interest [28,29].
MSI experiments typically require 10-20 μm thick tissue sections [30]. Embedding tissues in supporting media such as gelatin [12,13] or sucrose [31] allows for easy handling and precise sectioning of tissue samples without introducing interferences to mass spectrometer, whereas polymer-containing material, such as optimal cutting temperature (OCT) compound, Tissue-tek and carboxymethylcellulose (CMC), should be avoided [26].
Tissue sections are then transferred and attached onto a stainless steel conductive plate or indium-tin-oxide (ITO)-coated conductive glass slides [32,33] by thaw-mounting [26]. ITO-coated glass slides are more routinely used to analyze biomedical samples these days since it allows microscopic analyses of the tissue in MSI experiments with clinical diagnostic purpose.
Washing tissue sections with organic solvents is another recommended step to increase ion yields of protein/peptide signal by fixing tissues and removing ion-suppressing salts and lipids [34]. Through comparison with various solvents [35,36], graded ethanol has become a routine procedure for MSI of clinical proteomics by enhancing protein signals effectively and reproducibly. In contrast, SIMS and DESI-MS require relatively simple sample preparation, mainly involving tissue sectioning, due to their limited applicability to low molecular weight (MW) analytes instead of peptides and proteins.
2.2.1.2 Sample preparation for FFPE tissues
Although many successes have been achieved using fresh frozen tissues for MSI, the most commonly used preservation technique for clinical tissue specimens is formaldehyde-fixed paraffin-embedding (FFPE). FFPE allows long-term storage at room temperature and is a preferred tissue preservation technique by clinical researchers. Nevertheless, the use of formalin leads to protein/peptide cross-linking and tissue embedding in paraffin wax brings severe interferences to MS analysis [37]. Caprioli et al. reported a novel technique to re-gain access to the proteins through a process of antigen retrieval by heating [38]. This process first denatures proteins, thus allowing for enzymatic digestion of these accessible proteins to non-crosslinked peptides that can be analyzed by MS. The peptides of interest are then isolated for MS/MS sequencing and searched against a database to retrieve the parent protein. With this approach, the distribution information of proteins can be obtained from FFPE tissues and correlated to the histology. This method opens up new possibilities of discovering disease biomarkers by analyzing a larger pool of samples from tissue bank, compared to available fresh frozen tissues, for statistical confidence. Unfortunately, FFPE tissues are not amenable to analyses of lipids, which are involved in various biological and disease processes, with current technologies, due to the repetitive washing of the tissues.
2.2.1.3 Laser capture microdissection
A major challenge of MSI is the lack of selectivity of the analytes during the ionization process and thus the ion suppression effect could impact the detection of low-level analytes of interest. One solution to this problem is to purify cells of interest from the entire tissue section by laser capture microdissection (LCM) prior to MSI. LCM is a technique that allows for isolation of samples down to single cell scale from thin tissue sections by irradiating a focused laser beam to the target region that is adhered to a heat-sensitive polymer film and subsequently removed upon irradiation [39]. The incorporation of LCM into MSI pipeline for spatially-resolved sampling has been demonstrated by mapping of proteins in the mouse epididymis [32], ocular lens [40], human breast tissue [41] and rat kidneys [42], etc.
2.2.1.4 In situ tryptic digestion
Although MALDI-MSI is powerful in mapping proteins throughout tissue sections, it is significantly more difficult to identify the potential proteins of interest via MALDI-MS alone due to limited fragmentation obtainable from the singly-charged, high-mass ions produced by MALDI-MS. Therefore, the ability to digest the proteins and perform MS/MS sequencing directly on the tissue allows us to identify proteins in situ with high confidence without losing spatial resolution, thus facilitating the discovery of protein disease biomarkers. Groseclose et al. carried out in situ digestion by robotically spotting trypsin solution onto a coronal rat brain section in a well-defined microspotted array followed by automatic deposition of matrix [43]. Subsequent collection of MS and MS/MS spectra enables sequencing of tryptic fragments and thereby protein identification. This technique is fairly flexible in the workflow of MSI, compatible with upstream preparation like FFPE-tissue processing [44] or downstream coupling to ion mobility analysis [45,46].
2.2.2 Single-cell MSI
While MSI offers a powerful tool for clinical diagnosis and disease prognosis evaluation, single-cell MSI presents unique merits and challenges that deserve special attention. One limiting factor of single-cell MSI is instrument sensitivity [47], since the amount of analytes decreases as the square of the spatial resolution, as reviewed by McDonnell [48]. Induction-based fluidics (IBF) reported by Tu et al. alleviated this issue by using a charged capillary tip to charge the nL-sized matrix solution followed by launching the droplet onto the specimen [49]. This technique is amenable to single-cell MSI since the analyte diffusion is minimized and the resulting intensity is increased by 10-fold compared to conventional manual spotting by pipette.
An adaptable and cost-effective setup, the stretched sample method, was developed by Zimmerman et al. to improve the spatial resolution for single-cell scale MSI [50]. Briefly, dissected, individual cells, like neurons, were placed on a stretchable substrate, Parafilm, embedded with a monolayer of bead array. Then Parafilm was manually stretched and placed on conductive ITO glass slides for further MALDI-MSI experiments [50,51]. The MS images of the cells were registered to their original locations based on the optical image taken prior to stretching.
The Sweedler group further improved the spatial resolution by reducing the incremental movement of the laser beam to smaller than the laser diameter [52]. They introduced this setup as an “oversampling” method by desorbing/ionized from a much smaller area with each incremental step after sample material is ablated away at the initial spot. Such oversampling procedures are relatively simple and do not need any additional hardware. Nevertheless, this method flaws in that the total sample has to be consumed, excluding the possibility to re-analyze the samples.
On the other hand, several custom-designed mass spectrometers permit single-cell resolution. For example, Spengler et al. implemented special confocal-type objectives, which were furthered modified by Rompp and Guenther et al., offering sub-micron spatial resolution [53-55]. This ion optical and laser setup was then coupled to a high-end Q-Exactive Orbitrap Fourier transform mass spectrometer (FTMS) by Schober et al., delivering high resolution MS images of metabolites and lipids in Hela cells [56]. High spatial resolution down to 7 μm was reported by Chaurand et al. by introducing a coaxial laser illumination ion source to a MALDI-TOF [33]. Moreover, a proprietary smartbeam-II MALDI laser developed by Bruker Daltonics allows beam diameters to be focused down to 10 μm and permits routine MSI analyses to be performed at the cellular level [57].
An alternative approach to increasing single-cell spatial resolution is “microscope” MALDI-MSI by employing a setup that integrates a defocused UV laser, high-quality ion optical and a position-sensitive detector to record the position of the stigmatic projection of ions [58, 59]. Luxembourg et al. reached a pixel size of 500 nm and a resolving power of 4 μm using this configuration [58, 60]. However, the use of this method is limited due to technical constraints like the specialized ion optics, a fast detector necessitated to achieve high magnification and computing software to reconstruct ion images.
MSI has been extensively used in differentiation between healthy and diseased tissues with the purpose of clinical prognostics and diagnostics. Its cutting-edge applications to single-cell analyses will provide detailed biochemical information at a cellular scale for mechanistic understanding and, ultimately, development of therapeutic treatments. The recent advances of single-cell MALDI-MSI and its clinical impact are presented by Boggio et al.[47].
2.3 Matrix Application
Choosing a matrix and its application method are critical to MSI results. Other than conventional matrices such as α-cyano-4-hydroxycinnamic acid (CHCA) and 2,5-dihydroxy benzoic acid (DHB) [12, 13, 26], ionic matrices made by mixing conventional matrices with organic bases are also widely used and reported to improve spectral quality, crystallization and vacuum stability [61, 62].
Matrix application is another area under continuous innovation. Several types of matrix application apparatus exist by depositing matrix either as homogeneous layers (spray coating) or discrete spots (microspotting). Pneumatic spray device such as pneumatic sprayer, airbrush, TM sprayer system from HTX imaging and thin layer chromatography sprayer, and vibrational spray apparatus like ImagePrep Device from Bruker Daltonics are all capable of applying a uniform layer of small to medium-sized matrix droplets [12,13,63,64], whereas microspotters like CHIP manufactured by Shimadzu deposit pL-sized droplets of matrix according to a predefined array and require multiple rounds of spotting for sufficient matrix coverage. In comparison to wet application, solvent free methods like sublimation or dry coating yield very fine crystals amenable to high-spatial-resolution-MSI. However, it suffers from relatively low sensitivities due to the limited analyte-matrix interactions [65]. Deutskens et al. modified this procedure by rehydrating the sections following dry-coating and improved the detection sensitivity of proteins from rat cerebellum sections [63].
2.4 MS Instrumentation
An ion source can be couple to a majority of the current state-of-the-art mass analyzers for MSI capabilities. Time-of-flight (TOF) mass analyzers are most extensively used in MALDI-MSI applications. In TOF analyzers, desorbed ions are accelerated to the same kinetic energy and the m/z is determined by the time the ions take to travel through the TOF tube [66]. This design provides high sensitivity, a wide mass range (2~30 kDa) and fast analysis speed, thereby favored by the applications of MSI to clinical proteomics. The detection sensitivity of higher mass ions was further improved by Leinweber et al. who observed that the number of high MW proteins ( > 20 kDa) detected from tissues by MALDI-TOF was significantly increased by doping Triton X-100 into the matrix [67]. Nevertheless, the characterization of peptides and low MW compounds necessitate the addition of another TOF tube, allowing for accurate measurement of peptides’ and small molecules’ masses and tandem mass fragmentation for sequence validation [68].
The integration of ion mobility (IM) with MALDI-TOF is a breakthrough to MALDI-MS-based analyses. IM-TOF is a 2-dimensional gas-phase separation technique that discriminates ions based on their m/z and collisional cross-section. Upon desorption/ionization from the tissue surface in MALDI source, ions travel inside an IM drift cell, which is equipped with an applied electric field and a carrier buffer gas that opposes ion motion. The ion’s mass, charge, size and shape combinatorially determine its migration time inside the drift cell, consequently managing to discriminate isobaric ions according to their collisional cross-section [69]. For instance, IM-TOF has proven its usefulness in MSI of in situ digested proteins. Stauber et al. successfully separated and identified isobaric ions of tubulin and ubiquitin peptide fragments at m/z 1039 directly from a rat brain section based on different drift times, whereas the MS images of the two peptides would be merged without additional separation by IM [46]. The benefit of performing IM separation prior to MS measurements is enormous and promises to find extensive applicability to MALDI-MSI-based clinical research.
Compared to the relatively low resolving power offered by a TOF/TOF mass analyzer, Fourier transform ion cyclotron resonance (FT-ICR) mass analyzers provide superior resolution and accuracy for unambiguous discrimination of analytes without sacrificing spatial resolution [70]. Moreover, it also provides unique ion trapping and storage capabilities, which are utilized by Kutz et al. for in-cell accumulation [71] and by Bruker Daltonics for continuous accumulation of selected ions (CASI) [72] to improve the detection sensitivity of trace-level analytes. The primary drawback of FT-ICR is the slow scan speed, which lowers the throughput for serial MSI experiments. A more powerful magnetic field could partially alleviate this problem by increasing the ion cyclotron frequency without affecting spectral quality. In addition, the Orbitrap manufactured by Thermo Fisher Scientific provides comparable resolution to an FT-ICR instrument. Orbitrap determines the m/z by measuring the axial oscillation frequency of ions back and forth along a spindle-like electrode within an electrostatic field [73], which is proportional to the square root of the electrical-field strength, whereas the cyclotron frequency measured by FT-ICR is related to the magnetic field strength [74]. To obtain high resolving power on an FT-ICR, high-field magnets are needed, concomitantly at a high cost. Predictably, the Orbitrap is gaining more popularity in future MSI applications due to its excellent performance and relatively low cost compared with FT-ICR.
2.5 Histology-guided MSI
One of MSI’s advantages is that spatial localization of analytes within a tissue is preserved in comparison to homogenization. It allows for differentiation of tissue regions based on molecular features. Therefore, MSI can be integrated with histology to directly target for diseased regions and correlate the peaks that are up-regulated/down-regulated in the specific regions with the reference information provided by histology. To date, histology-guided imaging has become the standard for applying MSI to clinical diagnostics [47,75-79]. Hematoxylin and eosin (H&E) staining is commonly performed on clinical tissue samples. This technique allows the visualization of cells with bright field microscopy by labeling the nucleus and cytoplasm of the cells [47]. A trained pathologist can then use the H&E stained tissue to establish a diagnosis [75]. Methods of histology guided MSI have been developed that allow both the histological features and the MS image of the tissue to be observed. In such methods, the histological stain is applied to a serial section of the tissue that is to be analyzed by MSI, which dissipates the concern for mass spectrometry-friendly stains [75, 76]. One downfall to this method is that it can be difficult to obtain serial sections, reproducibly, without tearing or folding the tissue on the cryostat [77]. This can cause a misalignment of the sections, causing a loss of correlation of small features. Another method is to apply the histological stain directly to the tissue that is to be analyzed by MSI. This method requires MS-compatible stains, such as cresyl violet or methylene blue, or performing imaging first and washing off the matrix before applying the histological stain [47,77]. The downfall of this method is that H&E staining is not compatible with MSI. Therefore the standard workflow would have to be changed in order to include non-standard histological stains and non-routine evaluation of the results [77]. The future of MSI in clinical diagnostics relies on incorporating it into the current diagnostic workflow, and must be compared and correlated with histological results [78].
2.6 Data Analysis
MSI data analysis software, such as BioMap (http://www.maldi-msi.org, available for free downloading) and proprietary programs for MSI systems (e.g., FlexImaging from Bruker Daltonics, ImageQuest from Thermo Fisher Scientific and TissueView from Applied Biosystems/MDS), are mostly employed to produce distribution maps for selected analytes. These software packages allow the user to adjust color scales, overlay ion density maps, and integrate MS images with acquired histological pictures.
Other than image processing, software that provides statistical support are also applied to MSI data analysis. For example, biomarker discovery studies usually involve comparing MSI data sets sampled from a control group with those from a treated group. Data-mining software, like ClinProTools by Bruker Daltonics, capable of performing principal component analysis (PCA) and hierarchical clustering of multiple MSI data sets to extract differentially expressed or distributed molecules, are widely used for identification of potential disease biomarker candidates.
3 Clinical Applications
Over the past decade, MSI has become a powerful tool that has been extensively applied to various clinical applications. In this section, we present current advances of this technique and its novel applications in clinical setting, including drug response measurement (3.1), lipid and protein biomarker discovery (3.2) and several other novel applications (3.3) as summarized in Figure 1.
3.1 Measuring Drug Response and Metabolites
Histology guided MSI has been extensively applied to the study of drug response and distribution [80-83]. Drexler et al. [80] combined quantitative whole-body autoradiography (QWBA) and imaging mass spectrometry to simulate an in vivo phototoxicity study of a proprietary drug candidate (Bristol-Myers Squibb Company, Princeton, NJ) within ocular tissue. QWBA reveals quantitative and tissue distribution information of radiolabeled analytes [84]. However, QWBA cannot be used to obtain information about the drug and its metabolites or degradation products. MSI is notorious for giving poor quantitative data, but can reveal information about the analyte and its products; therefore it was advantageous to combine QWBA and MSI for this study. The radio-labeled and non-labeled drug candidate, BMS-X, was dosed to two different test groups of rats. Two hours after the dose was administered, the animals were sacrificed for QWBA and MALDI-MSI experiments. The QWBA experiment showed that radio-labeled analyte(s) preferentially distributed to the back of the eye, not the cornea or lens, but revealed no information about the molecular species of the radio-labeled analyte(s). The MSI experiments detected only BMS-X, again in the back of the eye, therefore suggesting that there were no metabolites or degradation products of the drug present in the tissue. Employing complementary technologies of MALDI-MSI and QWBA, Drexler et al. were able to assess the distribution of a new drug candidate and its potential metabolites which is essential for phototoxicity studies of pharmaceuticals before they can be prescribed in a clinical setting.
For pharmacokinetic and toxicity studies, the quantitative analysis of the in vivo distribution of a drug after it has been administered is essential. One of the major limitations to MSI is the lack of ability for successful quantification, as mentioned previously. A quantitative research method using MSI is greatly desired and could be very insightful for the pharmaceutical industry and for clinical diagnostics. Takai et al. [81] developed a quantitative MALDI-MSI approach to analyze the drug, raclopride (RCP), in multiple mouse organs and study drug distribution/ accumulation. RCP (m/z 347) is a dopamine D2 receptor-selective antagonist. RCP was initially spotted onto tissue sections and MS/MS experiments were performed in order to choose a fragment ion to monitor for the quantitation studies. The daughter ion m/z 129 was analyzed for the quantitation study. RCP was dosed to mice intravenously. The mice were sacrificed and whole-body sections of the mice were imaged with MALDI-MSI at different time points post-dosage:10 minutes, 30 minutes, and 60 minutes post-dosage. The average signal intensities (n=3) of RCP were calculated for each organ at each time point. For more precise quantification, Takai et al. used a normalization method by using the signal intensity of DHB peaks as an internal standard. The RCP signal intensities were divided by the DHB signal intensity within the specific organ of interest in order to normalize the results. Additionally, RCP was administered to mice and the concentrations of the drug in each organ studied (liver, heart, spleen, brain, and plasma) were determined by LC/MS/MS for comparison with the quantitative MSI procedure. There was a strong correlation observed between the concentrations determined from the LC/MS/MS and the intensities calculated in the MSI method suggesting the future potential of MSI for quantitative analysis in multiple organs simultaneously.
Ambient ionization methods have gained interest and popularity for clinical MSI [82]. Wiseman et al. utilized DESI MS, an ambient ionization technique, to image drugs and their metabolites in histological tissue sections. Their study includes monitoring the antipsychotic drug, clozapine (m/z 327.1) and its dominant, N-desmethylclozapine, metabolite (m/z 313.1) in rat brain, lung, kidney, and testis tissue samples. By using their developed DESI-MSI method, and imaging in MS/MS mode, Wiseman et al. were able to detect clozapine not only in brain tissue, but also the accumulation of the drug in lung, kidney, and testis tissue sections, which was previously unknown. The research team demonstrated the ability of DESI-MSI to detect small molecule pharmaceuticals and their metabolites simultaneously, directly from histological tissue sections. They were able to rapidly detect the drug and metabolite and display spatial distributions as well as provide relative quantitation with minimal sample preparation, giving this ambient ionization technique potential for use in the clinical setting.
There have been many reports of drug and metabolite imaging in the literature [21, 80, 82, 83, 85-93]. Using multi-faceted approaches to distinguish between the drug and its metabolite provides more detailed and useful information for potential use in clinical studies. The ability to quantify active drugs simultaneously in multiple organs provides another key layer of information for toxicology studies for new potential drug candidates.
3.2 Biomarker Discovery and Validation
The discovery and validation of biomarkers, along with other laboratory and clinical evaluations, contribute to the assessment of disease severity, disease progression, and treatment response [94]. Gene expression profiling was the prominent source of biomarker discovery, but more recently, protein expression profiling has exploded in this field [95], and lipidomics has been gaining popularity [96].
3.2.1 Protein Biomarkers
Changes in protein levels in a tissue can correlate with disease state. These protein levels can be monitored in order to reveal, not only the type of disease, but also the severity of the disease. MSI has emerged as a powerful tool for biomarker discovery largely because of its ability to probe the proteomics of the tissue while maintaining the spatial distribution, which allows for direct comparison with histology [75]. MSI can be applied simultaneously to multiple tissues, allowing a more in depth study of the target disease and its potential biomarkers [78].
Six common types of cancer (Barrett’s cancer, breast cancer, colon cancer, hepatocellular carcinoma, gastric cancer, and thyroid carcinoma) have been probed with MALDI-MSI by Meding et al. [95]. Patient diagnosis begins with tumor origin identification and classification and when a primary tumor cannot be identified, the sample is diagnosed as cancer of unknown primary (CUP) [95]. The researchers used MALDI-MSI to establish distinct protein biomarkers for each type of known cancer. Cancer cell specific spectra were extracted and classified based on their proteomic differences with high confidence. These data were applied toward the identification of cancer from CUP samples. It is extremely important for the development of an individual patient treatment regimen that the secondary tumor (metastasis) be identified even if the primary tumor cannot be found. Meding et al. introduced colon cancer liver metastases into the study in order to test the ability of MALDI-MSI to distinguish between colon cancer primary tumors, colon cancer liver metastases, and hepatocellular carcinomas (liver cancer). Their results indicated that MSI can be used to distinguish between, and classify these closely related cancer entities. This research team generated a classifier system based on MALDI-MSI methods that provides accurate tumor classification with high confidence levels. This method could become a valuable addition to the workflow for clinical tumor diagnostics.
Not only are biomarkers useful for diagnosing and classifying different types of cancer, they are also being used to distinguish between diseases with similar histological characteristics. One such example is Spitz Nevi and Spitzoid malignant melanoma which is found primarily in children [97]. Spitz Nevi is a benign skin lesion, whereas Spitzoid malignant melanoma requires surgery and chemotherapy [98]. These 2 diseases are very difficult to distinguish and can result in misdiagnoses when relying on histological criteria alone [97, 98]. Lazova et al. performed a study using MALDI-MSI to compare the protein profiles of Spitz Nevi and Spitzoid malignant melanoma [98]. They found 5 peptide peaks (m/z 976.49, 1060.18, 1336.72, 1410.74, and 1428.77) that were able to best discriminate between the two diseases, and a total of 12 discriminatory peaks that were used to build a classification model. Similarly to Dill et al., as mentioned previously, Lazova et al. developed their model with a training group, and validated it with a test group. The classification method for the tumor showed a sensitivity of 97% and specificity of 90%. Interestingly, the research team was able to correctly classify 28 of 31 samples based solely on the proteomic information found in the dermis and not the tumor itself. These results show great promise for improving the diagnostic accuracy of these diseases in conjunction with standard histological evaluations.
Biomarkers can also show signs of effective treatment of a disease, not just identify the disease. Kim et al. utilized MSI to study drug distribution and potential biomarkers of response to therapy in prostate cancer [99]. The research team sought to study the spatial pharmacokinetics of prostate tumors treated with a novel tyrosine kinase inhibitor, AEE788. AEE788 potentially inhibits VEGFR and EGFR in nanomolar concentrations, which could enhance radiotherapy. Mice with prostate tumor xenografts were split into 4 groups: treated with AEE788, treated with radiation therapy, treated with AEE788 and radiation therapy, or untreated. The tumors were imaged for drugs or potential biomarker proteins. The drug and protein distributions were altered when the tumors were treated with irradiation. One potential biomarker was found at m/z 7765.4 which was present in the tumors treated with AEE788, but not when treated with radiation therapy alone. Radiation treated tumors had increased expression of multiple proteins that were not present in the AEE788 treated tumors. These could be potential biomarkers for treated vs. untreated tumors, which could provide means for evaluating efficacy of different therapeutics and optimizing individual patient treatment regimes.
3.2.2 Lipid Biomarkers
Lipidomics has recently gained popularity in clinical applications. Similarly to protein biomarker discovery, lipidomics aims to characterize lipid molecules in tissues and relate this information to disease states, among other applications [96]. Proteomics aims to characterize and quantify the cellular performers reflecting gene expression, whereas lipidomics (a subset of metabolomics) studies the molecular products of metabolism and is closest to patient phenotype, making lipidomics an important field of study [100]. Traumatic brain injuries can have a great effect on the lipid distribution and composition of the brain. Cox et al. used MSI to study brain lipids after cortical impact injury [18]. For this study, rats were subjected to controlled cortical impact injury and the brains were removed at different time points of 24 hours, 3 days, and 7 days post injury. The distribution of lipid species in the brain showed large variation in the MS images, demonstrating time dependent changes in the lipid profile of the injured areas after traumatic injury. Not only do these results show lipid biomarker distributions that can reveal the extent and time post-injury, which can be used to monitor patient recovery after treatment, but also the observed changes in lipid abundances shed light on the mechanisms of injury and repair on the molecular level.
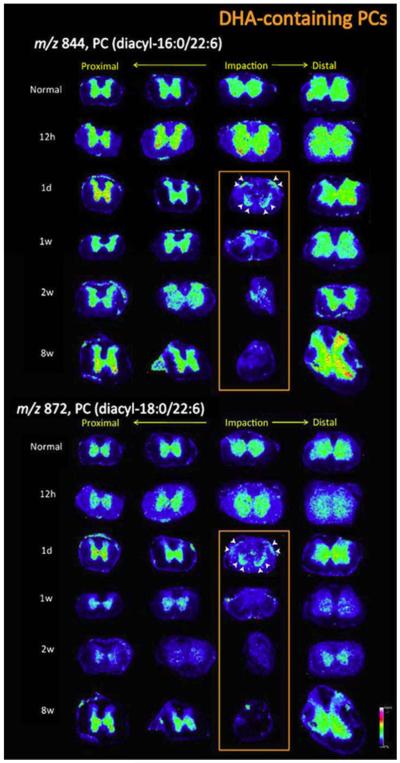
On a related note, Hanada et al. studied the alteration of phospholipids and prostaglandins after a spinal cord injury [101]. Alterations in lipid metabolism may play a key role in neurological disorders; therefore it is important to study these alterations to the lipid profiles during the occurrence and progression of a spinal cord injury. For this study, rats with applied spinal cord injuries were used, and the lipid distributions were studied after 12 h, 1 day, or 1, 2, or 8 weeks post-injury. Unique distribution patterns were observed for different types of phospholipids as the tissue damage resulting from the spinal cord injury progressed. Spatiotemporal changes in phosphatidylcholines (PCs) were examined with MSI. Both temporal and irreversible alterations in distinct PC species were observed, especially in species that contained 3-docosahexaenoic acid (DHA). Figure 2 shows the spatio-temporal images of two different DHA-containing PCs. At the impact site, an irreversible change in DHA-containing PCs was observed over time. This observation may result from an irreversible deficit of the neurons and could lead to motor dysfunction, meaning that DHA-PC reduction could be a potential indicator of the pathology of spinal cord injuries to be used in clinical settings.
Figure 2.
DHA-containing PCs exhibited impact site-specific irreversible reductions from 1 day to 8 weeks post-SCI. The MSI results for DHA-containing PCs, i.e.: PC(diacyl-16:0/22:6) and PC (diacyl-18:0/22:6), are detailed. In particular, the 24 ion images for each DHA-PC from sections of normal (sham-operated) and SCI-treated samples at five different time points are shown. The distribution of DHA-PCs was unaltered at 12 h post-SCI in comparison with the control. The primary reduction was observed around the central canal and gray commissure region to a severe extent at 1 d post-SCI, whereas the decreases at the anterior and posterior horns were moderate (arrowheads). However, at 1 week post-SCI, DHA-PCs were also lost from these tissue regions, and these reductions evolved at later time points and the DHA-PCs had almost disappeared by 8 weeks post-SCI [103]. Reprinted with permission from Hanada M, Sugiura Y, Shinjo R, et al. Spatiotemporal alteration of phospholipids and prostaglandins in a rat model of spinal cord injury. Anal Bioanal Chem 2012; 403:1873-1884.
3.2.3 Small Molecule Biomarkers
Ambient ionization methods can also be used to study and identify biomarkers in cancerous tissues [18,102,103]. Dill et al. used DESI-MSI to study human bladder cancer and develop a statistical method for distinguishing between cancerous and non-cancerous tissue samples in conjunction with standard histological identification methods. The researchers used a training set of samples to establish the best predictive features and a test/validation set to evaluate the performance of their statistical model on representative samples. There were 20 pairs of cancerous and adjacent normal human bladder tissue samples used in this study. DESI-MSI data showed significant changes in glycerophosphoinositols, glycerophosphoserines, and fatty acids between the cancerous and normal tissue samples. A series of DESI-MSI images were used to visually characterize the distribution of particular molecular species across the set of tissue samples. These images were visually compared to optically scanned images of the H&E-stained histological sections. Their results agreed with the pathological diagnosis of cancer and normal tissue for 15 of the tissue pairs. There was excellent agreement between the H&E-stained sections and the DESI images. Interestingly, for one sample, a border of tumor was detected on the normal tissue section, demonstrating the utility of DESI-MSI for determining the margins of the tumor before surgery. Overall, these results are very encouraging for the development of a method that could be used in a clinical setting for the diagnosis of cancer.
3.3 Unique Applications for Clinical Diagnostics
Apart from drug distribution and biomarker discovery, MSI has been used for several other clinical applications [104-107]. Osteoporosis is a well-known disease for which biomarkers have been extensively studied [97,108-114], but little attention has been devoted to bone material quality [106]. Routine clinical fracture risk assessments do not consider the quality of the bone mineral matrix. Zoehrer et al. used SIMS-MSI to investigate the spatial distribution and relationship of phosphorus and calcium in bone [106]. Calcium (Ca) and phosphate (P) are the main elements of the bone mineral building block, hydroxyapatite, and therefore support bone tissue mineralization. Bone material properties of the femoral head in male patients, age 65-80 y, with fragility fractures were compared to male, age-matched, non-fracture controls. SIMS-MSI clearly showed a greater frequency of areas on high Ca ion intensity in the tissue samples with fragility fractures, when compared to the control group. In the fragility fracture samples, a distinct, ~25 μm wide line of Ca ions was observed along the surface of the endosteum portion of the trabecular bone. In the control group, this line of Ca ions was distributed more diffusely, over a larger area of ~50 μm. Regions of high P ion intensity were observed in the control samples. The overall results show that a significant decrease in the P levels is associated with fragility fractures and the Ca/P ratio and distribution on the trabecular bone could be valuable parameters to consider during therapeutic and diagnostic trials.
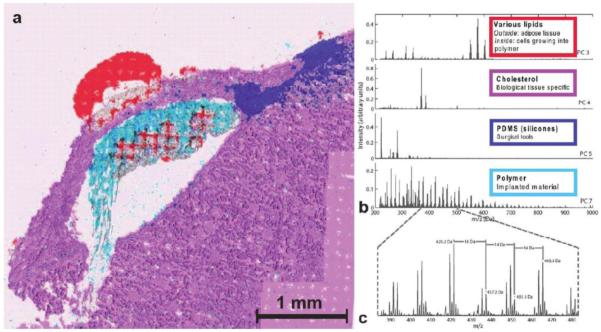
Assessing the compatibility of treatments with the body is a key step necessary to ensure the safety of the patients. Biodegradable polymers are of high interest in the medical field because of their potential applications in the field of tissue engineering and as drug delivery carriers [105]. Supramolecular polymers have gained much interest as potential drug delivery carriers because of their compatibility with weak water soluble compounds and their potential for controlled drug release [19,115-121]. In drug delivery systems, these polymers are designed to locally deliver active pharmaceutical ingredients to a localized place in the body, that needs the drug, and expose other parts of the body at a much lower dose. Klerk et al. used SIMS-MSI to elucidate molecular distributions in and around such polymer. A supramolecular polymeric hydrogel (containing no drugs) was implanted into rat renal tissue and the rat kidneys were harvested 15 days after implantation. Half of the kidneys were prepared for MSI and half were used for histological comparison. The MSI images show lipids from the kidneys, the polymer capsule, and cholesterol distribution around the implantation site, as well as silicone contaminants from the surgical tools. The images show that lipids from the kidney tissue entered into the polymer capsule, as shown in Figure 3, demonstrating biological activity within the polymer. When the study is continued with a polymer implant that does contain active pharmaceutical ingredients, the researchers will have the opportunity to study drug release by imaging the drug distribution in the surrounding tissue at various time points. Overall, this research shows the great potential of SIMS-MSI for understanding controlled drug distribution.
Figure 3.
(a) Large area image of the hydrogel implant under the renal capsule of a rat, 15 days after implantation. Various localizations are indicated, based on PCA+VARIMAX results (see spectra in (b)) and in the overlay with an optical microscope image. The presence of lipids inside the polymer area shows cellular infiltration in the drug delivery carrier. Some smearing artifact is visible at the bottom region of the polymer. The respective spectral results are given in (b). The first and second PCs gave non-informative distributions. PC 3 shows signal for various lipids, including Diacyl glycerols (m/z 550-620), PC 4 shows cholesterol ((M-OH)+ at m/z 369.4 and M+ at m/z 385.4), PC 5 shows silicone contamination (C7H21O2Si3+ at m/z 221.1, further identified from low-mass peaks in the corresponding region), PC 7 shows the polymer distribution, readily recognize from the m/z 44 spacing between the peaks, which exactly corresponds to the mass of one PEG unit. (c) Shows the PEG distribution in detail with characteristic 16 Da (K+ and Na+ difference or O loss) and 14 Da (CH2 loss) intervals. The change from 0.2 to 0.3 values is due to binning down and, thus, rounding to 0.1 Da [105]. Reprinted with permission from Klerk LA, Dankers PYW, Popa ER, et al. TOF-secondary ion mass spectrometry imaging of polymeric scaffolds with surrounding tissue after in vivo implantation. Anal Chem 2010; 82:4337-4343. Copyright 2010 American Chemical Society
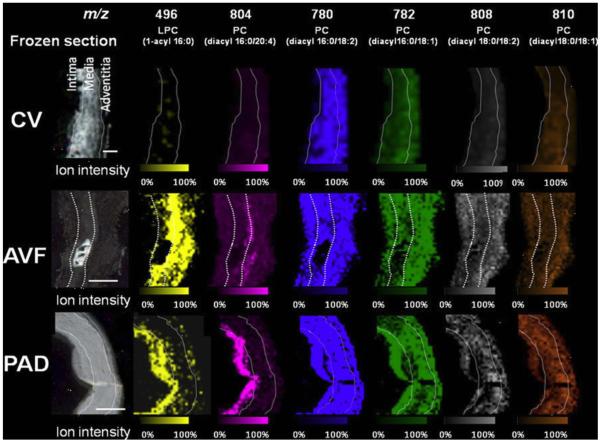
Biodegradable polymers are also being applied to the field of tissue engineering, requiring extensive study of the compatibility of these materials with the body. In tissue engineering, scaffolds made of biodegradable polymers can be designed to be slowly degraded by the body as their function is taken over by newly generated tissue. During renal failure, it may be necessary for a patient to undergo hemodialysis, which requires the doctors to surgically form an arteriovenous fistulae (AVF), using vein grafts [107]. In this procedure, the patients artery and vein are sutured together with a graft connecting them, which allows for the blood to bypass the capillaries and the vein to be enlarged so that it can accommodate the volume of blood being transferred during hemodialysis [122]. These grafts are typically made, not with polymers, but with the patient’s existing veins. Although the grafts come from the patient themselves, the veins in and around the graft degrade over time due to biological changes in the tissue [107]. Previous studies have shown that graft deformation is associated with abnormal lipid metabolism in hemodialysis patients [123, 124]. Tanaka et al. used MALDI-MSI to study the lipid profile of AVF tissues [107]. In this study, there were three types of vein samples: AVF tissues, control veins (CV), and peripheral artery occlusive disease (PAD) tissue. PAD is a disease that causes veins to deteriorate and is caused naturally, rather than through surgery like AVF. The control veins were segmental cephalic vein tissues taken from patients who underwent AVF creation, but the CV tissues were not part of the AVF. MALDI-MSI was performed on all three tissue samples, and the distributions of lysophosphatidylcholine (LPC) and phosphatidylcholine (PC) were determined. The results show that the distributions of LPC and PC differed between the 3 tissue types as shown in Figure 4. LPC was localized in the intima of the CV and PAD tissues, but distributed through the media and adventitia of the AVF tissue. The intensities of PC were much higher in the AVF and PAD tissues than it was in the CV tissue, suggesting an abnormal accumulation of these lipid molecules in both tissues. These results were compared with histological data, and both revealed complementary patterns of lipid accumulation in the AVF and PAD tissue samples. Overall, the MALDI-MSI results of this study provided the first evidence of the characteristic lipid distribution of AVF tissue on the molecular level which suggests an association between molecule-induced inflammation and tissue degeneration. This research provides valuable insight into the cause of vein degradation that can be applied to future procedures developed for hemodialysis or for the continued monitoring of patients with AVF’s already in place.
Figure 4.
Imaging mass spectrometry and optical images of frozen section (8 μm) of the control vein (CV), arteriovenous fistula (AVF), and peripheral artery occlusive disease (PAD) samples Scale bar=200 μm. Ion intensity was normalized by total ion current. MSI of human AVF revealed the characteristic distribution of phospholipid molecules in the intima and media compared with that in the CV [107]. Reprinted with permission from Tanaka H, Zaima N, Yamamoto N, et al. Distribution of phospholipid molecular species in autogenous access grafts for hemodialysis analyzed using imaging mass spectrometry. Anal Bioanal Chem 2011; 400:1873-1880.
4. Future Perspectives
4.1 3D MSI
One of the more exciting, recent advances in the field of clinical MSI is 3D MSI. 3D MSI grants the ability to study a broad mass range of molecular species by creating a lateral and vertical distribution map of select compounds [125]. This technique serves as a powerful discovery tool for pathologists and to the pharmaceutical industry by allowing a more complete visualization of tissue samples, which improves the ability for identification of distinct molecular signatures and drug distribution throughout the entire tissue. Advances involving imaging acquisition speed, image resolution, and data processing are always on-going in the biological sciences [126]. By adding the third dimension to the traditional 2D MSI method, there is a greater need for improvements in sample preparation, data acquisition, and data file transfer [125]. MSI generates extremely large data files; advances in computational tools will need to be made in order to improve the efficiency of data transfer and processing that will be much more time consuming when collecting a third dimension of imaging data. 3D imaging shows great promise for advancing clinical diagnostics, but there are still many areas for potential improvements before it becomes a widespread technique.
4.2 Alternative to Formalin-Fixed and Paraffin-Embedded (FFPE) Tissues
Formalin-fixation and paraffin-embedding is the standard procedure for preserving biological tissue samples for histological analysis and for long term storage at room temperature [127]. This technique preserves the cellular and morphological details of the tissue. The analysis of human tissue specimen is key to the identification of novel biomarkers that can be used to create more specific therapies and treatments. Recent progress has been made in the analysis of FFPE tissues, but proteomic analysis from these tissues is still very difficult because formaldehyde causes protein-protein cross-linking. PreAnalytiX (Hilden, Germany) developed an alcohol-based, formalin-free tissue fixation system, PAXgene that is commercially available for research use. The PAXgene system is a two-step approach composed of the PAXgene Tissue Fixation Reagent and the PAXgene Tissue Stabilization Reagent. The fixation is carried out without the cross-linking of biomolecules, allowing the stabilization of proteins and nucleic acids while still retaining the histo-morphology of the tissue. Ergin et al. compared MALDI-MSI capabilities on FFPE-fixed, cryopreserved, and PAXgene-fixed tissue samples [127]. The protein recovery efficiency of these three types of tissue fixation methods was compared, and the results showed that PAXgene fixation allows for high quantity of protein from mouse and human tissue samples. The protein pattern of these three tissue fixation methods was also examined, and the results showed that PAXgene and cryo-preserved tissue samples revealed similar proteomic signatures when examined by MALDI-MSI, and no protein peaks were observed from the FFPE tissue samples. Overall, these data demonstrate the potential of the PAXgene fixation system to become an integral part of protein biomarker discovery, which will facilitate advancing the clinical applications of MSI, if it becomes more widely used.
4.3 Quantitative Imaging
Quantitative imaging mass spectrometry is of great interest to the field of mass spectrometry and to the application of MSI in the clinical setting. Advances are slowly being made on the development of a reproducible, quantitative MSI method. Takai et al. [81], as mentioned previously, have developed a quantitative MALDI-MSI approach to analyze the concentration of a drug in mouse tissue after specific time points. This method is based on generating a calibration curve for the drug by spiking different concentrations of the drug of interest directly onto a tissue section and analyzing it with MSI. Their results were compared to trusted quantitation results obtained by LC/MS/MS, and had good agreement. Drexler et al. obtained quantitative information by combining MSI with QWBA. The downfall of this approach is that the analyte must be synthesized to contain radiolabels for QWBA, which is often time consuming and costly that are undesirable in the early stages of drug development. Several other groups combine MSI with a variety of other techniques in order to obtain quantitative data along with high spatial resolution images [128,129]. There is no universal technique for obtaining high resolution images and robust quantitative information, but progress is being made on this front, and there is potential for future improvements that would greatly impact the field of clinical diagnostics as well as many other scientific fields.
4.4 Incorporation of Ambient Ionization Techniques
MSI had been mainly viewed as an invasive process until the development of ambient ionization techniques, such as DESI. Ambient ionization techniques allow for direct analysis in real time and also remove the limitation imposed from requiring vacuum pressure conditions in traditional MSI [125]. DESI and other ambient techniques, which can be performed on untreated histological samples in the open lab environment, have the potential to promote in situ analysis in the clinical setting in the near future [24]. Miniature mass spectrometers have been developed that are capable of performing DESI-MSI for disease diagnostics and paper-spray ionization MS for therapeutic drug monitoring [24]. These techniques have significant potential to apply real-time diagnostic information in order, for example, to guide surgery. With the current advancements in MSI technology in the areas of miniaturization and ambient sampling, future improvements may involve configuring the instrument in the most efficient and useful way for use by doctors and surgeons in the clinic. The ability to perform in situ analysis and the convenience of the portable mass spectrometers suggests the potential role of DESI-MSI and other ambient ionization techniques in guiding therapy in parallel with standard histological methods in the clinical setting [24].
Conclusions
Over the past decade, MSI has obtained increasing attention from biologists and become more routinely employed to map various classes of biomolecules from biological specimens. Its novel applications to biomarker discovery in clinical settings have provided valuable knowledge regarding disease mechanisms and related reparative processes. These studies offer significant insight in clinical studies and hold promise for our continuous search for effective treatment of diseases. Moreover, exciting technical advances in areas such as sample preparation and instrumentation are further improving this powerful analytical tool and its clinical applications and impacts. We anticipate that MSI in clinical applications will lead to a better understanding of the biology of diseases and improvements in clinical diagnostics.
Highlights.
Current advances of MSI and its emerging clinical applications are reviewed.
Novel clinical applications of MSI are reviewed.
Future perspectives of MSI in the clinical setting are discussed.
Acknowledgements
Preparation of this manuscript was supported in part by National Science Foundation (CHE- 0957784) and National Institutes of Health through grant 1R01DK071801. L.L. acknowledges an H. I. Romnes Faculty Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Teng PN, Hood BL, Sun M, Dhir R, Conrads TP. Differential proteomic analysis of renal cell carcinoma tissue interstitial fluid. J Proteome Res. 2011;10:1333–1342. doi: 10.1021/pr101074p. [DOI] [PubMed] [Google Scholar]
- [2].Lowenthal MS, Mehta AI, Frogale K, et al. Analysis of albumin-associated peptides and proteins from ovarian cancer patients. Clin Chem. 2005;51:1933–1945. doi: 10.1373/clinchem.2005.052944. [DOI] [PubMed] [Google Scholar]
- [3].Hu S, Arellano M, Boontheung P, et al. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res. 2008;14:6246–6252. doi: 10.1158/1078-0432.CCR-07-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Craig-Schapiro R, Perrin RJ, Roe CM, et al. YKL-40: a Novel prognostic fluid biomarker for preclinical Alzheimer’s disease. Biol Psychiat. 2010;68:903–912. doi: 10.1016/j.biopsych.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Flint MS, Hood BL, Sun M, Stewart NA, Jones-Laughner J, Conrads TP. Proteomic analysis of the murine liver in response to a combined exposure to psychological stress and 7,12-Dimethylbenz(a)anthracene. J Proteome Res. 2010;9:509–520. doi: 10.1021/pr900861j. [DOI] [PubMed] [Google Scholar]
- [6].Ishimura R, Ohsako S, Kawakami T, Sakaue M, Aoki Y, Tohyama C. Altered protein profile and possible hypoxia in the placenta of 2,3,7,8-tetrachlorodibenzo-p-dioxin-exposed rats. Toxicol Appl Pharm. 2002;185:197–206. doi: 10.1006/taap.2002.9539. [DOI] [PubMed] [Google Scholar]
- [7].Ruepp SU, Tonge RP, Shaw J, Wallis N, Pognan F. Genomics and proteomics analysis of acetaminophen toxicity in mouse liver. Toxicol Sci. 2002;65:135–150. doi: 10.1093/toxsci/65.1.135. [DOI] [PubMed] [Google Scholar]
- [8].Li TA, Xu SL, Oses-Prieto JA, et al. Proteomics analysis reveals post-translational mechanisms for cold-induced metabolic changes in Arabidopsis. Mol Plant. 2011;4:361–374. doi: 10.1093/mp/ssq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cazares LH, Troyer D, Mendrinos S, et al. Imaging mass spectrometry of a specific fragment of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase 2 discriminates cancer from uninvolved prostate tissue. Clin Cancer Res. 2009;15:5541–5551. doi: 10.1158/1078-0432.CCR-08-2892. [DOI] [PubMed] [Google Scholar]
- [10].Deininger SO, Ebert MP, Futterer A, Gerhard M, Rocken C. MALDI imaging combined with hierarchical clustering as a new tool for the interpretation of complex human cancers. J Proteome Res. 2008;7:5230–5236. doi: 10.1021/pr8005777. [DOI] [PubMed] [Google Scholar]
- [11].Murphy RC, Hankin JA, Barkley RM. Imaging of lipid species by MALDI mass spectrometry. J Lipid Res. 2009;50:S317–S322. doi: 10.1194/jlr.R800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chen R, Hui L, Sturm RM, Li L. Three dimensional mapping of neuropeptides and lipids in crustacean brain by mass spectral imaging. J Am Soc Mass Spectr. 2009;20:1068–1077. doi: 10.1016/j.jasms.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].DeKeyser SS, Kutz-Naber KK, Schmidt JJ, Barrett-Wilt GA, Li L. Mass spectral imaging of neuropeptides in crustacean nervous tissue by MALDI TOF/TOF. J Proteome Res. 2007;6:1782–1791. doi: 10.1021/pr060603v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Schwamborn K, Caprioli RM. Molecular imaging by mass spectrometry--looking beyond classical histology. Nat Rev Cancer. 10:639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- [15].Suckau D, Resemann A, Schuerenberg M, Hufnagel P, Franzen J, Holle A. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal Bioanal Chem. 2003;376:952–965. doi: 10.1007/s00216-003-2057-0. [DOI] [PubMed] [Google Scholar]
- [16].Fletcher JS, Lockyer NP, Vickerman JC. Developments in molecular SIMS depth profiling and 3D imaging of biological systems using polyatomic primary ions. Mass Spectrom Rev. 30:142–174. doi: 10.1002/mas.20275. [DOI] [PubMed] [Google Scholar]
- [17].Jones EA, Lockyer NP, Vickerman JC. Mass spectral analysis and imaging of tissue by ToF-SIMS - The role of buckminsterfullerene, C-60(+), primary ions. Int J Mass Spectrom. 2007;260:146–157. [Google Scholar]
- [18].Dill AL, Eberlin LS, Costa AB, et al. Multivariate statistical identification of human bladder carcinomas using ambient ionization imaging mass spectrometry. Chem-Eur J. 2011;17:2897–2902. doi: 10.1002/chem.201001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gao YH, Yang CH, Liu X, Ma RJ, Kong DL, Shi LQ. A multifunctional nanocarrier based on nanogated mesoporous silica for enhanced tumor-specific uptake and intracellular delivery. Macromol Biosci. 2012;12:251–259. doi: 10.1002/mabi.201100208. [DOI] [PubMed] [Google Scholar]
- [20].Laskin J, Heath BS, Roach PJ, Cazares L, Semmes OJ. Tissue imaging using nanospray desorption electrospray ionization mass spectrometry. Anal Chem. 2012;84:141–148. doi: 10.1021/ac2021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Vismeh R, Waldon DJ, Teffera Y, Zhao ZY. Localization and quantification of drugs in animal tissues by use of desorption electrospray ionization mass spectrometry imaging. Anal Chem. 2012;84:5439–5445. doi: 10.1021/ac3011654. [DOI] [PubMed] [Google Scholar]
- [22].Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- [23].Ifa DR, Wu CP, Ouyang Z, Cooks RG. Desorption electrospray ionization and other ambient ionization methods: current progress and preview. Analyst. 2010;135:669–681. doi: 10.1039/b925257f. [DOI] [PubMed] [Google Scholar]
- [24].Cooks RG, Manicke NE, Dill AL, et al. New ionization methods and miniature mass spectrometers for biomedicine: DESI imaging for cancer diagnostics and paper spray ionization for therapeutic drug monitoring. Faraday Discuss. 2011;149:247–267. doi: 10.1039/c005327a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jehl B, Bauer R, Dorge A, Rick R. The use of propane-isopentane mixtures for rapid freezing of biological specimens. J Microsc-Oxford. 1981;123:307–309. doi: 10.1111/j.1365-2818.1981.tb02475.x. [DOI] [PubMed] [Google Scholar]
- [26].Schwartz SA, Reyzer ML, Caprioli RM. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J Mass Spectrom. 2003;38:699–708. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- [27].Che FY, Lim J, Pan H, Biswas R, Fricker LD. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol Cell Proteomics. 2005;4:1391–1405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- [28].Svensson M, Boren M, Skold K, et al. Heat stabilization of the tissue proteome: a new technology for improved proteomics. J Proteome Res. 2009;8:974–981. doi: 10.1021/pr8006446. [DOI] [PubMed] [Google Scholar]
- [29].Rountree CB, Van Kirk CA, You HN, et al. Clinical application for the preservation of phosphoproteins through in-situ tissue stabilization. Proteome Sci. 2010;8 doi: 10.1186/1477-5956-8-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Crossman L, McHugh NA, Hsieh YS, Korfmacher WA, Chen JW. Investigation of the profiling depth in matrix-assisted laser desorption/ionization imaging mass spectrometry. Rapid Commun Mass Sp. 2006;20:284–290. doi: 10.1002/rcm.2259. [DOI] [PubMed] [Google Scholar]
- [31].Verhaert PDEM, Pinkse MWH, Strupat K, Conaway MCP. Imaging of similar mass neuropeptides in neuronal tissue by enhanced resolution MALDI MS with an ion trap-orbitrap(TM) hybrid instrument. Mass spectrometry imaging: principles and protocols. 2010;656:433–449. doi: 10.1007/978-1-60761-746-4_25. [DOI] [PubMed] [Google Scholar]
- [32].Chaurand P, Fouchecourt S, DaGue BB, et al. Profiling and imaging proteins in the mouse epididymis by imaging mass spectrometry. Proteomics. 2003;3:2221–2239. doi: 10.1002/pmic.200300474. [DOI] [PubMed] [Google Scholar]
- [33].Chaurand P, Schriver KE, Caprioli RM. Instrument design and characterization for high resolution MALDI-MS imaging of tissue sections. J Mass Spectrom. 2007;42:476–489. doi: 10.1002/jms.1180. [DOI] [PubMed] [Google Scholar]
- [34].Kaletas BK, van der Wiel IM, Stauber J, et al. Sample preparation issues for tissue imaging by imaging MS. Proteomics. 2009;9:2622–2633. doi: 10.1002/pmic.200800364. [DOI] [PubMed] [Google Scholar]
- [35].Lemaire R, Wisztorski M, Desmons A, et al. MALDI-MS direct tissue analysis of proteins: improving signal sensitivity using organic treatments. Anal Chem. 2006;78:7145–7153. doi: 10.1021/ac060565z. [DOI] [PubMed] [Google Scholar]
- [36].Seeley EH, Oppenheimer SR, Mi D, Chaurand P, Caprioli RM. Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections. J Am Soc Mass Spectr. 2008;19:1069–1077. doi: 10.1016/j.jasms.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Scicchitano MS, Dalmas DA, Boyce RW, Thomas HC, Frazier KS. Protein extraction of formalin-fixed, paraffin-embedded tissue enables robust proteomic profiles by mass spectrometry. J Histochem Cytochem. 2009;57:849–860. doi: 10.1369/jhc.2009.953497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Casadonte R, Caprioli RM. Proteomic analysis of formalin-fixed paraffin-embedded tissue by MALDI imaging mass spectrometry. Nat Protoc. 2011;6:1695–1709. doi: 10.1038/nprot.2011.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].EmmertBuck MR, Bonner RF, Smith PD, et al. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- [40].Wang Z, Han J, Schey KL. Spatial differences in an integral membrane proteome detected in laser capture microdissected samples. J Proteome Res. 2008;7:2696–2702. doi: 10.1021/pr700737h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Xu BGJ, Caprioli RM. Direct analysis of laser capture microdissected cells by MALDI mass spectrometry. J Am Soc Mass Spectr. 2002;13:1292–1297. doi: 10.1016/S1044-0305(02)00644-X. [DOI] [PubMed] [Google Scholar]
- [42].Xu BJ, Shyr Y, Liang XB, et al. Proteomic patterns and prediction of glomerulosclerosis and its mechanisms. J Am Soc Nephrol. 2005;16:2967–2975. doi: 10.1681/ASN.2005030262. [DOI] [PubMed] [Google Scholar]
- [43].Groseclose MR, Andersson M, Hardesty WM, Caprioli RM. Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J Mass Spectrom. 2007;42:254–262. doi: 10.1002/jms.1177. [DOI] [PubMed] [Google Scholar]
- [44].Ronci M, Bonanno E, Colantoni A, et al. Protein unlocking procedures of formalin-fixed paraffin-embedded tissues: Application to MALDI-TOF imaging MS investigations. Proteomics. 2008;8:3702–3714. doi: 10.1002/pmic.200701143. [DOI] [PubMed] [Google Scholar]
- [45].Djidja MC, Francese S, Loadman PM, et al. Detergent addition to tryptic digests and ion mobility separation prior to MS/MS improves peptide yield and protein identification for in situ proteomic investigation of frozen and formalin-fixed paraffin-embedded adenocarcinoma tissue sections. Proteomics. 2009;9:2750–2763. doi: 10.1002/pmic.200800624. [DOI] [PubMed] [Google Scholar]
- [46].Stauber J, MacAleese L, Franck J, et al. On-tissue protein identification and imaging by MALDI-ion mobility mass spectrometry. J Am Soc Mass Spectr. 2010;21:338–347. doi: 10.1016/j.jasms.2009.09.016. [DOI] [PubMed] [Google Scholar]
- [47].Boggio KJ, Obasuyi E, Sugino K, Nelson SB, Agar NYR, Agar JN. Recent advances in single-cell MALDI mass spectrometry imaging and potential clinical impact. Expert Rev Proteomic. 2011;8:591–604. doi: 10.1586/epr.11.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].McDonnell LA, Corthals GL, Willems SM, van Remoortere A, van Zeijl RJM, Deelder AM. Peptide and protein imaging mass spectrometry in cancer research. J Proteomics. 2010;73:1921–1944. doi: 10.1016/j.jprot.2010.05.007. [DOI] [PubMed] [Google Scholar]
- [49].Tu T, Sauter AD, Sauter AD, Gross ML. Improving the signal intensity and sensitivity of MALDI mass spectrometry by using nanoliter spots deposited by induction-based fluidics. J Am Soc Mass Spectr. 2008;19:1086–1090. doi: 10.1016/j.jasms.2008.03.017. [DOI] [PubMed] [Google Scholar]
- [50].Zimmerman TA, Monroe EB, Sweedler JV. Adapting the stretched sample method from tissue profiling to imaging. Proteomics. 2008;8:3809–3815. doi: 10.1002/pmic.200800331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zimmerman TA, Rubakhin SS, Sweedler JV. MALDI mass spectrometry imaging of neuronal cell cultures. J Am Soc Mass Spectr. 2011;22:828–836. doi: 10.1007/s13361-011-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jurchen JC, Rubakhin SS, Sweedler JV. MALDI-MS imaging of features smaller than the size of the laser beam. J Am Soc Mass Spectr. 2005;16:1654–1659. doi: 10.1016/j.jasms.2005.06.006. [DOI] [PubMed] [Google Scholar]
- [53].Koestler M, Kirsch D, Hester A, Leisner A, Guenther S, Spengler B. A high-resolution scanning microprobe matrix-assisted laser desorption/ionization ion source for imaging analysis on an ion trap/Fourier transform ion cyclotron resonance mass spectrometer. Rapid Commun Mass Sp. 2008;22:3275–3285. doi: 10.1002/rcm.3733. [DOI] [PubMed] [Google Scholar]
- [54].Römpp A, Guenther S, Schober Y, et al. Histology by Mass Spectrometry: Label-free tissue characterization obtained from high-accuracy bioanalytical imaging. Angew Chem Int Edit. 2010;49:3834–3838. doi: 10.1002/anie.200905559. [DOI] [PubMed] [Google Scholar]
- [55].Guenther S, Rompp A, Kummer W, Spengler B. AP-MALDI imaging of neuropeptides in mouse pituitary gland with 5 mu m spatial resolution and high mass accuracy. Int J Mass Spectrom. 2011;305:228–237. [Google Scholar]
- [56].Schober Y, Guenther S, Spengler B, Rompp A. Single cell matrix-assisted laser desorption/ionization mass spectrometry imaging. Anal Chem. 84:6293–6297. doi: 10.1021/ac301337h. [DOI] [PubMed] [Google Scholar]
- [57].Schafer R. ultrafleXtreme: redefining MALDI mass spectrometry performance. LC GC N Am. 2009:14–15. [Google Scholar]
- [58].Luxembourg SL, Mize TH, McDonnell LA, Heeren RMA. High-spatial resolution mass spectrometric imaging of peptide and protein distributions on a surface. Anal Chem. 2004;76:5339–5344. doi: 10.1021/ac049692q. [DOI] [PubMed] [Google Scholar]
- [59].Klerk LA, Altelaar AFM, Froesch M, McDonnell LA, Heeren RMA. Fast and automated large-area imaging MALDI mass spectrometry in microprobe and microscope mode. Int J Mass Spectrom. 2009;285:19–25. [Google Scholar]
- [60].Altelaar AFM, Taban IM, McDonnell LA, et al. High-resolution MALDI imaging mass spectrometry allows localization of peptide distributions at cellular length scales in pituitary tissue sections. Int J Mass Spectrom. 2007;260:203–211. [Google Scholar]
- [61].Fitzgerald JJD, Kunnath P, Walker AV. Matrix-enhanced secondary ion mass spectrometry (ME-SIMS) using room temperature ionic liquid matrices. Anal Chem. 2010;82:4413–4419. doi: 10.1021/ac100133c. [DOI] [PubMed] [Google Scholar]
- [62].Lemaire R, Tabet JC, Ducoroy P, Hendra JB, Salzet M, Fournier I. Solid ionic matrixes for direct tissue analysis and MALDI Imaging. Anal Chem. 2006;78:809–819. doi: 10.1021/ac0514669. [DOI] [PubMed] [Google Scholar]
- [63].Deutskens F, Yang JH, Caprioli RM. High spatial resolution imaging mass spectrometry and classical histology on a single tissue section. J Mass Spectrom. 2011;46:568–571. doi: 10.1002/jms.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ye H, Greer T, Li L. Probing neuropeptide signaling at the organ and cellular domains via imaging mass spectrometry. J Proteomics. 2012;75:5014–5026. doi: 10.1016/j.jprot.2012.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Thomas A, Charbonneau JL, Fournaise E, Chaurand P. Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1,5-diaminonapthalene deposition. Anal Chem. 2012;84:2048–2054. doi: 10.1021/ac2033547. [DOI] [PubMed] [Google Scholar]
- [66].Cohen MZ. A historical overview of the phenomenologic movement. Image J Nurs Sch. 1987;19:31–34. doi: 10.1111/j.1547-5069.1987.tb00584.x. [DOI] [PubMed] [Google Scholar]
- [67].Leinweber BD, Tsaprailis G, Monks TJ, Lau SS. Improved MALDI-TOF imaging yields increased protein signals at high molecular mass. J Am Soc Mass Spectr. 2009;20:89–95. doi: 10.1016/j.jasms.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Vestal ML, Campbell JM. Tandem time-of-flight mass spectrometry. Methods Enzymol. 2005;402:79–108. doi: 10.1016/S0076-6879(05)02003-3. [DOI] [PubMed] [Google Scholar]
- [69].Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH. Ion mobility-mass spectrometry. J Mass Spectrom. 2008;43:1–22. doi: 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- [70].Marshall AG, Hendrickson CL, Jackson GS. Fourier transform ion cyclotron resonance mass spectrometry: A primer. Mass Spectrom Rev. 1998;17:1–35. doi: 10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- [71].Kutz KK, Schmidt JJ, Li LJ. In situ tissue analysis of neuropeptides by MALDI FTMS in-cell accumulation. Anal Chem. 2004;76:5630–5640. doi: 10.1021/ac049255b. [DOI] [PubMed] [Google Scholar]
- [72].Fuchser JCS, Becker M. High resolution molecular imaging of pharmaceuticals at theraputic levels. Bruker Daltonik GmbH. 2008 [Google Scholar]
- [73].Makarov A. Electrostatic axially harmonic orbital trapping: a high-performance technique of mass analysis. Anal Chem. 2000;72:1156–1162. doi: 10.1021/ac991131p. [DOI] [PubMed] [Google Scholar]
- [74].Hu QZ, Noll RJ, Li HY, Makarov A, Hardman M, Cooks RG. The Orbitrap: a new mass spectrometer. J Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- [75].Seeley EH, Caprioli RM. Imaging mass spectrometry: towards clinical diagnostics. Proteom Clin Appl. 2008;2:1435–1443. doi: 10.1002/prca.200800013. [DOI] [PubMed] [Google Scholar]
- [76].Seeley EH, Caprioli RM. MALDI imaging mass spectrometry of human tissue: method challenges and clinical perspectives. Trends Biotechnol. 2011;29:136–143. doi: 10.1016/j.tibtech.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Rauser S, Deininger SO, Suckau D, Hofler H, Walch A. Approaching MALDI molecular imaging for clinical proteomic research: current state and fields of application. Expert Rev Proteomic. 2010;7:927–941. doi: 10.1586/epr.10.83. [DOI] [PubMed] [Google Scholar]
- [78].Cazares LH, Troyer DA, Wang BH, Drake RR, Semmes OJ. MALDI tissue imaging: from biomarker discovery to clinical applications. Anal Bioanal Chem. 2011;401:17–27. doi: 10.1007/s00216-011-5003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Meding S, Nitsche U, Balluff B, et al. Tumor classification of six common cancer types based on proteomic profiling by MALDI imaging. J Proteome Res. 2012;11:1996–2003. doi: 10.1021/pr200784p. [DOI] [PubMed] [Google Scholar]
- [80].Drexler DM, Tannehill-Gregg SH, Wang LF, Brock BJ. Utility of quantitative whole-body autoradiography (QWBA) and imaging mass spectrometry (IMS) by matrix-assisted laser desorption/ionization (MALDI) in the assessment of ocular distribution of drugs. J Pharmacol Tox Met. 2011;63:205–208. doi: 10.1016/j.vascn.2010.10.003. [DOI] [PubMed] [Google Scholar]
- [81].Takai N, Tanaka Y, Inazawa K, Saji H. Quantitative analysis of pharmaceutical drug distribution in multiple organs by imaging mass spectrometry. Rapid Commun Mass Sp. 2012;26:1549–1556. doi: 10.1002/rcm.6256. [DOI] [PubMed] [Google Scholar]
- [82].Wiseman JM, Ifa DR, Zhu YX, et al. Desorption electrospray ionization mass spectrometry: Imaging drugs and metabolites in tissues. P Natl Acad Sci USA. 2008;105:18120–18125. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Greer T, Sturm R, Li LJ. Mass spectrometry imaging for drugs and metabolites. J Proteomics. 2011;74:2617–2631. doi: 10.1016/j.jprot.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Markowitz L, Anis SI, Linehan ST. QWBA - current methodologies and future advancements positioning of image. J Labelled Compd Rad. 2010;53:330–331. [Google Scholar]
- [85].Cornett DS, Frappier SL, Caprioli RM. MALDI-FTICR imaging mass spectrometry of drugs and metabolites in tissue. Anal Chem. 2008;80:5648–5653. doi: 10.1021/ac800617s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Leinweber BD, Tsaprailis G, Monks TJ, Lau SS. Improved MALDI-TOF imaging yields increased protein signals at high molecular mass. J Am Soc Mass Spectr. 2009;20:89–95. doi: 10.1016/j.jasms.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Lorey DR, Morrison GH, Chandra S. Dynamic secondary ion mass spectrometry analysis of boron from boron neutron capture therapy drugs in co-cultures: Single-cell imaging of two different cell types within the same ion microscopy field of imaging. Anal Chem. 2001;73:3947–3953. doi: 10.1021/ac0103266. [DOI] [PubMed] [Google Scholar]
- [88].Prideaux B, Staab D, Stoeckli M. Applications of MALDI-MSI to Pharmaceutical Research. Methods Mol Biol. 2010;656:405–413. doi: 10.1007/978-1-60761-746-4_23. [DOI] [PubMed] [Google Scholar]
- [89].Trim PJ, Francese S, Clench MR. Imaging mass spectrometry for the assessment of drugs and metabolites in tissue. Bioanalysis. 2009;1:309–319. doi: 10.4155/bio.09.33. [DOI] [PubMed] [Google Scholar]
- [90].Wittig A, Arlinghaus HF, Kriegeskotte C, et al. Laser postionization secondary neutral mass spectrometry in tissue: a powerful tool for elemental and molecular imaging in the development of targeted drugs. Mol Cancer Ther. 2008;7:1763–1771. doi: 10.1158/1535-7163.MCT-08-0191. [DOI] [PubMed] [Google Scholar]
- [91].Lagarrigue M, Lavigne R, Guevel B, Com E, Chaurand P, Pineau C. Matrix-assisted laser desorption/ionization imaging mass spectrometry: a promising technique for reproductive research. Biol Reprod. 2012;86 doi: 10.1095/biolreprod.111.094896. [DOI] [PubMed] [Google Scholar]
- [92].Nilsson A, Fehniger TE, Gustavsson L, et al. Fine mapping the spatial distribution and concentration of unlabeled drugs within tissue micro-compartments using imaging mass spectrometry. Plos One. 2010;5 doi: 10.1371/journal.pone.0011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Trim PJ, Henson CM, Avery JL, et al. Matrix-assisted laser desorption/ionization-ion mobility separation-mass spectrometry imaging of vinblastine in whole body tissue sections. Anal Chem. 2008;80:8628–8634. doi: 10.1021/ac8015467. [DOI] [PubMed] [Google Scholar]
- [94].Young SP, Piraud M, Goldstein JL, et al. Assessing disease severity in Pompe disease: the roles of a urinary glucose tetrasaccharide biomarker and imaging techniques. Am J Med Genet C. 2012;160C:50–58. doi: 10.1002/ajmg.c.31320. [DOI] [PubMed] [Google Scholar]
- [95].Meding S, Nitsche U, Balluff B, et al. Tumor classification of six common cancer types based on proteomic profiling by MALDI imaging. J Proteome Res. 2012;11:1996–2003. doi: 10.1021/pr200784p. [DOI] [PubMed] [Google Scholar]
- [96].Postle AD. Lipidomics. Curr Opin Clin Nutr. 2012;15:127–133. doi: 10.1097/MCO.0b013e32834fb003. [DOI] [PubMed] [Google Scholar]
- [97].Walsh N, Crotty K, Palmer A, McCarthy S. Spitz nevus versus spitzoid malignant melanoma: an evaluation of the current distinguishing histopathologic criteria. Hum pathol. 1998;29:1105–1112. doi: 10.1016/s0046-8177(98)90421-x. [DOI] [PubMed] [Google Scholar]
- [98].Lazova R, Seeley EH, Keenan M, Gueorguieva R, Caprioli RM. Imaging mass spectrometry-a new and promising method to differentiate Spitz Nevi From Spitzoid malignant melanomas. Am J Dermatopath. 2012;34:82–90. doi: 10.1097/DAD.0b013e31823df1e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Kim DW, Huamani J, Reyzer ML, Mi D, Caprioli RM, Hallahan DE. Imaging mass spectrometry to map distribution of radiation enhancing vasculature targeted drug and protein biomarkers of response to therapy in prostate cancer. Int J Radiat Oncol. 2006;66:S554–S554. [Google Scholar]
- [100].Thomas A, Lenglet S, Chaurand P, et al. Mass spectrometry for the evaluation of cardiovascular diseases based on proteomics and lipidomics. Thromb Haemostasis. 2011;106:20–33. doi: 10.1160/TH10-12-0812. [DOI] [PubMed] [Google Scholar]
- [101].Hanada M, Sugiura Y, Shinjo R, et al. Spatiotemporal alteration of phospholipids and prostaglandins in a rat model of spinal cord injury. Anal Bioanal Chem. 2012;403:1873–1884. doi: 10.1007/s00216-012-5900-3. [DOI] [PubMed] [Google Scholar]
- [102].Weaver CM, Liebman M. Biomarkers of bone health appropriate for evaluating functional foods designed to reduce risk of osteoporosis. Brit J Nutr. 2002;88:S225–S232. doi: 10.1079/BJN2002687. [DOI] [PubMed] [Google Scholar]
- [103].Gamez-Pozo A, Sanchez-Navarro I, Calvo E, et al. PTRF/Cavin-1 and MIF proteins are identified as non-small cell lung cancer biomarkers by label-free proteomics. Plos One. 2012;7 doi: 10.1371/journal.pone.0033752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Tanaka H, Zaima N, Yamamoto N, et al. Imaging mass spectrometry reveals unique lipid distribution in primary varicose veins. Eur J Vasc Endovasc. 2010;40:657–663. doi: 10.1016/j.ejvs.2010.08.001. [DOI] [PubMed] [Google Scholar]
- [105].Klerk LA, Dankers PYW, Popa ER, et al. TOF-secondary ion mass spectrometry imaging of polymeric scaffolds with surrounding tissue after in vivo implantation. Anal Chem. 2010;82:4337–4343. doi: 10.1021/ac100837n. [DOI] [PubMed] [Google Scholar]
- [106].Zoehrer R, Perilli E, Kuliwaba JS, Shapter JG, Fazzalari NL, Voelcker NH. Human bone material characterization: integrated imaging surface investigation of male fragility fractures. Osteoporosis Int. 2012;23:1297–1309. doi: 10.1007/s00198-011-1688-9. [DOI] [PubMed] [Google Scholar]
- [107].Tanaka H, Zaima N, Yamamoto N, et al. Distribution of phospholipid molecular species in autogenous access grafts for hemodialysis analyzed using imaging mass spectrometry. Anal Bioanal Chem. 2011;400:1873–1880. doi: 10.1007/s00216-011-4850-5. [DOI] [PubMed] [Google Scholar]
- [108].Johnson RB, Gilbert JA, Cooper RC, Parsell DE, Streckfus CF, Boring JG. Isolation of osteoporosis biomarkers from saliva. Faseb J. 2001;15:A716–A716. [Google Scholar]
- [109].Garnero P. Biomarkers for osteoporosis management - utility in diagnosis, fracture risk prediction and therapy monitoring. Mol Diagn Ther. 2008;12:157–170. doi: 10.1007/BF03256280. [DOI] [PubMed] [Google Scholar]
- [110].Bhattacharyya S, Siegel E, Jennings S, Khosla S, Suva LJ. The discovery of new biomarkers for the diagnosis of osteoporosis: Is SELDI the answer? J Bone Miner Res. 2005;20:S224–S225. [Google Scholar]
- [111].Holland SA, Bateman KP, Desmarais S, Chauret N, Percival MD, Seto C. Detection of bone resorption biomarkers in an osteoporosis animal model. Drug Metab Rev. 2003;35:164–164. [Google Scholar]
- [112].Liu JH, Tsang R, Gass M, Kao L, Muse K. Identification of women at risk for osteoporosis with bone biomarkers during early menopause. J Bone Miner Res. 1996;11:T577–T577. [Google Scholar]
- [113].Maddali KK, Starks CI, McDonough C, Dharmadhikari J, Litzenberger BA. Bone biomarkers significantly enhances the predictability of preclinical study outcomes and translation to first in man studies targeted towards osteoporosis. Clin Chem. 2009;55:A158–A158. [Google Scholar]
- [114].Moayyeri A, Hammond CJ, Spector TD. Novel metabolomic biomarkers for osteoporosis and longitudinal bone loss. Bone. 2012;50:S45–S45. [Google Scholar]
- [115].Chytil P, Etrych T, Kostka L, Ulbrich K. Hydrolytically degradable polymer micelles for anticancer drug delivery to solid tumors. Macromol Chem Phys. 2012;213:858–867. [Google Scholar]
- [116].Ooya T, Mori H, Terano M, Yui N. Synthesis of a biodegradable polymeric supramolecular assembly for drug-delivery. Macromol Rapid Comm. 1995;16:259–263. [Google Scholar]
- [117].Salmaso S, Bersani S, Scomparin A, Mastrotto F, Caliceti P. Supramolecular bioconjugates for protein and small drug delivery. Isr J Chem. 2010;50:160–174. [Google Scholar]
- [118].Nakayama M, Okano T. Intelligent thermoresponsive polymeric micelles for targeted drug delivery. J Drug Deliv Sci Tec. 2006;16:35–44. [Google Scholar]
- [119].Sant VP, Smith D, Leroux JC. Novel pH-sensitive supramolecular assemblies for oral delivery of poorly water soluble drugs: preparation and characterization. J Control Release. 2004;97:301–312. doi: 10.1016/j.jconrel.2004.03.026. [DOI] [PubMed] [Google Scholar]
- [120].Vergaro V, Scarlino F, Bellomo C, et al. Drug-loaded polyelectrolyte microcapsules for sustained targeting of cancer cells. Adv Drug Deliver Rev. 2011;63:847–863. doi: 10.1016/j.addr.2011.05.007. [DOI] [PubMed] [Google Scholar]
- [121].Gillies ER, Frechet JMJ. Development of acid-sensitive copolymer micelles for drug delivery. Pure Appl Chem. 2004;76:1295–1307. [Google Scholar]
- [122].Lin CH, Mardini S, Lin YT, Yeh JT, Wei FC, Chen HC. Sixty-five clinical cases of free tissue transfer using long arteriovenous fistulas or vein grafts. J Trauma. 2004;56:1107–1117. doi: 10.1097/01.ta.0000114637.29779.ab. [DOI] [PubMed] [Google Scholar]
- [123].Zhu Y, Lin JHC, Liao HL, Verna L, Stemerman MB. Activation of ICAM-1 promoter by lysophosphatidylcholine: Possible involvement of protein tyrosine kinases. Bba-Lipid Lipid Met. 1997;1345:93–98. doi: 10.1016/s0005-2760(96)00169-5. [DOI] [PubMed] [Google Scholar]
- [124].Rong JX, Berman JW, Taubman MB, Fisher EA. Lysophosphatidylcholine stimulates monocyte chemoattractant protein-1 gene expression in rat aortic smooth muscle cells. Arterioscl Throm Vas. 2002;22:1617–1623. doi: 10.1161/01.atv.0000035408.93749.71. [DOI] [PubMed] [Google Scholar]
- [125].Ye H, Greer T, Li LJ. From pixel to voxel: a deeper view of biological tissue by 3D mass spectral imaging. Bioanalysis. 2011;3:313–332. doi: 10.4155/bio.10.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Jones EA, van Zeijl RJM, Andren PE, Deelder AM, Wolters L, McDonnell LA. High speed data processing for imaging MS-based molecular histology using graphical processing units. J Am Soc Mass Spectr. 2012;23:745–752. doi: 10.1007/s13361-011-0327-1. [DOI] [PubMed] [Google Scholar]
- [127].Ergin B, Meding S, Langer R, et al. Proteomic analysis of PAXgene-fixed tissues. J Proteome Res. 2010;9:5188–5196. doi: 10.1021/pr100664e. [DOI] [PubMed] [Google Scholar]
- [128].Hattori K, Kajimura M, Hishiki T, et al. Paradoxical ATP elevation in ischemic penumbra revealed by quantitative imaging mass spectrometry. Antioxid Redox Sign. 2010;13:1157–1167. doi: 10.1089/ars.2010.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Wang HAO, Grolimund D, Van Loon LR, et al. Quantitative chemical imaging of element diffusion into heterogeneous media using laser ablation inductively coupled plasma mass spectrometry, synchrotron micro-X-ray fluorescence, and extended X-ray absorption fine structure spectroscopy. Anal Chem. 2011;83:6259–6266. doi: 10.1021/ac200899x. [DOI] [PubMed] [Google Scholar]