Abstract
Bapineuzumab is a humanized antibody developed by Pfizer and Johnson & Johnson targeting the amyloid (Aβ) plaques that underlie Alzheimer's disease neuropathology. Here we report the crystal structure of a Fab-Aβ peptide complex that reveals Bapineuzumab surprisingly captures Aβ in a monomeric helical conformation at the N-terminus. Microscale thermophoresis suggests that the Fab binds soluble Aβ(1–40) with a KD of 89 (±9) nM. The structure explains the antibody's exquisite selectivity for particular Aβ species and why it cannot recognize N-terminally modified or truncated Aβ peptides.
Alzheimer's disease (AD) is a common and devastating age related disease with no effective disease-modifying treatments1. Approximately, 34 million people worldwide are currently afflicted by AD and its prevalence is expected to triple over the next 40 years as people live longer2. Various antibodies targeting proteins implicated in AD are being developed as immunotherapies and are considered amongst the most promising approaches for the treatment and prevention of AD and related diseases3.
Bapineuzumab is a humanized monoclonal antibody (Mab) that targets the neurotoxic amyloid-beta (Aβ) peptide, an early biomarker of Alzheimer's disease pathology and the major component of plaques found in AD brains4. Large scale phase three clinical trials of Bapineuzumab in patients with mild to moderate Alzheimer's disease were halted in August 2012 when the drug failed to arrest cognitive decline. Despite the disappointing results, Bapineuzumab was effective in stabilizing amyloid plaque burden and lowering phosphorylated-tau levels in cerebrospinal fluid, two biomarkers of AD pathology (http://www.alzforum.org/new/detail.asp?id=3268). Immunotherapeutics are now being considered as prophylaxes for patients with mild cognitive impairment, many of whom go on to develop Alzheimer's disease.
The murine antibody 3D6, parent of the humanized monoclonal antibody Bapineuzumab, was raised in mice against Aβ (residues 1–5) conjugated to sheep anti-mouse immunoglobulin5. Recent evidence suggests that N-terminal Aβ directed antibodies interact with both soluble and insoluble forms of Aβ6. Here we describe the crystal structure of the humanized antigen binding fragment (Fab) in complex with Aβ (residues 1 to 28) that reveals Bapineuzumab recognizes the N-terminal end of the Aβ peptide in a helical conformation. The structure provides a molecular basis for Aβ recognition that may prove useful in informing the outcomes of possible future clinical trials with Bapineuzumab and other anti-AD therapeutic antibodies.
Results
Binding of Aβ
Amyloid-β peptides are conformationally sensitive to their environment. DeMattos et al. report binding data for the parent murine intact antibody 3D6 to Aβ with a KD of ∼5 nM to soluble and insoluble forms of Aβ7. We used Microscale Thermophoresis (MST)8 to measure the affinity of the humanized 3D6 antibody Fab fragment to synthetic wild type Aβ40, Aβ28, Aβ8 and N-terminally biotinylated Aβ40 under equivalent experimental conditions. Under these solution conditions the Fab binds wild type Aβ40 with a KD of 89 (±9) nM (see Supplementary Fig. S1 online), and the affinity is reduced approximately 65 fold when Aβ is N-terminally biotinylated, indicating that the free N-terminus of Aβ is critical for binding. Under the same solution conditions, the affinity of the Fab drops marginally to 151 (±12) nM for Aβ28, and quite significantly for Aβ8, showing low μM affinity.
We wanted to identify exactly how these N-terminal Aβ (1–5) specific antibodies engage the Aβ peptide. To that end we crystallized a recombinant humanized 3D6 antibody Fab fragment complexed to the Aβ peptide (residues 1 to 28) and determined the complex structure to a resolution of 2.2 Å (Fig. 1). Data refinement and model statistics are given in Table 1.
Figure 1. Structure of the humanized 3D6 Fab-Aβ peptide complex.
Both panels show Aβ nestled in the surface of the Fab CDRs. The peptide is shown in green sticks with the light chain in light blue surface and heavy chain in a darker blue surface. (a) A 2Fo - Fc electron density map in the vicinity of the peptide contoured at 1.5σ. (b) Intra-Aβ hydrogen bonding, shown as dashed lines, stabilizes the helical conformation of the peptide.
Table 1. Data collection and refinement statistics.
| Data collection | |
|---|---|
| Space group | P21221 |
| Cell dimensions | |
| a, b, c (Å) | 59.3, 83.0, 91.2 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 2.20 (2.28–2.20) * |
| Rmerge (%) | 16.3 (78.8) |
| I/σI | 11.2 (2.8) |
| Completeness (%) | 94.4 (75.7) |
| Redundancy | 12.1 (5.8) |
| Refinement | |
| Resolution (Å) | 2.2 |
| No. reflections | 22098 |
| Rwork/Rfree (%) | 17.0/22.0 |
| No. atoms | 3615 |
| Protein | 3324 |
| Ligand/ion | 54 |
| Water | 237 |
| B-factors | |
| Protein | 20.6 |
| Ligand/ion | 19.9 |
| Water | 23.8 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 0.98 |
*Data are from a single crystal.
Structure of Aβ
Fig. 1 shows the Aβ peptide in the antibody binding site. We observed strong electron density across Aβ residues 1–5 (DAEFR), weaker density for His 6, and no electron density for building amino acids further into the peptide sequence (Fig. 1a), presumably because they have high mobility outside the confines of the antibody. The structure shows that Aβ is captured by Bapineuzumab in a helical conformation stabilized by five putative intramolecular hydrogen bonds (Fig. 1b).
Three of the five intra-Aβ hydrogen bonds involve Glu 3, first between its backbone amide a side-chain carboxyl, and two bonds between the side-chain carboxyls and the free N-terminal amine of Asp 1 (Fig. 1b). Further hydrogen bonding is observed between the amide of Phe 4 and the main-chain carbonyl of Asp 1, and between the main-chain amide of Arg 5 and the carbonyl of Ala 2.
To our surprise, the Aβ structure seen in the antibody complementarity determining regions (CDRs) (Fig. 2a) is very similar with TFE (2,2,2-trifluoroethylalcohol)-stabilized solution structures of Aβ determined by NMR9 (0.8 Å rms deviation on Cα atoms when the lowest energy NMR structure is superimposed) (Fig. 2b). TFE is commonly used as a co-solvent to promote intramolecular hydrogen bonding and stabilizes secondary structure in intrinsically disordered peptides such as Aβ by mimicking the solvent-deprived core of folded proteins and membranes10. Preferential binding of Bapineuzumab for plaque deposited Aβ11 suggests that this helical conformation at the N-terminus is either enriched or exists in an equilibrium of conformational states in dense plaque deposits. X-ray diffraction of Aβ oligomers, protofibrils and fibrils suggest that the N-terminal region of the peptide is free to adopt structure independent of the core cross-β structure12,13.
Figure 2. Different conformations of the Aβ peptide.
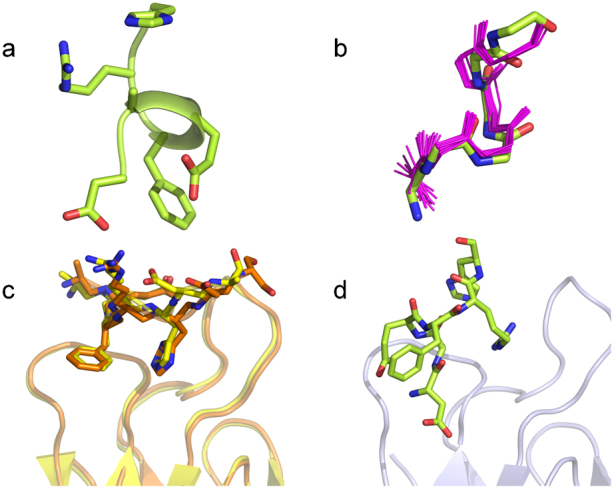
(a) The helical conformational epitope of Aβ recognized by Bapineuzumab highlighted in green ribbon. (b) Superposition of the main-chain heavy atoms of TFE-stabilized Aβ (residues 1 to 6) NMR structures (9) (in purple) with those of Aβ as recognized by Bapineuzumab (in green). (c), (d) Superposition over light chain of Fab-Aβ complexes with murine antibody Fabs in (c) WO2-Aβ is in orange ribbon, PFA1-Aβ in yellow and (d) Bapineuzumab related Fab in grey with Aβ in green sticks.
CDR engagement of Aβ
How humanized 3D6 engages the Aβ peptide is described in detail in Fig. 3. The interface surface area between the antibody and the antigen is 580 Å2. Eleven possible hydrogen bonds are observed between the antibody and Aβ residues Asp 1 (3 bonds), Glu 3 (4 bonds) and Arg 5 (4 bonds). In addition, there are five water-mediated hydrogen bonds. Numerous van der Waals contacts are made between antibody and Aβ: nine residues from the light chain and ten residues from the heavy chain. Not surprisingly, the hydrophobic Aβ residues are particularly involved in van der Waals contacts: Ala 2 interacts with five residues, all from light chain CDR 3, while Phe 4 is completely enveloped by seven contacting residues (six from the heavy chain).
Figure 3. Aβ-Fab interactions.
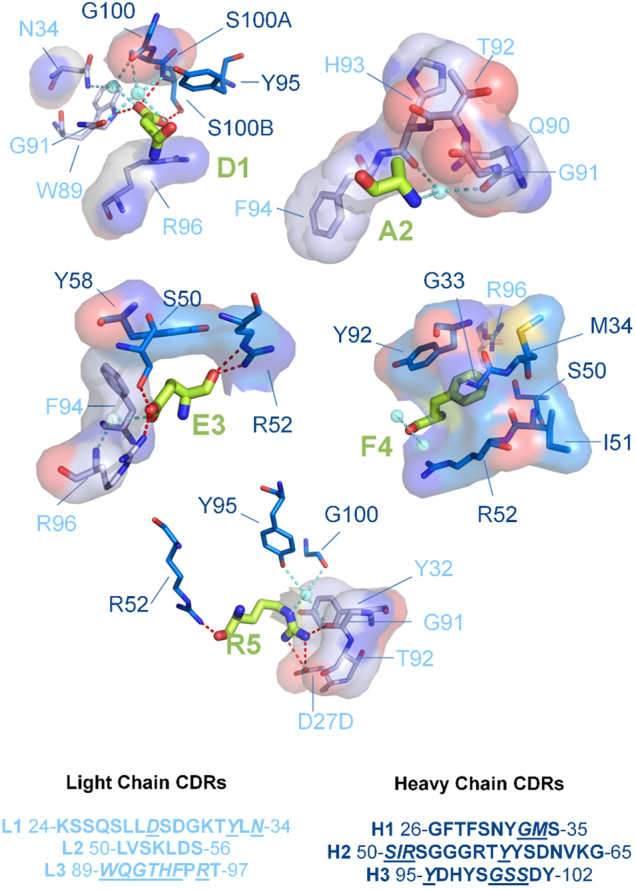
The Aβ residues are shown as green sticks. Amino acid sequences corresponding to the CDRs of Bapineuzumab are shown in light blue and darker blue for light and heavy chains respectively. Amino acids involved in Fab binding to Aβ are underlined and italicized in the CDR sequences common to 3D6 antibodies including Bapineuzumab. Direct polar contacts between the Fab and Aβ are shown graphically as red dashed lines. Waters involved in the hydrogen bonding network are shown as aquamarine spheres, and their putative hydrogen bonds are shown as aquamarine dashed lines. Fab residue labels and carbon atoms are colored by chain (shades of blue), whereas nitrogen, oxygen and sulfur atoms are shown in dark blue, red and yellow respectively. Surfaces represent non-polar contacts to Fab residues. Intra-chain contacts have been omitted for clarity. Figure produced using LigPlus30.
All hydrogen bonds stabilizing helical conformation in Aβ involve the first three residues, as do most of the ligand-Fab interactions. A high proportion, perhaps 60% of Aβ molecules in brain deposits are N-terminally truncated14 and N-terminal truncation of Aβ to any extent is likely to have deleterious effects on the stability of this helical fold and affinity for Bapineuzumab. Neurotoxic N-terminal modifications like pyro-Glu 3 (AβpE3) found in amyloid deposits15 would completely disrupt this fold and consequently presumably not be captured by Bapineuzumab. The buried free N-terminus of Aβ explains the lack of cross reactivity of Bapineuzumab with the amyloid precursor protein, which is proteolytically processed to yield Aβ.
Bapineuzumab is dissimilar to other N-terminal Aβ antibodies
This structure is unique amongst published anti-N-terminal-Aβ structures. Gardberg and co-workers published the first structure of a Fab-Aβ complex, murine-derived PFA1 in 200716 which was quickly followed by our own structure of a Fab derived from the murine anti-N-terminal-Aβ-antibody, WO2, bound to Aβ17,18. There are differences between CDR sequences of WO2 and PFA1, but they share conserved amino acids and engage the Aβ N-terminal region in essentially the same way (Fig. 2c). Both report structures for Aβ residues Ala 2 to Ser 8 with the core residues Phe 4, Arg 5 and His 6 buried in the CDR loops. Asp 1 is absent in each structure as are residues C-terminal to Ser 8 owing to high mobility. Contrast the two extended Aβ structures shown in Fig. 2c (1.3 Å rms deviation on Aβ alpha carbons upon superposition of the antibodies) with the helical Aβ structure as captured by Bapineuzumab, shown in Fig. 2d.
In 2010, Basi et al. reported structures for the highly homologous antibodies 12A11, 10D5, and 12B4, that recognize Aβ in much the same way as WO2 and PFA1/PFA219. In contrast, Bapineuzumab CDR loops share little sequence similarity with any of these antibodies. The WWDDD motif conserved between PFA1 and PFA2, and YWDDD in WO2, is not found in the Bapineuzumab CDR-H2, where the corresponding sequence is RSGGG. Only Tyr 32 of the light chain is involved in Aβ binding in all these structures. PFA1 and WO2 binding pockets have Tyr 32L pi-stacked against the aromatic side chain of His 6 in Aβ. In Bapineuzumab, Tyr 32L pi-stacks against the Aβ Arg 5 side-chain. The structure of the anti-AD immunotherapy Gantenerumab was reported in 2012 showing a longer extended coil structure for residues 1-11 of Aβ running across the antigen binding site of the antibody, but coordinates are not available from the PDB for examination20. Given the distinct structure of Aβ in Bapineuzumab and the lack of any consensus binding motif, it is clear that Bapineuzumab recognizes the overlapping binding epitope at the N-terminus in an entirely unique fashion.
Discussion
The recent setbacks in clinical trials of immunotherapies targeting Aβ (Bapineuzumab, Solanezumab and Ponezumab) in patients with mild to moderate AD have been disappointing and expensive but very informative. In the case of Bapineuzumab, the antibody was shown to be doing what it was designed to do: promoting clearance of brain amyloid with the downstream effect of lowering phosphorylated-tau levels in the cerebrospinal fluid. And in the case of Solanezumab, there was a small but significant cognitive improvement in a cohort of patients suffering “mild” AD. Proponents of the amyloid hypothesis of AD now believe that disease-modifying drugs may need to be administered early, in asymptomatic AD candidate patients before the disease causes its irretrievable effects21 and Bapineuzumab is being considered as one of the candidates in such trials (http://www.alzforum.org/new/detail.asp?id=3268).
We observe a lower affinity of the humanized 3D6 antibody for Aβ than the binding affinity reported by De Mattos et al. for the intact IgG murine antibody7. Our binding studies of truncated Aβ peptides suggest a more complex picture than simple antibody recognition of a linear epitope. Our MST data suggest that the antibody does not co-opt the peptide into the helical conformation but likely binds to a population of peptide that already adopts a helical structure as seen in the crystal structure. The minimal epitope containing peptide Aβ8 appears to sample this helical conformation less than longer peptides under our experimental conditions. Aβ peptides are highly pleiomorphic, with their conformation and oligomeric states exquisitely sensitive to their environment. Hence it was important that our measurements of the different peptides were done under the same solution conditions. An absolute KD value can therefore be misleading, and a nuanced approach must be taken in applying these data to an in vivo model.
The work reported here is part of a program to determine the structural basis of how clinically relevant antibodies recognize the conformationally variable Aβ peptide with the aim of aiding the interpretation of clinical trial outcomes, and for the development of more potent antibodies as elegantly demonstrated by Zahnd and co-workers where introduced mutations achieved a 500 fold improvement in antibody affinity for a helical peptide ligand22.
Methods
Protein expression, purification and crystallization will be published in detail elsewhere (Crespi, G.A.N., Ascher, D.B., Parker, M.W. and Miles, L.A., submitted) so only a brief description is presented here.
Humanized 3D6 Fab DNA constructs (variable light chain (VL) Seq ID NO:3 and variable heavy chain (VH) Seq ID NO:4, respectively, in (23)) were synthesized and cloned into pcDNA3.1 expression plasmids (Genscript). Heavy (C-terminally hexa-histidine tagged) and light chain constructs were co-transfected into FreeStyleTM 293-F cells (Invitrogen). Cell culture supernatants were harvested by centrifugation and concentrated by tangential flow filtration (Millipore, Proflux M12). Fab was purified with Ni-NTA resin (Qiagen) followed by size exclusion chromatography, dialyzed extensively against Buffer A (20 mM HEPES pH 7.5 and 50 mM NaCl), and finally concentrated to 5 mg/mL (measured by absorbance at 280 nm) and stored in small aliquots at −80°C until required for crystallization.
Peptides corresponding to the wild type amyloid-β sequence (DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVV) were purchased from GenicBio (residues 1–8, 95% purity), and the corresponding 1–28 and 1–40 peptides (≥95% purity) from AnaSpec. N-terminally biotinylated 1–40 peptide was a generous gift from laboratory of A/Prof. Kevin J. Barnham (Department of Pathology, the University of Melbourne). Lyophilized peptides, quantified by amino acid analysis, were resuspended in TFE and aliquoted to give 100 μg per Eppendorf tube. All aliquots were freeze-dried for 4 hours and stored at −80°C until required. TFE-treated, lyophilized peptides were taken up in 5 μL of 10 mM NaOH and diluted two fold with Buffer A (20 mM HEPES pH 7.5, 50 mM NaCl) to a final concentration of 10 mg/mL. Peptide was added to Fab in a Fab:Aβ molar ratio of 1∶5.
Solution MST binding studies between Fab and Aβ peptides (Aβ40, Aβ28 and Aβ8) were performed using standard protocols on a Monolith NT.115 (Nanotemper Technologies). Briefly, purified Fab was labeled using the RED-NHS (Amine Reactive) Protein Labelling Kit (Nanotemper Technologies). Lyophilized Aβ peptide was taken up in 5 μL of 10 mM NaOH and diluted in PBS and 0.05%v/v Tween-20. Labeled Fab antibody was mixed with Aβ with a final buffer condition of PBS and 0.05% Tween-20. Each replicate contained a 16 step of 1∶1 serial dilution series starting from 119 μM of Aβ40, 380 μM of Aβ8 and 151 μM of Aβ28. The protein concentration was chosen such that the observed fluorescence was approximately 600 units at 50% LED power. The samples were loaded into standard capillaries and heated for 30 sec, followed by 5 sec cooling at 40% laser power for Aβ40 and Aβ28, and 80% laser power for Aβ8. All experiments were performed with a minimum of 3 independent replicates. Affinity, KD, is quantified by analyzing the change in normalized fluorescence (Fnorm = fluorescence after thermophoresis/initial fluorescence) as a function of the concentration of the titrated peptide. The fraction of Fab bound (ΔFnorm/amplitude) was plotted against the concentration of peptide and the curves were analyzed using Graphpad Prism (Version 5, GraphPad, San Diego, CA, USA).
The first crystals obtained were the Fab complexed to the minimal epitope peptide Aβ8 with a well solution composed of 0.1 M HEPES pH 7.5, 25% (w/v) PEG 6000 (or 25% (w/v) PEG 8000). These crystals were used to promote crystallization of a Fab:Aβ28 complex via microseed matrix screening24. The best crystals obtained for Fab:Aβ28 were grown with reservoir solution containing 0.2 M sodium formate and 20% (w/v) PEG 3350. Crystals were harvested after 2 days for diffraction studies, requiring the addition of glycerol (to 10% (v/v)) as a cryo-protectant before being flash-frozen in liquid nitrogen.
X-ray diffraction data were acquired on the MX2 beamline at the Australian Synchrotron, Clayton, Victoria, Australia. The data collection was controlled using Blue-Ice software25. Data sets of 720 images were acquired at a single wavelength of 0.9537 Å, in a nitrogen cryostream (100 K), each with 0.5° rotation per frame. The best data set was indexed, integrated and scaled in point group P222 using the HKL2000 software package26. Data collection statistics are shown in Table 1.
The initial structure was determined by molecular replacement with Phaser from the Phenix software suite27 in the P21221 space group. A successful molecular replacement solution was achieved with a probe model derived from an antibody Fab structure with PDB entry code 3SOB, identified in a Protein Data Bank search for sequence similarities to humanized 3D6. 10% of reflections were set aside for the free R set by the program Phenix Refine prior to the first round of refinement. Refinement and rebuilding was done with Coot28 and Phenix Refine and structure validation monitored with MolProbity29. Refinement statistics are shown in Table 1. The final model has 98.0% of residues in favoured regions and 2.0% of residues in allowed regions of the Ramachandran plot with no outliers.
Author Contributions
G.A.N.C. expressed, purified and crystallized the Fab/Mab complexes and assisted in data acquisition. G.A.N.C. and L.D. designed and executed the MST experiments. L.A.M. acquired synchrotron diffraction data, refined data, solved structure and built model. L.A.M. and M.W.P. designed, supervised the study and analyzed the results. All authors wrote and revised the manuscript.
Supplementary Material
Supplementary Material
Acknowledgments
We thank David Ascher for his help in developing in-house MST protocols. This research was partly undertaken on the MX1 and MX2 beamlines at the Australian Synchrotron, Victoria, Australia. This work was supported by funding from a National Health and Medical Research Council of Australia (NHMRC) Project Grant (APP1021935) and grants from the JO & JR Wicking Trust, The Mason Foundation and The Bethlehem Griffith Research Foundation to MWP and LAM. The Australian Cancer Research Foundation provided substantial funding support for equipment critical for this work. Infrastructure support from the NHMRC Independent Research Institutes Infrastructure Support Scheme and the Victorian State Government Operational Infrastructure Support Program are gratefully acknowledged. MWP is an NHMRC Research Fellow.
References
- Selkoe D. J. Preventing Alzheimer's disease. Science 337, 1488–1492 (2012). [DOI] [PubMed] [Google Scholar]
- Barnes D. E. & Yaffe K. The projected effect of risk factor reduction on Alzheimer's disease prevalence. Lancet Neurol. 10, 819–828 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A. Sting of Alzheimer's failures offset by upcoming prevention trials. Nat. Rev. Drug Discov. 11, 657–660 (2012). [DOI] [PubMed] [Google Scholar]
- Salloway S. et al. A phase 2 multiple ascending dose trial of bapineuzumab in mild to moderate Alzheimer disease. Neurology 73, 2061–2070 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Wood K. et al. Amyloid precursor protein processing and Aβ42 deposition in a transgenic mouse model of Alzheimer disease. Proc. Natl. Acad. Sci. USA. 94, 1550–1555 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zago W. et al. Neutralization of soluble, synaptotoxic amyloid-β species by antibodies is epitope specific. J. Neurosci. 32, 2696–2702 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demattos R. B. et al. A plaque-specific antibody clears existing β-amyloid plaques in Alzheimer's disease mice. Neuron 76, 3523–3530 (2012). [DOI] [PubMed] [Google Scholar]
- Lippok S. et al. Direct detection of antibody concentration and affinity in human serum using microscale thermophoresis. Anal. Chem. 84, 3523–3530 (2012). [DOI] [PubMed] [Google Scholar]
- Zirah S. et al. Structural changes of region 1–16 of the Alzheimer disease amyloid beta-peptide upon zinc binding and in vitro aging. J. Biol. Chem. 281, 2151–2161 (2006). [DOI] [PubMed] [Google Scholar]
- Roccatano D., Colombo G., Fioroni M. & Mark A. E. Mechanism by which 2,2,2-trifluoroethanol/water mixtures stabilize secondary-structure formation in peptides: a molecular dynamics study. Proc. Natl. Acad. Sci. USA 99, 12179–12184 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seubert P. et al. Antibody capture of soluble Abeta does not reduce cortical Abeta amyloidosis in the PDAPP mouse. Neurodegener. Dis. 5, 65–71 (2008). [DOI] [PubMed] [Google Scholar]
- Colletier J. P. et al. Molecular basis for amyloid-beta polymorphism. Proc. Natl. Acad. Sci. USA 108, 16938–16943 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud J. C., Liu C., Teng P. K. & Eisenberg D. Toxic fibrillar oligomers of amyloid-β have cross-β structure. Proc. Natl. Acad. Sci. USA 109, 7717–7722 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant N. et al. Truncated beta-amyloid peptide species in pre-clinical Alzheimer's disease as new targets for the vaccination approach. J. Neurochem. 85, 1581–1591 (2003). [DOI] [PubMed] [Google Scholar]
- Nussbaum J. M. et al. Prion-like behaviour and tau-dependent cytotoxicity of pyroglutamylated amyloid-β. Nature 485, 651–655 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardberg A. S. et al. Molecular basis for passive immunotherapy of Alzheimer's disease. Proc. Natl. Acad. Sci. USA 104, 15659–15664 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles L. A. et al. Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope. J. Mol. Biol. 377, 181–192 (2008). [DOI] [PubMed] [Google Scholar]
- Wun K. S. et al. Crystallization and preliminary X-ray diffraction analysis of the Fab fragment of WO2, an antibody specific for the Abeta peptides associated with Alzheimer's disease. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64, 438–441 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basi G. S. et al. Structural correlates of antibodies associated with acute reversal of Amyloid β-related behavioral deficits in a mouse model of Alzheimer disease. J. Biol. Chem. 285, 3417–3427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohrmann B. et al. Gantenerumab: a novel human anti-Aβ antibody demonstrates sustained cerebral amyloid-β binding and elicits cell-mediated removal of human amyloid-β. J. Alzheimers Dis. 28, 49–69 (2012). [DOI] [PubMed] [Google Scholar]
- Callaway E. Alzheimer's drugs take a new tack. Nature 489, 13–14 (2012). [DOI] [PubMed] [Google Scholar]
- Zahnd C., Spinelli S., Luginbühl B., Amstutz P., Cambillau C. & Plückthun A. Directed in vitro evolution and crystallographic analysis of a peptide-binding single chain antibody fragment (scFv) with low picomolar affinity. J. Biol. Chem. 279, 18870–18877 (2004). [DOI] [PubMed] [Google Scholar]
- Schroeter S. & Games K. D. Prevention and treatment of cerebral amyloid angiopathy. US Patent Application 20080292625. (2008).
- Obmolova G., Malia T. J., Teplyakov A., Sweet R. & Gilliland G. L. Promoting crystallization of antibody-antigen complexes via microseed matrix screening. Acta Crystallogr. D Biol. Crystallogr. 66, 927–933 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhillips T. M. et al. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 9, 401–406 (2002). [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. & Minor W. Processing of X-ray diffraction data collected in the oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- Adams P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W. G. & Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R. A. & Swindells M. B. J. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. Chem. Inf. Model. 51, 2778–2786 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material



