Abstract
Purpose.
CaBP4 is a neuronal Ca2+-binding protein that is expressed in the retina and in the cochlea, and is essential for normal photoreceptor synaptic function. CaBP4 is phosphorylated by protein kinase C zeta (PKCζ) in the retina at serine 37, which affects its interaction with and modulation of voltage-gated Cav1 Ca2+ channels. In this study, we investigated the potential role and functional significance of protein phosphatase 2A (PP2A) in CaBP4 dephosphorylation.
Methods.
The effect of protein phosphatase inhibitors, light, and overexpression of PP2A subunits on CaBP4 dephosphorylation was measured in in vitro assays. Pull-down experiments using retinal or transfected HEK293 cell lysates were used to investigate the association between CaBP4 and PP2A subunits. Electrophysiologic recordings of cotransfected HEK293 cells were performed to analyze the effect of CaBP4 dephosphorylation in modulating Cav1.3 currents.
Results.
PP2A inhibitors, okadaic acid (OA), and fostriecin, but not PP1 selective inhibitors, NIPP-1, and inhibitor 2, block CaBP4 dephosphorylation in retinal lysates. Increased phosphatase activity in light-dependent conditions reverses phosphorylation of CaBP4 by PKCζ. In HEK293 cells, overexpression of PP2A enhances the rate of dephosphorylation of CaBP4. In addition, inhibition of protein phosphatase activity by OA increases CaBP4 phosphorylation and potentiates the modulatory effect of CaBP4 on Cav1.3 Ca2+ channels in HEK293T cells.
Conclusions.
This study provides evidence that CaBP4 is dephosphorylated by PP2A in the retina. Our findings reveal a novel role for protein phosphatases in regulating CaBP4 function in the retina, which may fine tune presynaptic Ca2+ signals at the photoreceptor synapse.
In the retina, protein phosphatase 2A (PP2A) reverses phosphorylation of CaBP4 by protein kinase C zeta in light conditions. Dephosphorylation of CaBP4 by PP2A suppresses its modulatory effects on Cav1.3 Ca2+ channels.
Introduction
CaBP4 is a member of a subfamily of calmodulin-like neuronal Ca2+-binding proteins (CaBP1-8).1–4 In addition to modulating voltage-gated Cav Ca2+ channels,5–13 CaBP family members modulate transient receptor potential (TRP) channels and inositol 1,4,5-trisphosphate (IP3) receptors,14–17 CaBP4 is localized in photoreceptor synaptic terminals and in cochlear inner hair cells.7,10,12,18 CaBP4 is essential for photoreceptor synaptic function through enhanced activation of Cav1.4 L-type voltage-gated Ca2+ channels and transmitter release.10,13 CaBP4-knockout mice (Cabp4−/−) possess morphologically and functionally deficient synapses,10,19 similar to that observed in mice lacking the pore-forming Cav1.4 subunit (α1 1.4) or the auxiliary Cav β2 subunit.20–22 Moreover, human mutations in the Cabp4 gene are associated with autosomal recessive incomplete congenital stationary night blindness and cone–rod synaptic disorder.23–26
Reversible phosphorylation of proteins is an essential mechanism regulating many cellular functions. Multiple photoreceptor proteins are regulated by light-dependent phosphorylation.27–34 Phosphorylation of Ca2+-binding proteins, including calmodulin, can affect their Ca2+-binding capability and interaction with target proteins.15,35–38 Similar to calmodulin, an analogous residue (Serine 120) in CaBP1 is phosphorylated by casein kinase 2, which weakens the effect of CaBP1 in inhibiting Ca2+ release by the IP3 receptor.15 We have shown that CaBP4 undergoes light-dependent phosphorylation by protein kinase C zeta (PKCζ) at serine 37 both in vitro and in the retina.39 In electrophysiologic recordings of transfected HEK293 cells, phosphorylation of CaBP4 at serine 37 enhances the effect of CaBP4 in prolonging the opening of Cav1.3 Ca2+ channels, which have been reported to be present in photoreceptors.40–43 In contrast, Ca2+-binding to CaBP4 weakens modulation of Cav1.3 channels.39 These findings suggest that phosphorylation as well as Ca2+ regulate the synaptic function of CaBP4 in the retina.
In the present study, we investigated the mechanisms underlying reversible phosphorylation of CaBP4. We identified protein phosphatase 2A (PP2A) as a phosphatase that dephosphorylates CaBP4 in the retina and in transfected cells. Although PKCζ activity dictates the level of CaBP4 phosphorylation under light-adapted conditions, PP2A activity is also higher in light-adapted than dark-adapted retinas. In HEK293 cells, we characterized PP2A subunits involved in CaBP4 dephosphorylation and show that dephosphorylation of CaBP4 by PP2A inhibits its modulation of Cav1.3 activity.
Methods
Antibodies
Commercially available antibodies were: alkaline phosphatase-conjugated anti-mouse and anti-rabbit (Promega Corp., Madison, WI); rabbit anti-PP2A sampler kit (anti-PP2A A [α/β], PP2A B [PR55, Bα], and PP2A C [α/ β] subunits [for specificity, see #9780 online at Cell signaling technology, Danvers, MA]); anti-DYKDDDDK epitope (FLAG) (Sigma-Aldrich, St. Louis, MO); anti–c-myc, anti-influenza hemagglutinin epitope (HA) (Roche Applied Science, Indianapolis, IN). The specificity of anti-tag antibodies was confirmed by Western blot analysis using untransfected and epitope-tagged PP2A transfected HEK293 cell lysates (data not shown). The development of the anti-CaBP4 antibody and demonstration of its specificity was described previously.10
Cloning, Bacterial Expression, and Purification of Active Nuclear Inhibitor of Protein Phosphatase-1 (NIPP-1) and Inhibitor2
Mouse NIPP-1 and inhibitor 2 were amplified by PCR with Pfx polymerase (Life Technologies, Carlsbad, CA) from a mouse retina cDNA library with specific primers (Table) and cloned into pentr-D-TOPO vector (Life Technologies). After sequencing, the cDNAs were transferred by recombination into the pDest17 vector using the Gateway Technology System (Life Technologies) for fusion to a His-tag and expression in bacteria. The His-fusion proteins were expressed in BL21(DE3)pLysS Escherichia coli after induction with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and purified on Ni2+-NTA or glutathione column according to the manufacturer's protocol. Protein concentration was determined by quantification of the corresponding band on Coomassie-Blue–stained SDS gels (Coomassie Blue R250; EM Science, Gibbstown, NJ) relative to known amounts of BSA. The intensity of each band on the scanned gels was quantified using ImageJ (National Institutes of Health, Bethesda, MD). The concentration of purified NIPP-1 was estimated at 3.4 μM and 7.5 μM for inhibitor 2.
Table. .
PCR Primers Used to Amplify PP2A Subunits and PP1 Inhibitors by PCR
|
mRNA / Gene Symbol* |
PCR Primer Sequence† |
mRNA Accession Number |
Primer Location on the mRNA Sequence |
| NIPP-1 / Ppp1r8 (+) | 5′- CACC ATGGCGGCAGCCGTGAACTCCG-3′ | NM_146154 | 58–79 |
| NIPP-1 / Ppp1r8 (−) | 5′-CCACCCTCTCCATAACCAAAATATC-3′ | NM_146154 | 1136–1112 |
| Inhibitor 2 / Ppp1r2 (+) | 5′- CACC ATGGCGGCCTCAACGGCCTCG-3′ | NM_025800.3 | 308–328 |
| Inhibitor 2 / Ppp1r2 (−) | 5′-CGACCGCATTGTTGAACATGTTTCT-3′ | NM_025800.3 | 954–930 |
| PP2A Aα / Ppp2r1a (+) | 5′-AAGATGGCAGCTGCCGACGGTGACG−3′ | NM_016891.3 | 50–74 |
| PP2A Aα / Ppp2r1a (−) | 5′-TCAGGCAAGAGAGAGAACAGTCAG-3′ | NM_016891.3 | 1822–1799 |
| PP2A Cα / Ppp2ca (+) | 5′-ATCATGGACGAGAAGTTGTTCACC-3 | NM_019411.4 | 223–246 |
| PP2A Cα / Ppp2ca (−) | 5′-TTACAGGAAGTAGTCTGGGGTAC-3′ | NM_019411.4 | 1155–1133 |
| PP2A Cβ / Ppp2cb (+) | 5′-GCCATGGACGACAAGGCGTTCACCAAG−3′ | NM_017374.3 | 294–320 |
| PP2A Cβ / Ppp2cb (−) | 5′-TTACAGGAAGTAGTCTGGGGTGC−3′ | NM_017374.3 | 1226–1204 |
| PP2A Bα / Ppp2r2a (+) | 5′-GACATGGCAGGAGCTGGAGGAGGGA-3′ | NM_028032.3 | 373–397 |
| PP2A Bα / Ppp2r2a (−) | 5′-CTAATTCACTTTGTCTTGAAATATATAC−3′ | NM_028032.3 | 1719–1692 |
|
Epitope-Tagged PP2A Subunits |
PCR Primer Sequence |
||
| HA-tagged PP2A Aα (+) | 5′- ACCATGTACCCATACGATGTTCCAGATTACGCTCTT ATGGCAGCTGCCGACGGTGACG-3′ | ||
| c-myc-tagged PP2A Cα (+) | 5′- ACCATGGAGCAAAAGCTAATTAGCGAGGAGGACCTT ATGGACGAGAAGTTGTTCACC-3′ | ||
| c-myc-tagged PP2A Cβ (+) | 5′- ACCATGGAGCAAAAGCTAATTAGCGAGGAGGACCTT ATGGACGACAAGGCGTTCACCAAG-3′ | ||
| FLAG-tagged PP2A Bα (+) | 5′- ACCATGGATTACAAGGATGACGACGATAAGCTT ATGGCAGGAGCTGGAGGAGG-3′ | ||
(+) and (−) indicate forward and reverse primer, respectively.
Start and Stop codons are underlined. Added sequence and tag are italicized.
Phosphorylation/Dephosphorylation Assay
Mouse retinas were obtained from C57Bl/6J mice. Animals were treated in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. All the procedures for the maintenance and use of animals were approved by the Institutional Animal Care and Use Committee of the University of Washington. When specified, dark-adapted retinas were obtained from mice kept in darkness for 4 hours and dissected under a dim red light. For all other experiments, retinas were obtained from light-adapted mice. The assay was performed as described previously but with different phosphatase inhibitors.39 Briefly, retinas were dissected and homogenized in 100 μL of 10 mM bis-Tris-propane (BTP), pH 7.4 containing 150 mM NaCl and a mixture of proteinase inhibitors (Sigma-Aldrich). The absorbance at 280 nm of a 20 × dilution of our retinal homogenates was approximately 0.5/cm. Phosphorylation assays involved native CaBP4 in retinal lysates, recombinant glutathione S-transferase (GST)-tagged CaBP4 (2 μg) or GST-tagged CaBP4-S37A (2 μg), prepared as described previously,39 and were carried out in a 20 μL reaction using 5 μL of retinal homogenate and 3 μCi of [γ-32P]ATP in 10 mM BTP, pH 7.4, 2 mM MgCl2, 100 μM ATP and 0.5 mM dithiothreitol (DTT) for 10 minutes at 30°C. After inhibition of the endogenous PKCζ with 10 μM Bisindolymaleimide I (Bis; Calbiochem, La Jolla, CA), the reactions were incubated for another 45 minutes at 30°C with or without protein phosphatase inhibitors: 1 μM calyculin A (Calbiochem), 1 μM okadaic acid (OA; Sigma-Aldrich), 1 μM cyclosporin A (Calbiochem), or 300 nM NIPP-1. Reactions were terminated with the addition of SDS-PAGE sample buffer. The proteins were separated on SDS-PAGE gels followed by transfer onto immobilon-P membrane. 32P-labeled CaBP4 was detected by autoradiography and equivalent input and transfer of CaBP4 were confirmed by red Ponceau staining of immobilon membrane (See Supplementary Material and Supplementary Fig. S1, http://www.iovs.org/content/54/2/1214/suppl/DC1) or Western blot analysis using anti-CaBP4 antibody. The contrast and brightness of the scanned images was increased equally for all lanes to improve band visibility.
For assays measuring dose-dependence of phosphatase inhibitors with respect to CaBP4 dephosphorylation and for the comparative assays involving recombinant PP1 or purified PP2A, recombinant CaBP4 was first phosphorylated with recombinant PKCζ (Calbiochem) in the presence of [γ-32P]ATP for 45 minutes at 30°C in a master mix. 32P-labeled (phosphorylated) CaBP4 was then added to 5 μL of mouse retinal homogenate along with 0.1 units of purified PP2A (Millipore, Billerica, MA) or 0.1 units of recombinant PP1 (New England Biolabs, Ipswich, MA) in a 15-μL reaction. The reactions were carried out further for 45 minutes at 30°C and analyzed on SDS-PAGE gels as described above. In experiments with NIPP-1, inhibitor 2, and fostriecin (Calbiochem), retinal lysate, rPP1, and purified PP2A were pre-incubated at 30°C for 10 minutes with the inhibitors.
For assays using HEK293 cell lysate, cells collected from a 10 cm2 plate of confluent HEK293 cells were sonicated in 10 mM BTP, pH7.2 (500 μL) containing 5 mM benzamidin. After centrifugation for 30 minutes at 10,000g, the supernatant was collected and processed as described above for mouse retinal homogenate.
Analysis of Phosphorylated CaBP4 in Dark- and Light-Adapted Mouse Retina
For those experiments repeated three times, native retinal CaBP4 from two dark- and two light-adapted mouse retinas was labeled with [γ-32P] ATP as described above in the presence or absence of OA. Reactions were then diluted twice with PBS and were cleared by incubation for 1 hour at 4°C with protein G-magnetic beads. After removal of the beads, samples were incubated for 1 hour at 4°C with affinity purified anti-CaBP4 polyclonal antibodies (10 μg). Protein G-magnetic beads were then added and incubated overnight at 4°C. Beads were washed 5 times with the same buffer, and proteins were eluted with 0.1% glycine, pH 2.8. Phosphorylated CaBP4 from dark/light-adapted retinas were resolved on SDS-PAGE and analyzed by autoradiography and Western blotting using anti-CaBP4 antibody. The intensity of each band was quantified using ImageJ.
Analysis of PP2A Activity in Dark- and Light-Adapted Mouse Retina Using Recombinant Phosphorylated CaBP4
Recombinant CaBP4 was phosphorylated with recombinant PKCζ in the presence of [γ-32P]ATP for 45 minutes at 30°C in a master mix as described above. After inactivation of PKC by addition of Bis (10 μM), aliquots containing 1 μg of labeled CaBP4 were then used as substrate for phosphatase. Phosphorylated proteins were added to 10 μL of dark- or light-adapted retinas homogenized in 10 mM BTP pH 7.4 and incubated further for 15 or 45 minutes at 30°C. An aliquot incubated for 45 minutes in the presence of OA (5 μM) was used as an internal control wherein both the kinase and phosphatase activity should be inhibited and, therefore, should be similar to results obtained immediately after addition of retinal extract (at time [T] = 0). Reactions were stopped by addition of SDS-PAGE sample buffer, and proteins were separated on SDS-PAGE followed by transfer onto immobilon membrane. 32P-labeled CaBP4 was detected by autoradiography and amount of CaBP4 was confirmed by Ponceau staining. The intensity of each band on the autoradiography was quantified using ImageJ.
Analysis of PP2A Expression Using Western Blot Analysis
Mouse retina or HEK293 cells were homogenized in 100 μL of PBS. Homogenate was centrifuged at 100,000g for 1 hour. After centrifugation, the supernatant was collected and the pellet was resuspended in 100 μL of PBS. Both samples were mixed with SDS-PAGE sample buffer and proteins were separated by SDS-PAGE follow by transfer onto immobilon. Membranes saturated with 5% milk in Phosphate buffer saline containing 0.1% Triton X-100 (PBST) were incubated for 1 hour at room temperature with polyclonal antibodies raised against PP2A A, PP2A B, or PP2A C. After extensive washing with PBST and 1 hour incubation with alkaline phosphatase-conjugated secondary antibodies, the labeled bands were detected colorimetrically by developing in 5-bromo-4-chloro-3-indolyl-phosphate (BCIP)/Nitro Blue Tetrazolium Chloride (NBT) solution.
Cloning of Retina PP2A Subunits
Mouse PP2A subunits (Aα, Cα, Cβ, and Bα) were cloned after amplification by PCR with Pfx polymerase from a mouse retina cDNA library with primers listed in the Table. Epitope tags were added by PCR to the cloned PP2A subunits using primers containing the HA, c-myc, or FLAG tag (Table). After sequencing of all cloned subunits, the tagged-subunits were subcloned into the pcDNA3.1 (–) vector for expression in mammalian cells.
GST Pull-Down Assay
HEK293 cells were cotransfected with equal amounts of pcDNA-PP2A Aα, pcDNA-PP2A Cα, or Cβ and pcDNA-PP2A Bα plasmids using Lipofectamine (Life Technologies) and collected 2 days after transfection. Cell lysates (∼5–7 mg/mL) were prepared by sonication of a 10 cm2 plate of HEK293 cells in 1 mL of PBS containing 0.1% Triton X-100 and 5 mM benzamidine, and centrifuged for 30 minutes at 10,000g. GST (negative control) and GST-CaBP4 (5 μg), immobilized on glutathione magnetic beads, were incubated for 3 hours at 4°C with 0.5 mL of cell lysate. The beads were washed five times with the same buffer and the proteins were eluted by boiling in 100 μL SDS-PAGE sample buffer. Samples were probed by Western blot analysis using anti-tag or anti-PP2A subunit antibodies. All samples were loaded on the same gel. When indicated, the development of the Western blot with NBT/BCIP colorimetric reagent was stopped after 1 minute for lanes loaded with the cell lysate compared with 5 minutes for the eluted proteins of the pull-down assay; a black line was added to the Western blot to delineate the cut membrane. The contrast of the Western blot images was increased equally across the whole image in Photoshop CS3 (Adobe Systems, San Jose, CA) to improve band visibility. For pull-down assays using retinas, we used bovine retinas, which provided a larger amount of tissue than is possible with mouse retina. Two bovine retinas (In Vision BioResources, Seattle, WA) were homogenized and sonicated in 4 mL of PBS containing 0.1% Triton X-100 and 5 mM benzamidine and centrifuged for 30 minutes at 10,000g. Retina supernatant was used in pull-down assays as described above.
Analysis of Native and Recombinant PP2A Activity in HEK293 Cell Lysates
Cells collected from a 10 cm2 plate of nontransfected cells or cells transfected with PP2A A, B, and C were sonicated in 500 μL of 10 mM BTP pH 7.2 containing 5 mM benzamidin. After centrifugation for 30 minutes at 10,000g, the total protein concentration of the supernatants was determined and equal amounts of proteins (∼25 μg) were used in the assay. A master mix for eight samples containing recombinant CaBP4 (8 × 2 μg), was added to 40 μL (8 × 5 μL) of cell extracts in the presence of [γ-32P]ATP and 10 mM BTP pH7.2, 2 mM MgCl2, 100 μM cold ATP, 0.5 mM DTT, 5 mM benzamidin in a 160 μL (8 × 20 μL ) volume, and incubated for 15 minutes at 30°C. OA (5 μM) was added to one aliquot and further incubated for 10 minutes at 30°C. One aliquot was taken and the reaction stopped as T equals 0 by addition of SDS-PAGE sample buffer. Bis (10 μM) was added to the rest of the reaction and further incubated at 30°C. Aliquots (20 μL) were taken after 5, 15, 30, 60, or 90 minutes and stopped by addition of SDS-PAGE sample buffer. The reactions were loaded on SDS-PAGE and transferred to immobilon membrane followed by Ponceau staining and autoradiography. The intensity of each band on the autoradiography was quantified using ImageJ.
Electrophysiologic Recordings
Patch clamp recordings were performed as described previously.10 Briefly, HEK293T cells were plated on 35-mm culture dishes and transiently transfected with approximately 3-μg total DNA (α11.3, β, α2δ subunits ± CaBP4) using Fugene transfection reagent according to manufacturer instructions. The plasmid pEGFP-N1 (0.1 μg) was included for fluorescent detection of transfected cells. Ca2+ currents were recorded at room temperature using the whole cell patch clamp technique 48 to 72 hours after transfection. The internal recording solution contained (in mM): 140 N-methyl-D-glucamine (NMDG), 5 EGTA, 10 HEPES, 2 MgCl2, 2 Mg-ATP, pH 7.3 (adjusted with methanesulfonate), and 290 milliosmole (mOsmol). The external solution contained the following (in mM): 130 Tris, 2 MgCl2, 20 CaCl2, pH 7.35 (adjusted with methanesulfonate), and 310 mOsmol. Dimethyl sulfoxide (DMSO), (0.04% final concentration) or OA (200 nM final concentration) was included in the internal solution. Electrode resistances were 3 to 5 megaohm (MΩ). Whole cell currents were acquired under voltage clamp with a EPC-9 amplifier (HEKA Elektronik, Lambrecht, Germany) controlled by PULSE software (HEKA Elektronik). Currents were filtered at 2 kHz and sampled at 10 kHz. A P/4 protocol was used to subtract the leak and capacitive transients. Electrophysiologic data were analyzed with IGOR Pro software (Wavemetrics, Portland, OR). Current-voltage relations were fit with the equation:
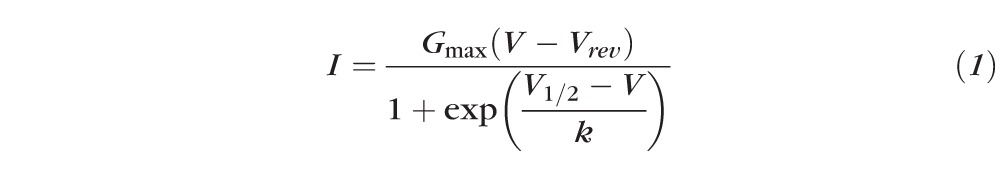 |
where Vrev is reversal potential, V1/2 is the half-maximal activation voltage, and k is the slope factor. Average data are expressed as mean ± SEM. ANOVA or t-test was used for statistical analysis.
Results
CaBP4 Phosphorylation Is Increased by PP2A Inhibitors in Retina Lysates
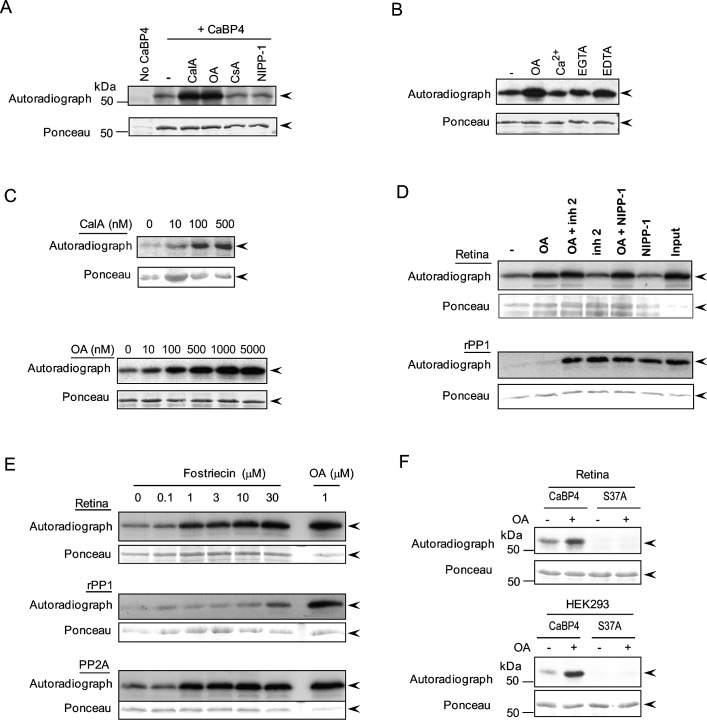
To investigate which phosphatase is involved in CaBP4 dephosphorylation in the retina, we first tested inhibitors of major protein serine/threonine phosphatases, PP1, PP2A, and PP2B (calcineurin). Recombinant GST-tagged CaBP4 was first labeled in retinal extracts in the presence of [γ-32P]ATP with the endogenous protein kinase in the retina, which we showed previously to be PKCζ.39 PKC was then inhibited with Bis and the endogenous retinal phosphatase activity assayed in the presence or absence of protein phosphatase inhibitors. Inhibition of a specific protein phosphatase involved in CaBP4 dephosphorylation should result in increased level of phosphorylated CaBP4 compared to that in the absence of that inhibitor. Without phosphatase inhibitors, phosphorylated CaBP4 was relatively low, due to strong dephosphorylation by the endogenous retinal phosphatase (Fig. 1A, lane 2). CaBP4 dephosphorylation was largely reversed in the presence of calyculin A, an inhibitor of PP1 and PP2A (Fig. 1A, lane 3). Compared with a retinal extract without inhibitor (Fig. 1A, lane 2), neither NIPP-1, a potent and specific inhibitor of PP1, nor cyclosporin A, a strong inhibitor of PP2B, had a significant effect on CaBP4 dephosphorylation (Fig. 1A, lanes 5, 6). In contrast, limited CaBP4 dephosphorylation was observed with OA, a stronger inhibitor of PP2A than PP1 and PP2B (Fig. 1A, lane 4).44,45 Unlike for PP2B and PP2C, the activity of PP2A is cation-independent. Therefore, CaBP4 dephosphorylation by PP2A should not be affected by the presence or absence of divalent cations. Consistent with this prediction, the level of CaBP4 phosphorylation was not substantially increased by the inclusion of Ca2+or EGTA; however, there was a slight increase in the level of CaBP4 phosphorylation with EDTA (Fig. 1B). Dose-dependent inhibition of CaBP4 dephosphorylation by calyculin A and OA further supported a role for PP2A in CaBP4 dephosphorylation (Fig. 1C).
Figure 1. .
CaBP4 phosphorylation by mouse retinal lysates is inhibited by PP2A inhibitors. Dephosphorylation of GST-CaBP4 due to the endogenous phosphatase in lysates from light-adapted mouse retinas (A–F) or HEK293 cells (F) was determined by autoradiography (upper panel) and equivalent input and transfer of CaBP4 in all reactions was confirmed by Ponceau staining (lower panel). The data are representative results of three separate experiments. Arrowheads indicate the band corresponding to GST-CaBP4. (A) Identification of the serine/threonine phosphatase involved in dephosphorylation of CaBP4 in retinal extracts. GST-CaBP4 was phosphorylated with the endogenous kinase in mouse retinal extracts in the presence of [γ-32P]ATP. After quenching of kinase activity, the reaction was further incubated with or without calyculin A (CalA, 1 μM), OA (1 μM), cyclosporin A (CsA, 1 μM), or NIPP-1 (300 nM). (B) Effect of Ca2+ on retinal phosphatase activity. GST-CaBP4 was first phosphorylated with recombinant PKCζ in the presence of [γ-32P]ATP. After inactivation of PKCζ, 32P-CaBP4 was added to an extract of mouse retinas without or with OA (5 μM), 5 mM CaCl2, 5 mM EGTA, or 5 mM EDTA as indicated and the incubation was carried on for 45 minutes at 30°C. (C) Dose-dependent inhibition of CaBP4 dephosphorylation by calyculin A (upper panel) and OA (lower panel) in mouse retina extract. CaBP4 dephosphorylation was analyzed in retinal extracts as described in (A) with or without inhibitors as indicated. (D) Comparison of retinal phosphatases and rPP1 activities on 32P-labeled CaBP4 in the presence or absence of phosphatase inhibitors. Phosphatase assays were carried out with radioactively labeled CaBP4 with or without 300 nM OA, 300 nM inhibitor 2 or 300 nM NIPP-1, or a combination of OA and inhibitor-2 or NIPP-1 (300 nM each). (E) Dose-dependent inhibition of CaBP4 dephosphorylation by fostriecin. GST-CaBP4 dephosphorylation assays were carried out as in (D) using mouse retinal lysate, rPP1, or purified PP2A, but with various concentrations of fostriecin or OA (1 μM). (F) Phosphorylated CaBP4-Ser37 is the substrate for the endogenous phosphatase in retina and HEK293 cells. GST-CaBP4 or GST-CaBP4-S37A mutant were incubated with equivalent amounts of retina or HEK293 extract in the presence of [γ-32P]ATP. After quenching of PKC activity with Bis, reactions were incubated further in the presence or absence of OA (1 μM).
However, the level of CaBP4 phosphorylation was maximal at concentration of OA (∼300 nM) that was high enough to affect the activity of PP1 as well as PP2A.45 Indeed, we found that recombinant PP1 (rPP1) could promote CaBP4 dephosphorylation in a cell free assay, and was inhibited by OA at slightly higher concentrations than those required to inhibit dephosphorylation of CaBP4 by purified PP2A (see Supplementary Material and Supplementary Figs. S2A, S2B, http://www.iovs.org/content/54/2/1214/suppl/DC1). To exclude the possibility that PP1 mediates CaBP4 dephosphorylation in retinal extracts, we tested the effect of PP1-specific inhibitors, inhibitor 2, and NIPP-1.45–48 Neither of these inhibitors affected dephosphorylation of CaBP4 in retinal lysates at a concentration (>100 nM; Fig. 1D) that fully inhibited the ability of rPP1 to dephosphorylate CaBP4 (see Supplementary Material and Supplementary Figs. S2C, S2D, http://www.iovs.org/content/54/2/1214/suppl/DC1). In addition, OA fully inhibited the CaBP4 retinal phosphatase at a concentration (300 nM) that did not affect rPP1 dephosphorylation of CaBP4, and there were no additive effects of inhibitor 2 and NIPP-1 to those of OA (Fig. 1D). Because OA inhibits not only PP2A but also PP4, PP5, and PP6, we also analyzed whether CaBP4 phosphorylation is promoted by fostriecin, which is a more selective inhibitor of PP2A.49 CaBP4 dephosphorylation in retinal extracts was strongly inhibited by fostriecin concentrations that do not inhibit CaBP4 dephosphorylation by rPP1 (Fig. 1E). Taken together, these results suggest that the properties of the phosphatase that dephosphorylates CaBP4 in the retina are consistent with PP2A.
We next tested the involvement of serine 37 as the residue in mouse CaBP4 that is dephosphorylated by PP2A. This residue was shown to be phosphorylated by PKC in HEK293 cells and by PKCζ in the retina, which is important for Cav1 channel modulation.39 If PP2A dephosphorylates serine 37, inhibiting PP2A should not affect the phosphorylation status of CaBP4 lacking this phosphorylation site. Consistent with this prediction, phosphorylation of wild-type (WT) CaBP4 but not CaBP4-S37A was increased with OA (Fig. 1F). These results suggest that PP2A can dephosphorylate CaBP4 at serine 37.
Interaction of CaBP4 with PP2A Subunits
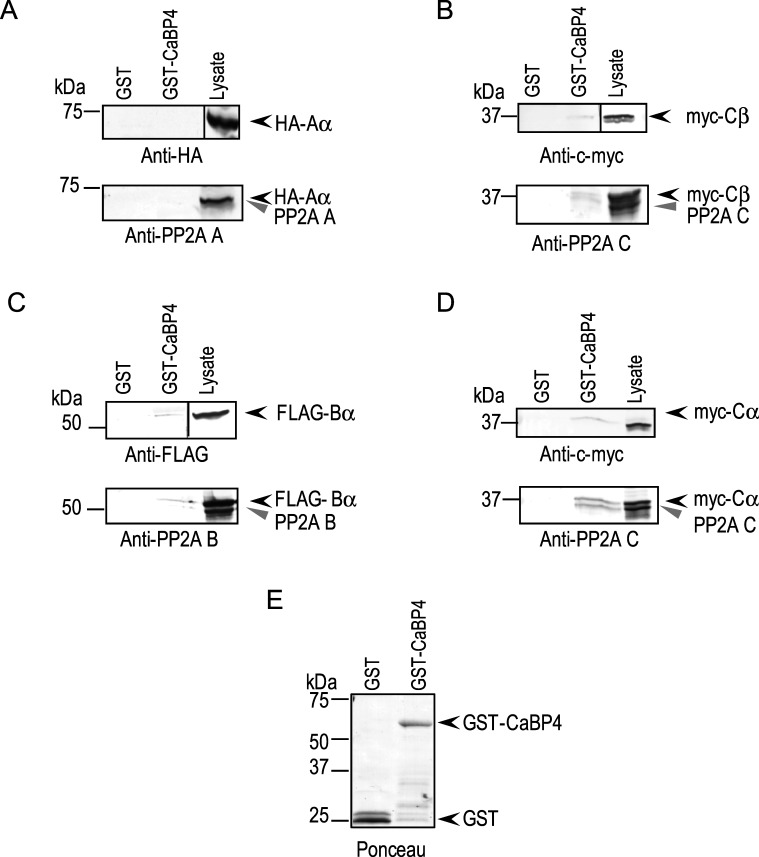
Two structural (Aα, β) and 2 catalytic (Cα, β) subunits of PP2A have been characterized. In addition, there are 16 PP2A regulatory (B) subunits divided across four gene families (B, B', B'', and B'''), which help define substrate specificity.50–53 To gain insights as to which PP2A subunits may regulate CaBP4 in the retina, we performed pull-down assays using GST-tagged CaBP4 (GST-CaBP4) and lysate from bovine retinas or HEK293 cells. Unlike for the CaBP4 phosphorylation assays, in which mouse retinal lysate was used, larger amounts of starting material are required for pull-down assays, which is why we turned to lysates from bovine retinas and HEK293 cells. Consistent with previous results,54,55 the A, B, and C subunits of PP2A were detected in extracts of retina by Western blotting with anti-PP2A (Aα, β), anti-PP2A (Bα), and anti-PP2A (Cα, β) antibodies (Fig. 2A). In pull-down assays, GST-CaBP4 interacted with A, B, and C subunits that were endogenously expressed in retina (Fig. 2B). By affinity chromatography and mass spectrometry of bovine retinal lysates, peptide sequences corresponding to PP2A Aα and common to particular PP2A subunits (Cα, and Cβ) were identified among the proteins interacting with CaBP4 (data not shown). In addition, each of these PP2A subunits (Table) found to interact with CaBP4 could be amplified by PCR from mouse retina cDNA, consistent with their potential role in regulating CaBP4 dephosphorylation in the retina. These results provide further support for a role of PP2A in CaBP4 dephosphorylation in the retina.
Figure 2. .
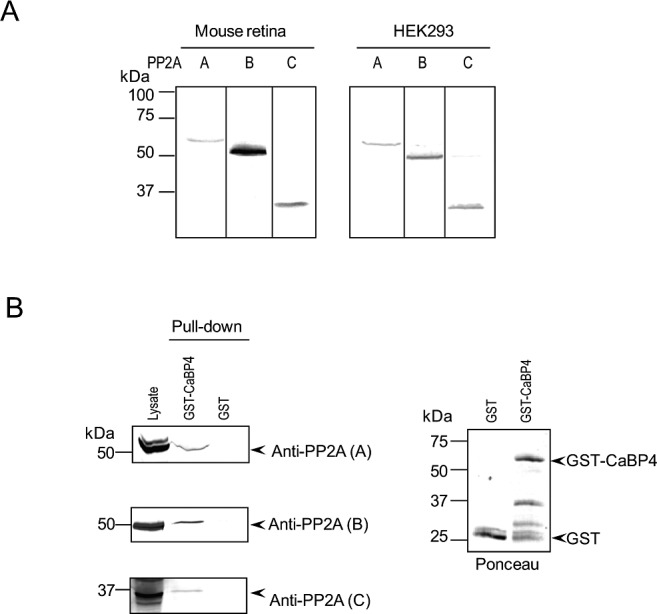
PP2A subunits are expressed in the retina and interact with CaBP4. (A) Detection of PP2A subunits A, B, and C in mouse retina and HEK293 cells. Mouse retinas or HEK293 cells were homogenized and analyzed by SDS-PAGE followed by transfer onto immobilon-P membrane. The membrane was cut in three sections that were analyzed separately by Western blot using antibodies raised against PP2A subunits: A (α/β), B (PR55, Bα), or C (α/β). Both in retinal extracts and HEK293 cell extracts, each anti-PP2A antibody detected one single band consistent in size with the A (∼65 kDa), B (∼55 kDa), or C (∼35 kDa) subunit. (B) Pull-down of retinal PP2A subunits by GST-CaBP4. Lysate of bovine retinas was incubated with either GST or GST-CaBP4 immobilized on glutathione-coupled magnetic beads. Western blot analysis was done with the same antibodies used in panel A and detected bands in retinal lysate (lane 1), and in reactions incubated with GST-CaBP4 (lane 2), but not GST (lane 3). Levels of GST proteins used in the assay were visualized by Ponceau staining (right).
To confirm the interaction of CaBP4 with these specific PP2A subunits, epitope-tagged PP2A subunits (Aα, Cα, Cβ, and Bα) were expressed in HEK293 cells and tested for binding to GST-CaBP4. Cα, Cβ, and Bα, but not Aα, subunits interacted specifically with GST-CaBP4 as shown by Western blot analysis using anti-tag antibodies (Figs. 3A–D, top panels). Western blotting using antibodies against A, B, and C subunits detected two bands corresponding to the epitope-tagged PP2A Cα, Cβ, and Bα (Figs. 3B–D, lower panels, upper band) and the endogenous B and C subunits, which are expressed in HEK293 cells (Figs. 3B–D, lower panels, lower band). Our inability to detect interaction of PP2A Aα subunit with GST-CaBP4 in these experiments with HEK293 cell lysates suggested interactions through other retina-specific proteins, which could account for the pull-down of PP2A A subunit by GST-CaBP4 from retinal lysates (Fig. 2B). Thus, we conclude that CaBP4 interacts, either directly or indirectly, with PP2A subunits Aα, Cα, Cβ, and Bα in the retina.
Figure 3. .
Recombinant PP2A B and C subunits interact with CaBP4. GST or GST-CaBP4 immobilized on glutathione magnetic beads was incubated with lysates of HEK293 cells cotransfected with epitope-tagged PP2A subunits: HA-tagged Aα (A), c-myc–tagged Cβ (B), FLAG-tagged Bα (C), or c-myc–tagged Cα (D). Bound PP2A subunits were detected by Western blot analysis with antibodies against epitope tags (upper panels) or PP2A subunits (bottom panels). Bands corresponding to the epitope-tagged and endogenous untagged PP2A subunits are indicated in the lysates with black and grey arrowheads, respectively. Ponceau staining indicated the amounts of GST or GST-CaBP4 used in the pull-down assay (E).
Overexpression of PP2A Enhances the Rate of Dephosphorylation of CaBP4 in Transfected HEK293 Cells
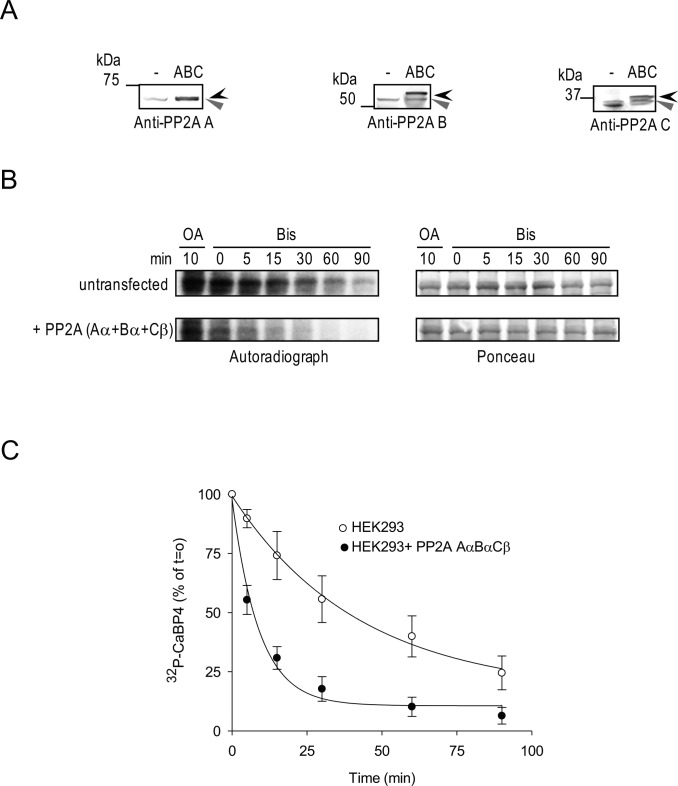
To verify the importance of PP2A in regulating CaBP4 dephosphorylation, we tested the consequences of varying expression levels of PP2A on CaBP4 phosphorylation in HEK293 cells. Because we were unable to achieve knock-down of PP2A subunit expression in HEK293 cells with siRNA, we assessed instead the effect of increasing PP2A by overexpression on CaBP4 dephosphorylation in HEK293 cells. For these experiments, we cotransfected HEK293 cells with PP2A subunits (Aα, Bα, Cβ; Fig. 4A), which interacted with CaBP4 (Figs. 2, 3), and compared the effects of lysates from untransfected and PP2A-transfected cells on CaBP4 dephosphorylation. Recombinant GST-CaBP4 was first prephosphorylated by the endogenous kinase in HEK293 cells and similar levels of kinase activity were confirmed in untransfected and PP2A-transfected cells upon phosphatase inhibition with OA (Fig. 4B, lane 1). We then analyzed the time course of CaBP4 dephosphorylation by PP2A after addition of the PKC inhibitor, Bis. The rate of CaBP4 dephosphorylation for PP2A-transfected cells (11.8 ± 2.5%/min, P = 0.009 by t-test) was significantly greater than that in untransfected cells (1.8 ± 0.7%/min). These findings confirm that PP2A can efficiently dephosphorylate CaBP4.
Figure 4. .
Recombinant PP2A subunits enhance CaBP4 dephosphorylation in HEK293 cells. HEK293 cells were not transfected (lane 1) or cotransfected with rPP2A Aα, Bα, and Cβ (lane 2). (A) Western blot analysis of the recombinant epitope-tagged (black arrowhead) or endogenous untagged (grey arrowhead) PP2A subunits in HEK293 cell lysates used in (B) using anti-PP2A subunit antibodies. (B) Time course of CaBP4 dephosphorylation by PP2A subunits. Lysates of HEK293 cells were incubated with GST-CaBP4 in the presence of [γ-32P]ATP. Similar levels of kinase activity between groups was confirmed upon further incubation with OA (5 μM) for 10 minutes to inhibit PP2A activity. Dephosphorylation of radiolabeled CaBP4 by PP2A was analyzed between 0 and 90 minutes after addition of Bis (10 μM) to inhibit PKC activity. Autoradiograph (left) and Ponceau staining (right) shows levels of phosphorylated CaBP4 and equivalent amounts of CaBP4 in each reaction, respectively. (C) Quantitative analysis of time course of CaBP4 dephosphorylation by PP2A subunits. The intensity of each band of the autoradiography as shown in (B) was quantified using ImageJ and normalized to that of the reaction at T = 0 minutes after quenching of PKC activity. Points represent mean ± SEM, n = 4. The data were fit with a single exponential function. The rate for untransfected cells (1.8 ± 0.7%/min) is significantly slower than that for PP2A-transfected cells (11.8 ± 2.5%/min, P = 0.009 by t-test).
CaBP4 Dephosphorylation by PP2A Inhibits Modulation of Cav1.3
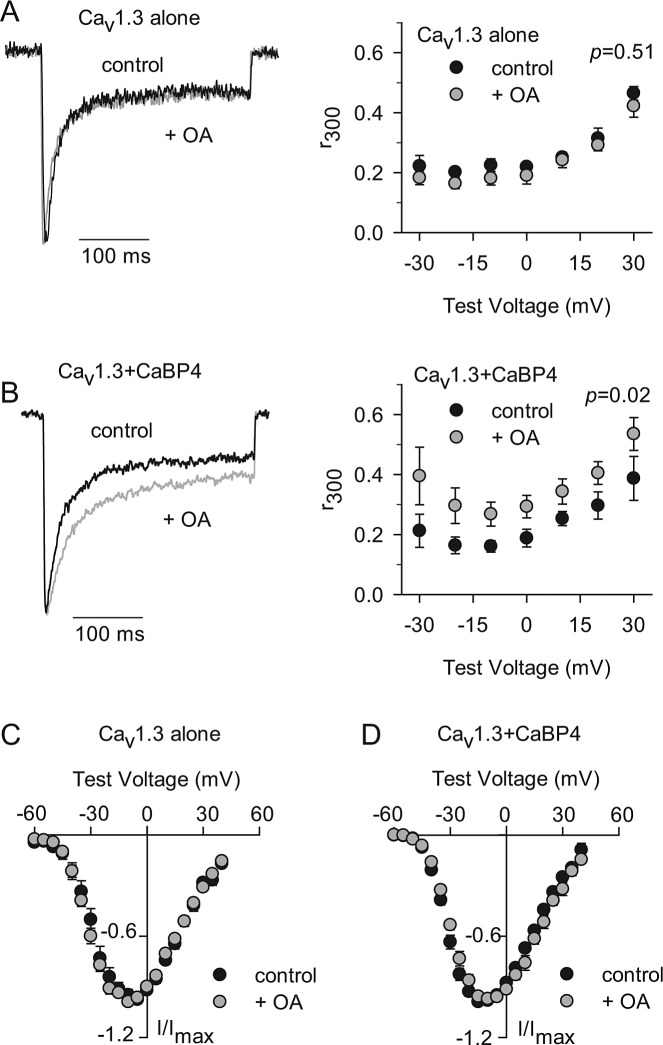
To address the functional consequences of CaBP4 dephosphorylation by PP2A, we used an assay that measures the modulation of Cav1 channels by CaBP4 in transfected HEK293T cells.39 While we have shown that CaBP4 enhances voltage-dependent activation of Cav1.4 channels, Cav1.4 Ca2+ currents (ICa) are relatively small in amplitude compared with Cav1.3 currents, which complicates analysis of CaBP4 modulation. Therefore, we have used Cav1.3 channels to characterize CaBP4 modulation by phosphorylation. CaBP4 has a modest effect on slowing inactivation of ICa in HEK293T cells cotransfected with Cav1.3, which is prevented by the S37A mutation that prevents phosphorylation of CaBP4.39 If PP2A dephosphorylation of CaBP4 affects the function of CaBP4, then OA should enhance the effect of CaBP4 on slowing Cav1.3 inactivation. To test this hypothesis, we compared ICa in HEK293T cells cotransfected with Cav1.3 and CaBP4 (Fig. 5). Inactivation was measured as the ratio of the current amplitude at the end of a 300 ms test pulse and the peak current amplitude (r300, Figs. 5A, B). In cells cotransfected with CaBP4 and Cav1.3, ICa inactivation was significantly weaker (∼22%–43% across the voltage range tested, Fig. 5B) when exposed to OA compared with control solution (P = 0.02, by two-way ANOVA). By contrast, OA had no effect on ICa inactivation in cells transfected with Cav1.3 alone (P = 0.58, Fig. 5A). These results confirm that CaBP4 dephosphorylation can negatively regulate functional interactions with targets such as Cav1.3. Although PP2A inhibition can directly affect activation of Cav1 currents,56 we found no effect of OA on the voltage-dependent activation in cells transfected with Cav1.3 alone (Fig. 5C) or cotransfected with Cav1.3 and CaBP4 (Fig. 5D). Together with our biochemical data, these results suggest that CaBP4 dephosphorylation by PP2A can inhibit CaBP4 modulation of Cav1.3 inactivation.
Figure 5. .
OA potentiates modulation of Cav1.3 channels by CaBP4. (A, B) Left panels: Representative traces showing Ca2+ currents (ICa) evoked by a 300 ms test pulse from −90 mV to −20 mV for cells with Cav1.3 alone (A) or cotransfected with CaBP4 (B). Recordings were with 0.04% DMSO (control; black) or with 200 nM OA included in the patch pipette (+OA, grey). Control and OA traces were normalized for comparison. Right panels: Inactivation was measured as the amplitude of ICa at the end of the 300 ms depolarization normalized to the peak current amplitude (r300) and plotted against test voltage for Cav1.3 alone (n = 5 for control, n = 4 for +OA) or for Cav1.3 + CaBP4 (n = 4 for control, n = 4 for +OA). P values were based on two-way ANOVA. (C, D) Current-voltage (I-V, left panels) relationships for cells transfected and recorded as in (A, B). Currents were evoked by 20 ms test pulses from −90 mV to various test potentials ranging from −70 mV to +40 mV for cells with Cav1.3 alone (C) or cotransfected with CaBP4 (D). Peak currents were normalized to Imax for each cell (I/Imax) and plotted against voltage. Parameters from Boltzmann fits of IV curves in (C, D) were: for Cav1.3 alone (control: V1/2 = −15.3 ± 1.4 mV; k = −7.5 ± 0.3; n = 6; + OA: V1/2 = −18.4 ± 1.6 mV; k = −7.4 ± 0.5; n = 6), and for Cav1.3 + CaBP4 (control: V1/2 = −21.1 ± 1.1 mV; k = −6.6 ± 0.4; n = 7; +OA: V1/2 = −17.9 ± 1.3 mV; k = −7.4 ± 0.6; n = 7). OA had no significant effect on any of the parameters (Cav1.3 alone: V1/2: P = 0.17; k: P = 0.76; Vrev: P = 0.90, by t-test; Cav1.3 + CaBP4: V1/2: P = 0.10; k: P = 0.17, Vrev: P = 0.27).
Dephosphorylation of CaBP4 by PP2A is Greater in Light-Adapted than Dark-Adapted Retina
CaBP4 phosphorylation by the atypical Ca2+-independent PKCζ is greater in light-adapted conditions when Ca2+ levels are low than in dark-adapted mouse retinas when Ca2+ levels are comparatively high.39,57–59 Therefore, the phosphorylation state of CaBP4 could be determined by Ca2+-dependent dephosphorylation in darkness or Ca2+-independent enhancement of CaBP4 phosphorylation in light-adapted retina. The former possibility is unlikely since CaBP4 dephosphorylation by PP2A is unaffected by Ca2+ (Fig. 1B). Nevertheless, we compared the effects of PP2A inhibition on CaBP4 phosphorylation in light- and dark-adapted retina. In these experiments, PP2A inhibition by OA only slightly increased CaBP4 phosphorylation in retina extract from dark-adapted mice (Figs. 6A, B). Since an effect of PP2A may have been missed due to the low level of CaBP4 phosphorylation under dark-adapted conditions, we compared the effect of OA on recombinant CaBP4 that was prephosphorylated with recombinant PKCζ to the same levels prior to the addition of extract of dark- or light-adapted mouse retinas. Under these conditions, the same initial level of phosphorylated CaBP4 should be then regulated by the endogenous phosphatase activity in light- or dark-adapted retinal lysates. By analyzing samples taken at different time points, we found that dephosphorylation of CaBP4 was faster in light- compared with dark-adapted retinas in that the level of phosphorylated CaBP4 was significantly lower in light-adapted than in dark-adapted retinas (Figs. 6C, D; T = 15 minutes, P < 0.015, T = 45 minutes, P < 0.005, t-test). Thus, PP2A activity is actually greater under light-adapted than dark-adapted conditions. These results were not due to a light-dependent alteration in the subcellular distribution of PP2A subunits, which was confirmed by Western blot analysis of soluble and membrane fractions of light- and dark-adapted retinal extracts (data not shown). While Ca2+-independent activity of PKCζ may dictate the level of light-dependent CaBP4 phosphorylation, activation of PP2A in light conditions may be necessary to balance the activity of PKCζ in controlling the phosphorylation state of CaBP4.
Figure 6. .
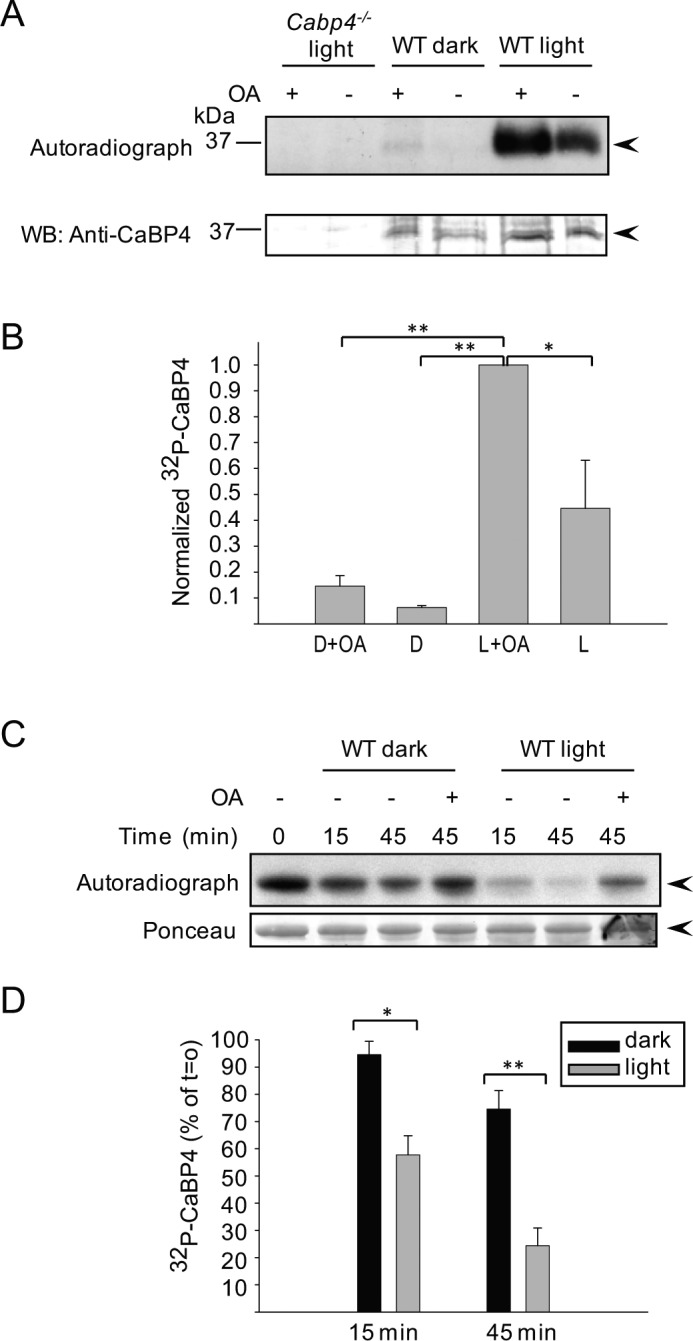
CaBP4 dephosphorylation is greater in light-adapted retina than dark-adapted retinas. (A) Extracts from dark- and light-adapted mouse retinas were incubated under red dim light or in the light, respectively, with [γ-32P]ATP. Samples were further incubated with or without OA and subjected to immunoprecipitation with anti-CaBP4 antibodies. Level of phosphorylated CaBP4 was revealed by autoradiography (upper panel) and amount of immunoprecipitated CaBP4 in each sample was detected by Western blot analysis (WB, bottom panel) with anti-CaBP4 antibodies. CaBP4 knockout mouse retinas were used as a negative control (lanes 1, 2). The data are representative results of three independent experiments. Arrowheads indicate the band corresponding to CaBP4. (B) Quantitative analysis of 32P-CaBP4 in dark- and light-adapted mouse retina in the presence and absence of OA as shown in (A). The intensity of each band of the autoradiograph was quantified and normalized to the intensity of each band in the anti-CaBP4 Western blot. The values were then normalized to the intensity of the peak signal (i.e., in light condition with OA). D = dark-adapted retinas. L = light-adapted retinas. Data represent mean ± SEM (n = 3, *P < 0.05, **P < 0.001, t-test). (C) Recombinant CaBP4 was phosphorylated with recombinant PKCζ in the presence of [γ-32P]ATP. After inactivation of PKCζ with Bis (T = 0 minutes), radioactively labeled CaBP4 was added to an extract of dark- or light-adapted mouse retinas and the incubation was carried out for 15 minutes and 45 minutes at 30°C. A reaction carried out with OA provided a negative control in which both the native kinase and phosphatase activity should be inhibited. The level of 32P-CaBP4 is revealed by autoradiography (upper panel) and equivalent amount of the protein in each reaction was confirmed by Ponceau staining (bottom panel). (D) Quantitative analysis of 32P-CaBP4 dephosphorylation in dark- and light-adapted mouse retina as shown in (B). The intensity of each band of the autoradiograph in (B) was quantified using ImageJ and normalized to that of the control reaction carried out in the presence of OA. Points represent mean ± SEM (n = 4, *T = 15 minutes, P < 0.015, **T = 45 minutes, P < 0.005, t-test).
Discussion
Reversible phosphorylation of Ca2+ binding proteins such as calmodulin (CaM) has emerged as an important mechanism for the dynamic regulation of effectors. Small-conductance Ca2+-activated potassium channels (SK channels) are gated by Ca2+ ions through the association of CaM directly with the SK channel protein.60 Casein kinase 2 phosphorylation of CaM at threonine 80 inhibits Ca2+ activation of SK channels, which may be reversed by PP2A dephosphorylation.37 In this study, we demonstrate that PP2A dephosphorylates CaBP4 in the retina. CaBP4 dephosphorylation is enhanced by light in the retina and limits CaBP4 potentiation of Cav1.3 activity in transfected HEK293T cells.
Although multiple serine/threonine protein phosphatases have been identified in the retina,61–65 PP2A inhibitors, OA, and fostriecin, but not PP2B or PP1 selective inhibitors, block CaBP4 dephosphorylation in retinal lysates (Fig. 1). Higher concentrations of fostriecin and OA (hundreds of nanomolar) were required to fully inhibit CaBP4 dephosphorylation both in the retinal extract and by purified PP2A than would be predicted based on previous reports (∼low nanomolar concentrations)45,49 (Figs. 1C, 1D, 1F and see Supplementary Material and Supplementary Fig. S2, http://www.iovs.org/content/54/2/1214/suppl/DC1). However, variation among IC50 values of OA and fostriecin in cell-free assays may depend on the amount of phosphatase45 as well as the substrate employed.66 Although inhibition of PP1 is independent of phosphatase concentration, the concentration of OA required for inhibition of PP2A increases with increasing PP2A levels.45 Therefore, it is possible that CaBP4 as a PP2A substrate and/or high phosphatase levels in our assay account for the higher concentration of OA and fostriecin needed to inhibit fully CaBP4 dephosphorylation.
The PP2A-like phosphatases, PP4, PP5, and PP6, are also inhibited by OA concentrations close to those that inhibit PP2A.50,67,68 However, fostriecin inhibited CaBP4 dephosphorylation at concentrations that inhibit PP2A, but not PP1 and PP549 (Fig. 1E), arguing against a role for PP1 or PP5 in CaBP4 dephosphorylation in retinal extracts. These data alone cannot rule out the involvement of PP4, since there are currently no selective inhibitors, including fostriecin, that distinguish between PP2A and PP4. The sensitivity of PP6 to fostriecin has not been characterized, although predicted to be comparable to that of PP2A based on conserved amino acids between the PP2A and PP6 fostriecin-binding domain.49 However, PP6 is inhibited by 10 times higher concentration of OA than PP2A.50 Therefore, PP6 is unlikely to be involved in CaBP4 dephosphorylation in the retina.
As expected for PP2A, phosphatase activity was not changed by addition of Ca2+ or EGTA; however, a small decrease of CaBP4 dephosphorylation was observed in the presence of EDTA compared with EGTA (Fig. 1B). In contrast to EGTA, which preferentially binds Ca2+, EDTA chelates all divalent cations. PP2A activity does not require cations but can be stimulated by cations including Mn2+.69 Reversible methylation of PP2A is also significantly stimulated by divalent cations, especially Mn2+.70 Methylation of PP2A C is important for regulating the formation of PP2A heterotrimers containing B-type subunits,71–73 and, therefore, may indirectly regulate PP2A activity.
Considering the role of PP2A B subunits in regulating various aspects of substrate specificity,50–53 the association of the PP2A Bα subunit with CaBP4 (Figs. 2B, 3C) may mediate specific dephosphorylation by the heterotrimeric PP2A holoenzyme. PP2A B subunits have been shown to recruit PP2A to substrates such as calcium/calmodulin-dependent protein kinase IV,74 and may similarly facilitate CaBP4 dephosphorylation at photoreceptor synapses by the PP2A holoenzyme. Crystallization of a PP2A holoenzyme including the Bα subunit revealed that a highly acidic groove contributes to the putative substrate binding site that associates with the cytoskeletal tau protein.75 The two peptide sequences in tau that were identified as interacting with Bα were highly basic, with 11 lysine residues. The 40 amino acids surrounding the CaBP4 phosphorylation site (serine 37) are also highly basic, with nine positively charges residues, which may similarly contribute to a PP2A Bα-binding region. The faster CaBP4 dephosphorylation in HEK293 cells overexpressing PP2A Bα, as well as Aα and Cβ (Figs. 4B, 4C), support the participation of PP2A Bα in PP2A complexes involved in CaBP4 dephosphorylation. However, it is possible that other regulatory B subunits in addition to Bα contribute to heterotrimeric PP2A complexes that recognize CaBP4 as a substrate.
PP2A complexes containing the B' subunit, B56 epsilon, translocate to the cytosolic fraction upon exposure to light, which stimulates light-dependent dephosphorylation of phosducin and downregulation of visual transduction.54 However, light-dependent translocation of the Bα subunit was not observed54 (and data not shown). Therefore, formation of PP2A A, Bα, C heterotrimers that may interact with CaBP4 most likely occurs at the membrane and does not require light-dependent redistribution. In PP2A A, Bα, C, the Bα subunit was also shown to promote targeting of PP2A to the microtubule cytoskeleton.53,76 It is possible that light-driven changes in PP2A ABαC association with substrates, such as CaBP4, may underlie enhanced PP2A activity in light-adapted retinas (Fig. 6).
In photoreceptors, activation of the phototransduction cascade by light results in hyperpolarization of the membrane potential that is transmitted from the outer segment to the synaptic terminals and causes closure of synaptic Cav1 voltage-gated Ca2+ channels. Because neurotransmitter release is proportional to Ca2+ influx, Cav1 Ca2+ channels are key players in transforming the change in membrane potential into a change in neurotransmitter release.58,59,77 In photoreceptor terminals, CaBP4 interacts with and potentiates the activity of Cav1.4 Ca2+ channels, primarily by enhancing voltage-dependent activation.10 The analysis of CaBP4 modulation of Cav1.4 in transfected HEK293 cells is greatly hindered by the low open probability and unitary conductance of Cav1.4,78 and the inhibitory effects of CaBP4 on Cav1 channel expression.7 Therefore, we have used the prolongation of Cav1.3 Ca2+ currents, due to effects of CaBP4 in antagonizing inactivation, as a metric for CaBP4 modulation. Using this approach, we showed that phosphorylation of serine 37 in CaBP4 facilitates low affinity interactions with Cav1.3 and enhances the effect of CaBP4 in prolonging Cav1.3 Ca2+ currents in HEK293 cells.39 The functional importance of CaBP4 reversible phosphorylation is supported by our present data showing that inhibition of PP2A by OA enhances the effect of CaBP4 on Cav1.3 inactivation in HEK293 cells (Fig. 5).
While CaBP4 does not affect voltage-dependent activation of Cav1.3, it strongly suppresses Ca2+dependent inactivation (CDI) of Cav1.3.39 Cav1.4 channels show little CDI independent of CaBP4 due to an inhibitory C-terminal domain (CTD) in the pore-forming Cav1.4 α1 subunit.79,80 Deletion of the CTD restores CDI to Cav1.4, which can then be suppressed by CaBP4.13 Splice variants of the Cav1.4 α1 subunit lacking the CTD have been detected in human retina.81 Therefore, while our present data indicate effects of CaBP4 dephosphorylation on Cav1.3 CDI, they are likely relevant for regulation of CDI of these Cav1.4 splice variants. It was previously reported that in retinal explants, OA increases Ca2+ influx in a time- and dose-dependent manner, which is prevented by inhibitors of Cav1 Ca2+ channels and PKC.82 These findings are consistent with our results that OA inhibits inactivation of Cav1.3 channels by enhancing the modulatory effects of phosphorylated CaBP4.
Photoreceptors are able to respond to a wide range of light intensity. Thus, Cav1 channel activity must be highly regulated to transform any change in light intensity into a precise change in the rate of transmitter release. In addition to Ca2+ binding, reversible phosphorylation provides an additional mechanism to control the function of CaBP4 and consequently Cav1 activity. In light conditions, CaBP4 phosphorylation may maintain Cav1-mediated Ca2+ influx to sustain neurotransmitter release at various light intensities. However, prolonged CaBP4 phosphorylation would cause excessive Ca2+ influx and neurotransmitter release, which would likely degrade visual signal transmission. Therefore, the light-dependent enhancement in PP2A activity may be important for terminating the PKCζ-dependent boost in Cav1 Ca2+ signaling, so as to preserve the fidelity of photoreceptor responses to light stimuli.
Supplementary Material
Acknowledgments
The authors thank Sharona Gordon and the members of her group for helpful discussions and the use of lab equipment, and Bhuvana Parampalli for assistance with figure preparation.
Footnotes
Supported by grants from National Institutes of Health/National Eye Institute (R01-EY020850, [FH]), National Institutes of Health (R01 DC009433 and R01 HL087120 [AL]; DA015040 and K12/GM00068 [FDG]), and the Carver Research Program of Excellence (AL).
Disclosure: F. Haeseleer, None; I. Sokal, None; F.D. Gregory, None; A. Lee, None
References
- 1. Haeseleer F, Imanishi Y, Sokal I, Filipek S, Palczewski K. Calcium-binding proteins: intracellular sensors from the calmodulin superfamily. Biochem Biophys Res Commun. 2002; 290: 615–623 [DOI] [PubMed] [Google Scholar]
- 2. Haeseleer F, Paczewski K. Calmodulin and Ca2+-binding proteins (CaBPs): variations on a theme. Adv Exp Med Biol. 2002; 514: 303–317 [DOI] [PubMed] [Google Scholar]
- 3. Haeseleer F, Sokal I, Verlinde C, et al. Five members of a novel Ca2+-binding protein (CABP) subfamily with similarity to calmodulin. J Biol Chem. 2000; 275: 1247–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McCue HV, Haynes LP, Burgoyne RD. Bioinformatic analysis of CaBP/calneuron proteins reveals a family of highly conserved vertebrate Ca2+-binding proteins. BMC Res notes. 2010; 3: 118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou H, Kim SA, Kirk EA, et al. Ca2+-binding protein-1 facilitates and forms a postsynaptic complex with Ca(v)1.2 (L-Type) Ca2+ channels. J Neurosci. 2004; 24: 4698–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee A, Westenbroek RE, Haeseleer F, Palczewski K, Scheuer T, Catterall WA. Differential modulation of Ca(v)2.1 channels by calmodulin and Ca2+-binding protein 1. Nat Neurosci. 2002; 5: 210–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang PS, Alseikhan BA, Hiel H, et al. Switching of Ca2+-dependent inactivation of Ca(V)1.3 channels by calcium binding proteins of auditory hair cells. J Neurosci. 2006; 26: 10677–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Findeisen F, Minor DL. Structural basis for the differential effects of CaBP1 and calmodulin on Ca(v)1.2 calcium-dependent inactivation. Structure. 2010; 18: 1617–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Few AP, Nanou E, Scheuer T, Catterall WA. Molecular determinants of Ca(v)2.1 channel regulation by calcium-binding protein-1. J Biol Chem. 2011; 286: 41917–41923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haeseleer F, Imanishi Y, Maeda T, et al. Essential role of Ca2+-binding protein 4, a Ca(v)1.4 channel regulator, in photoreceptor synaptic function. Nat Neurosci. 2004; 7: 1079–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou H, Yu K, McCoy KL, Lee A. Molecular mechanism for divergent regulation of Ca(v)1.2 Ca2+ channels by calmodulin and Ca2+-binding protein-1. J Biol Chem. 2005; 280: 29612–29619 [DOI] [PubMed] [Google Scholar]
- 12. Cui G, Meyer A, Calin-Jageman I, et al. Ca2+-binding proteins tune Ca2+ -feedback to Cav1.3 Ca 2+ channels in auditory hair cells. J Physiol. 2007; 585: 791–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shaltiel L, Paparizos C, Fenske S, et al. Complex regulation of voltage-dependent activation and inactivation properties of retinal voltage-gated Cav1.4 L-type Ca2+ channels by Ca2+-binding protein 4 (CaBP4). J Biol Chem. 2012; 287: 36312–36321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kinoshita-Kawada M, Tang JS, Xiao R, Kaneko S, Foskett JK, Zhu MX. Inhibition of TRPC5 channels by Ca2+-binding protein 1 in Xenopus oocytes. Pflugers Archiv-Eur J Physiol. 2005; 450: 345–354 [DOI] [PubMed] [Google Scholar]
- 15. Kasri NN, Holmes AM, Bultynck G, et al. Regulation of InsP(3) receptor activity by neuronal Ca2+-binding proteins. Embo J. 2004; 23: 312–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang J, McBride S, Mak DOD, et al. Identification of a family of calcium sensors as protein ligands of inositol trisphosphate receptor Ca2+ release channels. Proc Natl Acad Sci U S A. 2002; 99: 7711–7716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haynes LP, Tepikin AV, Burgoyne RD. Calcium-binding protein 1 is an inhibitor of agonist-evoked, inositol 1,4,5-trisphosphate-mediated calcium signaling. J Biol Chem. 2004; 279: 547–555 [DOI] [PubMed] [Google Scholar]
- 18. Lee SW, Briklin O, Hiel H, Fuchs P. Calcium-dependent inactivation of calcium channels in cochlear hair cells of the chicken. J Physiol. 2007; 583: 909–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maeda T, Lem J, Palczewski K, Haeseleer F. A critical role of CaBP4 in the cone synapse. Invest Ophthalmol Vis Sci. 2005; 46: 4320–4327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ball SL, Powers PA, Shin HS, Morgans CW, Peachey NS, Gregg RG. Role of the beta(2) subunit of voltage-dependent calcium channels in the retinal outer plexiform layer. Invest Ophthalmol Vis Sci. 2002; 43: 1595–1603 [PubMed] [Google Scholar]
- 21. Chang B, Heckenlively JR, Bayley PR, et al. The nob2 mouse, a null mutation in Cacnalf: anatomical and functional abnormalities in the outer retina and their consequences on ganglion cell visual responses. Vis Neurosci. 2006; 23: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mansergh F, Orton NC, Lalonde MR, et al. Mutation of the calcium channel gene Cacna1f disrupts calcium signaling, synaptic transmission and cellular organization in mouse retina. Hum Mol Genet. 2005; 14: 3035–3046 [DOI] [PubMed] [Google Scholar]
- 23. Aldahmesh MA, Al-Owain M, Alqahtani F, Hazzaa S, Alkuraya FS. A null mutation in CABP4 causes Leber's congenital amaurosis-like phenotype. Mol Vis. 2010; 16: 207–212 [PMC free article] [PubMed] [Google Scholar]
- 24. Littink KW, van Genderen MM, Collin RWJ, et al. A novel homozygous nonsense mutation in CABP4 causes congenital cone-rod synaptic disorder. Invest Ophthalmol Vis Sci. 2009; 50: 2344–2350 [DOI] [PubMed] [Google Scholar]
- 25. Zeitz C, Labs S, Lorenz B, et al. Genotyping microarray for CSNB-associated genes. Invest Ophthalmol Vis Sci. 2009; 50: 5919–5926 [DOI] [PubMed] [Google Scholar]
- 26. Khan AO, Alrashed M, Alkuraya FS. Clinical characterisation of the CABP4-related retinal phenotype [published online ahead of print October 31 2012]. Brit J Ophthalmol. doi:10.1136/bjophthalmol-2012-302186 [DOI] [PubMed] [Google Scholar]
- 27. BoeszeBattaglia K, Kong FS, Lamba OP, Stefano FP, Williams DS. Purification and light-dependent phosphorylation of a candidate fusion protein, the photoreceptor cell peripherin/rds. Biochemistry. 1997; 36: 6835–6846 [DOI] [PubMed] [Google Scholar]
- 28. Hu G, Jang GF, Cowan CW, Wensel TG, Palczewski K. Phosphorylation of RGS9-1 by an endogenous protein kinase in rod outer segments. J Biol Chem. 2001; 276: 22287–22295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee RH, Brown BM, Lolley RN. Light-induced dephosphorylation of a 33k protein in rod outer segments of rat retina. Biochemistry. 1984; 23: 1972–1977 [DOI] [PubMed] [Google Scholar]
- 30. Kuhn H, Dreyer WJ. Light dependent phosphorylation of rhodopsin by ATP. Febs Lett. 1972; 20: 1–6 [DOI] [PubMed] [Google Scholar]
- 31. Rajala RVS, McClellan ME, Ash JD, Anderson RE. In vivo regulation of phosphoinositide 3-kinase in retina through light-induced tyrosine phosphorylation of the insulin receptor beta-subunit. J Biol Chem. 2002; 277: 43319–43326 [DOI] [PubMed] [Google Scholar]
- 32. Roof DJ, Hayes A, Adamian M, Chishti AH, Li TS. Molecular characterization of abLIM, a novel actin-binding and double zinc finger protein. J Cell Biol. 1997; 138: 575–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trojan P, Giessl A, Pulvermuller A, Hofmann KP, Wolfrum U. Light-dependent phosphorylation of Centrins in vertebrate retina. Eur J Cell Biol. 2003; 82: 76–76 [Google Scholar]
- 34. Hayashi F, Matsuura I, Kachi S, et al. Phosphorylation by cyclin-dependent protein kinase 5 of the regulatory subunit of retinal cGMP phosphodiesterase II. Its role in the turnoff of phosphodiesterase in vivo. J Biol Chem. 2000; 275: 32958–32965 [DOI] [PubMed] [Google Scholar]
- 35. Benaim G, Villalobo A. Phosphorylation of calmodulin - functional implications. Eur J Biochem. 2002; 269: 3619–3631 [DOI] [PubMed] [Google Scholar]
- 36. Allen D, Fakler B, Maylie J, Adelman JP. Organization and regulation of small conductance Ca2+-activated K+ channel multiprotein complexes. J Neurosci. 2007; 27: 2369–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bildl W, Strassmaier T, Thurm H, et al. Protein kinase CK2 is coassembled with small conductance Ca2+-activated K+ channels and regulates channel gating. Neuron. 2004; 43: 847–858 [DOI] [PubMed] [Google Scholar]
- 38. Quadroni M, L'Hostis EL, Corti C, et al. Phosphorylation of calmodulin alters its potency as an activator of target enzymes. Biochemistry. 1998; 37: 6523–6532 [DOI] [PubMed] [Google Scholar]
- 39. Lee A, Jimenez A, Cui G, Haeseleer F. Phosphorylation of the Ca2+-binding protein CaBP4 by protein kinase C zeta in photoreceptors. J Neurosci. 2007; 27: 12743–12754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kersten FFJ, van Wijk E, van Reeuwijk J, et al. Association of whirlin with Ca(v)1.3 (alpha1D) channels in photoreceptors, defining a novel member of the usher protein network. Invest Ophthalmol Vis Sci. 2010; 51: 2338–2346 [DOI] [PubMed] [Google Scholar]
- 41. Morgans CW. Calcium channel heterogeneity among cone photoreceptors in the tree shrew retina. Eur J Neurosci. 1999; 11: 2989–2993 [DOI] [PubMed] [Google Scholar]
- 42. Morgans CW, El Far O, Berntson A, Wassle H, Taylor WR. Calcium extrusion from mammalian photoreceptor terminals. J Neurosci. 1998; 18: 2467–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xiao HL, Chen XM, Steele EC. Abundant L-type calcium channel Ca-v 1.3 (alpha 1D) subunit mRNA is detected in rod photoreceptors of the mouse retina via in situ. Mol Vis. 2007; 13: 764–771 [PMC free article] [PubMed] [Google Scholar]
- 44. Cohen P, Holmes CFB, Tsukitani Y. Okadaic acid- a new probe for the study of cellular regulation. Trends Biochem Sci. 1990; 15: 98–102 [DOI] [PubMed] [Google Scholar]
- 45. Cohen P, Klumpp S, Schelling DL. An improved procedure for identifying and quantitating protein phosphatases in mammalian tissues. Febs Lett. 1989; 250: 596–600 [DOI] [PubMed] [Google Scholar]
- 46. Foulkes JG, Cohen P. The regulation of glycogen metabolism - purification and properties of protein phosphatase inhibitor-2 from rabbit skeletal muscle. Eur J Biochem. 1980; 105: 195–203 [DOI] [PubMed] [Google Scholar]
- 47. Helps NR, Street AJ, Elledge SJ, Cohen PTW. Cloning of the complete coding region for human protein phosphatase inhibitor-2 using the 2-hybrid system and expression of inhibitor-2 in Escherichia coli. Febs Lett. 1994; 340: 93–98 [DOI] [PubMed] [Google Scholar]
- 48. Vaneynde A, Wera S, Beullens M, et al. Molecular cloning of NIPP-1, a nuclear inhibitor of protein phosphatase 1, reveals homology with polypeptides involved in RNA processing. J Biol Chem. 1995; 270: 28068–28074 [DOI] [PubMed] [Google Scholar]
- 49. Swingle M, Ni L, Honkanen RE. Small-molecule inhibitors of ser/thr protein phosphatases: specificity, use and common forms of abuse. Methods Mol Biol. 2007; 365: 23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cohen PTW. Novel protein serine/threonine phosphatases: variety is the spice of life. Trends Biochem Sci. 1997; 22: 245–251 [DOI] [PubMed] [Google Scholar]
- 51. Shi YG. Serine/threonine phosphatases: mechanism through structure. Cell. 2009; 139: 468–484 [DOI] [PubMed] [Google Scholar]
- 52. Slupe AM, Merrill RA, Strack S. Determinants for substrate specificity of protein phosphatase 2A. Enzyme Res. 2011; 398751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sontag E. Protein phosphatase 2A: the Trojan Horse of cellular signaling. Cell Signal. 2001; 13: 7–16 [DOI] [PubMed] [Google Scholar]
- 54. Brown BM, Carlson BL, Zhu XM, Lolley RN, Craft CM. Light-driven translocation of the protein phosphatase 2A complex regulates light/dark dephosphorylation of phosducin and rhodopsin. Biochemistry. 2002; 41: 13526–13538 [DOI] [PubMed] [Google Scholar]
- 55. Liu WB, Li Y, Zhang L, et al. Differential expression of the catalytic subunits for PP-1 and PP-2A and the regulatory subunits for PP-2A in mouse eye. Mol Vis. 2008; 14: 762–773 [PMC free article] [PubMed] [Google Scholar]
- 56. Shi J, Gu PY, Zhu ZH, et al. Protein phosphatase 2A effectively modulates basal L-type Ca2+ current by dephosphorylating Ca(v)1.2 at serine 1866 in mouse cardiac myocytes. Biochem Biophys Res Commun. 2012; 418: 792–798 [DOI] [PubMed] [Google Scholar]
- 57. Choi SY, Jackman S, Thoreson WB, Kramer RH. Light regulation of Ca(2+) in the cone photoreceptor synaptic terminal. Vis Neurosci. 2008; 25: 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Krizaj D, Copenhagen DR. Calcium regulation in photoreceptors. Front Biosci. 2002; 7: D2023–D2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mercer AJ, Thoreson WB. The dynamic architecture of photoreceptor ribbon synapses: cytoskeletal, extracellular matrix, and intramembrane proteins. Vis Neurosci. 2011; 28: 453–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xia XM, Fakler B, Rivard A, et al. Mechanism of calcium gating in small-conductance calcium-activated potassium channels. Nature. 1998; 395: 503–507 [DOI] [PubMed] [Google Scholar]
- 61. Cooper NGF, McLaughlin BJ, Tallant EA, Cheung WY. Calmodulin-dependent protein phosphatase - immunocytochemical localization in chick retina. J Cell Biol. 1985; 101: 1212–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fowles C, Akhtar M, Cohen P. Interplay of phosphorylation and dephosphorylation in vision - protein phosphatases of bovine rod outer segments. Biochemistry. 1989; 28: 9385–9391 [DOI] [PubMed] [Google Scholar]
- 63. Klumpp S, Selke D, Fischer D, Baumann A, Muller F, Thanos S. Protein phosphatase type-2C isozymes present in vertebrate retinae: purification, characterization, and localization in photoreceptors. J Neurosci Res. 1998; 51: 328–338 [DOI] [PubMed] [Google Scholar]
- 64. Palczewski K, Hargrave PA, McDowell JH, Ingebritsen TS. The catalytic subunit of phosphatase 2A dephosphorylates phosphoopsin. Biochemistry. 1989; 28: 415–419 [DOI] [PubMed] [Google Scholar]
- 65. Selke D, Anton H, Klumpp S. Serine/threonine protein phosphatases type 1, 2A and 2C in vertebrate retinae. Acta Anatomica. 1998; 162: 151–156 [DOI] [PubMed] [Google Scholar]
- 66. Maki K, Motoki R, Fujii K, et al. Catalyst-controlled asymmetric synthesis of fostriecin and 8-epi-fostriecin. J Am Chem Soc. 2005; 127: 17111–17117 [DOI] [PubMed] [Google Scholar]
- 67. Hastie CJ, Cohen PTW. Purification of protein phosphatase 4 catalytic subunit: inhibition by the antitumour drug fostriecin and other tumour suppressors and promoters. Febs Lett. 1998; 431: 357–361 [DOI] [PubMed] [Google Scholar]
- 68. Prickett TD, Brautigan DL. The alpha 4 regulatory subunit exerts opposing allosteric effects on protein phosphatases PP6 and PP2A. J Biol Chem. 2006; 281: 30503–30511 [DOI] [PubMed] [Google Scholar]
- 69. Ingebritsen TS, Stewart AA, Cohen P. The protein phosphatases involved in cellular regulation. 6. Measurement of type-1 and type-2 protein phosphatases in extracts of mammalian tissues - an assessment of their physiological roles. Eur J Biochem. 1983; 132: 297–307 [DOI] [PubMed] [Google Scholar]
- 70. Kowluru A, Metz SA. Purine nucleotide- and sugar phosphate-induced inhibition of the carboxyl methylation and catalysis of protein phosphatase-2A in insulin-secreting cells: protection by divalent cations. Bioscience Rep. 1998; 18: 171–186 [DOI] [PubMed] [Google Scholar]
- 71. Tolstykh T, Lee J, Vafai S, Stock JB. Carboxyl methylation regulates phosphoprotein phosphatase 2A by controlling the association of regulatory B subunits. Embo J. 2000; 19: 5682–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yu XX, Du X, Moreno CS, et al. Methylation of the protein phosphatase 2A catalytic subunit is essential for association of B alpha regulatory subunit but not SG2NA, striatin, or polyomavirus middle tumor antigen. Mol Biol Cell. 2001; 12: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bryant JC, Westphal RS, Wadzinski BE. Methylated C-terminal leucine residue of PP2A catalytic subunit is important for binding of regulatory B alpha subunit. Biochem J. 1999; 339: 241–246 [PMC free article] [PubMed] [Google Scholar]
- 74. Reece KM, Mazalouskas MD, Wadzinski BE. The B alpha and B delta regulatory subunits of PP2A are necessary for assembly of the CaMKIV.PP2A signaling complex. Biochem Biophys Res Commun. 2009; 386: 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xu YH, Chen Y, Zhang P, Jeffrey PD, Shi YG. Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation. Mol Cell. 2008; 31: 873–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sontag E, Nunbhakdicraig V, Bloom GS, Mumby MC. A novel pool of protein phosphatase 2A is associated with microtubules and is regulated during the cell-cycle. J Cell Biol. 1995; 128: 1131–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Choi SY, Borghuis B, Rea R, Levitan ES, Sterling P, Kramer RH. Encoding light intensity by the cone photoreceptor synapse. Neuron. 2005; 48: 555–562 [DOI] [PubMed] [Google Scholar]
- 78. Doering CJ, Hamid J, Simms B, McRory JE, Zamponi GW. Ca(v)1.4 encodes a calcium channel with low open probability and unitary conductance. Biophys J. 2005; 89: 3042–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Singh A, Hamedinger D, Hoda JC, et al. C-terminal modulator controls Ca2+-dependent gating of Ca(v)1.4 L-type Ca2+ channels. Nat Neurosci. 2006; 9: 1108–1116 [DOI] [PubMed] [Google Scholar]
- 80. Wahl-Schott C, Baumann L, Cuny H, Eckert C, Griessmeier K, Biel M. Switching off calcium-dependent inactivation in L-type calcium channels by an autoinhibitory domain. Proc Natl Acad Sci U S A. 2006; 103: 15657–15662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tan GMY, Yu DJ, Wang JJ, Soong TW. Alternative splicing at C terminus of Ca(V)1.4 calcium channel modulates calcium-dependent inactivation, activation potential, and current density. J Biol Chem. 2012; 287: 832–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Adao-Novaes J, Guterrres CDB, Linden R, Sholl-Franco A. Rod photoreceptor cell death is induced by okadaic acid through activation of PKC and L-type voltage-dependent Ca(2+) channels and prevented by IGF-1. Neurochem Int. 2010; 57: 128–135 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.