Abstract
The genetic etiology of complex human diseases has been commonly viewed as a process that involves multiple genetic variants, environmental factors, as well as their interactions. Statistical approaches, such as the multifactor dimensionality reduction (MDR) and generalized MDR (GMDR), have recently been proposed to test the joint association of multiple genetic variants with either dichotomous or continuous traits. In this paper, we propose a novel Forward U-Test to evaluate the combined effect of multiple loci on quantitative traits with consideration of gene-gene/gene-environment interactions. In this new approach, a U-Statistic-based forward algorithm is first used to select potential disease-susceptibility loci and then a weighted U statistic is used to test the joint association of the selected loci with the disease. Through a simulation study, we found the Forward U-Test outperformed GMDR in terms of greater power. Aside from that, our approach is less computationally intensive, making it feasible for high-dimensional gene-gene/gene-environment research. We illustrate our method with a real data application to Nicotine Dependence (ND), using three independent datasets from the Study of Addiction: Genetics and Environment. Our gene-gene interaction analysis of 155 SNPs in 67 candidate genes identified two SNPs, rs16969968 within gene CHRNA5 and rs1122530 within gene NTRK2, jointly associated with the level of ND (p-value = 5.31e-7). The association, which involves essential interaction, is replicated in two independent datasets with p-values of 1.08e-5 and 0.02, respectively. Our finding suggests that joint action may exist between the two gene products.
Keywords: gene-gene interaction, Forward U-Test, Nicotine Dependence
INTRODUCTION
The genetic etiology of common complex human diseases has been of tremendous interest to clinical and basic science researchers as well as to the general public. In the past few years, the radical breakthrough of biotechnologies has enabled us to generate almost unlimited genotypic data with great accuracy [Schuster 2008]. Testing the association between these genetic variants and complex traits provides an unprecedented opportunity to unravel the hidden secret of gene functions, which would be crucial for a better understanding of the disease etiology. Meanwhile, the rapid growth of the data dimensionality also presents daunting challenges to statistical modeling and hypothesis testing.
Most of the first generation genome wide association studies have tested the association between genetic variants and disease outcome on a single-locus basis [Barrett, et al. 2008; Easton, et al. 2007; Zeggini, et al. 2008]. Though a substantial number of genetic variants have been identified to be associated with the development of many complex diseases, such as diabetes and Crohn’s disease, the current findings have accounted for only a small proportion of heritability [Manolio, et al. 2009]. One possible reason is that most of the complex human diseases are polygenic in nature. Multiple genetic variants, each conferring a small or moderate effect, may contribute to the disease development [Moore 2003; Nagel 2005]. In addition, the effect of one genetic variant could be suppressed or enhanced by the existence of others, which is termed epistasis [Bateson and Mendel 1909]. Whereas epistasis per se cannot account for missing additive heritability, it may often lead to lack of power to identify association when loci are examined individually without considering their potential interactions [Chatterjee, et al. 2006].
Considering the probable polygenic nature of many human diseases, statistical approaches for multi-locus association analysis have been recently developed. Lin et al. [Lin and Wu 2006] proposed a sequence interaction model in a multivariate regression framework for quantitative traits. Several studies have modeled multi-locus interactions through haplotype analysis [Li, et al. 2010a; Tzeng, et al. 2006; Zhang, et al. 2003]. Schaid et al. proposed a U-statistic-based score test that can simultaneously examine the association of multiple genetic variants with dichotomous traits [Schaid, et al. 2005]. Wei et al. further extended this approach for quantitative traits by using data-adaptive weights for different variants [Wei, et al. 2008]. These approaches comprise the commonly used single-locus methods, providing powerful alternatives for genetic association analysis. Their limitation is, however, that they are less suitable for handling a large number of genetic variants and for considering interactions, especially high order interactions.
Another group of methods uses a different strategy, by first selecting a subset of genetic variants from the totality of genotyped variants and then conducting an association test to assess the combined effect of the selected loci with disease. The subset is usually selected to best describe the risk of binary disease outcomes or the variation of quantitative traits. For example, Ritchie et al. proposed a Multifactor Dimensionality Reduction (MDR) method for balanced case-control studies [Ritchie, et al. 2001]. It pools multi-locus genotypes into high-risk and low-risk groups, and hence reduces the data dimensions to one. This method has been widely used and further extended in a series of articles. Martin et al. extended the MDR method for family-based designs [Martin, et al. 2006]. Lou et al. derived a generalized MDR (GMDR) method that can be applied to both dichotomous and quantitative traits [Lou, et al. 2007]. The GMDR method is not limited to studies with a balanced design and has the advantage of allowing for covariate adjustment.. It maps the phenotypic traits into residual scores through certain link functions under a generalized linear model framework, and then conducts SNP selection and association testing based on the residual scores. The extension of GMDR can also be applied for family-based designs, referred to as pedigree-based GMDR (PGMDR) [Lou, et al. 2008]. These approaches have now been commonly used to search for gene-gene/gene-environment interactions. They are generally non-parametric and model free.
These advantages aside, the above methods commonly use an exhaustive search algorithm. When the number of genetic variants is large, the chances are that a model of irrelevant combination may outperform the real disease model simply due to sample randomness. Therefore, when dealing with hundreds of thousands of genetic variants and environmental factors, an exhaustive search may suffer from loss of power due to the substantial increase in the feature space [Wu and Zhao 2009]. In addition, an exhaustive search may not be computationally feasible for high order interaction, especially at a genome-wide scale. As discussed by Cordell and Marchini et al., high order epistasis beyond pair-wise interactions would not be computationally affordable and can be pursued only after single-locus-based filtering [Cordell 2009; Marchini, et al. 2005].
As an alternative approach, the forward or sequential selection algorithm has received growing attention for its computational efficiency [Brem, et al. 2005; Lu and Elston 2008; Storey, et al. 2005]. The algorithm starts with a null feature set and sequentially adds the best feature that satisfies certain criteria. Real data applications and simulation studies have also suggested that forward search may have greater power than exhaustive search [Storey, et al. 2005; Wu and Zhao 2009]. In this paper, we propose a U-statistic-based multi-locus testing approach for quantitative traits. It searches a relatively large number of SNPs for joint gene-gene action through forward selection. It has the following advantages: (1) It tests the overall association for multiple genetic variants together with any interaction effects, including high-order interactions; (2) It is a non-parametric method that makes no assumptions about the trait distribution; and (3) It is computationally efficient and can be applied to high-dimensional data.
METHODS
We first introduce notation and the hypothesis of interest. Suppose the study has N subjects. Let Yi denote the quantitative trait for the ith subject, i=1,2,......,N; and let Xi = (Xi1, Xi2 ...... XiK) denote K independent SNP genotypes, each taking a value from one of the three possible genotypes Xij ∈ {AA,Aa,aa} j = 1,2,......,K. The hypothesis is whether these K SNPs, or a subset of them, are associated with the quantitative trait Y. To test this hypothesis, we 1) first select k out of the K SNPs that best describe the variation of Y, where k ≤ K; and 2) test whether these selected SNPs are jointly associated with the trait Y. In what follows, we explain our method that conducts the selection and test simultaneously.
U-Statistics
Since the foundational work of Hoeffding [Hoeffding 1948], U-Statistics have been widely used in both theoretical and applied statistical research. They have been recently used to build test statistics for multiple genetic variants [Schaid, et al. 2005; Wei, et al. 2008]. However, while considering multiple genetic variants simultaneously, these approaches calculated the global U-Statistic by assuming additive across multiple genetic variants, and thus did not consider the gene-gene interaction. We here introduce a new U-Statistic to test joint association of multiple genetic variants with the consideration of possible gene-gene interactions. In this new method, we measure the difference of the quantitative trait between two individuals i and j as:
Suppose we have k selected SNPs, which comprise L multi-SNP genotypes, denoted by G1,G2,......,GL. A multi-SNP genotype,Gl, is defined as a vector of k single-SNP genotypes that an individual carries (e.g., {g1, g2,…, gk}). The k SNPs and L multi-SNP genotypes are selected sequentially out of a total of K genotyped SNPs (See Section below for details). We denote by Sl={i,Xi=Gl} the group of subjects carrying multi-SNP genotype Gl, l=1,2, ......,L and m=|Sl| the number of subjects in group Sl. We define the between-group U-statistic for group l and group l′ as:
Ul,l′ is the summation of all possible pair-wise trait differences for any two subjects from Sl and Sl′. In the presence of association, we would expect different trait values for individuals carrying different multi-SNP genotype (e.g., those carrying high risk multi-SNP genotype have higher trait values than those carrying low risk multi-SNP genotype). We assume that the expected quantitative trait value of the L multi-SNP genotypes decreases with l (i.e., E(YS1) ≥ E(YS2) ≥ ...... ≥ E(YSL)). Practically, we can sort the multi-SNP genotypes according to their average trait values (i.e., ȲS1 ≥ ȲS2 ≥ ...... ≥ ȲSL). Based on Ul,l′, we further define the global U-statistic for L groups as:
Here, the weight parameter ω is chosen to account for the number of subjects in each genotype group. The above U-Statistic is defined to measure the overall trait differences among a total of L multi-SNP genotype groups.
U-Statistic Based Forward Selection Algorithm
When dealing with a large number of SNPs, it is likely that a significant proportion of the SNPs are not disease-related, and thus conducting a model selection will be necessary. We here introduce a computationally efficient U-Statistic-based forward selection algorithm that is capable of searching among a large number of SNPs for disease-susceptibility loci that best describe the variation of the quantitative trait. We start by taking all individuals as a single genotype group. In the first step, each SNP j can form two single-SNP genotypes,{ }, in three possible ways, denoted as and { }. This leads to a total of 3K possible partitions that can be represented by{ }, where denotes the lth multi-SNP genotype at step s. We calculate the U-Statistic for each partition { }. The SNP with the largest value of this U-statistic is selected, and the corresponding partition is recorded. In the second step, based on the first selected SNP, a second SNP j′ is chosen to form four two-SNP genotypes, denoted by{ }. It should be noted that, if the same SNP from step one is chosen in step two, only three single-SNP genotypes will be formed, denoted by { }. We screen all SNPs and calculate the U-statistic for each of these partitions. The SNP that increases the U-statistic the most is chosen, together with its corresponding partition. As the algorithm moves forward, the overall U-Statistic is expected to increase until all the genotype groups are separated. The largest number of possible genotype groups will be 3K.
We use a 10-fold cross-validation (CV) procedure to decide when the selection algorithm should be stopped. In this procedure, all the subjects are randomly divided into 10 subgroups. Nine of the ten subgroups are used as a training set, while the remaining one is used as the testing set. The process is repeated 10 times to make sure all subgroups have served as a testing set. The U-statistics are calculated and averaged over the testing sets based on the selected model determined from the corresponding training sets. The selection algorithm is stopped when the overall U-statistic averaged over the testing sets ceases to increase. After the number of forwarding steps is determined, an overall U-statistic is calculated on the whole dataset including all the subjects. An empirical p-value, which accounts for inflated Type I Error due to model selection and genotype groups ordering, can also be obtained by sampling the permutation distribution. In a replication study when the sequence of genotype partitions is pre-determined from an initial study, under the null, the corresponding global U-Statistic is expected to have a zero mean and asymptotically follow a normal distribution. The significance level of the association could be tested by using the asymptotic distribution of the U-Statistic. For simplicity, we denote , and its variance is estimated as:
where Var (Yi) = σ2 for any 1≤i≤N. The derivation is described in Appendix.
Note that, although the illustration above is specified for joint gene-gene action, the same procedure is also valid for joint gene-environment action. Similar to genetic variables, environmental factors with categorical or ordinal levels can be directly analyzed. For continuous environmental factors, however, we need to first cluster them into different levels and then put them into the model as discrete variables.
RESULTS
Simulation Results
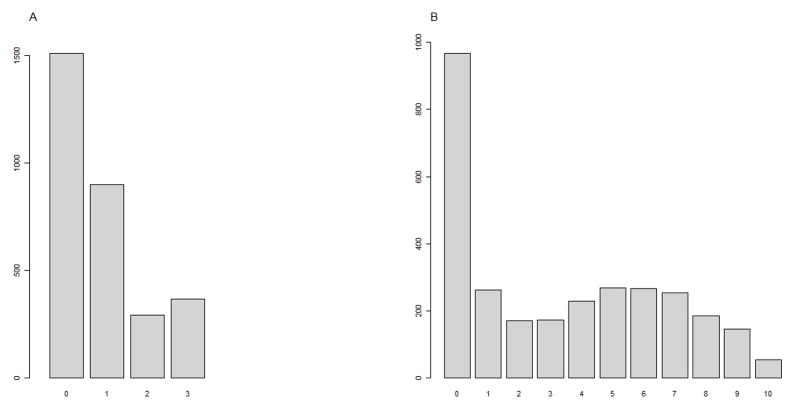
We conducted two sets of simulations to evaluate the performance of our proposed method, and compared it with a commonly used approach, GMDR. The first set of simulations compared the performance of the two approaches under various underlying disease models. The second set of simulations evaluated the performance of the two approaches when the distribution of the quantitative trait is unknown. The quantitative traits for the second set of simulations were simulated according to the distributions of two traits from the Study of Addiction: Genetics and Environment (SAGE) dataset. The two traits used were ‘number of cigarettes smoked per day’ and ‘lifetime Fagerström Test for Nicotine Dependence (FTND) score’. The trait distributions in SAGE are illustrated in Figure 1.
Figure 1. Trait distribution in Simulation II.
A: the distribution of the number of cigarette smoked per day;
B: the distribution of Participants’ life-time score of FTND.
Simulation I
In the first set of simulations, we considered a variety of underlying disease models, starting with three types of two-locus SNP models (Table 1) introduced by Marchini et al (i.e., multiplicative-effect model, additive-effect model and threshold-effect model) [Marchini, et al. 2005]. We are here assuming only one SNP in each locus. To mimic more complex disease scenarios, we also simulated two three-locus models and two four-locus models. The two three-locus models, which are extensions of the two-locus models to three loci, were simulated with multiplicative and additive effects, respectively. Each of the four-locus models comprises two two-locus models (i.e., two two-way joint actions). We simulated the two-locus models of the first and second four-locus models with multiplicative and addictive effects, respectively. We further assumed the effects between the two two-locus models for the first and second four-locus models follow an addictive model and a multiplicative model, respectively. The multi-SNP genotypes were simulated under the assumption of joint Hardy-Weinberg Equilibrium (HWE). For the two-locus models, the minor allele frequencies for the risk loci were set at 0.4 and 0.3. For the three-locus models, they were set at 0.4, 0.5 and 0.3. For the four-locus models, they were set at (0.4, 0.3) and (0.3, 0.4) for each of the two-locus models, respectively. The allele frequencies remained fixed in this study unless specified otherwise. Noise loci were also introduced to mimic real data application. The minor allele frequencies of the SNPs at the noise loci were simulated from a uniform distribution ranging from 0.1 to 0.9. The number of noise loci was adjusted to ensure the total number of SNPs was always ten. A total of L multi-locus SNP genotypes were formed from the simulated SNPs at the ten loci,{G1,G2,......GL}, corresponding to different levels of the quantitative trait. Assuming multi-locus group l had an expected trait value of μl, calculated based on the simulated setting (e.g., additive-effect model), we simulated quantitative traits for a reference population of one million subjects as:
where εi ~ N(0,1) and I{·} is an indicator function. The Forward U-Test and GMDR were applied to 1000 subjects randomly selected from the reference population. For each underlying disease model, the simulation was repeated 1000 times with 1000 permutations. For both methods, the association was significant if the test statistic exceeded the 95-th percentile of the corresponding permutation distribution. The power was then calculated as the probability to detect the overall association based on 1000 replicates. In a similar manner, we calculated the type I error by only considering non-causal loci in the model.
Table 1.
Average trait values for two-locus joint action models
| Two-locus joint action with multiplicative effects | Two-locus joint action with additive effects | Two-locus joint action with a threshold effect | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| bb | Bb | BB | bb | Bb | BB | bb | Bb | BB | |||
| aa | α | α(1+θ21) | α(1+θ21)(1+θ22) | aa | α | α+θ21 | α+θ22 | aa | α | α | α |
| Aa | α(1+ θ11) | α(1+θ11)(1+θ22) | α(1+θ11)(1+θ22) | Aa | α+θ11 | α+θ11+θ22 | α+θ11+θ22 | Aa | α | α (1+θ) | α (1+θ) |
| AA | α(1+ θ11)(1+θ12) | α(1+θ12)(1+θ21) | α(1+θ12)(1+θ22) | AA | α+θ12 | α+θ12+θ21 | α+θ12+θ22 | AA | α | α (1+θ) | α (1+θ) |
The simulation results are summarized in Table 2. We report power, Type I error, sensitivity and specificity. The sensitivity (specificity) was calculated as the probability of selecting (not selecting) a causal (non-causal) SNP. U-statistics and Testing Balanced Accuracy (default in GMDR) were used as test statistics to examine the significance level of the two methods. For GMDR, since the quantitative traits were simulated under a normal distribution, an identity link was used to calculate the scores statistics. The simulation results has showed that, compared to GMDR, the Forward U-Test significantly increased the test power under multiplicative and additive models, while properly controlling the Type I error. For the threshold effect model, GMDR and the Forward U-Test had comparable power. In terms of selection accuracy, the sensitivities of GMDR tended to be higher than those of Forward U-Test, with a few exceptions when both of the causal SNPs have strong marginal effects. However, the specificities of Forward U-Test were consistently higher than those of GMDR, and were greater than 0.95 in all scenarios. On the other hand, the specificities of GMDR were significantly reduced when the effect size decreased or the complexity of disease model increased. This result indicated that Forward U-Test had a low false positive rate for SNP selection, which could partially explain the increase rather than loss of testing power over GMDR despite of the relatively lower sensitivities, because less noise was selected into the final model. The power increased in most scenarios could also be explained by allowing for more than two risk groups in the model. As illustrated in Table 2, Forward U-Test attained significant power increase over GMDR under disease models with more than two risk groups, with the exception of the two-locus threshold model containing only two risk groups.
Table 2.
Comparison of the Forward U-Test with GMDR under different disease models
| Disease Model | Forward U-Test | GMDR | Disease Model | Forward U-Test | GMDR | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Two-locus Multiplicative
|
Power | 0.947 | 0.781 | Two-locus Additive
|
Power | 0.991 | 0.824 | ||||||
| Type I Err. | 0.026 | 0.038 | Type I Err. | 0.038 | 0.045 | ||||||||
| Average trait 1a | 1 | 1.1 | 1.2 | Sensitivity | 0.564 | 0.604 | Average trait 1 | 1 | 1.3 | 1.4 | Sensitivity | 0.915 | 0.817 |
| Average trait 2 | 1 | 1.3 | 1.4 | Specificity | 0.968 | 0.921 | Average trait 2 | 1 | 1.3 | 1.4 | Specificity | 0.978 | 0.878 |
|
| |||||||||||||
| Two-locus Multiplicative
|
Power | 0.878 | 0.462 | Two-locus Additive
|
Power | 0.762 | 0.267 | ||||||
| Type I Err. | 0.042 | 0.051 | Type I Err. | 0.074 | 0.061 | ||||||||
| Average trait 1 | 1 | 1.2 | 1.3 | Sensitivity | 0.794 | 0.780 | Average trait 1 | 1 | 1.2 | 1.3 | Sensitivity | 0.749 | 0.758 |
| Average trait 2 | 1 | 1.2 | 1.3 | Specificity | 0.970 | 0.843 | Average trait 2 | 1 | 1.2 | 1.3 | Specificity | 0.964 | 0.803 |
|
| |||||||||||||
| Two-locus Multiplicative
|
Power | 0.623 | 0.342 | Two-locus Additive
|
Power | 0.474 | 0.098 | ||||||
| Type I Err. | 0.059 | 0.048 | Type I Err. | 0.06 | 0.044 | ||||||||
| Average trait 1 | 1 | 1.1 | 1.2 | Sensitivity | 0.589 | 0.695 | Average trait 1 | 1 | 1.1 | 1.2 | Sensitivity | 0.562 | 0.672 |
| Average trait 2 | 1 | 1.2 | 1.3 | Specificity | 0.961 | 0.789 | Average trait 2 | 1 | 1.2 | 1.3 | Specificity | 0.954 | 0.746 |
|
| |||||||||||||
| Two-locus Threshold Model
|
Power | 0.808 | 0.871 | Two-locus Threshold Model
|
Power | 0.412 | 0.385 | ||||||
| Type I Err. | 0.049 | 0.058 | Type I Err. | 0.057 | 0.031 | ||||||||
| RAFb | 0.4 | 0.4 | Sensitivity | 0.609 | 0.992 | RAF | 0.5 | 0.5 | Sensitivity | 0.554 | 0.897 | ||
|
|
|
||||||||||||
| Average trait | 1.5 | Specificity | 0.984 | 0.935 | Average trait | 1.3 | Specificity | 0.972 | 0.838 | ||||
|
| |||||||||||||
| Three-locus Multiplicative
|
Power | 0.612 | 0.198 | Three-locus Additive
|
Power | 0.672 | 0.228 | ||||||
| Type I Err. | 0.042 | 0.051 | Type I Err. | 0.054 | 0.045 | ||||||||
| Average trait 1 | 1 | 1.1 | 1.1 | Sensitivity | 0.408 | 0.609 | Average trait 1 | 1 | 1.1 | 1.2 | Sensitivity | 0.454 | 0.610 |
| Average trait 2 | 1 | 1.1 | 1.2 | Specificity | 0.960 | 0.741 | Average trait 2 | 1 | 1.2 | 1.2 | Specificity | 0.967 | 0.799 |
| Average trait 3 | 1 | 1.2 | 1.2 | Average trait 3 | 1 | 1.2 | 1.3 | ||||||
|
| |||||||||||||
| Two-locus × Two-locus Multiplicative/Additive
|
Power | 0.871 | 0.438 | Two-locus × Two-locus Additive/Multiplicative
|
Power | 0.838 | 0.483 | ||||||
| Type I Err. | 0.046 | 0.045 | Type I Err. | 0.059 | 0.050 | ||||||||
| Average trait 1-1 | 1 | 1.1 | 1.2 | Sensitivity | 0.474 | 0.568 | Average trait 1-1 | 1 | 1.1 | 1.2 | Sensitivity | 0.423 | 0.533 |
| Average trait 1–2 | 1 | 1.2 | 1.3 | Specificity | 0.978 | 0.840 | Average trait 1–2 | 1 | 1.2 | 1.3 | Specificity | 0.976 | 0.840 |
| Average trait 2-1 | 1 | 1.2 | 1.3 | Average trait 2-1 | 1 | 1.1 | 1.3 | ||||||
| Average trait 2-2 | 1 | 1.1 | 1.2 | Average trait 2-2 | 1 | 1.1 | 1.2 | ||||||
Average trait value for genotype AA, Aa, aa of 1st causal SNP.
Risk allele frequency for causal SNPs.
Simulation II
For common complex diseases, the trait distribution is commonly unknown or hard to determine. We conducted a second set of simulations to compare the performance of the methods when the trait distribution is unknown. Two quantitative traits were simulated according to the distributions of the two variables ‘number of cigarettes smoked per day’ and ‘lifetime Fagerstrom Test for Nicotine Dependence (FTND) score’ in SAGE. For each trait, two-SNP disease models with three types of joint action effects, multiplicative, additive and threshold, were used for the comparison. Because of the unknown trait distribution, various link functions were used to calculate the residual scores for GMDR, including zero inflated Poisson, Poisson, negative binomial, and Gamma. The residual score for zero inflated Poisson was calculated with the package ‘pscl’ in R [Zeileis, et al. 2008].
The simulation results are summarized in Table 3 and Table 4. For both traits, the Forward U-Test attained greater power than the GMDR, especially under two-SNP models with additive or multiplicative effect. For the given trait distributions, GMDR had its best performance on assuming zero-inflated Poisson. When the underlying disease model was the threshold model, GMDR with a zero-inflated Poisson link could reach the same power as the Forward U-Test. However, the power could be significantly reduced for GMDR on using an inappropriate link function. In all scenarios, the specificities of Forward U-Test were greater than 0.95 and were consistently higher than those of GMDR. In terms of sensitivity, the performance largely varied, depending on the underlying disease model, effect size and link functions.
Table 3.
Comparison of the Forward U-Test with GMDR when the quantitative traits are simulated from the distribution of number of cigarette smoked per day
| Disease Model | Forward U-Test | GMDR (Zero Infl. Poisson) | GMDR (Poisson) | GMDR (Negative Binomial) | GMDR (Gamma) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Two-locus Multiplicative
|
Power | 0.930 | 0.540 | 0.289 | 0.217 | 0.355 | |||
| Type I Err. | 0.056 | 0.056 | 0.079 | 0.061 | 0.052 | ||||
| Relative Risk 1 | 1 | 1.1 | 1.2 | Sensitivity | 0.562 | 0.634 | 0.546 | 0.511 | 0.659 |
| Relative Risk 2 | 1 | 1.3 | 1.4 | Specificity | 0.957 | 0.870 | 0.910 | 0.938 | 0.841 |
|
| |||||||||
| Two-locus Threshold
|
Power | 0.952 | 0.924 | 0.526 | 0.291 | 0.843 | |||
| Type I Err. | 0.050 | 0.054 | 0.063 | 0.074 | 0.064 | ||||
| RAF | 0.6 | 0.6 | Sensitivity | 0.754 | 0.982 | 0.781 | 0.623 | 0.952 | |
|
|
|||||||||
| Relative Risk | 1.5 | Specificity | 0.967 | 0.944 | 0.918 | 0.943 | 0.931 | ||
|
| |||||||||
| Two-locus Additive
|
Power | 0.948 | 0.694 | 0.343 | 0.247 | 0.579 | |||
| Type I Err. | 0.056 | 0.069 | 0.066 | 0.086 | 0.063 | ||||
| Relative Risk 1 | 1 | 1.3 | 1.4 | Sensitivity | 0.749 | 0.789 | 0.646 | 0.570 | 0.764 |
| Relative Risk 2 | 1 | 1.3 | 1.4 | Specificity | 0.966 | 0.847 | 0.920 | 0.932 | 0.870 |
Table 4.
Comparison of the Forward U-Test with GMDR when the quantitative traits are simulated from the distribution of life-time FTND scores
| Disease Model | Forward U-Test | GMDR (Zero Infl. Poisson) | GMDR (Poisson) | GRMD (Negative Binomial) | GMDR (Gamma) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Two-locus Multiplicative
|
Power | 0.875 | 0.624 | 0.421 | 0.126 | 0.297 | |||
| Type I Err. | 0.039 | 0.064 | 0.046 | 0.048 | 0.047 | ||||
| Relative Risk 1 | 1 | 1.1 | 1.2 | Sensitivity | 0.547 | 0.611 | 0.648 | 0.530 | 0.560 |
| Relative Risk 2 | 1 | 1.3 | 1.4 | Specificity | 0.951 | 0.885 | 0.860 | 0.963 | 0.929 |
|
| |||||||||
| Two-locus Threshold
|
Power | 0.779 | 0.780 | 0.107 | 0.45 | 0.064 | |||
| Type I Err. | 0.048 | 0.055 | 0.061 | 0.057 | 0.068 | ||||
| MAF | 0.4 | 0.4 | Sensitivity | 0.609 | 0.984 | 0.496 | 0.465 | 0.462 | |
|
|
|||||||||
| Relative Risk | 1.5 | Specificity | 0.981 | 0.904 | 0.907 | 0.972 | 0.955 | ||
|
| |||||||||
| Two-locus Additive
|
Power | 0.971 | 0.657 | 0.583 | 0.160 | 0.369 | |||
| Type I Err. | 0.036 | 0.060 | 0.073 | 0.063 | 0.081 | ||||
| Relative Risk 1 | 1 | 1.3 | 1.4 | Sensitivity | 0.853 | 0.813 | 0.803 | 0.556 | 0.664 |
| Relative Risk 2 | 1 | 1.3 | 1.4 | Specificity | 0.980 | 0.853 | 0.871 | 0.964 | 0.916 |
Application to Nicotine Dependence
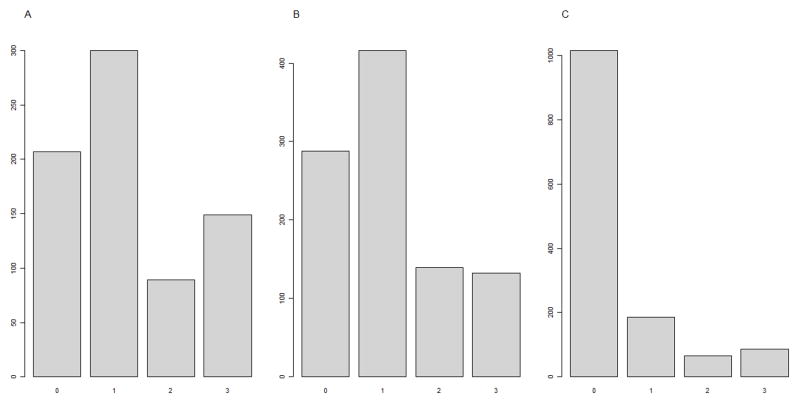
We applied the proposed method to the Study of Addiction: Genetics and Environment (SAGE) GWAS dataset, searching for potential joint gene-gene actions among 155 known ND-associated SNPs. The participants of the SAGE were unrelated individuals selected from three large, complementary studies: the Family Study of Cocaine Dependence (FSCD), the Collaborative Study on the Genetics of Alcoholism (COGA), and the Collaborative Genetic Study of Nicotine Dependence (COGEND). The trait of primary interest was the level of addiction to cigarettes, assessed by the answer to the question ‘How many cigarettes do you smoke per day?’. It had four ordinal levels: 0 (10 cigarettes or less), 1 (11–20 cigarettes), 2 (21–30 cigarettes) and 3 (31 cigarettes or more). 760, 799 and 1356 subjects in FSCD, COGA and COGEND, respectively, had the trait information and were used in the analysis. The trait distributions are shown in Figure 2. From the literature, we selected 155 SNPs across 67 candidate genes that had been reported for potential association with ND. Among those, genotypes for 128 SNPs are available in the SAGE dataset, and genotypes for the remaining 27 SNPs were imputed by using PLINK [Frazer, et al. 2007; Marchini, et al. 2007]. The HapMap phase III founders of the CEU and ASW populations were used in the imputation as the reference panels for the white and black subjects [Altshuler, et al. 2010].
Figure 2. Trait Distributions in FSCD, COGA and COGEND.
A: the distribution of trait in FSCD;
B: the distribution of trait in COGA;
C: the distribution of trait in COGEND.
We applied the Forward U-Test to FSCD for an initial association test and then replicated the initial findings in COGA and COGEND. Two SNPs, rs16969968 (A/G) and rs1122530 (C/T), were identified to be significantly jointly associated with the trait with a nominal p-value of 5.31e-7 in FSCD. Permutation test was also conducted and the empirical p-value was p<0.001. The two SNPs are located in genes CHRNA5 and NTRK2, respectively. Evaluation of the association finding in COGA (p-value=1.08e-5) and COGEND (p-value=0.02) showed the association remained significant at the 0.05 level (Table 5). The two SNPs together formed four two-SNP genotypes:G1={{AA or AG} & {CC or CT}}, G2 ={{AA or AG} & {TT}}, G3={{GG}& {CC or CT}}, G4={{GG} & {TT}}. In order to study any potential interaction between the two SNPs, we calculated the average trait level in each genotype groups. From the FSCD we found that the effect of rs16969969 was modified differently by the genotypes of rs1122530, indicating a potential interaction effect between the two SNPs (Figure 3). A similar trend was observed in COGA and COGEND (Figure 3). In particular, it should be noted that this interaction is “essential” [Wu, et al. 2009] and not completely removable by a monotonic transformation of the data.
Table 5.
Summary of two SNPs identified in FSCD and replicated in COGA and COGEND
| SNP | Allele | Chro | Position | Gene | Grouping | p-values |
|---|---|---|---|---|---|---|
| rs16969968 | A/G | 15 | 78882925 | CHRNA5 | {AA,AG},{GG} | FSCD : 5.31e-7 COGA : 1.08e-5 |
| rs1122530 | C/T | 9 | 87464352 | NTRK2 | {CC,CT},{TT} | COGEND :0.02 |
Figure 3. Joint Effects of Two SNPs Show Potential Statistical Interaction.
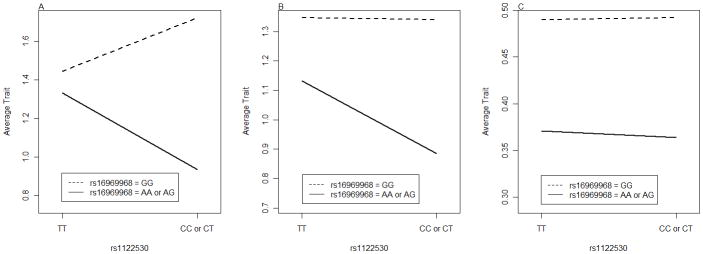
A: average trait by genotype groups in FSCD;
B: average trait by genotype groups in COGA;
C: average trait by genotype groups in COGEND;
We also applied GMDR on the same datasets. For the initial association study on FSCD, the disease models were searched with up to 3-way joint actions, and a zero inflated Poisson link was assumed. The results showed that the model with two SNPs performed best in terms of Testing Balance Accuracy, CV consistency, and sign test p-value (Table 6). Whereas GMDR identified rs16969968 (A/G) that overlapped with the results of Forward U-Test, it also picked up a different SNP, rs573400 (A/G), which located in gene GABRA2. Examination of the two SNPs in the other two datasets showed, the association remained significant in COGA (p-value=0.0001), but was not significant in COGEND (p-value=0.6230). We used linear regression models to fit the trait values with the groupings identified by both methods and examined the goodness of fit with R-Squares (Table 7). The results showed that the SNPs identified by GMDR had a better fit than the SNPs identified by Forward U-Test in FSCD, but not in COGA and COGEND. Both methods may indicate plausible joint gene-gene actions. Although the findings of both methods can’t be directly compared, the results from the association and goodness-of-fit analyses suggested that the finding of Forward U-Test may be more robust across different studies.
Table 6.
Analysis result of GMDR in FSCD and replication in COGA and COGEND
| Study | Model | Allele | Gene | Training Bal. Acc | Testing Bal. Acc | Sign Test (p) | CV | |
|---|---|---|---|---|---|---|---|---|
| FSCD | 1 | rs2836823 | 0.5944 | 0.5511 | 7 (0.1719) | 7/10 | ||
| 2 | rs16969968 rs573400 |
A/G A/G |
CHRNA5 GABRA2 |
0.6448 | 0.6369 | 10 (0.001) | 10/10 | |
| 3 | rs16969968 rs573400 rs9321013 |
0.6764 | 0.5803 | 10 (0.001) | 3/10 | |||
|
| ||||||||
| COGA | rs16969968 rs573400 |
0.6093 | 0.6107 | 10 (0.001) | 10/10 | |||
|
| ||||||||
| COGEND | rs16969968 rs573400 |
0.5511 | 0.4840 | 5 (0.6230) | 10/10 | |||
Table 7.
Goodness of Fit with the SNPs identified by Forward U-Test and GMDR
| Study | R-Squares
|
|
|---|---|---|
| Forward U-Test | GMDR | |
| FSCD | 0.0567 | 0.0656 |
| COGA | 0.0348 | 0.0165 |
| COGEND | 0.0051 | 0.0033 |
DISCUSSION
Complex traits are expected to be caused by the interplay of multiple genetic variants and environmental factors through complicated mechanisms. If two genes are jointly involved in producing the variability of a phenotype whether additively or not, biological interaction between them or their products must be involved [Wang, et al. 2010a]. In addition, there may be statistical interaction that may or may not be removable by a transformation of the data. Thus, statistical approaches that consider gene-gene/gene-environment interactions, including high order interaction, are more likely to take this complexity into account and could improve the discovery process of identifying important genetic variants. In this paper, we proposed a Forward U-Test for joint association of multiple genetic variants, with consideration of possible gene-gene interaction. Through simulation, we have shown that our method has a better performance than GMDR under various scenarios, whether or not statistical interaction exists. This improvement can be explained by the following reasons: 1) Our method is an entirely non-parametric approach and makes no assumption about the trait distribution, while the GMDR is based on a generalized linear model and implicitly specifies the link function with an assumption of the trait distribution. 2) Similar to MDR, GMDR assumes two levels of the quantitative traits by clustering multi-locus genotypes into a high-risk group and a low-risk group. Our method measures the differences of traits on genotype group levels without constraining the genotype groups to two levels, which may gain more strength from the quantitative variation of the trait. 3) Unlike MDR and GMDR, which select a set of candidate models for each model size, the Forward U-Test uses the cross-validation procedure to choose the most parsimonious model, making it easy for interpretation and replication. 4) Our method uses a forward search instead of an exhaustive search as does GMDR. The forward selection can substantially reduce the search space of the interaction combinations. As discussed by Wu et al [Wu and Zhao 2009], the performance of the selection strategies depends on the underlying disease model. Our results indicated that, under additive and multiplicative models, forward selection outperformed exhaustive selection. However, we expect power to decrease for forward selection if none of the genetic variants has any marginal effect. In this specific case, exhaustive selection will perform better than forward selection.
Besides the potential improvement of testing power, forward selection is also less computational intensive than exhaustive selection. When the number of loci increases, the computation time and memory use for the exhaustive search increase exponentially, while those increase only quadratic for the forward selection algorithm. This makes it computationally feasible for testing high-order interaction on high-dimensional data (e.g., whole genome-wide data). High-order epistasis may play an important role in gene networks. The early evidence in plant has shown that the aggressiveness of the isolate of phytophthora capsici is influence by high order epistasis [Bartual, et al. 1993]. A recent study has also detected a significant three-locus interaction that is associated with the development of inflammatory bowel disease (IBD) in human [Wang, et al. 2010b]. Furthermore, in our study, we illustrated the proposed method with a moderate number of SNPs. For genome-wide association studies with millions of SNPs, Li et al. recently proposed a two-step analysis framework by integrating a trait preconditioning procedure with the feature selection [Li, et al. 2011]. This approach first predicts ‘preconditioned’ trait by a linear combination of features that are strongly correlated with the trait, and further applies the feature selection to the ‘preconditioned’ trait. It has been shown that the preconditioning can improve the performance of feature selection, especially for interaction effects. Such a strategy may also be helpful to detect genetic interactions by combing trait preconditioning with the proposed forward selection procedure.
We also compared the power of the proposed method with the stepwise linear regression method. The stepwise linear regression model was performed using the glm and step function in R. During the stepwise regression process, the SNPs were selected forwardly into the model and the most parsimonious model was determined based on Akaike information criterion (AIC). Through simulation, we found forward U-Test outperformed linear regression. For instance, under the two-locus multiplicative model with the largest marginal effect in the simulation (first scenario in Table 2), the power of stepwise linear regression is 0.16 without considering the interaction effects and 0.152 if interaction effects are considered, which are much lower than the power of the forward U-Test. We also applied stepwise linear regression to SAGE data. Due to a large number of parameters required for modeling interactions, we applied stepwise linear regression with only considering marginal effects. By applying the stepwise linear regression to the initial data of FSCD, 26 SNPs were selected. Further evaluation of these 26 SNPs in COGA and COGEND showed these SNPs were not significantly associated with the trait. This result may indicate that the parametric methods, such as linear regression, were less robust when a large number of SNPs were considered.
The Forward U-Test also differs from existing U-Statistic based methods: 1) It calculates the global U-Statistic by a summation over the U-Statistics of multi-SNP genotype groups instead of each single SNP, which implicitly considers the joint gene-gene action that is additive or not; 2) It searches for the multi-SNP genotypes by a forward selection algorithm, which is important for high-dimensional data with a large number of non-functional SNPs. The size of the model selected by the forward selection algorithm may depend on the study sample size. The larger the sample size, the more complex the model with the possibility of high-order interactions, the approach can find. In addition, the choice of the weight parameter ω can also have an impact on the performance of the approach. Different weights can be used in the proposed method (e.g. ωll′ = 1, for all l,l′), but we chose in our study because it appeared to have the best testing power.
In the real data application, we identified two SNPs, located in CHRNA5 and NTRK2, jointly associated with ND. Both CHRNA5 and NTRK2 have been suggested to be functionally related to ND. SNP rs16969968, a non-synonymous coding SNP in exon 5 of CHRNA5, was first reported to be ND-related with a significance level of 0.00064 [Saccone, et al. 2007], and has been replicated in several other studies [Berrettini, et al. 2008; Caporaso, et al. 2009; Grucza, et al. 2008; Schuckit, et al. 2008; Spitz, et al. 2008; Stevens, et al. 2008]. Studies have also suggested that CHRNA5 may interact with CHRNA3 and CHRNB4 to affect ND and lung cancer [Li, et al. 2010b; Li, et al. 2010c; Schlaepfer, et al. 2008; Weiss, et al. 2008]. SNP rs1122530, a non-coding SNP in NTRK2, has been found to be associated with ND in a haplotype analysis with two other SNPs (rs1659400 and rs1187272) of NTRK2 [Beuten, et al. 2007]. A previous study has found evidence of joint action between NTRK2 and multiple functional genes for ND, such as CHRNA4, CHRNB2, and BDNF [Li, et al. 2008]. However, to our knowledge, no joint action has been previously reported for CHRNA5 and NTRK2. Although the joint association of CHRNA5 and NTRK2 with ND, involving statistical interaction, reached a statistically significant level and replicated in independent studies, further study would be necessary to further replicate and investigate the statistical interaction.
Acknowledgments
This work was supported by start-up funds from Michigan State University. We thank James C. (Jim) Anthony for advice supported by his NIDA K05 Senior Scientist Award (K05DA015799). We also want to thank two anonymous references for helpful suggestions that improve the manuscript.
APPENDIX
Estimation of the variance of the U-Statistic under the null hypothesis
Suppose we have a study sample of N subjects. We assume their quantitative traits are independent and have the same variance, denoted as Var (Yi) = σ2. For simplicity, we denote .
The variance of the U-statistic can be expressed as
For all 1≤l≤l′≤L, we estimate the group-wise variance for the U-Statistic as:
The covariance between group-wise U-Statistics is estimated according to different scenarios:
- l1 = l2 = l,
- ,
- ,
is equivalent to 4)
Contributor Information
Ming Li, Email: liming@msu.edu.
Chengyin Ye, Email: cye@epi.msu.edu.
Wenjiang Fu, Email: fuw@msu.edu.
Robert C. Elston, Email: rce2@case.edu.
Qing Lu, Email: qlu@epi.msu.edu.
References
- Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Bonnen PE, de Bakker PI, Deloukas P, Gabriel SB, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–8. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40(8):955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartual R, Lacasa A, JM, JE Epistasis in the resistance of pepper to phytophthora stem blight (Phytophthora capsici L.) and its significance in the prediction of double cross performances. Euphytica. 1993;72:149–152. [Google Scholar]
- Bateson W, Mendel G. Mendel’s principles of heredity. Cambridge: At the University press; 1909. [Google Scholar]
- Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, Waterworth D, Muglia P, Mooser V. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13(4):368–73. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuten J, Ma JZ, Payne TJ, Dupont RT, Lou XY, Crews KM, Elston RC, Li MD. Association of specific haplotypes of neurotrophic tyrosine kinase receptor 2 gene (NTRK2) with vulnerability to nicotine dependence in African-Americans and European-Americans. Biol Psychiatry. 2007;61(1):48–55. doi: 10.1016/j.biopsych.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Brem RB, Storey JD, Whittle J, Kruglyak L. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature. 2005;436(7051):701–3. doi: 10.1038/nature03865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso N, Gu F, Chatterjee N, Sheng-Chih J, Yu K, Yeager M, Chen C, Jacobs K, Wheeler W, Landi MT, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS One. 2009;4(2):e4653. doi: 10.1371/journal.pone.0004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, Kalaylioglu Z, Moslehi R, Peters U, Wacholder S. Powerful multilocus tests of genetic association in the presence of gene-gene and gene-environment interactions. Am J Hum Genet. 2006;79(6):1002–16. doi: 10.1086/509704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10(6):392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grucza RA, Wang JC, Stitzel JA, Hinrichs AL, Saccone SF, Saccone NL, Bucholz KK, Cloninger CR, Neuman RJ, Budde JP, et al. A risk allele for nicotine dependence in CHRNA5 is a protective allele for cocaine dependence. Biol Psychiatry. 2008;64(11):922–9. doi: 10.1016/j.biopsych.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffding W. A Class of Statistics with Asymptotically Normal Distribution. Annals of Mathematical Statistics. 1948;19(3):293–325. [Google Scholar]
- Li J, Das K, Fu G, Li R, Wu R. The Bayesian lasso for genome-wide association studies. Bioinformatics. 2011;27(4):516–23. doi: 10.1093/bioinformatics/btq688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Romero R, Fu WJ, Cui Y. Mapping haplotype-haplotype interactions with adaptive LASSO. BMC Genet. 2010a;11:79. doi: 10.1186/1471-2156-11-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Lou XY, Chen G, Ma JZ, Elston RC. Gene-gene interactions among CHRNA4, CHRNB2, BDNF, and NTRK2 in nicotine dependence. Biol Psychiatry. 2008;64(11):951–7. doi: 10.1016/j.biopsych.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Xu Q, Lou XY, Payne TJ, Niu T, Ma JZ. Association and interaction analysis of variants in CHRNA5/CHRNA3/CHRNB4 gene cluster with nicotine dependence in African and European Americans. Am J Med Genet B Neuropsychiatr Genet. 2010b;153B(3):745–56. doi: 10.1002/ajmg.b.31043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD, Yoon D, Lee JY, Han BG, Niu T, Payne TJ, Ma JZ, Park T. Associations of variants in CHRNA5/A3/B4 gene cluster with smoking behaviors in a Korean population. PLoS One. 2010c;5(8):e12183. doi: 10.1371/journal.pone.0012183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Wu RL. Detecting sequence-sequence interactions for complex diseases. Current Genomics. 2006;7(1):59–72. [Google Scholar]
- Lou XY, Chen GB, Yan L, Ma JZ, Mangold JE, Zhu J, Elston RC, Li MD. A combinatorial approach to detecting gene-gene and gene-environment interactions in family studies. Am J Hum Genet. 2008;83(4):457–67. doi: 10.1016/j.ajhg.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, Li MD. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am J Hum Genet. 2007;80(6):1125–37. doi: 10.1086/518312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Elston RC. Using the optimal receiver operating characteristic curve to design a predictive genetic test, exemplified with type 2 diabetes. Am J Hum Genet. 2008;82(3):641–51. doi: 10.1016/j.ajhg.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005;37(4):413–7. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genetics. 2007;39(7):906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Martin ER, Ritchie MD, Hahn L, Kang S, Moore JH. A novel method to identify gene-gene effects in nuclear families: the MDR-PDT. Genet Epidemiol. 2006;30(2):111–23. doi: 10.1002/gepi.20128. [DOI] [PubMed] [Google Scholar]
- Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum Hered. 2003;56(1–3):73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- Nagel RL. Epistasis and the genetics of human diseases. C R Biol. 2005;328(7):606–15. doi: 10.1016/j.crvi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69(1):138–47. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Human Molecular Genetics. 2007;16(1):36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, McDonnell SK, Hebbring SJ, Cunningham JM, Thibodeau SN. Nonparametric tests of association of multiple genes with human disease. Am J Hum Genet. 2005;76(5):780–93. doi: 10.1086/429838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer IR, Hoft NR, Collins AC, Corley RP, Hewitt JK, Hopfer CJ, Lessem JM, McQueen MB, Rhee SH, Ehringer MA. The CHRNA5/A3/B4 gene cluster variability as an important determinant of early alcohol and tobacco initiation in young adults. Biol Psychiatry. 2008;63(11):1039–46. doi: 10.1016/j.biopsych.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA, Danko GP, Smith TL, Bierut LJ, Bucholz KK, Edenberg HJ, Hesselbrock V, Kramer J, Nurnberger JI, Jr, Trim R, et al. The prognostic implications of DSM-IV abuse criteria in drinking adolescents. Drug Alcohol Depend. 2008;97(1–2):94–104. doi: 10.1016/j.drugalcdep.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster SC. Next-generation sequencing transforms today’s biology. Nat Methods. 2008;5(1):16–8. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- Spitz MR, Amos CI, Dong Q, Lin J, Wu X. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008;100(21):1552–6. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens VL, Bierut LJ, Talbot JT, Wang JC, Sun J, Hinrichs AL, Thun MJ, Goate A, Calle EE. Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiol Biomarkers Prev. 2008;17(12):3517–25. doi: 10.1158/1055-9965.EPI-08-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Akey JM, Kruglyak L. Multiple locus linkage analysis of genomewide expression in yeast. PLoS Biol. 2005;3(8):e267. doi: 10.1371/journal.pbio.0030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng JY, Wang CH, Kao JT, Hsiao CK. Regression-based association analysis with clustered haplotypes through use of genotypes. Am J Hum Genet. 2006;78(2):231–42. doi: 10.1086/500025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Elston RC, Zhu X. The Meaning of Interaction. Hum Hered. 2010a;70(4):269–277. doi: 10.1159/000321967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Liu T, Lin Z, Hegarty J, Koltun WA, Wu R. A general model for multilocus epistatic interactions in case-control studies. PLoS One. 2010b;5(8):e11384. doi: 10.1371/journal.pone.0011384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Li M, Rebbeck T, Li H. U-statistics-based tests for multiple genes in genetic association studies. Ann Hum Genet. 2008;72(Pt 6):821–33. doi: 10.1111/j.1469-1809.2008.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RB, Baker TB, Cannon DS, von Niederhausern A, Dunn DM, Matsunami N, Singh NA, Baird L, Coon H, McMahon WM, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4(7):e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Zhang H, Liu X, Dewan A, Dubrow R, Ying Z, Yang Y, Hoh J. Detecting essential and removable interactions in genome-wide association studies. Stat Interface. 2009;2(2):161–170. doi: 10.4310/sii.2009.v2.n2.a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Zhao H. Statistical power of model selection strategies for genome-wide association studies. PLoS Genet. 2009;5(7):e1000582. doi: 10.1371/journal.pgen.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40(5):638–45. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeileis A, Kleiber C, Jackman S. Regression models for count data in R. Journal of Statistical Software. 2008;27(8):1–25. [Google Scholar]
- Zhang J, Liang F, Dassen WR, Veldman BA, Doevendans PA, De Gunst M. Search for haplotype interactions that influence susceptibility to type 1 diabetes, through use of unphased genotype data. Am J Hum Genet. 2003;73(6):1385–401. doi: 10.1086/380417. [DOI] [PMC free article] [PubMed] [Google Scholar]