SUMMARY
The angiosperm extracellular matrix, or cell wall, is composed of a complex array of cellulose, hemicelluose, pectins and proteins, the modification and regulated synthesis of which are essential for cell growth and division. The wall associated kinases (WAKs) are receptor-like proteins that have an extracellular domain that bind pectins, the more flexible portion of the extracellular matrix, and are required for cell expansion as they have a role in regulating cellular solute concentrations. We show here that both recombinant WAK1 and WAK2 bind pectin in vitro. In protoplasts pectins activate, in a WAK2-dependent fashion, the transcription of vacuolar invertase, and a wak2 mutant alters the normal pectin regulation of mitogen-activated protein kinases. Microarray analysis shows that WAK2 is required for the pectin activation of numerous genes in protoplasts, many of which are involved in cell wall biogenesis. Thus, WAK2 plays a major role in signaling a diverse array of cellular events in response to pectin in the extracellular matrix.
Keywords: cell wall, kinase, wall associated kinase
INTRODUCTION
Plant cell walls are composed of a complex array of cellulose, hemicelluose, pectins and proteins that surround each cell. This matrix must enlarge and be modified during cell growth and division, and the regulation and monitoring of its production and modification is critical to plant form and function (Ridley et al., 2001; Willats et al., 2001; Bosch and Hepler, 2005; Bacic, 2006; Kohorn et al., 2006a,b). Pectin is thought to be the more flexible portion of the cell wall, and many have predicted that its movement, cleavage and modification by esterification is regulatory, and may be monitored by receptors that mediate cellular responses (Willats et al., 2001; Bosch and Hepler, 2005; Bacic, 2006). Pectin fragments generated by pathogens, wounding or perhaps during developmental programs have long been hypothesized to trigger numerous cellular responses, and a physiological and developmental change to the plant (Ridley et al., 2001; Willats et al., 2001; Bacic, 2006).
Pectins are synthesized in the Golgi in a methyl-esterified form, but upon exit into the cell wall can undergo de-esterification that allows ionic interactions via the exposed carboxyl group with other similar pectins, carbohydrates and proteins (Ridley et al., 2001; Willats et al., 2001; Bosch and Hepler, 2005). These interactions can change the plasticity of the cell wall and thereby regulate cell growth and shape.
The wall-assocaited kinases (WAKs) bind to pectins and are required for cell expansion, in part because of their regulation of invertase transcriptional activity, and hence solute concentrations, within the cell (Kohorn, 2000; Lally et al., 2001; Wagner and Kohorn, 2001; Kohorn et al., 2006a). WAKs are receptor-like proteins that in addition to their association with the cell wall, contain a cytoplasmic serine/threonine protein kinase domain (Kohorn, 2000; Lally et al., 2001; Wagner and Kohorn, 2001; Kohorn et al., 2006a,b). WAKs are encoded by five tightly linked and highly similar genes in Arabidopsis thaliana, and are expressed in leaves, meristems and cells undergoing expansion (He et al., 1996, 1999; Wagner and Kohorn, 2001). WAK gene expression is also induced by a variety of agents, including pathogens, wounding and numerous stresses (He et al., 1998, 1999; Wagner and Kohorn, 2001; Sivaguru et al., 2003).
Expression of an inducible antisense WAK2 or WAK4 in Arabidopsis leaf cells leads to a 50% reduction in WAK protein levels and a subsequent loss of cell elongation, and hence lead to dwarf plants (Lally et al., 2001; Wagner and Kohorn, 2001). An insertion mutation of WAK2, wak2-1, also leads to an arrest of cell expansion and a reduction of invertase expression, and subsequently activity, in the vacuole (Kohorn et al., 2006a). This provides a theoretical basis for a loss of cell expansion.
WAK1 has been reported to bind to a glycine-rich protein (GRP) in the yeast two-hybrid assay, and possibly forms a 450-kDa complex with GRP in Arabidopsis leaf extracts (Park et al., 2001). The significance of the finding that a thylakoid lumen protein OEC 32 is phosphorylated upon GRP-WAK binding in protoplasts remains to be determined (Yang et al., 2003).
The extracellular domains of WAKs indeed can bind to pectin in vitro, and this is enhanced by a calcium-induced bridging of the pectin (Decreux and Messiaen, 2005; Decreux et al., 2006). The WAK-pectin binding may be charge dependent, as de-esterified pectin binds WAK1 more than does esterified pectin. The requirement for a charge interaction is partly confirmed by substitution of arginine and lysine residues for glutamines and threonines, respectively, in the WAK1 extracellular domain that results in reduced binding to pectin (Decreux et al., 2006).
A large number of pharmacological studies provide intriguing yet conflicting results of how pectin fragments might stimulate cytoplasmic events. Although some suggest that in intact tissues pectins are antagonistic to the effects of auxin, others indicate pectins can be stimulatory (Ridley et al., 2001), and the mechanism by which pectins mediate cellular responses, including those to pathogens, remain yet to be fully characterized.
Wall-associated kinases have also been implicated in the plant’s response to pathogens, as their NPR1-dependent expression is required during the pathogen response, and the induced expression of a WAK kinase domain can lead to resistance to otherwise toxic levels of salicylate (SA) (He et al., 1999). In Arabidopsis, a larger family of at least 35 members encoding WAK-like proteins contains extracellular epidermal growth factor repeats and a cytoplasmic kinase domain. The similarity to WAKs is confined to the kinase and EGF repeats, such that the large extracellular domains are quite distinct. The WAK-like genes (WAKL) are also clustered in several tandem repeated arrays (Verica and He, 2002; Verica et al., 2003), yet although these receptors are clearly on the cell surface, their association with the cell wall remains uncharacterized. A dominant allele of the WAKL22 locus was identified by its ability to confer resistance to Fusarium oxysporum (Diener and Ausubel, 2005), indicating that this class of receptor has a role in pathogenesis. Also noteworthy is the large expansion of the WAK-like family in crop plants relative to the Arabidopsis genome. Such an increase in size of tandemly repeated and clustered gene families is correlated with a role in disease resistance and response to environmental stresses (Richter and Ronald, 2000; Lespinet et al., 2002).
Thus, the WAKs bind to pectin, are required for cell expansion and are induced by a variety of environmental stimuli, including pathogens and wounding. As their position in the cell provides a mechanism for relaying the presence and modification of pectin, it was of interest to determine if there was a direct link between pectin, WAKs and cellular responses. We show here that pectins activate both mitogen-activated protein kinase (MAPK) and multiple gene expression changes associated with cell wall biogenesis and pathogen response in a WAK2-dependent fashion, indicating that WAK2 plays a major role in signaling a diverse array of cellular events in response to pectin in the extracellular matrix.
RESULTS AND DISCUSSION
Pectin binding
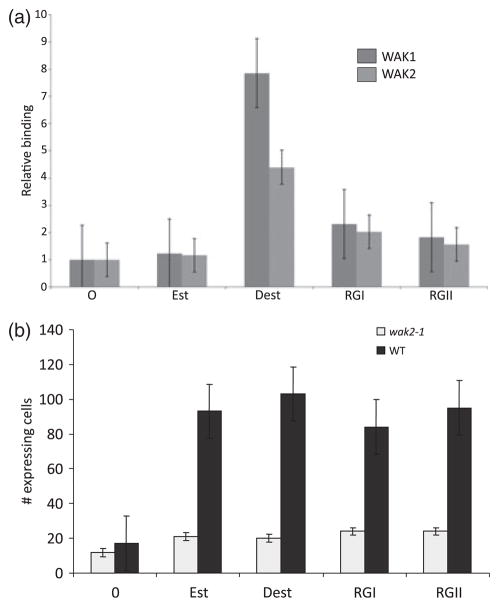
The association of pectin with the extracellular domain of WAK in Arabidopsis is in part covalent, as it survives strong denaturing and ionic conditions (He et al., 1996; Kohorn, 2000). The purified extracellular domain of WAK1 can bind to pure pectin fragments in vitro, and this appears to be dependent upon interaction between charged residues in WAK1 and charged oxygen groups in pectin (Decreux and Messiaen, 2005; Decreux et al., 2006). Previous work had shown that the extracellular domain of WAK1 binds to esterified pectin (homogalacturonan), but more binds to de-esterified pectin, where there is a larger charge on the homo-galacturonan backbone (Decreux and Messiaen, 2005; Decreux et al., 2006). The in vitro binding of pectin to the extracellular domain of WAK1 and WAK2 expressed and purified from Saccharomyces cereviseae was tested. Figure 1(a) shows that the WAK2 extracellular domain also has a similar preference, but less binds to pectin than to WAK1. The pectin preparation used in the previous studies and here is a mixed-length polymer purified from citrus, and is 89% esterified. Neither WAK1 nor WAK2 bound to the branched pectin varieties RGI or RGII. In this experiment these molecules were esterified on the galacturonan and rhamnogalacturonan backbones, and have few charged groups on their varied oligosaccharide side chains (Bacic, 2006). The results are consistent with both WAK1 and WAK2 binding the de-esterified, charged galacturonic acid backbone common to pectins. Galacturonic acid did not bind the WAK extracellular domain (Decreux and Messiaen, 2005) (data not shown), indicating that a polymer backbone is required for binding.
Figure 1.
(a) Binding of the purified wall-assocaited kinase 1 (WAK1) and WAK2 extracellular domain to pectin. The indicated pectin was adhered to a microtiter well, followed by incubation with WAK-haemaglutanin (HA) protein, which was subsequently detected with anti-HA antiserum. The y-axis indicates -fold binding over background binding of β-galactosidase-HA. (b) Pectin, WAK2-dependent, invertase expression. A 100-μg protein equivalent of protein from wild-type or wak2-1 protoplasts was transformed with 30 μg of both Invp-RFP and 35S-GFP plasmids, and treated with 4 μg ml−1 of the indicated pectin. The y-axis indicates the total number of GFP-expressing protoplasts that also expressed RFP, as detected by fluorescent microscopy. The type of pectin added is indicated below each bar: 0, none; Est, esterified; Dest, de-esterified.
Pectin-induced activity
As pectins can bind to WAK, it was of interest to ask if pectins can induce WAK activity. Arabidopsis mutants with a loss of WAK2 function have a corresponding reduction in invertase mRNA levels (Kohorn et al., 2006a). This implies that WAK2 is required for the full activation of invertase gene (Inv1) expression. To serve as an indicator of invertase expression and thus indirectly indicate WAK activity, the Inv1 gene regulatory region 1.5-kb 5′ to the start of translation was fused to the red fluorescent protein-coding region (Inv-RFP) (Jian and Rogers, 1998), and transformed into Arabidopsis protoplasts. All cells were also co-transformed with a plasmid expressing GFP from the constitutive Cauliflower mosaic virus 35S promoter (Mindrinos et al., 1994). After 12 h, of all cells expressing GFP, few cells also expressed RFP. If cells were also incubated with pectin for the 12 h following transformation with Inv-RFP, then most of GFP-expressing cells also expressed the Inv-RFP gene (Figure 1b). Both esterified and de-esterified pectin activated Inv-GFP. However, this activation was not seen in protoplasts isolated from a wak2-1 mutant. After 24 h of incubation, all samples, including nontreated protoplasts, show equivalent activation of Inv-RFP (data not shown), and this may be because of sufficient production of new cell wall material, including pectin deposition (Kohorn et al., 2006b). Thus, pectin induces the Inv1 promoter in a WAK2-dependent fashion. Smaller oligogalacturonides of degree of polymerization 9–15 also activate Inv-RFP, yet galacturonic acid did not active Inv-RFP in the protoplasts (data not shown), indicating that a polymer is required, and this is consistent with the results reported for the in vitro binding (Decreux and Messiaen, 2005; Decreux et al., 2006). It was not expected that esterified pectin would activate gene expression through WAK2, as these pectins have low binding in vitro (Figure 1a), but this may be because of the presence of esterases in protoplasts that de-esterify pectin at the cell surface (Bacic, 2006; Pelloux et al., 2007). Indeed, using esterified pectin and the pH-sensitive dye methyl red (Hewezi et al., 2008), esterase activity is detected in both wild-type and wak2-1 protoplasts (data not shown), and has been reported in leaf tissues (Bacic, 2006), the source of the protoplasts used here. It does remain possible, however, that pectins do not require de-esterification to bind or activate WAK1 or WAK2 in vivo, and that the in vitro binding of purified proteins and pectins is missing critical co-factors, the functions of which are substituted by charged groups. The data heredosupport a role for pectin activation of gene expression that requires WAK2, and further studies on the native state of WAK-pectin binding will be required to resolve the issue.
MAPKs
The events that are induced downstream of plant receptor kinases are less well characterized than that of metazoan signal transduction pathways. MAPKs are key components of many pathways, and form a diversified cascade in hormone, stress and developmentally induced pathways in angiosperms (Colcombet and Hirt, 2008). MAPK3 and 6 in Arabidospis are known to mediate pathogen and stress responses, and small pectin fragments have been shown to activate MAPKs (Moscatiello et al., 2006; Wang et al., 2007; Colcombet and Hirt, 2008). We therefore determined if the larger pectin fragments of mixed lengths that induce invertase gene expression in a WAK2-dependent fashion, also induced MAPKs. Protoplasts from wild-type or wak2-1 mutants were treated with de-esterified pectin for 1 min, and then the cells were frozen, lysed and equal protein quantities of all extracts were analyzed in an in-gel kinase assay for MAPK activity (Suzuki and Shinshi, 1995). The results of one experiment are shown in Figure 2(a), and the quantification of three experiments is shown in Figure 2(b). In wild-type cells pectin induces MAPK3 activity by 17% relative to untreated cells (P < 0.01). The MAPK3 activity is not affected by the wak2-1 allele, as untreated wild-type and mutant protoplasts have similar levels of MAPK3 activity (P > 0.3), but the induction by pectin is abated in wak2-1 cells (wak2-1 versus wak2-1 pectin MAPK3 activities are not different, P > 0.3). The levels of MAPK6 are higher than those of MAPK3 in all samples, but are quite variable between treatments and experiments, and no consistent pattern is apparent. MAPK4, detected as the lowest molecular weight band, has very low activity in both wild-type and mutant untreated cells, and pectin treatment appears to reduce the activity to a level that is not detectable, even upon longer exposures. However, the levels of MAPK4 are too low to quantify in some experiments. The activity changes can not be attributed to changes in MAPK gene expression (see below and Figure S1).
Figure 2.
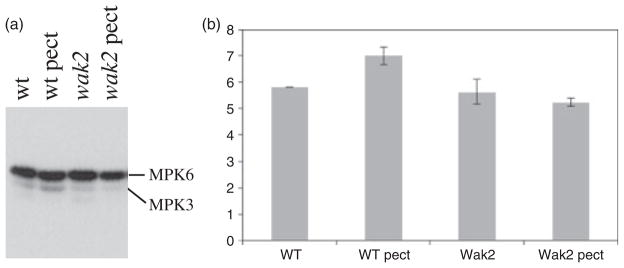
Pectin, wall-assocaited kinase (WAK)-induced, mitogen-activated protein kinase 3 (MAPK3) activity.
(a) Protoplasts from wild-type (+) or wak2-1 leaves were treated with 4 μg ml−1 de-esterified pectin, and then after 1 min were frozen in liquid nitrogen. Samples were run in a gel containing myelin basic protein, were incubated with [γ-32P]ATP, and were subjected to autoradiography as described by Wang et al. (2007). MAPK6 and MAPK3 were identified by their relative migration in the gel.
(b) Quantification of MAPK3 activities. The y-axis shows relative levels of activity.
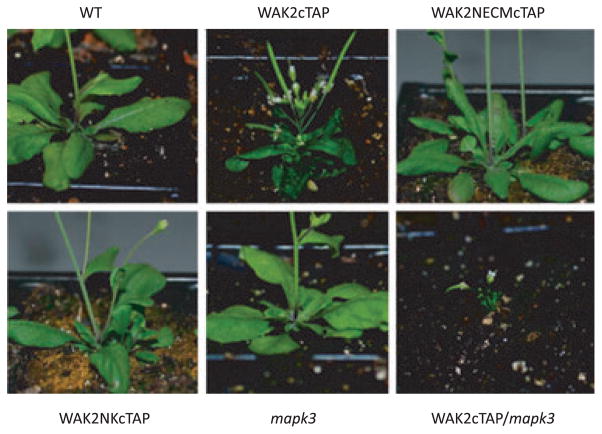
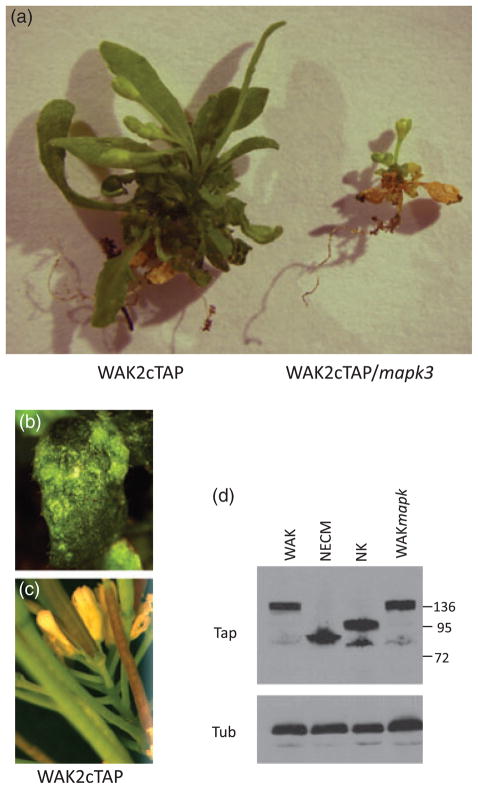
The results suggest that pectin activates a pathway that involves both WAK2 and MAPK3, and this predicts that double mutants of WAK2 and MAPK3 should provide informative phenotypes. The null wak2-1 allele provides a very weak phenotype in the whole plant (Kohorn et al., 2006a), and so we isolated a new stronger dominant negative allele of WAK2 that causes a 78% reduction in leaf area relative to the wild type (P < 0.01, Figure 3), curling of leaves (Figures 3 and 4a), necrosis of some of the leaves in the absence of pathogen (Figure 4b), and a dramatic shortening of floral stems such that flowers and siliques are clustered (Figures 3 and 4c). The allele was created by the fusion of the tandem affinity tag (TAP) onto the carboxyl terminus of WAK2, and expression using the constitutive 35S CaMV promoter in wild-type plants. Expression of a WAK2cTAP fusion that lacks the extracellular domain (amino acids 30–345, WAK2NECMcTAP), or that lacks the kinase domain (amino acids 365–731, WAK2NKcTAP), had no abnormal phenotype (Figure 3) and leaf areas similar to the wild type (P < 0.01), indicating that both of these domains are required for the dominant negative phenotype. Multiple plants for each transformation provided similar results, and the plants shown all express equivalent levels of the fusion proteins (Figure 4d).
Figure 3.
WAKcTAP/mapk3 double mutants. Arabidopsis expressing a WAK2cTAP fusion results in curled and stunted leaves, and altered floral stems (WAK2cTAP). A WAK2cTAP fusion lacking the WAK2 ECM (WAK2NEC-McTAP) or the kinase domain (WAK2NKcTAP) of WAK2 do not produce this phenotype. The WAK2cTAP phenotype is far stronger when in a mapk3 mutant background (WAK2cTAP/mapk3).
Figure 4.
(a) WAK2cTAP and WAK2cTAP/mapk3 plants.
(b) An example of leaf necrosis and curling in WAK2cTAP leaves.
(c) Floral stem shortening in WAK2cTAP plants.
(d) Western blot with anti-tandem affinity tag (anti-TAP; Kohorn et al., 2006b) to identify WAKcTAP fusions in total leaf extracts from the indicated plant. Western blot with tubulin antiserum (Tub) indicates the equal loading of samples. WAK, WAK2CTAP; NECM, WAK2NECMcTAP; NK, WAK2NKcTAP; WAKmapk3, WAK2cTAP/mapk3. Numbers on the right indicate molecular weights × 10−3.
The simplest interpretation of this phenotype is that WAK2cTAP is associating with some native signaling complex, which perhaps includes another WAK, and is inhibiting the activation of downstream events. We obtained a plant homozygous for a null MAPK3 allele that has no visible phenotype, because of a partial redundancy provided by MAPK6 (Bush and Krysan, 2007; Wang et al., 2007; Colcombet and Hirt, 2008), and transformed this with the WAK2c- TAP allele. We screened for plants that expressed equivalent levels of WAK2cTAP as the parental line (Figure 4d), and observed a more severe phenotype in the double mutant. The double mutant is shown in Figures 3 and 4(a): plants were far more stunted in the growth of all tissues and total leaf area is reduced to 4% (P < 0.01) of the wild type. If WAK2cTAP is partially inhibiting MAPK6 activity, then removal of the redundant MAPK3 should increase the severity of the effect of MAPK6 inhibition, which it indeed appears to do. As the MAPK assays (Figure 2) reveal that WAK2 also affects MAPK3 activity, the results support a model where WAK2 is upstream of both MAPK3 and MAPK6, but it is not clear which is stimulated more, or if additional kinases and related proteins are involved. Dual activation of these kinases appears to be common in many pathways (Colcombet and Hirt, 2008).
WAK2, pectin and gene expression
The initial screen for enzymes activated by WAK was limited to those involved in sugar metabolism, as the wak2-1 phenotype suggested an osmotic imbalance (Kohorn et al., 2006a). However, it remains likely that WAKs indirectly regulate numerous other genes, and hence regulate multiple physiological events. To gain an estimate of the breadth or the extent to which WAK is regulating cellular events, we performed a gene expression analysis using Arabidopsis Affymetrix expression arrays with RNA from wild-type or wak2-1 protoplasts treated or not treated with pectin.
Protoplasts were prepared from wild-type and wak2-1 leaves, and were incubated with or without de-esterified pectin for 12 h. Total RNA was then extracted and used to probe Affymetrix gene chips. The differences in gene expression between wild-type and wak2-1 samples in the presence and absence of pectin were analyzed with Gene-Spring GX 9.0.
In wild-type protoplasts pectin treatment resulted in a significant change of expression (more than twofold) of more than 200 genes (Figures S1 and 5). Of these genes, 50 were upregulated, and the rest were downregulated. The majority of the upregulated genes play a role in cell-wall synthesis, including oligogalacturonase-inhibiting protein, glycosyl hydrolase family protein, pectin esterase, a cellulose synthase family protein and a leucine-rich transmembrane kinase. Genes involved in plant defense, such as senescence-associated proteins, plant defensin, myrosinase-associated proteins and calreticulin 2 were also induced (Figure S1). The downregulated genes code for an array of proteins of multiple functions.
wak2-1 protoplasts displayed a distinct response to pectin, compared with the wild type. Only one of the 227 pectin-response genes identified in wild type, At4g30670 (unknown expressed protein), showed altered expression (1.96-fold upregulation) in wak2-1 protoplasts treated with pectin. Indeed, with this one exception, no genes were differentially expressed in pectin-incubated wak2-1 protoplasts, compared with untreated cells.
To further analyze the gene expression pattern of wak2-1 compared with the wild type, the expression of the 227 pectin-response genes in the untreated wak2-1 protoplasts was compared with that in the untreated wild type (Figure S1). Of the 50 pectin-induced genes in the wild type, 13 were significantly suppressed in the wak2-1 background (Figure S1a), whereas the other 37 showed expression levels similar to those in the wild type (Figure S1b). Of the downregulated genes, 20 demonstrated reduced expression in wak2-1 cells (Figure S1c), 24 were activated compared with the wild type (Figure S1d) and the rest demonstrated levels similar to the wild type (Figure S1e). These expression patterns enabled the distinction between genes regulated by WAK2 in the normal background (no pectin treatment) and genes that are normally independent of WAK2, but are WAK2-dependent in the pectin response.
WAK2 expression in wak2-1 protoplasts was 13.5-fold reduced compared with the wild type, and showed the most dramatic decrease in expression. As wak2-1 is a null mutation the result suggests that a reduction in expression levels higher than 13-fold effectively means that the gene is not transcribed. The presence of any WAK2 signal could result from the background of hybridization, or from cross-hybridization with other WAKs, because of the high sequence conservation among the members of the family. Both WAK1 and WAK2 were slightly upregulated in pectin-treated wild-type cells: 1.54- and 1.87-fold, respectively. The change in expression does not meet our criteria of significance (twofold change), but demonstrates a trend of pectin-induced activation of the two genes. This slight activation was not observed in wak2-1 protoplasts.
Comparison of the total gene expression between untreated wild-type and untreated wak2-1 protoplasts was also performed in order to identify how the lack of WAK2 affected gene expression. More than 300 genes were differentially expressed between untreated wak2-1 and wild-type protoplasts, out of which approximately half were upregulated compared with the wild type (Figure S2), and half were downregulated compared with the wild type (Figure S3). Many of the genes activated in wak2-1 compared with the wild type are heat-shock proteins (Figure 2). A total of 27 heat-shock proteins showed a higher than twofold increase of expression, and many of them were among the most upregulated genes in the mutant (3–5-fold higher than in the wild type). In the absence of WAK2 a number of cell wall-related genes were upregulated, including WAK3 (Figure S2). A high number of wall structure-related genes and defense response genes were downregulated in the wak2-1 mutant compared with the wild type (Figure S3).
Thus, pectin treatment of protoplasts altered the expression of more than 200 genes in the wild type, and this response was dramatically reduced in wak2-1 protoplasts. The result identifies WAK2 as an essential part of the pectin signaling pathway, as the absence of WAK2 in protoplasts is sufficient for negating or significantly reducing the transcriptional response to pectin. The WAK2-dependent pectin-response genes, both upregulated and downregulated, fall within four main categories: defense related, cell wall structure related, protein phosphorylation related and transcription factors. This pattern supports the previously established role of WAK2 in wall modification and expansion (Wagner and Kohorn, 2001; Kohorn et al., 2006a), and the upregulation of the protein during wounding and pathogen attack (He et al., 1998).
The observation that WAK2 is required for pectin response is surprising given that WAKs seem to have a somewhat redundant function in Arabidopsis. wak2-1 plants grown on soil do not display a visible phenotype different from the wild type, even though they have a short-root phenotype on minimal agar. As these mutants grow and develop normally on soil, pectin signaling and response must occur at comparatively normal levels. However, in protoplasts (plant cells with a newly growing wall) the redundancy of the pectin signaling seems to be reduced. It is possible that the digestion of the cell wall removes other wall-embedded pectin receptors, leaving predominately WAK2, which is known to be still present as a full-length receptor (He et al., 1996). Alternatively, non-WAK but important pathways may not be expressed in protoplasts. Protoplasts represent a subset of leaf cells, the mesophyll cells, and WAK2 is expressed in the mesophyll, whereas WAK1 is preferentially expressed in the leaf veins (Wagner and Kohorn, 2001). Therefore WAKs may not be redundant in certain tissues.
Other unexpected aspects of the data were both the small number of pectin-response genes that were identified in the wild type, and the moderate changes of expression that were observed. Indeed, none of the upregulated genes showed an activation of higher than threefold, and the most downregulated gene displayed a sevenfold reduction of transcription. In contrast, Moscatiello et al. (2006) identified more than 1200 genes in which expression was altered significantly after oligogalacturonide treatment. The origin of this discrepancy may lie both in the use of cultured Arabidopsis cells instead of protoplasts and in the treatment with purified, short pectin fragments of specific length. Furthermore, the analysis of plant material from whole leaves involves many different cell types, whereas protoplasts present a very specific subset of mesophyll cells, and their response provides a more narrow and specific reaction to pectin than that of the whole leaf.
We have shown here that WAKs, and in particular WAK2, mediate the plant’s response to large pectin fragments. Pectin appears to induce MAPK3, a kinase that has already been implicated in pathways regulating developmental, stress and pathogen responses. Genetic analysis indicates that WAK2 activates an MAPK-related event. Pectin also induces and represses the expression of numerous genes involved in cell wall integrity and environmental response, and this too is WAK2 dependent. The relationship between the large pectin fragments used in this study and the native pectin structures in the cell wall remains to be determined, and future studies will need to elucidate how pectin is arranged in the complex architecture that includes cellulose and cell wall proteins, which may also bind to WAK (Anderson et al., 2001; Park et al., 2001). As pectin comprises the more flexible portion of the wall, and is dynamic and modified during growth and environmental responses (Bacic, 2006), WAK2 clearly plays a fundamental role in relaying pectin-based signals from the cell wall. This study explored pectin activation in protoplasts that probably represent a developing cell as they re-synthesize the cell wall. Notably, most pectin-activated genes were WAK2 dependent, indicating that WAK2 plays a significant role in this cell type. Whole-plant phenotypes of single wak mutants are, however, subtle, and thus additional receptors are likely to be involved in pectin sensing.
EXPERIMENTAL PROCEDURES
Arabidopsis thaliana col. plants were grown in soil in a growth chamber, with a 16-h light/8-h dark photoperiod.
Pectin binding and protein expression
WAK expression
The extracellular domain of WAK1 and WAK2 (amino acids 30–345) were cloned by PCR into the yeast expression vector pYES (Invitrogen, http://www.invitrogen.com), expressed with a C-terminal HA tag in S. cereviseae and purified exactly as described by Decreux and Messiaen (2005). Plate binding assays were as described by Decreux and Messiaen (2005), where microtiter wells were coated first with pectin, then with purified WAK protein and then with mouse anti-HA antiserum (Sigma-Aldrich, http://www.sigmaaldrich.com), followed by anti-mouse conjugated peroxidase (Sigma-Aldrich). Levels of binding were measured colourometircally with a plate reader, and are reported as those relative to background binding to pectin of purified β-galactosidase-HA purified from yeast (Decreux and Messiaen, 2005).
Pectin was de-esterified by the addition of NaOH to 50 mM for 60 min on ice, followed by neutralization with 1 N HCl. Pectin was purchased from Sigma-Aldrich (Sigma P9561): a homogalacturonan purified from citrus as 89% esterified, and of mixed length. When specified, HPLC-purified oligogalacturonides, RGI and RGII, were provided by Malcolm O’Niel (University of Georgia).
Protoplasts
Protoplast preparation was as described by Kohorn et al. (2006b; http://genetics.mgh.harvard.edu/sheenweb/). In short, leaves from 2–3-week-old plants were sliced and placed in activated enzyme solution [0.4 M mannitol, 20 mM KCl, 20 mM 2-(N-morpholine)-ethanesulphonic acid (MES), pH 5.7, 9 mM CaCl2, 1 mg ml−1 BSA, 10 mg ml−1 cellulase, 2 mg ml−1 macerozyme]. Vacuum was applied for 5 min, followed by incubation for 3–4 h under constant shaking at 40 rpm. The supernatant containing the protoplasts was filtered through 200-μm nylon mesh, centrifuged at 100 g for 2 min and the enzyme solution was replaced with W5 (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, pH 5.7). The final concentration of protoplasts was equilibrated by diluting the samples to a standard chlorophyll concentration of 0.06 at OD660. Pectin was added to a final concentration of 4 μg ml−1 and samples were incubated overnight in Eppendorf tubes under constant light.
For protoplast transformation, protoplasts were resuspended in MMG buffer (0.4 M mannitol, 15 mM MgCl2, 4 mM MES, pH 5.7). A 100-μl portion of 5 × 105 ml−1 protoplasts was transformed with 30 μg of the appropriate plasmids by adding DNA and 100 μl of PEG solution (40% PEG 4000, 200 mM mannitol, 100 mM CaCal2) for 30 min at room temperature (25°C). Cells were diluted with 440 μl of W5 buffer, centrifuged at 100 g for 3 min, and resuspended in W5, and pectin if appropriate. The Inv1 promoter RFP was amplified by PCR from Arabidopsis genomic DNA using the forward primer 5′-TGTGATATGATCAAGTCCTATAAAAGTTTGAAATGATGATC-3′ and the reverse primer 5′-GTGCTCGCCATGGATCCTTTCTTGTTCGTTTTG-3′, and then cloned into HindIII and BamHI sites 5′ to the RFP coding region in JR900 (Jian and Rogers, 1998), which contains an in-frame RFP coding region. The 35S GFP was expressed from pSM. Cells were imaged with a BX51 Olympus fluorescent microscope under a 40× objective (http://www.olympus-global.com), and images were captured with a QImaging RetigaRX digital camera (http://www.qimaging.com), using Open Lab. ImageJ was used for quantification.
cTAP
35S:WAK2cTAP, WAK2NECMcTAP and WAK2NKcTAP were generated by PCR of the appropriate WAK coding regions, and were cloned into p289CTAP (Rohila et al., 2006). In short, the WAK2 gene was generated by PCR from Arabidopsis genomic DNA, thereby placing an XhoI site at the 5′ end of the signal sequence coding region, a SalI at the end of the signal sequence region, a SalI at the 5′ end of the transmembrane (TM) coding region, a SpeI at the 3′ end of the transmembrane coding region and a SpeI at the 3′ end of the kinase coding region. The various gene constructs were then combined using the XhoI-SalI-SpeI ligated pieces inserted into pSK+ and then p289cTAP. Primers for each domain: signal sequence forward, 5′-AAGGCAATTCTCGAGTTCATATTTTGAGCGTG-3′; reverse, 5′-GTCAAGGGGGTCGACCAACCTCGCAAG-3′ (compliment was made); TM forward, 5′-CATCGGCTTCGTCGACGTTATCATGC-3′, reverse 5′-GAGGCATGTTGATAACTAGTCAGCGAGTCTC-3′ (compliment was made); kinase reverse 5′-GAAGCTGGCCGTACTAGTGATATTAATGCTC-3′ (compliment was made). Plant transformation was via Agrobacterium tumefaciens into Arabidopsis by floral dipping (Clough and Bent, 1998). The C-terminal TAP tag was detected using a 1:2500 dilution of peroxidase-anti-peroxidase complex (PAP) antiserum (Sigma-Aldrich) on western blots as described by Kohorn et al. (2006b). MAPK in-gel assays were performed exactly as described by Wang et al. (2007), courtesy of Shuqun Zhang, University of Missouri. In short, equal quantities of protein extract were run in a denaturing gel containing myelin basic protein. The protein in the gel was renatured and incubated with [γ-32P]ATP, washed and then subjected to autoradiography. ImageJ was used for quantification.
Leaf size was measured by capturing an image using a QImaging RetigaRX digital camera and a Leica MZ3 dissecting microscope (http://www.leica.com). Open Lab software was used to digitize and measure the images, and ImageJ was used for quantification.
Total RNA extraction
Total RNA extraction was performed similarly on protoplasts and leaf tissue. Protoplasts were centrifuged, the W5 media was removed and they were immediately frozen in liquid nitrogen. Leaves (one or two, approximately 100 mg) were frozen in liquid nitrogen and homogenized with a motorized pestle. RNA was extracted using the RNeasy Plant Mini Kit (QIAGEN, http://www.qiagen.com).
Microarray preparation and hybridization
Frozen RNA samples were submitted to the Duke University Microarray Facility (Duke Institute for Genome Sciences and Policy, Durham, NC, USA). RNA quality assessments were performed on a Bioanalyzer, and chip hybridization was performed according protocols available at http://www.genome.duke.edu/cores/microarray/services/affymetrix/.
Differential gene expression analysis
Affymetrix gene chip files were analyzed with GeneSpring GX 9 using the Guided Workflow. The robust multiple chip average (RMA) algorithm and baseline transformation to the median of all samples were performed to obtain normalized data. To control for the quality of the samples and for any possible degradation, the 3′/5′ ratios for internal controls were calculated. The data was accepted if the ratios did not exceed 3. The quality of hybridization was controlled for with the hybridization controls, bioB, bioC, bioD and cre. These probes hybridize to artificial, non-Arabidopsis RNA added to each sample. Each is represented on the chip by four repeats of different concentrations. Data was used if the bioB signal was present at least 50% of the time, and if bioC, bioD and cre were present all the time, and appeared in increasing concentrations. All chips satisfied these quality standards. To minimize error, the lowest 20 percentile of the row intensity values was removed from further analysis. This excluded unreliable fluorescence levels from the analysis, ensuring that only real readings were included in further data interpretation.
Fold change analysis was performed on the normalized intensity values. The cut-off for significance that we used in our analysis was 2.0-fold. Gene onthology (GO) analysis was performed to identify genes of similar function or cellular localization within a large subset of genes. A P value identifies the significance of the GO term within the subset compared with the whole data set. The cut-off for the GO analysis that we used was 0.01, unless stated otherwise.
Supplementary Material
Figure S1. Genes differentially expressed in the wild type after pectin treatment, but not responsive to pectin in wak2-1.
Figure S2. Genes upregulated in untreated wak2-1 protoplasts compared with the untreated wild type.
Figure S3. Genes downregulated in untreated wak2-1 protoplasts compared with the untreated wild type.
Figure 5.
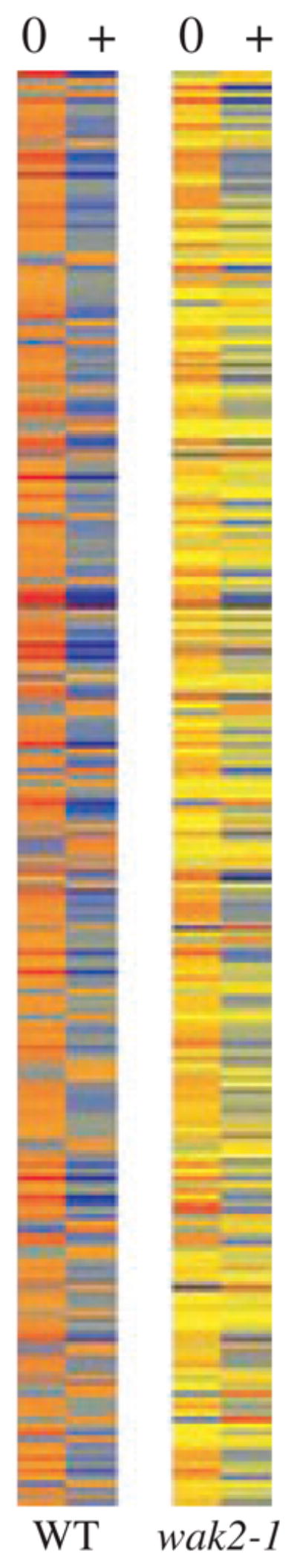
Heat map of 251 genes in which expression is activated by pectin and WAK2. Intensity of colors indicates the degree of induction: 0, no pectin, +, pectin; yellow, no change; red, relatively high; blue, relatively low.
Acknowledgments
We would like to thank Patrick Krysan (University of Wisconsin) for supplying the MAPK3 mutant plant, and Shuqun Zhang (University of Missouri) for running the protoplast extracts in his in-gel assay gels. RGI and RGII were a generous gift of Malcolm O’Niel (University of Georgia). This work is supported by an NSF grant MCB 0717983 to BDK. Students were supported in part by grants from the Howard Hughes Medical Institute, and NIH INBRI to Bowdoin College.
References
- Anderson CM, Wagner TA, Perret M, He ZH, He D, Kohorn BD. WAKs: cell wall-associated kinases linking the cytoplasm to the extracellular matrix. Plant Mol Biol. 2001;47:197–206. [PubMed] [Google Scholar]
- Bacic A. Breaking an impasse in pectin biosynthesis. Proc Natl Acd Sci. 2006;15:5639–5640. doi: 10.1073/pnas.0601297103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Hepler P. Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell. 2005;17:3219–3226. doi: 10.1105/tpc.105.037473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush ZS, Krysan PJ. Mutational evidence that the Arabidopsis MAP kinase MPK6 is involved in anther, inflorescence, and embryo development. J Exp Bot. 2007;58:2181–2191. doi: 10.1093/jxb/erm092. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium- mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Colcombet J, Hirt H. Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J. 2008;413:217–226. doi: 10.1042/BJ20080625. [DOI] [PubMed] [Google Scholar]
- Decreux A, Messiaen J. Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. J Plant Cell Physiol. 2005;46:268–278. doi: 10.1093/pcp/pci026. [DOI] [PubMed] [Google Scholar]
- Decreux A, Thomas A, Spies B, Brasseur R, Van Cutsem P, Messiaen J. In vitro characterization of the homogalacturonan-binding domain of the wall-associated kinase WAK1 using site-directed mutagenesis. Phytochemistry. 2006;67:1068–1079. doi: 10.1016/j.phytochem.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Diener AC, Ausubel FM. RESISTANCE TO FUSARIUM OXYSPORUM 1, a Dominant Arabidopsis Resistance Gene, is not Race Specific. Genetics. 2005;171:305–321. doi: 10.1534/genetics.105.042218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He ZH, Fujiki M, Kohorn BD. A cell wall-associated, receptor-like protein kinase. J Biol Chem. 1996;271:19789–19793. doi: 10.1074/jbc.271.33.19789. [DOI] [PubMed] [Google Scholar]
- He ZH, He D, Kohorn BD. Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 1998;14:55–63. doi: 10.1046/j.1365-313x.1998.00092.x. [DOI] [PubMed] [Google Scholar]
- He ZH, Cheeseman I, He D, Kohorn BD. A cluster of five cell wall-associated receptor kinase genes, Wak1-5, are expressed in specific organs of Arabidopsis. Plant Mol Biol. 1999;39:1189–1196. doi: 10.1023/a:1006197318246. [DOI] [PubMed] [Google Scholar]
- Hewezi T, Howe P, Maier TR, Hussey RS, Mitchum MG, Davis EL, Baum TJ. Cellulose binding protein from the parasitic nematode Heterodera schachtii interacts with Arabidopsis pectin methylesterase: cooperative cell wall modification during parasitism. Plant Cell. 2008;20:3080–3093. doi: 10.1105/tpc.108.063065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian L, Rogers JC. Integral membrane sorting to vacuoles in plant cells: evidence for two pathways. J Cell Bio. 1998;143:1183–1199. doi: 10.1083/jcb.143.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD. Plasma membrane-cell wall contacts. Plant Physiol. 2000;124:31–38. doi: 10.1104/pp.124.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD, Kobayashi M, Johansen S, Riese J, Huang L-F, Koch K, Dotson A, Byers N. An Arabidopsis cell wall associated kinase required for invertase activity and cell growth. Plant J. 2006a;46:307–316. doi: 10.1111/j.1365-313X.2006.02695.x. [DOI] [PubMed] [Google Scholar]
- Kohorn BD, Kobayashi M, Johansen S, Fischer A, Byers N. Wall associated kinase 1 is crosslinked in endomembranes and transport to the cell surface requires correct cell wall synthesis. J Cell Sci. 2006b;119:2282–2290. doi: 10.1242/jcs.02968. [DOI] [PubMed] [Google Scholar]
- Lally D, Ingmire P, Tong HY, He ZH. Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell. 2001;13:1317–1331. doi: 10.1105/tpc.13.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lespinet O, Wolf YI, Koonin EV, Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12:1048–1059. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu GL, Ausubel FM. The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell. 1994;78:1089–1099. doi: 10.1016/0092-8674(94)90282-8. [DOI] [PubMed] [Google Scholar]
- Moscatiello R, Mariani P, Sanders D, Maathuis FJ. Transcriptional analysis of calcium-dependent and calcium-independent signalling pathways induced by oligogalacturonides. J Exp Bot. 2006;57:2847–2865. doi: 10.1093/jxb/erl043. [DOI] [PubMed] [Google Scholar]
- Park AR, Cho SK, Yun UJ, Jin MY, Lee SH, Sachetto-Martins G, Park OK. Interaction of the Arabidopsis receptor protein kinase Wak1 with a glycine-rich protein, AtGRP-3. J Biol Chem. 2001;276:26688–26693. doi: 10.1074/jbc.M101283200. [DOI] [PubMed] [Google Scholar]
- Pelloux J, Rustérucci JC, Mellerowicz EJ. New insights into pectin methylesterase structure and function. Trends Plant Sci. 2007;12:267–277. doi: 10.1016/j.tplants.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Richter TE, Ronald PC. The evolution of disease resistance genes. Plant Mol Biol. 2000;42:195–204. [PubMed] [Google Scholar]
- Ridley BL, O’Neill MA, Mohnen D. Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry. 2001;57:929–967. doi: 10.1016/s0031-9422(01)00113-3. [DOI] [PubMed] [Google Scholar]
- Rohila JS, Chen M, Chen S, et al. Protein-protein interactions of tandem affinity purification-tagged protein kinases in rice. Plant J. 2006;46:1–13. doi: 10.1111/j.1365-313X.2006.02671.x. [DOI] [PubMed] [Google Scholar]
- Sivaguru M, Ezaki B, He Z-H, Tong H, Osawa H, Baluska F, Volkmann D, Matsumoto H. Aluminum induced gene expression and protein localization of cell wall-associated receptor kinase in Arabidopsis thaliana. Plant Physiol. 2003;132:2256–2266. doi: 10.1104/pp.103.022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Shinshi H. Transient activation and tyrosine phosphorylation of a protein kinase in tobacco cells treated with a fungal elicitor. Plant Cell. 1995;7:639–647. doi: 10.1105/tpc.7.5.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verica JA, He ZH. The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol. 2002;129:455–459. doi: 10.1104/pp.011028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verica JA, Chae L, Tong H, Ingmire P, He ZH. Tissue-specific and developmentally regulated expression of a cluster of tandemly arrayed cell wall associated kinase-like kinase genes in Arabidopsis. Plant Physiol. 2003;133:1732–1746. doi: 10.1104/pp.103.028530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner TA, Kohorn BD. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell. 2001;13:303–318. doi: 10.1105/tpc.13.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell. 2007;19:63–73. doi: 10.1105/tpc.106.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WG, McCartney L, Mackie W, Knox JP. Pectin: cell biology and prospects for functional analysis. Plant Mol Biol. 2001;47:9–27. [PubMed] [Google Scholar]
- Yang EJ, Oh YA, Lee ES, Park AR, Cho SK, Yoo YJ, Park O. Oxygen-evolving enhancer protein 2 is phosphorylated by glycine-rich protein 3/wall-associated kinase 1 in Arabidopsis. Biochem Biophys Res Commun. 2003;305:862–868. doi: 10.1016/s0006-291x(03)00851-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Genes differentially expressed in the wild type after pectin treatment, but not responsive to pectin in wak2-1.
Figure S2. Genes upregulated in untreated wak2-1 protoplasts compared with the untreated wild type.
Figure S3. Genes downregulated in untreated wak2-1 protoplasts compared with the untreated wild type.