Abstract
Glycogen Storage Disease type III (GSD III) is an autosomal recessive disorder in which a mutation in the AGL gene causes deficiency of the glycogen debranching enzyme. In childhood, it is characterized by hepatomegaly, keto-hypoglycemic episodes after short periods of fasting, and hyperlipidemia. In adulthood, myopathy, cardiomyopathy, and liver cirrhosis are the main complications. To determine the genotype of the GSD III patients (n = 14) diagnosed and treated in our center, mutation analysis was performed by either denaturing gradient gel electrophoresis or full gene sequencing. We developed, validated and applied both methods, and in all patients a mutation was identified on both alleles. Five novel pathogenic mutations were identified in seven patients, including four missense mutations (c.643G>A, p.Asp215Asn; c.655A>G, p.Asn219Asp; c.1027C>T, p.Arg343Trp; c.1877A>G, p.His626Arg) and one frameshift mutation (c.3911delA, p.Asn1304fs). The c.643G>A, p.Asp215Asn mutation is related with type IIIa, as this mutation was found homozygously in two type IIIa patients. In addition to five novel mutations, we present new genotype–phenotype relationships for c.2039G>A, p.Trp680X; c.753_756delCAGA, p.Asp251fs; and the intron 32 c.4260-12A>G splice site mutation. The p.Trp680X mutation was found homozygously in four patients, presenting a mild IIIa phenotype with mild skeletal myopathy, elevated CK values, and no cardiomyopathy. The p.Asp251fs mutation was found homozygously in one patient presenting with a severe IIIa phenotype, with skeletal myopathy, and severe symptomatic cardiomyopathy. The c.4260-12A>G mutation was found heterozygously, together with the p.Arg343Trp mutation in a severe IIIb patient who developed liver cirrhosis and hepatocellular carcinoma, necessitating an orthotopic liver transplantation.
Introduction
Glycogen storage disease type III (GSD III; OMIM no. 233400) is an autosomal recessive disorder in which mutations in the AGL gene cause deficiency of amylo-1,6-glucosidase and 1,4α-d-glucan 4-α-glycosyltransferase, also known as the glycogen debranching enzyme (GDE; EC no. 3.2.1.33 and 2.4.1.25). GDE catalyzes the last step in the conversion of glycogen to glucose, and GDE deficiency thus causes storage of an intermediate form of glycogen called limit dextrin (LD) (Smit et al. 2006). In the IIIa subtype, muscle and liver tissue are deficient in GDE, and this affects 85% of GSD III patients. Approximately 15% of the patients have type IIIb, in which only the liver is deficient in GDE (Shen et al. 1996). In the neonatal period and in infancy, the main features are hepatomegaly with elevated aspartate transaminase (ASAT) and alanine transferase (ALAT) values, keto-hypoglycemic episodes after relatively short periods of fasting, and hyperlipidemia. Untreated neonates and children have developmental delay, growth retardation, and delayed puberty. In puberty and early adulthood, myopathy becomes the predominant feature of GSD III; the disease presents as a slowly progressive muscle weakness in which the proximal muscles of the shoulder and hip joints are affected. Clinical muscle weakness in the upper and the lower limb muscles can develop in later adulthood, which may be worsened by the development of peripheral neuropathy (Hobson-Webb et al. 2010; Wolfsdorf and Weinstein 2003). LD can also be stored in heart muscle, which causes a form of cardiomyopathy that resembles idiopathic hypertrophic cardiomyopathy on an echocardiogram (Lee et al. 1997; Akazawa et al. 1997).
GDE is composed of 1,532 amino acid residues and has two catalytic centers (Bao et al. 1996; Liu et al. 1991). Before GDE starts to act, a phosphorylase enzyme separates four glucose molecules from the glycogen molecule to form LD (Newgard et al. 1989). Then the first catalytic center of GDE, 1,4-glucan-4-d-glucosyltransferase, transports the three outer glucose molecules of LD to another chain. The second catalytic center, amylo-1,6-glucosidase, then releases the last glucose molecule (Ding et al. 1990). GSD III can be diagnosed biochemically by measuring GDE activity in skin fibroblasts and/or leucocytes. GDE activity and LD content can also be measured in liver and/or muscle biopsies (Wolfsdorf and Weinstein 2003). LD content can be measured in erythrocytes.
The AGL gene is located on chromosome 1p22 and contains 35 exons covering 85 kb of DNA (Yang-Feng et al. 1992). The full cDNA is 7 kb with a 4,596 bp coding region. A total of six mRNA isoforms are created by alternative splicing. Isoform 1 is the major isoform and widely expressed, including in liver and muscle tissue (Bao et al. 1996). Isoforms 2, 3, and 4 are present in muscle and cardiac muscle and are formed by alternative splicing or because of the difference in transcription start points. Isoform 1 contains exons 1 and 3. Isoforms 2, 3, and 4 start with exon 2. Isoforms 1 through 4 all contain exon 3, which includes the normal initiation codon for protein translation. Exons 4 through 35 are present in all isoforms (Bao et al. 1996,1997). The glycogen-binding domain is encoded by exons 31–34. The 1,4α-d-glucan 4-α-glycosyltransferase catalytic site is encoded by exons 6, 13, 14, and 15. The amylo-1,6-glucosidase catalytic site is encoded by exons 26 and 27 (Shen and Chen 2002).
Molecular analyses of GSD III patients have been performed in several ethnic populations, and over 100 different AGL mutations have been described (Goldstein et al. 2010), but new mutations are still being reported (http://www.hgmd.org). No clear genotype-phenotype relationship has been established so far, although there is a relation between mutations in exon 3 and the IIIb subtype (Shen et al. 1996; Shen and Chen 2002). It is unclear, however, what mechanism enables patients with mutations in exon 3 to retain GDE activity in muscle tissue. A possible explanation was proposed by Goldstein et al. (2010) in which the exon 3 mutation is bypassed using a downstream start codon, thus creating an isoform without the exon 3 mutations.
To determine the genotype of the GSD III patients diagnosed and treated in our center (n = 14), the University Medical Centre Groningen (UMCG), the Netherlands, we performed mutation analysis by one of two methods. Denaturing gradient gel electrophoresis (DGGE) was applied on eight patients, and full gene sequencing was applied on six patients. We developed, validated, and applied both methods. Here we describe five novel mutations found in seven patients and their phenotypes, and evaluate the phenotype-genotype relationships in patients with previously described mutations.
Materials and Methods
Patients: For this study we analyzed 14 patients diagnosed with GSD III (seven females and seven males). They were diagnosed enzymatically by measuring GDE activity in leukocytes, fibroblasts, and/or liver tissue, and/or muscle tissue. All the patients were diagnosed and are being treated in the UMCG. Their clinical data was collected.
Mutation analysis by DGGE: Genomic DNA was isolated from EDTA blood (Miller et al. 1988). Primers were designed on the GenBank genomic reference sequence (NW_012865) to include at least 40 bp of intronic sequence on the front end of every exon and at least 20 bp at the back end. Analysis with NGRL Manchester’s SNP database (http://ngrl.manchester.ac.uk/SNPCheckV2/snpcheck.htm) confirmed that no known single nucleotide polymorphisms were situated under the primers. The primer specificity was checked and verified in complete genomic sequences with NCBI’s ePCR (http://www.ncbi.nlm.nih.gov/projects/e-pcr). We developed 50 primer sets to analyze 30 of the 33 coding exons by DGGE (primer sequences available upon request). The three remaining exons were sequenced directly, as designing DGGE primers with an appropriate melting curve was not possible. Per amplicon, the amplification mix consisted of 1.0 μl genomic DNA (40 ng/μl), 10 μl Amplitaq Gold ® Fast PCR Master Mix (Applied Biosystems, California, USA), 3.0 μl primer (3 pmol/μl), and 6.0 μl milliQ water; the reaction volume per sample was 20.0 μl. The samples were amplified by PCR in 96-well plates on a 96-well Gene Amp® 9700 PCR system (Applied Biosystems, California, USA).The PCR program used was as follows: 3 min at 96°C, 45 cycles of 1 min at 96°C, 1 min annealing at multiple temperatures, 1 min elongation at 72°C, and a final extension step at 72°C lasting 5 min. The annealing temperatures were 5 cycles at 60°C, 5 cycles at 56°C, 5 cycles at 52°C, and 30 cycles at 50°C. The PCR products were analyzed by DGGE (Hayes et al. 1999). Amplicons showing an aberrant banding pattern were sequenced on an ABI 3730 automated sequencer (Applied Biosystems, California, USA) using specific primers. DNA samples from 38 GSD III patients with 34 known mutations were used to validate the DGGE system – and all mutations were detected in our system.
Sequence analysis: We designed 33 primer sets according to the criteria used in conformation-sensitive capillary electrophoresis (CSCE, Table 1) and applied them for sequencing all coding exons of the gene including flanking intronic sequences. Per sample, the amplification mix consisted of 2 μl genomic DNA, 5 μl Amplitaq Gold® Fast PCR Master Mix (Applied Biosystems, California, USA), and 3.0 μl primer (150 fmol/μl). The reaction volume per sample was 10 μl; the samples were amplified by PCR in 384-well plates on a Veriti 384-well thermal cycler (Applied Biosystems, California, USA). The PCR program was as follows: 3 min at 94°C, 35 cycles of 1 min at 94°C, 1 min annealing at 60°C, 1 min at 72°C, and a final extension step at 72°C lasting 7 min, after which the samples were cooled down to 20°C. Five microliters of the PCR products were loaded with 5 μl loading buffer and run on a 2% agarose gel with a FastRuler Low Range DNA ladder (Fermentas, Vilnius, Lithuania) for comparison. The remaining PCR products were purified with ExoSAP-IT (Amersham Pharmacia Biotech, Piscataway, NY, USA) and subjected to direct sequencing on an ABI 3730 automated sequencer (Applied Biosystems, California, USA) using specific primers.
Table 1.
Sequences of the primer set designed according to the criteria of primer design used in conformation sensitive capillary electrophoresis (CSCE)a
| Amplicon | Forward primer sequence 5′-3′ | Reverse primer sequence 5′-3′ | Size bp |
|---|---|---|---|
| 3 | CGAACATGTAAGTGCCGCTGTCA | AGAACACAGCACCATCTTTGCACAA | 380 |
| 4 | GTAGTGCCAAAACAGCATTAGGTTTGC | GCACTGCCATGGTTCATACAGTAACAT | 457 |
| 5 | TTCCATTAAGTTTTGTTGCAAC | CTGCAATGAGAGAATGGACTAATACAC | 435 |
| 6 | TGAACCCAAGTGTTTGACCTCTTTTCC | CCTTTCTCTTATTTGTGTGTATATGTG | 432 |
| 7 | AACTTTTCCTGTAACAGTATCATCG | AATACAGGTTCTAAGTAATTTTCAACC | 429 |
| 8 | GCACTTTGGCGTTTCTCCTGTGA | GACGTTACCCAAAAGAGAGTTTTCCCT | 460 |
| 9 | GGGAGGAGGTAGGAGGATAC | CACATATAGAAACATGGCCCACACACA | 456 |
| 10 | CTGTGTGTGGGCCATGTTTCTATATGT | TTCCCAAAAGGCAATTAACTGCCTGAA | 409 |
| 11 | CTGCATTTCTCCATCTGCTCTAGCAA | ATTTAAGAAATGTACTGAACTCACATG | 440 |
| 12 | CATCCTGCTAGATTTACTCAAAAAGCC | ACCAATAGACTAATGGGGAAGAAAATC | 432 |
| 13 | TTAAAAACCAGTGTTTCCTTGAAG | AATGCTTGTGTCCAACTAGC | 381 |
| 14 | TATGTCAAATCATGCCTCCTTTTGTC | GAAATGAGGTATCTTACCCCAAAGTAG | 428 |
| 15 | CCATTTCTCCAGTTAAGTTATGGG | TGGGTATGATTGTGACCAAGTGTCAGA | 445 |
| 16 | GGTCACAATCATACCCATATACTTC | AAACCACTGAAATCTGGACAAAGG | 442 |
| 17 | CTATGGCATGTTGTGCTAGTGGAAGT | TCCACATACACCTGAGAAGCAGAAAGA | 433 |
| 18 | AGGAGCTTGGAGCCAAGGGTTT | CCATCATACCTGGCCAAGTTACCAAA | 447 |
| 19 | GATTTGAAACCACTTTAGCCTTCC | TGTGGCAACTCCAGCTTGTTTAAC | 340 |
| 20 | TGGGACTCTCATCTTACTACTGTG | GCATGTGGATCAAGACTAACTCTG | 340 |
| 22 | TTGAAAACTTGTCTCCAGGAAGTG | TGGACCGTACTTTGAGTAGCAAGGAT | 402 |
| 22 | GAATGCTGAGTTCCTAAAACATACAC | TGCAACCCAAGTAGGCATACTCTGA | 366 |
| 23 | TTGTGGACTGGGTAGCCCTTGT | GAAGGAAGGAGGAAAATGGTTCAGGTT | 397 |
| 24 | CCTCCTTCCTTCATCATCTTTCAG | CTATCCACCTACAAGCCTTTTCAG | 413 |
| 25 | TGGGTGAAATGAAAGCAGTTTTG | AAAATCTTGAGTAGCATTACAAGC | 458 |
| 26 | ACCCCAGGTTTAGAGTAACTGTTC | CTACCTAAAGAAAATACAGCTCCC | 323 |
| 27 | CAAAAGTGACTGGTTTTTGTCTTC | GGTGCCAAATCAATACTGACATTTG | 440 |
| 28 | CTGGCCTCACCCCAATTCCTATTTC | ATTATATCGTGAGGTTTGGCACAC | 352 |
| 29 | CAAACTGAGCTTTAGAGTGGTTGTCCT | AGATGAAGGGAAGAAGGCAGGGAAAT | 398 |
| 30 | TTCATTACAATTGTTTACCGAATGCCC | GGGTTTTCCGATATTAGCTGATAG | 301 |
| 31 | CACATCTCAATTCAGACTGGCCACAT | AACAAATGGGAATAAGGAACTAAGC | 441 |
| 32 | GGCTTTCCTAACTTCTACGGCCAAAA | AGATGGCATCTCCTTTTGTTGCCC | 407 |
| 33 | TGCCGAGCTTATTCTGTAGAAGAC | AGGCCACAGCCACTCCTAAAAAAG | 333 |
| 34 | TCACCAAGGACCTGTAAGAATTTC | CCTAGGGCATACAGAAATCAATTC | 350 |
| 35 | CACTAGAAGGCAAAAATCACCAGGTCT | AACTTGAGCCTGTGCATATAAGGCATT | 294 |
aPrimer and amplicon criteria: Optimal primer length 20bp (minimum 18bp; maximum 27bp), optimal annealing temperature 58°C (minimum 52°C; maximum 64°C), optimal GC% 55 (maximum 70), optimal amplicon size 400bp (minimum 200bp; maximum 464bp), maximal amplicon GC% 73. A PT1 tail was added to every forward primer sequence (5′-3′; TGTAAAACGACGGCCAGT), and a PT2 tail was added to every reverse primer sequence (5′-3′; CAGGAAACAGCTATGACC)
Analysis of the pathogenicity of novel mutations: The pathogenicity of novel mutations was assessed using six separate methods. One hundred control chromosomes from mixed ethnicity were checked for novel mutations. Conservation of the mutated nucleotide and amino acid was graded with Alamut Version 1.4 (©Interactive Biosoftware). The University of Harvard’s PolyPhen program (http://genetics.bwh.harvard.edu/pph/) predicted the impact of an amino acid substitution on the structure and function of a human protein. The SIFT program (http://blocks.fhcrc.org/sift/SIFT.html) predicted whether an amino acid substitution affects protein function based on sequence homology and the physical properties of amino acids. Finally, we assessed whether the mutation was located in an exon encoding the glycogen binding domain (exon 31-34), or encoding a catalytic site (exon 6, 14 16, 26-27), and measured the GDE activity.
Results
Patients: Of our 14 GSD III patients, 4 were related to one other patient (two sisters, and two brothers), all other patients were unrelated. Patient 7 was born to consanguineous parents. Nine patients had type IIIa, two patients had type IIIb, and three pediatric patients were too young to be subtyped based on their clinical presentation. The clinical and biochemical characteristics of the patients are presented in Table 2.
Table 2.
Demographic, clinical, and biochemical characteristics of the analyzed GSD III patients
| Patient no. | Age (years) | Sex | Subtype | Ethnic origin | GDE residual activity (%) | Liver complications | Cardiologic complications | Skeletal muscle complications | Most recent CK value (U/L) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 8 | M | IIIa | Caribbean | 0 | Hepatomegaly | None | Proximal myopathy | 1083 |
| 2 | 25 | F | IIIa | Caribbean | 0 | Hepatomegaly | None | None | 2232 |
| 3 | 26 | M | IIIa | Caribbean | 0 | Hepatomegaly | Septal hypertrophy | None | 893 |
| 4 | 40 | F | IIIa | Caribbean | 0 | Hepatomegaly | None | None | 3729 |
| 5 | 15 | F | IIIa | Mediterranean | 0 | Hepatomegaly | None | None | 662 |
| 6 | 20 | F | IIIa | Mediterranean | 0 | Hepatomegaly | None | None | 1342 |
| 7 | 32 | F | IIIa | Mediterranean | 0 | Hepatomegaly | Severe symptomatic left ventricular and septal hypertrophy | Exercise intolerance, distal myopathy | 1898 |
| 8 | 3 | M | Too young | Caucasian | 1 | Hepatomegaly | None | None | 133 |
| 9 | 30 | F | IIIa | Caucasian | 0 | None | None | Exercise intolerance, distal myopathy | 2257 |
| 10 | 30 | M | IIIb | Caucasian | 0 | Hepatomegaly | None | None | 68 |
| 11 | 41 | F | IIIa | Caucasian | 0 | Hepatomegaly | None | Exercise intolerance | 392 |
| 12 | 41 | F | IIIb | Caucasian | 0 | None, the patient is post-OLTc, pre-OLTc liver cirrhosis and hepatocellular carcinoma was present |
None | None | 70 |
| 13 | 3 | M | Too young | Caucasian | 0 | Hepatomegaly | None | None | 128 |
| 14 | 1 | M | Too young | Caucasian | 0 | Hepatomegaly | None | None | 466 |
M Male, F Female; OLT Orthotopic Liver Transplantation
Results of mutation analysis: The patients were fully analyzed and we detected two mutations in each patient (Table 3). Five novel mutations were identified in seven patients, including two sisters who were homozygous for c.643G>A, p.Asp215Asn and two brothers who were homozygous for c.3911delA, p.Asn1304fs. The other three patients were compound heterozygous with a novel missense mutation and a previously reported pathogenic mutation: c.655A>G, p.Asn219Asp in combination with c.4529dupA, p.Tyr1510X; c.1027C>T, p.Arg343Trp in combination with the splice mutation c.4260-12A>G; and c.1877A>G, p.His626Arg in combination with c.1222C>T, p.Arg408X.
Table 3.
Mutation analysis results in 14 GSD III patients
| Patient no. | Exon no. | Nucleotide change allele 1 | Amino acid change allele 1 | Mutation type | Exon no. | Nucleotide change allele 2 | Amino acid change allele 2 | Mutation type | Mutation analysis method |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 17 | c.2039G>A | p.Trp680X | Nonsense | 17 | c.2039G>A | p.Trp680X | Nonsense | DGGE |
| 2 | 17 | c.2039G>A | p.Trp680X | Nonsense | 17 | c.2039G>A | p.Trp680X | Nonsense | DGGE |
| 3 | 17 | c.2039G>A | p.Trp680X | Nonsense | 17 | c.2039G>A | p.Trp680X | Nonsense | DGGE |
| 4 | 17 | c.2039G>A | p.Trp680X | Nonsense | 17 | c.2039G>A | p.Trp680X | Nonsense | Sequencing |
| 5 | 6 | c.643G>A | p.Asp215Asn | Missense | 6 | c.643G>A | p.Asp215Asn | Missense | DGGE |
| 6 | 6 | c.643G>A | p.Asp215Asn | Missense | 6 | c.643G>A | p.Asp215Asn | Missense | DGGE |
| 7 | 7 | c.753_756delCAGA | p.Asp251fs | Frameshift | 7 | c.753_756delCAGA | p.Asp251fs | Frameshift | DGGE |
| 8 | 6 | c.655A>G | p.Asn219Asp | Missense | 35 | c.4529dupA | p.Tyr1510X | Nonsense | Sequencing |
| 9 | 35 | c.4529dupA | p.Tyr1510X | Nonsense | 35 | c.4529dupA | p.Tyr1510X | Nonsense | DGGE |
| 10 | 3 | c.16C>T | p.Gln6X | Nonsense | 3 | c.16C>T | p.Gln6X | Nonsense | DGGE |
| 11 | 11 | c.1222C>T | p.Arg408X | Nonsense | 15 | c.1877A>G | p.His626Arg | Missense | Sequencing |
| 12 | 9 | c.1027C>T | p.Arg343Trp | Missense | Intron 32 | c.4260-12A>G | Splice | Sequencing | |
| 13 | 30 | c.3911delA | p.Asn1304fs | Frameshift | 30 | c.3911delA | p.Asn1304fs | Frameshift | Sequencing |
| 14 | 30 | c.3911delA | p.Asn1304fs | Frameshift | 30 | c.3911delA | p.Asn1304fs | Frameshift | Sequencing |
Results of pathogenicity analysis: We consider the four new missense mutations to be pathogenic for several reasons. We did not detect the new mutations in 100 control chromosomes, and these mutations were not found in the NCBI SNP database. Highly conserved amino acids were mutated, and all mutations were predicted to affect protein function by the PolyPhen and SIFT programs. Additionally, three of the four new mutations (Asp215Asn, p.Asn219Asp, and p.His626Arg) were located in an exon encoding the 1,4α-d-glucan 4-α-glycosyltransferase catalytic site of GDE (Fig. 1). This makes a pathogenic effect of these mutations on GDE function very probable (Cheng et al. 2009). As the c.3911delA, p.Asn1304fs causes a frameshift with the new reading frame ending in a stop codon at position 10, this is considered to be a pathogenic mutation as well. Clinically, all the patients presented with a GSD III phenotype, and this was biochemically confirmed by enzymatic analysis that no GDE activity was measured to be present.
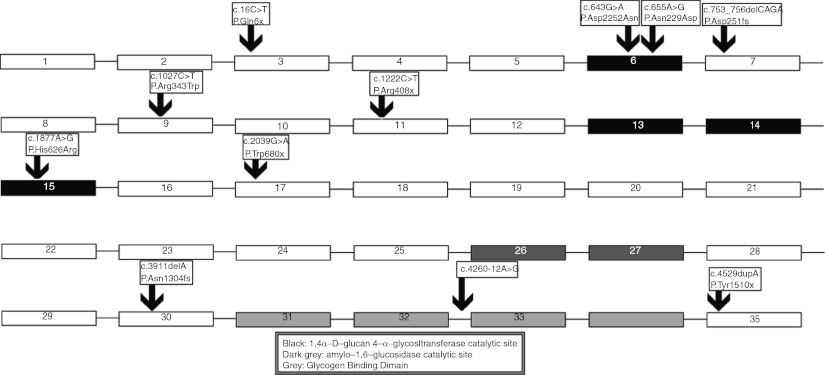
Fig. 1.
The location of all found mutations in the gene, the catalytic sites of GDE are depicted as well; three of the four novel mutations (Asp215Asn; p.Asn219Asp; and p.His626Arg) were located in exons encoding the 1,4α-d-glucan 4-α-glycosyltransferase catalytic site
Discussion
Here we describe five novel pathogenic mutations in the AGL gene, four of which are missense mutations, very likely causing GSD III in seven patients. Missense mutations causing GSD III are scarce, as truncating mutations compose the majority of the pathogenic mutations. However, pathogenic missense mutations have been described. Cheng et al. showed that missense mutations located in the active sites produce a GSD III phenotype in which there is total or partial abolishment of GDE activity depending on the location of the mutation (Cheng et al. 2009).
GSD III is characterized by clinical and genotypical heterogeneity, and clear genotype-phenotype correlations are rare. We therefore present the clinical characteristics of the patients with novel mutations, and discuss the correspondence between their clinical presentation and that described in the literature for the mutations that have been previously described.
Genotype-phenotype analysis: Patients 1 to 4 all had type IIIa and were homozygous for p.Trp680X. The phenotype was similar in all four patients, with hepatomegaly, no or mild cardiac involvement, and mild skeletal myopathy with elevated CK values. This suggests a link between the p.Trp680X mutation and the type IIIa phenotype. The p.Trp680X mutation was described previously in a compound heterozygote patient, in whom it was linked to the IIIb phenotype (Shen et al. 1996), which is in contrast to what we found in our patients. Interestingly, all our patients with the c.2039G>A, p.Trp680X mutation were from the same topographic region and ethnic origin, making a founder effect feasible, as seen for the c.1222C>T, p.Arg408X mutation on the Faroe Islands (Santer et al. 2001). However, in order to prove this, haplotyping for these patients would be required, which we have not done in this study.
Patient 5 (female, age 15 years) and patient 6 (female, age 20 years) were sisters (whose parents were from the Mediterranean) who had type IIIa, and were homozygous for the novel p.Asp215Asn mutation. Their phenotype was mild with hepatomegaly as the main finding, there was no myopathy or cardiomyopathy. In laboratory investigations ASAT, ALAT, and CK values were elevated, but there was no hypoglycemia or hyperlipidemia. The mild phenotype was probably due to their young age and the good metabolic control of these patients.
Patient 7 (female, age 32 years) had type IIIa and was found to be homozygous for p.Asp251fs. She had a severe IIIa phenotype, with hepatomegaly, and skeletal muscle involvement with severe exercise intolerance. There was also severe cardiac involvement with hypertrophy of the interventricular septum and left ventricle, necessitating pharmacological treatment and ICD placement. This mutation was previously described in a 3-year-old female, who was found to have hepatosplenomegaly and hypoglycemia but no myopathy or cardiomyopathy (Lucchiari et al. 2006).
In patient 8 (male, age 3 years), the novel mutation p.Asn219Asp was found to be heterozygous. The other mutation p.Tyr1510X was previously reported and associated with a severe IIIa phenotype (Shen et al. 1997). At first presentation (age 1.5 years), he had severe hepatomegaly extending 15 cm below the costal margin in the medioclavicular line, regular keto-hypoglycemic episodes and hyperlipidemia. These symptoms improved dramatically upon starting dietary treatment after diagnosis at the age of 1.5 years with frequent meals during the day and overnight gastric drip feeding. These findings are typical for GSD III patients in this age group, and the normal CK value does not exclude future muscle involvement, making it hard to assess the subtype or to determine the phenotype that goes with this novel mutation. Furthermore, the latter proves difficult because the patient was found to have compound heterozygous mutations.
Patient 9 (female, 30 years) had type IIIa and was found to be homozygous for p.Tyr1510X. She had a severe IIIa phenotype including distal myopathy and severe exercise intolerance with elevated CK (2257 U/L). Despite the absence of hepatomegaly, transaminases remained elevated (ASAT 237 U/L, ALAT 175 U/L). This is in concordance with a suggestion made by Shen et al. that this mutation is associated with a severe GSD IIIa phenotype (Shen et al. 1997).
Patient 10 (male, 30 years) had type IIIb and was homozygous for p.Gln6X. His phenotype was mild, with hepatomegaly, normal CK values, and no cardiac or skeletal muscle involvement. This mutation was previously described and is strongly linked with the GSD IIIb phenotype, as are other mutations in exon 3 (Shen et al. 1996; Shen and Chen 2002). Paradoxically, this patient first presented at the age of 2 years with proximal myopathy and severe hypotonia, but without cardiomyopathy or elevated CK values. His myopathy and hypotonia improved dramatically upon starting dietary treatment, and he has had no muscular symptoms since.
Patient 11 (female, age 41 years) had type IIIa and was compound heterozygous for the novel p.His626Arg mutation. Her other mutation, p.Arg408X, was previously described (Santer et al. 2001; Lam et al. 2004) and associated with the GSD IIIa phenotype. This corresponds with her phenotype: hepatomegaly and elevated transaminases. She has no proximal or distal myopathy but she does suffer from exercise intolerance and has elevated CK values.
Patient 12 (female, age 41 years) had type IIIb and was compound heterozygous for the novel p.Arg343Trp mutation and c.4260-12A>G in intron 32. c.4260-12A>G was previously described and associated with IIIb as well as IIIa (Okubo et al. 1998; Shaiu et al. 2000). This patients’ phenotype was severe IIIb and a case report on her was published after she received an orthotopic liver transplantation after being diagnosed with liver cirrhosis (Haagsma et al. 1997). Hepatocellular carcinoma was found upon pathological examination of the excised liver. She has never had any muscle involvement, and CK values have always been normal.
Patients 13 (male, age 3 years) and 14 (male, age 1 years) were brothers and homozygous for the novel mutation c.3911delA, p.Asn1304fs in exon 30. Except for prominent hepatomegaly and the need for appropriate dietary requirements, both patients were generally well. There was no clinical muscle involvement, even though patient 14 had elevated CK values. As these are very young pediatric patients, it is not yet possible to assess the subtype based on the clinical findings.
Conclusions: We identified two separate mutations in each of our 14 GSD III patients. Five were novel pathogenic mutations considered to be causal. As we also analyzed parts of the intron sequences (40 bp on the front end, and 20 bp at the back end of every exon), we assume that the chance that we missed other disease-causing mutations is slim. The novel c.643G>A, p.Asp215Asn mutation is related with type IIIa, as this mutation was found homozygously in two patients both clinically presenting as type IIIa. We also established new genotype-phenotype relationships between c.2039G>A, p.Trp680X and type IIIa; c.753_756delCAGA, p.Asp251fs and type IIIa; and the intron 32 c.4260-12A>G splice site mutation and type IIIb. The association between c.4529dupA, p.Tyr1510X and type IIIa complies with previous literature. However, as the GSD III subtype is not yet clear for every patient, it was not possible to establish a genotype-phenotype relationship for every novel mutation. There is still a large clinical and genotypical heterogeneity in GSD III, which makes establishing genotype-phenotype relationships for GSD III difficult. The fact that we found five new mutations in a relatively small number of GSD III patients further accentuates the need for more genotyping, and indicates that there are probably numerous unidentified mutations.
Acknowledgments
We thank Prof. René Santer of the University Clinic Hamburg, Germany, for providing the DNA samples for the validation of our methods.
Synopsis
GSD III is characterized by a large clinical and genotypical heterogeneity, which makes the establishment of clear genotype-phenotype relationships difficult.
Conflicts of Interest
None declared.
Footnotes
Competing interests: None declared
References
- Akazawa H, Kuroda T, Kim S, Mito H, Kojo T, Shimada K. Specific heart muscle disease associated with glycogen storage disease type III: clinical similarity to the dilated phase of hypertrophic cardiomyopathy. Eur Heart J. 1997;18:532–533. doi: 10.1093/oxfordjournals.eurheartj.a015283. [DOI] [PubMed] [Google Scholar]
- Bao Y, Dawson TL, Jr, Chen YT. Human glycogen debranching enzyme gene (AGL): complete structural organization and characterization of the 5’ flanking region. Genomics. 1996;38:155–165. doi: 10.1006/geno.1996.0611. [DOI] [PubMed] [Google Scholar]
- Bao Y, Yang BZ, Dawson TL, Jr, Chen YT. Isolation and nucleotide sequence of human liver glycogen debranching enzyme mRNA: identification of multiple tissue-specific isoforms. Gene. 1997;15:389–398. doi: 10.1016/S0378-1119(97)00291-6. [DOI] [PubMed] [Google Scholar]
- Cheng A, Zhang M, Okubo M, Omichi K, Saltiel AR. Distinct mutations in the glycogen debranching enzyme found in glycogen storage disease type III lead to impairment in diverse cellular functions. Hum Mol Genet. 2009;18:2045–2052. doi: 10.1093/hmg/ddp128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding JH, De Barsy T, Brown BI, Coleman RA, Chen YT. Immunoblot analyses of glycogen debranching enzyme in different subtypes of glycogen storage disease type III. J Pediatr. 1990;116:95–100. doi: 10.1016/S0022-3476(05)81652-X. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, Austin SL, Boyette K, Kanaly A, Veerapandiyan A, Rehder C, Kishnani PS, Bali DS. Molecular analysis of the AGL gene: identification of 25 novel mutations and evidence of genetic heterogeneity in patients with glycogen storage disease type III. Genet Med. 2010;12:424–430. doi: 10.1097/GIM.0b013e3181d94eaa. [DOI] [PubMed] [Google Scholar]
- Haagsma EB, Smit GPA, Niezen-Koning KE, Gouw AS, Meerman L, Slooff MJ. Type IIIb glycogen storage disease associated with end-stage cirrhosis and hepatocellular carcinoma. The Liver Transplant Group. Hepatology. 1997;25:537–540. doi: 10.1002/hep.510250307. [DOI] [PubMed] [Google Scholar]
- Hayes VM, Wu Y, Osinga J, Mulder IM, van der Vlies P, Elfferich P, Buys CH, Hofstra RM. Improvements in gel composition and electrophoretic conditions for broad-range mutation analysis by denaturing gradient gel electrophoresis. Nucleic Acids Res. 1999;27:e29. doi: 10.1093/nar/27.20.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson-Webb LD, Austin SL, Bali DS, Kishnani PS. The electrodiagnostic characteristics of glycogen storage disease type III. Genet Med. 2010;12:440–445. doi: 10.1097/GIM.0b013e3181cd735b. [DOI] [PubMed] [Google Scholar]
- Lam CW, Lee AT, Lam YY, Wong TW, Mak TW, Fung WC, Chan KC, Ho CS, Tong SF. DNA-based subtyping of glycogen storage disease type III: mutation and haplotype analysis of the AGL gene in Chinese. Mol Genet Metab. 2004;83:271–275. doi: 10.1016/j.ymgme.2004.07.017. [DOI] [PubMed] [Google Scholar]
- Lee PJ, Deanfield JE, Burch M, Baig K, McKenna WJ, Leonard JV. Comparison of the functional significance of left ventricular hypertrophy in hypertrophic cardiomyopathy and glycogenosis type III. Am J Cardiol. 1997;79:834–838. doi: 10.1016/S0002-9149(96)00885-5. [DOI] [PubMed] [Google Scholar]
- Liu W, Madsen NB, Braun C, Withers SG. Reassessment of the catalytic mechanism of glycogen debranching enzyme. Biochemistry. 1991;30:1419–1424. doi: 10.1021/bi00219a036. [DOI] [PubMed] [Google Scholar]
- Lucchiari S, Pagliarani S, Salani S, Filocamo M, Di Rocco M, Melis D, Rodolico C, Musumeci O, Toscano A, Bresolin N, Comi GP. Hepatic and neuromuscular forms of glycogenosis type III: nine mutations in AGL. Hum Mutat. 2006;27:600–601. doi: 10.1002/humu.9426. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1225. doi: 10.1093/nar/16.3.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard CB, Hwang PK, Fletterick RJ. The family of glycogen phosphorylases: structure and function. Crit Rev Biochem Mol Biol. 1989;24:69–99. doi: 10.3109/10409238909082552. [DOI] [PubMed] [Google Scholar]
- Okubo M, Horinishi A, Nakamura N, Aoyama Y, Hashimoto M, Endo Y, Murase T. A novel point mutation in an acceptor splice site of intron 32 (IVS32 A-12→G) but no exon 3 mutations in the glycogen debranching enzyme gene in a homozygous patient with glycogen storage disease type IIIb. Hum Genet. 1998;102:1–5. doi: 10.1007/s004390050646. [DOI] [PubMed] [Google Scholar]
- Santer R, Kinner M, Steuerwald U, Kjaergaard S, Skovby F, Simonsen H, Shaiu WL, Chen YT, Schneppenheim R, Schaub J. Molecular genetic basis and prevalence of glycogen storage disease type IIIa in the Faroe Islands. Eur J Hum Genet. 2001;9:388–391. doi: 10.1038/sj.ejhg.5200632. [DOI] [PubMed] [Google Scholar]
- Shaiu WL, Kishnani PS, Shen J, Liu HM, Chen YT. Genotype-phenotype correlation in two frequent mutations and mutation update in type III glycogen storage disease. Mol Genet Metab. 2000;69:16–23. doi: 10.1006/mgme.1999.2953. [DOI] [PubMed] [Google Scholar]
- Shen J, Bao Y, Liu HM, Lee PJ, Leonard JV, Chen YT. Mutations in Exon 3 of the glycogen debranching enzyme gene are associated with glycogen storage disease type III that is differentially expressed in liver and muscle. J Clin Invest. 1996;98:352–357. doi: 10.1172/JCI118799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Bao Y, Chen YT. A nonsense mutation due to a single base insertion in the 3'-coding region of glycogen debranching enzyme gene associated with a severe phenotype in a patient with glycogen storage disease type IIIa. Hum Mutat. 1997;9:37–40. doi: 10.1002/(SICI)1098-1004(1997)9:1<37::AID-HUMU6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Shen JJ, Chen YT. Molecular characterization of glycogen storage disease type III. Curr Mol Med. 2002;2:167–175. doi: 10.2174/1566524024605752. [DOI] [PubMed] [Google Scholar]
- Smit GPA, Rake JP, Akman HO, DiMauro S. The glycogen storage diseases and related disorders. In: Fernandes J, Saudubray J-M, van den Berghe G, Walter JH, editors. Inborn metabolic diseases: diagnosis and treatment. Heidelberg: Springer Medizin Verlag; 2006. pp. 103–116. [Google Scholar]
- Wolfsdorf JI, Weinstein DA. Glycogen storage diseases. Rev Endocr Metab Disord. 2003;4:95–102. doi: 10.1023/A:1021831621210. [DOI] [PubMed] [Google Scholar]
- Yang-Feng TL, Zheng K, Yu J, Yang BZ, Chen YT, Kao FT. Assignment of the human glycogen debrancher gene to chromosome 1p22. Genomics. 1992;13:931–934. doi: 10.1016/0888-7543(92)90003-B. [DOI] [PubMed] [Google Scholar]