Abstract
Integrons are genetic elements first described at the end of the 1980s. Although most integrons were initially described in human clinical isolates, they have now been identified in many non-clinical environments, such as water and soil. Integrons are present in ≈10% of the sequenced bacterial genomes and are frequently linked to mobile genetic elements (MGEs); particularly the class 1 integrons. Genetic linkage to a diverse set of MGEs facilitates horizontal transfer of class 1 integrons within and between bacterial populations and species. The mechanistic aspects limiting transfer of MGEs will therefore limit the transfer of class 1 integrons. However, horizontal movement due to genes provided in trans and homologous recombination can result in class 1 integron dynamics independent of MGEs. A key determinant for continued dissemination of class 1 integrons is the probability that transferred MGEs will be vertically inherited in the recipient bacterial population. Heritability depends both on genetic stability as well as the fitness costs conferred to the host. Here we review the factors known to govern the dissemination of class 1 integrons in bacteria.
Keywords: mobile genetic elements, class 1 integron, horizontal or lateral gene transfer, homologous recombination, transposition, transformation, conjugation, transduction, antibiotic resistance
Introduction
Horizontal gene transfer (HGT) is the transfer of genetic material between bacteria from the same generation. A successful HGT event does not rely only on the introduction of DNA into a recipient cell's cytoplasm, but also on the heritability of the transferred sequences in the recipient microorganism.1 HGT contributes to the genetic diversity and evolutionary trajectories of bacterial populations. For instance, HGT of mobile genetic elements (MGEs) is the major contributor to emergence, recombination and dissemination of multidrug resistance among bacterial pathogens.2,3 Due to their capacity to physically move between host genomes, MGEs are common elements in bacterial communities. A variety of MGEs have been described to date such as plasmids, bacteriophages, genomic islands (GIs), integrative and conjugative elements (ICEs), insertion sequences (ISs), transposons (Tns), integrons and miniature inverted repeat transposable elements (MITEs).4 Integrons are genetic elements that contain a site-specific recombination system able to integrate, express and exchange specific DNA elements, called gene cassettes.5 The complete integron is not considered to be a mobile element as such as it lacks functions for self-mobility. In contrast, the gene cassettes present in integrons are considered mobile, although the natural exchange of gene cassettes are rarely experimentally observed.6,7 Nevertheless, sequence similar integrons appear to be widespread among bacterial species and genetic backgrounds, suggesting that they are frequently exposed to mechanisms that allow them to disseminate horizontally through bacterial populations.8 The aim of this review is to examine how different vehicles and mechanisms of HGT enable the dissemination of integrons. We focus on the main type of integron involved in the spread of antibiotic resistance, the class 1 (C1) integrons.9
Integrons
Integrons were initially described at the end of the 1980s.8 Bioinformatics based analysis of partially or fully sequenced bacterial genomes show that integrons or integrase genes are present in approx. 10% and 17% of them, respectively.9,10 Most of the descriptions of integrons have focused on their presence in human clinical isolates,9,11,12 but they are also found in many non-clinical environments, such as in aquatic environment13-15 and soil.16,17
Integrons consists of three elements: the gene that encodes a tyrosine recombinase (integrase, encoded by the intI gene), needed for site-specific recombination within the integron; the adjacent recombination site (attI) that is recognized by the integrase; and the promoter (Pc), located upstream of the integration site, necessary for efficient transcription and expression of gene cassettes present in the integron. Although the Pc promoter is assumed to be part of all classes of integrons, its presence and activity has not been shown for all of them.10 Many of the integrase genes present in integrons carry LexA binding sites close to their promoter regions and can be controlled by the host LexA protein, which is a transcriptional repressor of the SOS response;6 as shown for a class 1 integrase and in an integrase present in a CI (chromosomal integron) of Vibrio cholorae.6,7 These studies suggest that SOS induction can lead to increased transcription of the integrase gene and increased integrase activity, including cassette rearrangements, in organisms harboring lexA alleles.6,7 The regulation of the integrase transcription in species where a LexA ortholog is absent is unknown.
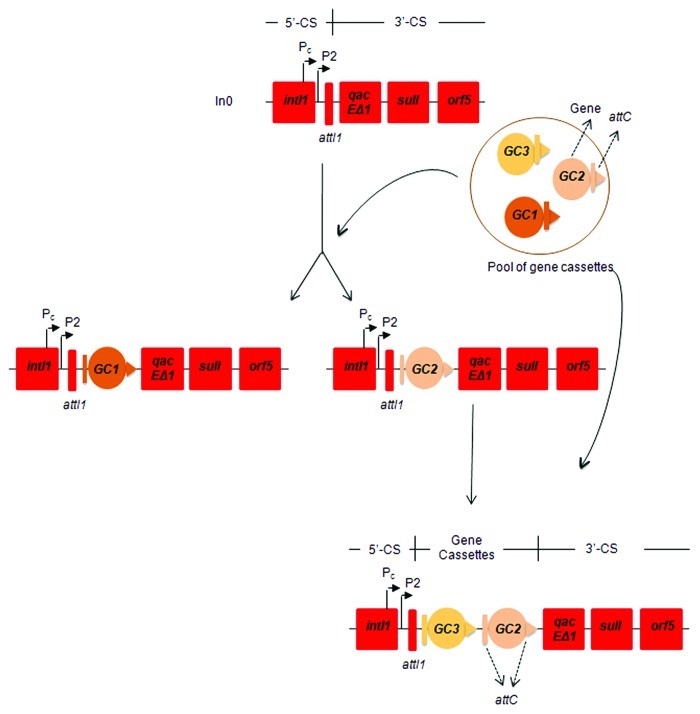
Integrons can incorporate one or more gene cassettes (Fig. 1).5 Usually integrons present in clinical bacterial strains carry less than five cassettes,18 although integrons with up to nine antibiotic resistance genes have been described.19 Numerous combinations of gene cassettes have been reported so far;20 a list of numbered C1 integrons is available from the INTEGRALL database (http://integrall.bio.ua.pt/).21
Figure 1. Schematic representation of class 1 (C1) integrons. In0 is the most basic integron that does not contain any gene cassettes. Different cassette arrays can be integrated in the variable region of the integron. Abbreviations: intI1, class 1 integrase gene; Pc and P2, gene cassette promoters; attI1, integron-associated recombination site; qacEΔ1, truncated version of a quaternary ammonium resistance gene; sulI, sulphonamide resistance gene; orf5, open reading frame; attC, recombination site of the gene cassette; GC, gene cassette; 5′-CS,5′ conserved segment of the integron; 3′-CS,3′ conserved segment of the integron.
Based on the genetic relatedness of the integrase intI gene sequence, the integrons were initially classified into a few classes, of which class 1, 2 and 3 have received most attention.10,16 More extensive DNA sequencing efforts have however now revealed more of the genetic diversity of the integrase gene and identified > 90 different gene variants, questioning the initial classification scheme.10,22 Phylogenetic analyses of sequenced intI genes suggest integrons belong to 3 broad groups, the soil/freshwater proteobacteria group, the marine γ-proteobacteria group and the inverted integrase group.10 C1 integrons have been reported as the most common and widespread, especially in clinical settings and are one of the main contributors to the problem of antibiotic resistance dissemination.9
The origin of C1 integrons is not clear, but it is assumed that they were present in bacteria before the “antibiotic era”.23 C1 integrons without antibiotic resistance genes found in environmental Betaproteobacteria have been suggested to be the original source of these genetic structures. These environmental integrons lack the Tn402 (also called Tn5090) features,22,23 and it is proposed that they were incorporated into a plasmid-borne Tn402 transposon, carrying a gene cassette that conferred a host advantage.22 For instance, the qac gene, which codes resistance to quaternary ammonium compounds, as such biocides have a long history of use in clinical practices;24 and the sulI gene, which confers resistance to sulphonamides, one of the first antibiotics introduced in the clinical practice.25 Another possible origin of the C1 integrons is the early association between the ancestor of the Tn402 transposon with a class 1 integrase and an attI1 site. This early association is supported by the observation that most C1 integrons contain the 5′-CS region at the same location.25 The 3′-CS region has been suggested to be the result of a fusion of the qacE gene of the Tn402 transposon with the sulI gene, with consequent partial deletion of the qacE gene; that occurred at the same time as a deletion event in the transposition functions of the Tn402 transposon, resulting in a structure unable of self-mobilization.25 In fact, most C1 integrons are defective transposons;26 though a few C1 integrons with a complete transposition module have been identified.27,28 After initial formation of C1 integrons, strong selection for resistance to antimicrobials has favored events of capture of antibiotic resistance gene cassettes resulting in the compositions of C1 integron as we know them today.4,18 There is also evidence that C1 integrons can capture gene cassettes from CIs (see description of CIs below).29
Integrons are also classified based on their context, which includes two different types of integrons, mobile integrons (MIs) and chromosomal integrons (CIs).9 CIs can carry a variable number of gene cassettes, ranging from zero to hundreds, which are usually not involved in antimicrobial resistance. CIs are considered to be sedentary, though movement of gene cassettes from CIs has been reported.9,10 In contrast, MIs, such as C1 integrons, carry a limited number of gene cassettes and are often involved in dissemination of antimicrobial resistance.9 This review focuses on mobile C1 integrons.
Gene cassettes are small mobile elements composed by a single and most often promoterless gene and a recombination site (attC). Gene cassettes are linear when integrated in the C1 integron and circular in the free form, after excision or before site-specific insertion.30 The attC site (previous called 59-base element or 59-be) is recognized by the integrase and recombination between this site and the attI1 site of the C1 integron backbone leads to the addition of the gene cassette into the integron structure.20,31,32 Due to the nature of the site-specific recombination, part of the attC site is located at the start of the gene cassette and part at the end.31,33 The majority of the 194 known gene cassettes encode antibiotic resistance; including resistance to frequently used aminoglycosides, β-lactams, quinolones, chloramphenicol and trimethoprim, among others.20 Transcription and posterior expression of gene cassettes depends on the promoter of the integron, as the gene cassettes themselves rarely possess a promoter;5 though a few exceptions have been reported.20 Thirteen different variants of the gene cassette promoter Pc, with different expression levels, have been described. As the Pc promoter is located within the intI1 coding sequence, the occurrence of different Pc leads to the existence of 10 different variants of the class 1 integrase IntI1.34 The Pc can be associated or not with a second gene cassette promoter, P2.35 The level of transcription and expression of gene cassettes present in the integron is also influenced by the distance to the promoter, where cassettes closer to the promoter are expressed at higher levels than more distal ones.35
Usually, C1 integrons have three distinct genetic regions of which two are highly conserved, the 5′-conserved segment (5′-CS) and the 3′-conserved segment (3′-CS), that flank the central variable region where the gene cassettes are located.8 The 5′-CS includes the integrase intI1 gene, the recombination site attI1 and the promoter Pc and P2 when present. The 3′-CS consists of the qacEΔ1 gene, which encodes an incomplete version of a protein that mediates resistance to certain detergents, the sulI gene, encoding resistance to sulphonamides and an open reading frame orf5, of unknown function.33 Although most of the C1 integrons have this classic structure, an increasing number of C1 integrons with different structure have been reported;36-38 these C1 integrons are associated with ISCR1 and therefore will be mentioned as ISCR1-linked C1 integrons throughout this review. These integrons are often referred to, in the scientific literature, as complex class 1 integrons. The ISCR1-linked C1 integrons frequently have non-cassette type resistance genes, a truncated version of 3′-CS region followed by an ISCR1 element and another variable region and or 3′-CS duplications.38,39
C1 integrons are mainly found in Gram-negative bacteria, although several studies have reported the presence of these mobile elements also in Gram-positive bacteria. In fact, Nandi and collaborators40 suggested that Gram-positives represent a larger reservoir of C1 integrons than Gram-negatives. Reports of C1 integrons in the Gram-positive genera Corynebacterium,40,41 Enterococcus,42 Staphylococcus,42-44 and Streptococcus,42 are available. Nonetheless, the majority of the reports of C1 integrons concerns Gram-negative genera, such as Acinetobacter,45,46 Aeromonas,13 Burkholderia,47 Enterobacter,46 Escherichia,13,46,48 Klebsiella,46,48 Morganella,13 Proteus,48 Pseudomonas,45,46 Salmonella,49,50 and Vibrio51 It should also be noted that C1 integron-carrying Gram-positive bacteria have mainly been isolated from human clinical samples, while C1 integron-carrying Gram-negative isolates have been collected from a wider range of environments, including hospitals,11,12,52,53 healthy humans,52,54 animals,52,55,56 food-products,53,57 soil17,58 and aquatic environments.13-15
The Genomic Locations of C1 integrons, Context and Mobility
Most of the descriptive studies of C1 integrons focus on the identification and characterization of various C1 integrons and their gene cassette arrays; fewer studies also aim to unravel the genetic context where they are inserted.57 When investigated, most of the C1 integrons are found genetically linked to and embedded in MGEs, such as MITEs, ISs, Tns, GIs and plasmids. Some of these MGEs contain self-mobility determinants, whereas others can only be mobilised by other elements or when some genes/elements are provided in trans (further information below). These elements confer mobility to the C1 integrons when part of the larger MGE. Thus, genetic linkage to various and sometimes several MGEs facilitates both intra and intergenomic transfers of integrons at frequencies determined by the mobility functions and transfer dynamics of the element(s).
Evidence of mobility of C1 integrons
There are many studies that suggest horizontal dissemination of C1 integrons, as the same C1 integron structure is found in different species, in different geographical places and isolated in different periods of time. For example, C1 integron In76, carrying a blaIMP-5 gene cassette, was isolated in Acinetobacter baumannii 65FFC from Coimbra University Hospital in 1998,59 and also in eight Pseudomonas aeruginosa isolates collected between 2001 and 2003 from São Bernardo Hospital, Setúbal, Santa Maria Hospital and Dona Estefânia Hospital, both from Lisbon;60 these latter isolates were not epidemiologically related.59,60 Similar is the case of 13 epidemiologically unrelated Acinetobacter baumannii isolates collected between 1989 and 2000 in six different Italian hospitals that carried a C1 integron with the cassette array aacC1-orfX-orfX'-aadA1a.12 The same C1 integron was described in an Acinetobacter baumannii collected in Spain in 2000.61 Interestingly, the same cassette array dfrA1-aadA1 was found in C1 integron in the Gram-negatives Escherichia coli and Aeromonas sp.,13 and in the Gram-positives Corynebacterium ammoniagenes and Corynebacterium casei.40
Genetic linkage to MGEs with consequences for mobility
The genetic linkage between C1 integrons and MGEs increases the potential of C1 integrons for dissemination by HGT. C1 integrons are frequently observed in larger MGEs with several layers of horizontal mobility and such combination of MGEs confers a broad horizontal dissemination potential to C1 integrons. Importantly, the mobility of C1 integrons embedded in MGEs emerges mainly from a complex interplay between various modes of transposition and plasmid-based conjugative processes. The fate of newly transferred integrons is further modulated by functional and selective constraints to the stable maintenance of the elements in new hosts. MGEs vary in their ability to enable intra- vs. inter-cellular movements of integrons. For instance GIs, MITEs, ISs and MTns (mobilisable Tns) allow transfer and genomic integration of C1 integrons within cells (see section 3.2.1 to 3.2.4) whereas conjugative plasmids, transduction and transformation contribute to the transfer of C1 integrons between cells (see section 3.2.5 to 3.2.7). Some bacteriophages, ICEs and some GIs can enable both intra- and intercellular transfers. In many other cases, horizontal dissemination of integrons should be viewed as a two-step process, the first step being dependent on mechanism allowing intercellular transfer. The second step can however draw on a much wider set of mechanisms allowing intracellular transfer and genetic relocalization.
Below we provide further considerations and examples of the horizontal mobility caused by the genetic linkage of C1 integrons with diverse MGEs; including combination of MGEs. Some of the studies referred to directly demonstrate HGT of the integron-associated MGE whereas others infer horizontal acquisition of C1 integron-containing elements, usually by transposition, due to the presence of target site duplications (TSDs). TSDs are short direct nucleotide repeats that can be found on both ends of the inserted mobile element. Table 1 shows some examples of the various MGEs associated with C1 integrons.
Table 1. Examples of mobile genetic elements (MGEs) associated with class1 integrons.
| MGE |
Species |
Isolation source |
Isolation country |
Reference |
|---|---|---|---|---|
| MITE | ||||
| MITE-like |
Acinetobacter johnsonii |
Prawn |
Australia |
69 |
| |
Acinetobacter baumannii |
Clinical |
Portugal |
68 |
| |
Acinetobacter bereziniae |
Clinical |
Portugal |
70 |
| IMU | Enterobacter cloacae | Clinical | Canada | 72 |
| IS | ||||
|---|---|---|---|---|
| IS6 family |
Escherichia coli |
Clinical |
Poland |
76 |
| |
Salmonella enterica serovar Newport |
Clinical |
France |
75 |
| |
Escherichia coli |
Clinical |
France |
19 |
| |
Escherichia coli |
Clinical |
Greece |
74 |
| |
Salmonella enterica serovar Typhimurium |
Clinical |
Albania |
74 |
| ISCR1 |
Aeromonas punctata |
Wastewater |
China |
80 |
| |
Citrobacter freundii |
Clinical |
Portugal |
78 |
| |
Klebsiella pneumoniae |
Clinical |
Argentina |
36 |
| |
Enterobacter cloacae |
Clinical |
Argentina |
36 |
| |
Escherichia coli |
Clinical |
Argentina |
36 |
| ISEcp1 |
Klebsiella pneumoniae |
Clinical |
Israel |
81 |
| |
Escherichia coli |
Clinical |
Israel |
81 |
| |
Enterobacter cloacae |
Clinical |
Israel |
81 |
| |
Proteus mirabilis |
Clinical |
Israel |
81 |
| Escherichia coli | Clinical | Tunisia | 82 |
| MTn | ||||
|---|---|---|---|---|
| Tn21-like |
Aeromonas salmonicida subsp. salmonicida |
Brown trout |
France |
93 |
| |
Escherichia coli |
Dead chicks and turkey poults |
China |
92 |
| |
Salmonella enterica serovar Brandenburg |
Clinical and foodborne |
Spain |
91 |
| |
Klebsiella oxytoca |
Clinical |
Spain |
90 |
| |
Enterobacter cloacae |
Clinical |
Spain |
90 |
| Tn5045 |
Pseudomonas aeruginosa |
Permafrost |
Russia |
96 |
| Tn6001 | Pseudomonas aeruginosa | Clinical | Taiwan | 98 |
| ICE | ||||
|---|---|---|---|---|
| SXT | Vibrio cholerae | Clinical | India, Sri Lanka, Bangladesh, Hong Kong | 151 |
| GI | ||||
|---|---|---|---|---|
| n.n. |
Pseudomonas aeruginosa |
Clinical |
Australia |
112 |
| n.n. |
Shigella flexneri |
Primate |
Japan |
111 |
| AbaR |
Acinetobacter baumannii |
Clinical |
Australia |
105 |
| |
Acinetobacter baumannii |
Clinical |
Belgium, Bulgaria, Czech Republic, Spain, Ireland, Italy, Netherlands, Poland, United Kingdom |
107 |
| SGI |
Salmonella enterica serovar Haifa and Newport |
Clinical |
France |
75 |
| Proteus mirabilis | Clinical | Palestine | 110 |
| Plasmid | ||||
|---|---|---|---|---|
| n.i. |
Enterococcus faecalis |
Clinical |
United States of America |
114 |
| n.i. |
Salmonella enterica serovar Typhimurium |
Foodborne |
Portugal |
53 |
| n.i. |
Acinetobacter baumannii |
Clinical |
Taiwan |
115 |
| n.i. |
Aeromononas sp |
Wastewater |
Portugal |
13 |
| n.i. |
Escherichia coli |
Wastewater |
Portugal |
13 |
| n.i. |
Enterobacter cloacae |
Clinical |
Spain |
90 |
| n.i. |
Klebsiella oxytoca |
Clinical |
Spain |
90 |
| n.i. |
Pseudomonas aeruginosa |
Clinical |
Spain |
90 |
| IncHI2 |
Enterobacter cloacae |
Clinical |
Spain |
90 |
| IncI1 |
Klebsiella pneumoniae |
Clinical |
Spain |
90 |
| Escherichia coli | Clinical | Spain | 90 |
Abbreviations: n.n., without attributed name; n.i., without Inc group determined.
Genetic linkage to MITEs
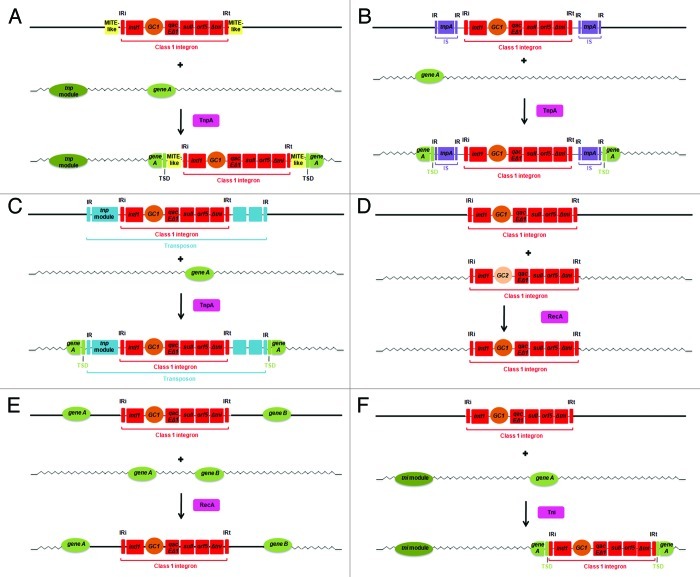
MITEs are non-autonomous mobile elements consisting of small repeat sequences, which do not encode proteins and are found in random locations in the genome of various bacteria.62,63 MITEs are quite abundant62,64,65 and their capacity of transposition has been shown in the genomes of archaea,66 eukaryotes67 and bacteria.63 Bacterial MITEs do not carry a transposase gene, but are suggested to transpose when a transposase is provided in trans. The movement of MITEs generates a TSD (Fig. 2A).63 To our awareness, there are only four reports of presence of MITE-like structures flanking C1 integrons. Identical copies of the same 439 bp MITE-like structure have been found flanking the C1 integron of a clinical Acinetobacter baumannii,68 a prawn-associated Acinetobacter johnsonii69 and in a clinical Acinetobacter bereziniae70 (GenBank accession number JX235356). TSDs were detected on both sides of the MITEs in these three isolates, indicating that the C1 integrons flanked by the MITEs were mobilized by transposition promoted by these genetic elements; in addition to the TSD, the fact that the host gene is interrupted by the single insertion of the MITE-integron-MITE structure in Acinetobacter baumannii68 and in Acinetobacter bereziniae (GenBank accession number JX235356) contribute to the assumption that it was acquired by transposition promoted by the MITEs. Transposition of the entire MITE-integron-MITE structures presented in the Acinetobacter baumannii71 and in the Acinetobacter johnsonii69 was not experimentally observed, suggesting that the transposase required for the movement is not present in the respective strains.
Figure 2. Representation of the possible mechanisms contributing to mobility of class 1 (C1) integrons (in red). (A) Transposition of C1 integrons due to linkage with MITEs and a transposase acting in trans; (B) Transposition of C1 integrons due to linkage with a composite transposon with IS elements; (C) Transposition of C1 integrons due to linkage with a transposon; (D) Exchange of gene cassettes by double homologous recombination between the conserved end regions of C1 integrons; (E) Integration of C1 integrons/gene cassettes by homologous recombination between two adjacent DNA regions in the chromosome; (F) Transposition-like movement of C1 integrons containing two Inverted Repeats and a tni module provided in trans. Abbreviations: IRi, inverted repeat at the 5′-CS outer end of the C1 integron; IRt, inverted repeat at the 3′-CS outer end of the C1 integron; IR, inverted repeat; tnp module, transposition module including the tnpA, tnpR and tnpM genes; TnpA, transposase enzyme; TSD, target site duplication; RecA, enzyme responsible for homologous recombination; tni module, transposition module including the tniA, tniB, tniQ and tniR genes; Tni, TniA, TniB and TniQ enzymes.
In the fourth study, two copies of a MITE-like structure, a 288 bp IMU (Integron Mobilization Unit) were found to flank a defective C1 integron in the plasmid of a clinical Enterobacter cloacae.72 In this case, TSD was not present, but the entire structure (IMU-integron-IMU) was shown to transpose and create a TSD when a transposase was experimentally provided in trans (Fig. 2A).72
Genetic linkage to ISs
ISs are one of the most basic and smallest mobile elements, consisting of a central region encoding a transposase, responsible for mobility that is flanked by terminal inverted repeats (IRs). Transposition of ISs also produce TSD.73 ISs are important for dissemination of antibiotic resistance genes. A DNA segment flanked on each side by the same IS is called a composite transposon, where at least one of the two IS copies must be intact to confer mobility.38 Only a few reports of C1 integrons flanked on each side by an IS can be found in the literature and these include mainly elements of the IS6 family.19,74-76 In some cases C1 integrons flanked by ISs are incorporated into even larger composite transposons.74,75
The C1 integron In53, which carry an unusual number of gene cassettes (9), is located in the pNLT-1 plasmid of Escherichia coli and is flanked on each side by IS26; the IS26-composite transposon, named Tn2000, is flanked by TSDs, suggesting acquisition of this element by transposition.19 The C1 integron carried by the p1658/97 plasmid of Escherichia coli is flanked by one complete and one incomplete IS26, followed by another intact IS26; the C1 integron is inserted in a 48 kb region flanked by different IS26 and similar TSDs, suggesting the acquisition of the entire region by transposition.76 One C1 integron of a Salmonella enterica serovar Newport isolate is flanked by one IS26 and one IS6100 and it is inserted in a larger IS26-composite transposon that was inserted into the genomic island SGI1 backbone by transposition, as suggested by the presence of TSDs.75
ISCRs (insertion sequence common regions) are an unusual group of IS that belongs to the IS91 family and are characterized by the lack of the inverted repeats and by dissimilar ends, an oriIS and a terIS. ISCR also transpose but by a mechanism different from other ISs. The mechanism is called rolling-circle (RC) and result in ‘one-ended transposition’. ISCRs do not generate TSD, which makes it challenging to identify DNA moved by this IS in sequencing analysis. The presence of a single copy in the genome is sufficient to mobilize adjacent DNA.38,39,77 ISCR1 is always found in ISCR1-linked C1 integrons,38,39 and have been reported in diverse bacterial species.36,37,78-80 Another IS conferring “one-ended transposition” is ISEcp1, which has been associated mainly with the dissemination of the blaCTX genes, which encode extended-spectrum β-lactamase (ESBL) enzymes, linked with C1 integrons.81,82
Genetic linkage to Tns
Tns are distinguished on the basis of transferability: some are mobilisable and others conjugative. Mobilisable Tns (MTns) do not encode functions for self-transfer, but can hitchhike with other MGEs. Conjugative Tns, also designated ICEs, have the ability to excise from the chromosome where they are located and promote their own transfer to a new host, where they integrate.83
Transposition of MTns relies on the action of a transposase, usually part of the MTn, which recognizes the IRs of the MTn. The insertion of a MTn in a new location also generates TSDs. Transposition of Tn402, a C1 integron that is also an active transposon has been observed.84,85 Another C1 integron containing a complete transposition module, named Tn6007, was detected in a human commensal strain of Enterobacter cloacae; Tn6007 with a single base pair mutation in the tniA gene, present in a fosmid clone, was shown to transpose in the presence of another tniA gene in trans—TSDs flanked both ends of the Tn6007 insert.86
With the exception of the Tn40284,87 and a few Tn402-like transposons,86,88 most C1 integrons are considered defective transposons, due to deletions present in DNA regions involved in transposition.22,26 Nevertheless, many C1 integrons are present in other functional and mobilisable Tns. For instance, C1 integron In2 is incorporated in Tn21;89 and various bacterial species carry Tn21-like transposons with different C1 integrons.90-93 Transposition of Tn21 and Tn21-like transposons has been widely observed, which contributes to the broad dissemination of C1 integrons.71,94,95 There are also reports of other MTns carrying C1 integrons, such as Tn5045,96 Tn508697 and Tn6001.98 The TSD flanking the C1 integron InC* and the TSD flanking the integron-carrying transposon Tn5045 suggest two transposition events in the generation and acquisition of this transposon by the chromosome of permafrost isolate Pseudomonas sp. Tik3.96 Another example of mobility of C1 integrons due to the linkage with a MTn includes the experimentally observed transposition of a C1 integron-carrying transposon, Tn5086, carried by an Escherichia coli isolate.97
Unlike MTns, ICEs do not generate TSD when inserted in a new genome;99 so HGT of these elements might be only inferred when comparing their nucleotide composition with the core genome. To date, there are no reports of C1 integrons carried in ICEs.
Genetic linkage to GIs
GIs are large chromosomal regions containing genes encoding a diverse range of functions and that display evidence of acquisition by HGT, such as a different GC content and codon usage from the core genome. When these islands encode resistance to antibiotics and heavy metals, they are denominated resistance islands.100 Similar GIs are usually inserted in defined sites on the chromosome of different host chromosomes.100,101 Most of the GIs are not self-mobile and transfer of GIs is supposed to occur by hitchhiking with other MGEs;100,102 however, self-transfer of a few GIs has recently been demonstrated.103
Several resistance islands containing C1 integrons have been described in diverse strains of Acinetobacter baumannii (AbaR islands),104-107 in several serovars of Salmonella enterica,108,109 in Proteus mirabilis,110 in Shigella flexneri111 and in Pseudomonas aeruginosa.112
A recent study demonstrated the mobilization in trans of the SGI1 island, containing the C1 integron In104, from two strains of Salmonella enterica serovar Agona to Escherichia coli by conjugation; only plasmids from the IncA/C incompatibility group were able to mobilise this GI.109
Genetic linkage to plasmids and horizontal transfer of C1 integrons by conjugation
Plasmids are ubiquitous vectors for HGT and often carry a variety of antimicrobial resistance genes; they have been identified in most bacterial species investigated and carriage of multiple plasmids is also common.18 Plasmids are termed conjugative, when they encode the functions needed for self-mobilization or mobilisable, when they rely on the help of other conjugative elements or functions to move between cells.18 Plasmids are the main vector for antibiotic resistance dissemination, including those resistances coded by C1 integrons.18,113 There are a high number of reports of plasmids carrying C1 integron in a wide range of bacterial species.13,41,52,53,90,114,115 These reports do not usually report on the incompatibility (Inc) group of the plasmid, which have important implications for the broader potential for dissemination of these elements. Plasmids that belong to the same Inc group utilize common mechanisms for replication or transfer and cannot co-exist in the same cell.113 One of the few studies that determined the Inc group of integron-carrying plasmids was done by Tato and collaborators.90 Plasmids belonging to the IncHI2 and IncI1 groups were detected in transconjugants, showing the capacity of these plasmids to horizontally move and disseminate C1 integrons.
Several other studies have reported on the transfer of C1 integrons by conjugation, due to the linkage with plasmids. These latter studies were done in several different species and genera.57,116-118 For instance, Meng and colleagues57 demonstrated a plasmid-mediated transfer of two C1 integrons, from Salmonella enterica serovar Derby and Salmonella enterica serovar Indiana strains, isolated from pork and chicken food, respectively, to Escherichia coli by conjugation. The conjugative plasmid transfer occurred at a frequency of 10−6 to 10−5. In another study, clinical isolates of Serratia marcescens were shown to carry C1 integron-borne plasmids; three different plasmids and respective C1 integron, were transferred to Escherichia coli by conjugation.117
A possible limitation in the interpretation of some of these conjugation studies is that the description of the genomic location of C1 integrons is usually limited to the plasmid; without further information being provided on the immediate flanking regions of the resistance determinants such as flanking ISs or Tns. The latter will likely also be a determinant for the mobility and exact genetic location of C1 integrons.
Conjugation has recently been shown to induce the SOS system and consequently to induce the integrase activity of the intI1 and intIA in integrons present in Escherichia coli and Vibrio cholerae, respectively, leading to recombination of gene cassettes.7
Horizontal transfer of C1 integrons by transduction
Viruses that infect bacteria, bacteriophages, are abundant forms of life and co-reside in environments with bacteria.119 The DNA of bacteriophages can exist in the host cell either in an extrachromosomal form or integrated in the host DNA. Bacteriophages can, during transduction, transfer fragments of the host DNA to the new infected bacterial cell.120 Therefore is expected that also DNA fragments with C1 integrons can be horizontally transferred to other cells by this mechanism. The amount of DNA that can be transferred during transduction depends on the size of the phage capsid, but can range from tens to hundreds of kbs.121 The ability to transduce host DNA seems to be limited to relatively large double-stranded DNA phages.120 Both chromosomal and plasmids are known to be transferrable by transduction.122
To our awareness there is only one report on the role of transduction as a mechanism of horizontal transfer of resistance genes embedded in C1 integrons; in Salmonella enterica serovar Typhimurium.123 It should however be noted that retrospective studies might not be able to identify transduction as a causal mechanism for intercellular DNA transfer as there is no specific genetic signature for events of DNA acquisition by transduction or events of natural transformation.
Horizontal transfer of C1 integrons by natural transformation
Natural transformation is the uptake and stabilization of extracellular DNA by a competent cell.3,124Any fragment of DNA can be taken up over the bacterial membrane of competent cells, except in species with sequence dependent uptake.125 However lack of DNA sequence similarity between recipient and donor DNA limits the probability of recombination-based integration of chromosomal DNA in the new host.3 Uptake of self-replicating plasmids by natural transformation has the potential to disseminate linked C1 integrons.57,126 The stable uptake of plasmid DNA by transformation is however less efficient than uptake of chromosomal DNA due to the complex steps necessary to reassemble a circular duplex molecule in the bacterial cytoplasm.3 For example, Meng and collaborators57 demonstrated the transfer of two integron-borne plasmids, one from a Salmonella enterica serovar Derby and one from a Salmonella enterica serovar Indiana strains, isolated from pork and chicken food, respectively, to the oral bacterium Streptococcus mutans UA159, via natural transformation; the transformation frequency was in the order of 10−6−10−7. Less is known about the transformation potential of integrons present in plasmids incompatible with the new host or when present in chromosomal locations. A recent study by Domingues et al.71 showed that natural transformation can contribute to the uptake of C1 integrons, gene cassettes and transposons in an Acinetobacter baylyi strain. Three different mechanism were found to be involved in the incorporation of C1 integrons in the chromosome of the recipient Acinetobacter baylyi BD413; transposition promoted by IS26 (Fig. 2B) or Tn21 (Fig. 2C) and homologous recombination (Fig. 2D and E). The acquisition of C1 integrons and transposons was shown to occur from related as well as unrelated host species. A previous study with synthetic circular cassettes and linear gene cassette arrays also demonstrated acquisition of gene cassettes by natural transformation in Pseudomonas stutzeri, which were integrated by site-specific recombination rather than by homologous or illegitimate recombination.127
Mobility of C1 integrons independent of genetic linkage to MGEs
Movement of C1 integrons supported by elements in trans
Some authors suggest that C1 integrons can move independently under appropriate circumstances, such as in the presence of a complete transposition (tni) module provided in trans and the presence of the two inverted repeats IRi and IRt flanking the C1 integron (Fig. 2F).26,128 However, only few studies have reported experimental evidence of horizontal transfer of C1 integrons without the genetic linkage to other MGEs. Transposon Tn2521, identified in clinical strains of Pseudomonas aeruginosa, has been shown to have the ability to transpose.129 This transposon was however later described to be a C1 integron, renamed In33 and lacking transposition genes.130 Further analysis of the recipient revealed the presence of a 5 bp direct TSD, which confirmed the transfer of In33 by transposition. It was suggested that the transposition event was supported by tni genes from another transposon present in the same cell, although this was not shown.130 In another study, experimental transposition of C1 integrons lacking a transposition module, In0 and In2, was observed when assisted by tni genes from transposon Tn502 provided in trans. Transposition was further confirmed by the presence of a 5 bp direct TSD flanking the C1 integrons in their new location.131
Movement of C1 integrons and gene cassettes by homologous recombination
As the conserved regions of C1 integrons provide sufficient stretches of DNA similarity for recombinational exchange between two C1 integron-containing bacteria, homologous recombination has been suggested to contribute to the replacement of the gene cassette arrays in C1 integrons.5 For example, such recombination could explain the loss of the gene cassette array of In3 when Tn1405 was acquired by the In3-containing R388 plasmid.130 The role of homologous recombination in the replacement of gene cassettes (Fig. 2D) and in the acquisition of C1 integrons (Fig. 2E) in new locations has been recently demonstrated.71 Although it has only been experimentally shown to occur after natural transformation, it is expected that stable integration of C1 integrons by homologous recombination also can occur after the uptake of DNA by conjugation or transduction.113
Limitations to the dissemination of C1 integrons
Genetic linkage to one or more MGEs enables C1 integrons to transfer horizontally within and between bacterial populations and species. The limiting factors to dissemination of C1 integrons will therefore in most cases be similar to those governing the dissemination of the MGE they are linked to. Typically, transfer by conjugation will be limited by mechanistic aspects of the conjugal machinery, including surface receptors of the recipient cell18 as well as compatibility of the plasmid with the new host cytoplasm and its restriction modification system.3 For transduction, key limiting factors are the host range of the transducing bacteriophages as well as the host restriction modification system.132 Moreover, not all bacterial species are known to undergo transduction. The host range of bacteriophages depends on the specificity of the interaction between the bacteriophage and bacterial receptor sites.1,132 For natural transformation, key limiting factors are the ability to develop competence for DNA uptake as well as the ability of incoming DNA to recombine with the genome of the transformed cell.3 In order to be vertically inherited, non-replicative DNA with C1 integrons transferred, taken up or injected into the bacterial cytoplasm must be able to recombine with the host genome. Such recombinational barriers are the same as those preventing HGT of any chromosomal DNA between the genomes of non-related bacteria.3,120
A general limiting factor, beyond the mechanistic aspects defining the probability of transfer to new hosts, is the likelihood that the acquired MGE will become vertically inherited in the larger bacterial population. It is known that initially rare events of HGT are unlikely to establish in a larger bacterial population if they do not confer a benefit to the host.133,134
Thus, in cases where the acquisition of C1 integron-carrying MGE will negatively affect the relative or absolute growth rate of the new host, rare HGT events will be lost from the larger population as they will be outgrown by more fit members of the population. At high transfer rates, a temporal equilibrium of C1 integron-carrying MGEs in the population can be established when the frequency of new successful HGT events equals the loss of non-fit members.135 However, several studies have shown that both the MGEs and the bacterial host can rapidly co-adapt; so that the initial high fitness costs to the host can, within weeks, be reduced or lost. Eventually, the MGE carrying hosts can be equally or more fit than the remaining bacterial population.135-137 This amelioration of fitness costs is due to compensatory mutations.138,139
In cases where the C1 integron-carrying MGE does not result in an initial fitness change in the new host,140 the frequency of successful transfer and random fluctuations among genotypes in the bacterial population (genetic drift) will determine the short-term-fate of the new C1 integron carrying host cells. C1 integrons present in such cells are not expected to remain in the bacterial population over evolutionary time in the absence of purifying selection, as spontaneous deletions will occur. However, in cases where co-adaptation takes place between the MGE/C1 integrons and the host genome, more complex population trajectories can be expected.141,142 Whereas the bacterial mutation rate is well understood,143 less is known about the various mechanisms leading to deletions of acquired DNA segments in the bacterial genome.
In cases where the C1 integron-containing MGE confers a growth advantage to the new host, even very few HGT events can be sufficient for establishing the C1 integron in the new host.140 For instance, the use of antimicrobial drugs in treatment of bacterial infections can effectively remove drug sensitive cells, so that even exceedingly low initial proportions of C1 integron-containing cells can rapidly expand their population sizes to become the dominant one. Dominant populations of resistance-carrying bacterial pathogens are a major public health issue.
Prohibiting the use of antibiotics is often considered as a strategy to reduce antimicrobial resistance, with the assumption that resistance genes impose a fitness cost to the host and the absence of the antibiotic pressure will lead to their loss. However, reversing resistance development is challenging,144,145 particularly because not all resistance genes confer initial or continual costs to their hosts.146,147 A recent study by Starikova and colleagues shows that this also applies to C1 integrons; although a C1 integron introduced in Acinetobacter baylyi by natural transformation conferred an initial fitness cost of 7%, the experimental evolution assay showed that the fitness of the transformant was completely restored.152
Other selection pressures on the MGE carrying host population can in some cases allow the maintenance of genes with negative host effects; selection on one resistance gene/trait will contribute to maintain adjacent and linked genes, even when those genes provide no beneficial effect to the host.148,149 This means that C1 integrons conferring a fitness cost to the host can be maintained when carried, for example, on a plasmid that confers other selective advantages to the host.
General Considerations and Summary
C1 integrons are usually found linked to various types of MGEs. C1 integrons will therefore move within and between bacterial genomes as part of the MGE(s) they reside in;150 a process called hitchhiking. Although C1 integrons embedded in transposons, IS or other replicating genetic elements are considered to have intragenomic mobility only, such elements can be transferred between cells when part of plasmids during conjugation or through direct DNA transfer by transduction or natural transformation. The conserved regions of C1 integrons also provide sufficient DNA sequence similarity for homologous recombination with the host genome to occur. Such recombination can exchange the gene cassettes present in the recombining parts of the C1 integrons also when present in unrelated bacterial species.71
The potential for dissemination of C1 integrons should be seen as product of the factors governing the linkage of C1 integrons to MGEs, the supply of MGEs into a given bacterial population, their transfer potential and heritable stability in new bacterial hosts and the larger opportunity for MGE carrying hosts to co-exist with or outcompete non-carrying members of the larger population. Thus, considerations of genetic linkage, dissemination rates and absolute and relative host fitness effects are essential to understand the fate of C1 integrons. It is further important to recognize that fitness will be a variable depending on both host and environmental factors; including continual changes in the genetic composition of the host and C1 integron-containing MGE. Further complicating factors in the understanding of dissemination potential are bacterial population structure and transmission dynamics, spatial and temporal variability in transfer rates (exposure dependency), variability in directional selection (e.g., temporal antibiotic usage) and randomness in most processes including host survival and dissemination; leading to non-uniform outcomes.
Most environmental factors can be controlled in experimental laboratory model systems providing an opportunity to conduct quantitative descriptions of C1 integrons and their dissemination characteristics. However, in clinical and other environments, variability and random fluctuations in the governing factors considered above rarely leads to opportunities for a precise quantitative understanding of the transfer dynamics of C1 integrons and the factors that determine current dissemination patterns.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
S.D. is supported by grant SFRH/BD/49061/2008 from the Fundação para a Ciência e a Tecnologia, Lisbon, Portugal. G.S. and K.M.N. acknowledge financial support from the Center of Pharmaceutical Sciences, Portugal and the University of Tromsø, Norway, respectively.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/22967
References
- 1.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura Y, Itoh T, Matsuda H, Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat Genet. 2004;36:760–6. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- 3.Thomas CM, Nielsen KM. Mechanisms of and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–21. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 4.Stokes HW, Gillings MR. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol Rev. 2011;35:790–819. doi: 10.1111/j.1574-6976.2011.00273.x. [DOI] [PubMed] [Google Scholar]
- 5.Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 6.Guerin E, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, Da Re S, et al. The SOS response controls integron recombination. Science. 2009;324:1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- 7.Baharoglu Z, Bikard D, Mazel D. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet. 2010;6:e1001165. doi: 10.1371/journal.pgen.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–83. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 9.Cambray G, Guerout AM, Mazel D. Integrons. Annu Rev Genet. 2010;44:141–66. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- 10.Boucher Y, Labbate M, Koenig JE, Stokes HW. Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol. 2007;15:301–9. doi: 10.1016/j.tim.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Lévesque C, Piché L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–91. doi: 10.1128/AAC.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gombac F, Riccio ML, Rossolini GM, Lagatolla C, Tonin E, Monti-Bragadin C, et al. Molecular characterization of integrons in epidemiologically unrelated clinical isolates of Acinetobacter baumannii from Italian hospitals reveals a limited diversity of gene cassette arrays. Antimicrob Agents Chemother. 2002;46:3665–8. doi: 10.1128/AAC.46.11.3665-3668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moura A, Henriques I, Ribeiro R, Correia A. Prevalence and characterization of integrons from bacteria isolated from a slaughterhouse wastewater treatment plant. J Antimicrob Chemother. 2007;60:1243–50. doi: 10.1093/jac/dkm340. [DOI] [PubMed] [Google Scholar]
- 14.Rosser SJ, Young HK. Identification and characterization of class 1 integrons in bacteria from an aquatic environment. J Antimicrob Chemother. 1999;44:11–8. doi: 10.1093/jac/44.1.11. [DOI] [PubMed] [Google Scholar]
- 15.Wright MS, Baker-Austin C, Lindell AH, Stepanauskas R, Stokes HW, McArthur JV. Influence of industrial contamination on mobile genetic elements: class 1 integron abundance and gene cassette structure in aquatic bacterial communities. ISME J. 2008;2:417–28. doi: 10.1038/ismej.2008.8. [DOI] [PubMed] [Google Scholar]
- 16.Nield BS, Holmes AJ, Gillings MR, Recchia GD, Mabbutt BC, Nevalainen KM, et al. Recovery of new integron classes from environmental DNA. FEMS Microbiol Lett. 2001;195:59–65. doi: 10.1111/j.1574-6968.2001.tb10498.x. [DOI] [PubMed] [Google Scholar]
- 17.Srinivasan V, Nam HM, Sawant AA, Headrick SI, Nguyen LT, Oliver SP. Distribution of tetracycline and streptomycin resistance genes and class 1 integrons in Enterobacteriaceae isolated from dairy and nondairy farm soils. Microb Ecol. 2008;55:184–93. doi: 10.1007/s00248-007-9266-6. [DOI] [PubMed] [Google Scholar]
- 18.Bennett PM. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br J Pharmacol. 2008;153(Suppl 1):S347–57. doi: 10.1038/sj.bjp.0707607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naas T, Mikami Y, Imai T, Poirel L, Nordmann P. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J Bacteriol. 2001;183:235–49. doi: 10.1128/JB.183.1.235-249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Partridge SR, Tsafnat G, Coiera E, Iredell JR. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009;33:757–84. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 21.Moura A, Soares M, Pereira C, Leitão N, Henriques I, Correia A. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics. 2009;25:1096–8. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- 22.Gillings M, Boucher Y, Labbate M, Holmes A, Krishnan S, Holley M, et al. The evolution of class 1 integrons and the rise of antibiotic resistance. J Bacteriol. 2008;190:5095–100. doi: 10.1128/JB.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stokes HW, Nesbø CL, Holley M, Bahl MI, Gillings MR, Boucher Y. Class 1 integrons potentially predating the association with tn402-like transposition genes are present in a sediment microbial community. J Bacteriol. 2006;188:5722–30. doi: 10.1128/JB.01950-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gillings MR, Xuejun D, Hardwick SA, Holley MP, Stokes HW. Gene cassettes encoding resistance to quaternary ammonium compounds: a role in the origin of clinical class 1 integrons? ISME J. 2009;3:209–15. doi: 10.1038/ismej.2008.98. [DOI] [PubMed] [Google Scholar]
- 25.Toleman MA, Walsh TR. Combinatorial events of insertion sequences and ICE in Gram-negative bacteria. FEMS Microbiol Rev. 2011;35:912–35. doi: 10.1111/j.1574-6976.2011.00294.x. [DOI] [PubMed] [Google Scholar]
- 26.Brown HJ, Stokes HW, Hall RM. The integrons In0, In2 and In5 are defective transposon derivatives. J Bacteriol. 1996;178:4429–37. doi: 10.1128/jb.178.15.4429-4437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juan C, Zamorano L, Mena A, Albertí S, Pérez JL, Oliver A. Metallo-beta-lactamase-producing Pseudomonas putida as a reservoir of multidrug resistance elements that can be transferred to successful Pseudomonas aeruginosa clones. J Antimicrob Chemother. 2010;65:474–8. doi: 10.1093/jac/dkp491. [DOI] [PubMed] [Google Scholar]
- 28.Marchiaro P, Viale AM, Ballerini V, Rossignol G, Vila AJ, Limansky A. First report of a Tn402-like class 1 integron carrying blaVIM-2 in Pseudomonas putida from Argentina. J Infect Dev Ctries. 2010;4:412–6. [PubMed] [Google Scholar]
- 29.Rowe-Magnus DA, Guerout AM, Mazel D. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol Microbiol. 2002;43:1657–69. doi: 10.1046/j.1365-2958.2002.02861.x. [DOI] [PubMed] [Google Scholar]
- 30.Collis CM, Hall RM. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol Microbiol. 1992;6:2875–85. doi: 10.1111/j.1365-2958.1992.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 31.Recchia GD, Hall RM. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–27. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 32.Loot C, Ducos-Galand M, Escudero JA, Bouvier M, Mazel D. Replicative resolution of integron cassette insertion. Nucleic Acids Res. 2012;40:8361–70. doi: 10.1093/nar/gks620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fluit AC, Schmitz FJ. Class 1 integrons, gene cassettes, mobility and epidemiology. Eur J Clin Microbiol Infect Dis. 1999;18:761–70. doi: 10.1007/s100960050398. [DOI] [PubMed] [Google Scholar]
- 34.Jové T, Da Re S, Denis F, Mazel D, Ploy MC. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet. 2010;6:e1000793. doi: 10.1371/journal.pgen.1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collis CM, Hall RM. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 1995;39:155–62. doi: 10.1128/AAC.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quiroga MP, Andres P, Petroni A, Soler Bistué AJ, Guerriero L, Vargas LJ, et al. Complex class 1 integrons with diverse variable regions, including aac(6′)-Ib-cr and a novel allele, qnrB10, associated with ISCR1 in clinical enterobacterial isolates from Argentina. Antimicrob Agents Chemother. 2007;51:4466–70. doi: 10.1128/AAC.00726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song JS, Jang SJ, Bae IK, Lee HJ, Jeong BC, Lee SH. New complex class 1 integron carrying an ISCR1 element in Escherichia coli clinical isolates harbouring the blaCMY-11 gene. J Med Microbiol. 2010;59:132–4. doi: 10.1099/jmm.0.012559-0. [DOI] [PubMed] [Google Scholar]
- 38.Toleman MA, Bennett PM, Walsh TRIS. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev. 2006;70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toleman MA, Bennett PM, Walsh TR. Common regions e.g. orf513 and antibiotic resistance: IS91-like elements evolving complex class 1 integrons. J Antimicrob Chemother. 2006;58:1–6. doi: 10.1093/jac/dkl204. [DOI] [PubMed] [Google Scholar]
- 40.Nandi S, Maurer JJ, Hofacre C, Summers AO. Gram-positive bacteria are a major reservoir of Class 1 antibiotic resistance integrons in poultry litter. Proc Natl Acad Sci U S A. 2004;101:7118–22. doi: 10.1073/pnas.0306466101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nesvera J, Hochmannová J, Pátek M. An integron of class 1 is present on the plasmid pCG4 from gram-positive bacterium Corynebacterium glutamicum. FEMS Microbiol Lett. 1998;169:391–5. doi: 10.1111/j.1574-6968.1998.tb13345.x. [DOI] [PubMed] [Google Scholar]
- 42.Shi L, Zheng M, Xiao Z, Asakura M, Su J, Li L, et al. Unnoticed spread of class 1 integrons in gram-positive clinical strains isolated in Guangzhou, China. Microbiol Immunol. 2006;50:463–7. doi: 10.1111/j.1348-0421.2006.tb03815.x. [DOI] [PubMed] [Google Scholar]
- 43.Xu Z, Li L, Shi L, Shirtliff ME. Class 1 integron in staphylococci. Mol Biol Rep. 2011;38:5261–79. doi: 10.1007/s11033-011-0676-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Z, Shi L, Alam MJ, Li L, Yamasaki S. Integron-bearing methicillin-resistant coagulase-negative staphylococci in South China, 2001-2004. FEMS Microbiol Lett. 2008;278:223–30. doi: 10.1111/j.1574-6968.2007.00994.x. [DOI] [PubMed] [Google Scholar]
- 45.Gu B, Tong M, Zhao W, Liu G, Ning M, Pan S, et al. Prevalence and characterization of class I integrons among Pseudomonas aeruginosa and Acinetobacter baumannii isolates from patients in Nanjing, China. J Clin Microbiol. 2007;45:241–3. doi: 10.1128/JCM.01318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu H, Su Z, Wang S, Dai X, Chen J, Kong F, et al. Four novel resistance integron gene-cassette occurrences in bacterial isolates from zhenjiang, china. Curr Microbiol. 2009;59:113–7. doi: 10.1007/s00284-009-9405-z. [DOI] [PubMed] [Google Scholar]
- 47.Crowley D, Daly M, Lucey B, Shine P, Collins JJ, Cryan B, et al. Molecular epidemiology of cystic fibrosis-linked Burkholderia cepacia complex isolates from three national referral centres in Ireland. J Appl Microbiol. 2002;92:992–1004. doi: 10.1046/j.1365-2672.2002.01612.x. [DOI] [PubMed] [Google Scholar]
- 48.White PA, McIver CJ, Rawlinson WD. Integrons and gene cassettes in the enterobacteriaceae. Antimicrob Agents Chemother. 2001;45:2658–61. doi: 10.1128/AAC.45.9.2658-2661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Antunes P, Machado J, Peixe L. Characterization of antimicrobial resistance and class 1 and 2 integrons in Salmonella enterica isolates from different sources in Portugal. J Antimicrob Chemother. 2006;58:297–304. doi: 10.1093/jac/dkl242. [DOI] [PubMed] [Google Scholar]
- 50.Krauland MG, Marsh JW, Paterson DL, Harrison LH. Integron-mediated multidrug resistance in a global collection of nontyphoidal Salmonella enterica isolates. Emerg Infect Dis. 2009;15:388–96. doi: 10.3201/eid1503.081131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falbo V, Carattoli A, Tosini F, Pezzella C, Dionisi AM, Luzzi I. Antibiotic resistance conferred by a conjugative plasmid and a class I integron in Vibrio cholerae O1 El Tor strains isolated in Albania and Italy. Antimicrob Agents Chemother. 1999;43:693–6. doi: 10.1128/aac.43.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang HY, Jeong YS, Oh JY, Tae SH, Choi CH, Moon DC, et al. Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J Antimicrob Chemother. 2005;55:639–44. doi: 10.1093/jac/dki076. [DOI] [PubMed] [Google Scholar]
- 53.Antunes P, Machado J, Sousa JC, Peixe L. Dissemination amongst humans and food products of animal origin of a Salmonella typhimurium clone expressing an integron-borne OXA-30 beta-lactamase. J Antimicrob Chemother. 2004;54:429–34. doi: 10.1093/jac/dkh333. [DOI] [PubMed] [Google Scholar]
- 54.Sepp E, Stsepetova J, Lõivukene K, Truusalu K, Kõljalg S, Naaber P, et al. The occurrence of antimicrobial resistance and class 1 integrons among commensal Escherichia coli isolates from infants and elderly persons. Ann Clin Microbiol Antimicrob. 2009;8:34. doi: 10.1186/1476-0711-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Machado E, Coque TM, Cantón R, Sousa JC, Peixe L. Antibiotic resistance integrons and extended-spectrum β-lactamases among Enterobacteriaceae isolates recovered from chickens and swine in Portugal. J Antimicrob Chemother. 2008;62:296–302. doi: 10.1093/jac/dkn179. [DOI] [PubMed] [Google Scholar]
- 56.Vo AT, van Duijkeren E, Fluit AC, Gaastra W. Characteristics of extended-spectrum cephalosporin-resistant Escherichia coli and Klebsiella pneumoniae isolates from horses. Vet Microbiol. 2007;124:248–55. doi: 10.1016/j.vetmic.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 57.Meng H, Zhang Z, Chen M, Su Y, Li L, Miyoshi S, et al. Characterization and horizontal transfer of class 1 integrons in Salmonella strains isolated from food products of animal origin. Int J Food Microbiol. 2011;149:274–7. doi: 10.1016/j.ijfoodmicro.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 58.Agersø Y, Sandvang D. Class 1 integrons and tetracycline resistance genes in alcaligenes, arthrobacter and Pseudomonas spp. isolated from pigsties and manured soil. Appl Environ Microbiol. 2005;71:7941–7. doi: 10.1128/AEM.71.12.7941-7947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Da Silva GJ, Correia M, Vital C, Ribeiro G, Sousa JC, Leitão R, et al. Molecular characterization of bla(IMP-5), a new integron-borne metallo-β-lactamase gene from an Acinetobacter baumannii nosocomial isolate in Portugal. FEMS Microbiol Lett. 2002;215:33–9. doi: 10.1016/S0378-1097(02)00896-0. [DOI] [PubMed] [Google Scholar]
- 60.Brízio A, Conceição T, Pimentel M, Da Silva G, Duarte A. High-level expression of IMP-5 carbapenemase owing to point mutation in the -35 promoter region of class 1 integron among Pseudomonas aeruginosa clinical isolates. Int J Antimicrob Agents. 2006;27:27–31. doi: 10.1016/j.ijantimicag.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 61.Ribera A, Vila J, Fernández-Cuenca F, Martínez-Martínez L, Pascual A, Beceiro A, et al. Spanish Group for Nosocomial Infection (GEIH) Type 1 integrons in epidemiologically unrelated Acinetobacter baumannii isolates collected at Spanish hospitals. Antimicrob Agents Chemother. 2004;48:364–5. doi: 10.1128/AAC.48.1.364-365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Delihas N. Small mobile sequences in bacteria display diverse structure/function motifs. Mol Microbiol. 2008;67:475–81. doi: 10.1111/j.1365-2958.2007.06068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Delihas N. Impact of small repeat sequences on bacterial genome evolution. Genome Biol Evol. 2011;3:959–73. doi: 10.1093/gbe/evr077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Filée J, Siguier P, Chandler M. Insertion sequence diversity in archaea. Microbiol Mol Biol Rev. 2007;71:121–57. doi: 10.1128/MMBR.00031-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han Y, Wessler SR. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 2010;38:e199. doi: 10.1093/nar/gkq862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blount ZD, Grogan DW. New insertion sequences of Sulfolobus: functional properties and implications for genome evolution in hyperthermophilic archaea. Mol Microbiol. 2005;55:312–25. doi: 10.1111/j.1365-2958.2004.04391.x. [DOI] [PubMed] [Google Scholar]
- 67.Yang G, Nagel DH, Feschotte C, Hancock CN, Wessler SR. Tuned for transposition: molecular determinants underlying the hyperactivity of a Stowaway MITE. Science. 2009;325:1391–4. doi: 10.1126/science.1175688. [DOI] [PubMed] [Google Scholar]
- 68.Domingues S, Nielsen KM, da Silva GJ. The blaIMP-5-carrying integron in a clinical Acinetobacter baumannii strain is flanked by miniature inverted-repeat transposable elements (MITEs) J Antimicrob Chemother. 2011;66:2667–8. doi: 10.1093/jac/dkr327. [DOI] [PubMed] [Google Scholar]
- 69.Gillings MR, Labbate M, Sajjad A, Giguère NJ, Holley MP, Stokes HW. Mobilization of a Tn402-like class 1 integron with a novel cassette array via flanking miniature inverted-repeat transposable element-like structures. Appl Environ Microbiol. 2009;75:6002–4. doi: 10.1128/AEM.01033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Domingues S, Duarte A, Nielsen KM, Da Silva GJ. Identification of a clinical isolate of Acinetobacter genomic species 10 with an integron-borne blaVIM-2 gene flanked by Miniature Inverted-Repeat Transposable Elements. Clin Microbiol Infect. 2012;18:S317. [Google Scholar]
- 71.Domingues S, Harms K, Fricke WF, Johnsen PJ, da Silva GJ, Nielsen KM. Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species. PLoS Pathog. 2012;8:e1002837. doi: 10.1371/journal.ppat.1002837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Poirel L, Carrër A, Pitout JD, Nordmann P. Integron mobilization unit as a source of mobility of antibiotic resistance genes. Antimicrob Agents Chemother. 2009;53:2492–8. doi: 10.1128/AAC.00033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–74. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miriagou V, Carattoli A, Tzelepi E, Villa L, Tzouvelekis LS. IS26-associated In4-type integrons forming multiresistance loci in enterobacterial plasmids. Antimicrob Agents Chemother. 2005;49:3541–3. doi: 10.1128/AAC.49.8.3541-3543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Doublet B, Praud K, Weill FX, Cloeckaert A. Association of IS26-composite transposons and complex In4-type integrons generates novel multidrug resistance loci in Salmonella genomic island 1. J Antimicrob Chemother. 2009;63:282–9. doi: 10.1093/jac/dkn500. [DOI] [PubMed] [Google Scholar]
- 76.Zienkiewicz M, Kern-Zdanowicz I, Gołebiewski M, Zyliñska J, Mieczkowski P, Gniadkowski M, et al. Mosaic structure of p1658/97, a 125-kilobase plasmid harboring an active amplicon with the extended-spectrum beta-lactamase gene blaSHV-5. Antimicrob Agents Chemother. 2007;51:1164–71. doi: 10.1128/AAC.00772-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tavakoli N, Comanducci A, Dodd HM, Lett MC, Albiger B, Bennett P. IS1294, a DNA element that transposes by RC transposition. Plasmid. 2000;44:66–84. doi: 10.1006/plas.1999.1460. [DOI] [PubMed] [Google Scholar]
- 78.Ferreira S, Paradela A, Velez J, Ramalheira E, Walsh TR, Mendo S. Carriage of qnrA1 and qnrB2, blaCTX-M15 and complex class 1 integron in a clinical multiresistant Citrobacter freundii isolate. Diagn Microbiol Infect Dis. 2010;67:188–90. doi: 10.1016/j.diagmicrobio.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 79.Bae IK, Lee YN, Lee WG, Lee SH, Jeong SH. Novel complex class 1 integron bearing an ISCR1 element in an Escherichia coli isolate carrying the blaCTX-M-14 gene. Antimicrob Agents Chemother. 2007;51:3017–9. doi: 10.1128/AAC.00279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xia R, Guo X, Zhang Y, Xu H. qnrVC-like gene located in a novel complex class 1 integron harboring the ISCR1 element in an Aeromonas punctata strain from an aquatic environment in Shandong Province, China. Antimicrob Agents Chemother. 2010;54:3471–4. doi: 10.1128/AAC.01668-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Navon-Venezia S, Chmelnitsky I, Leavitt A, Carmeli Y. Dissemination of the CTX-M-25 family beta-lactamases among Klebsiella pneumoniae, Escherichia coli and Enterobacter cloacae and identification of the novel enzyme CTX-M-41 in Proteus mirabilis in Israel. J Antimicrob Chemother. 2008;62:289–95. doi: 10.1093/jac/dkn182. [DOI] [PubMed] [Google Scholar]
- 82.Ben Slama K, Ben Sallem R, Jouini A, Rachid S, Moussa L, Sáenz Y, et al. Diversity of genetic lineages among CTX-M-15 and CTX-M-14 producing Escherichia coli strains in a Tunisian hospital. Curr Microbiol. 2011;62:1794–801. doi: 10.1007/s00284-011-9930-4. [DOI] [PubMed] [Google Scholar]
- 83.Roberts AP, Chandler M, Courvalin P, Guédon G, Mullany P, Pembroke T, et al. Revised nomenclature for transposable genetic elements. Plasmid. 2008;60:167–73. doi: 10.1016/j.plasmid.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shapiro JA, Sporn P. Tn402: a new transposable element determining trimethoprim resistance that inserts in bacteriophage lambda. J Bacteriol. 1977;129:1632–5. doi: 10.1128/jb.129.3.1632-1635.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kamali-Moghaddam M, Sundström L. Transposon targeting determined by resolvase. FEMS Microbiol Lett. 2000;186:55–9. doi: 10.1111/j.1574-6968.2000.tb09081.x. [DOI] [PubMed] [Google Scholar]
- 86.Labbate M, Roy Chowdhury P, Stokes HW. A class 1 integron present in a human commensal has a hybrid transposition module compared to Tn402: evidence of interaction with mobile DNA from natural environments. J Bacteriol. 2008;190:5318–27. doi: 10.1128/JB.00199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rådström P, Sköld O, Swedberg G, Flensburg J, Roy PH, Sundström L. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu and the retroelements. J Bacteriol. 1994;176:3257–68. doi: 10.1128/jb.176.11.3257-3268.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sajjad A, Holley MP, Labbate M, Stokes HW, Gillings MR. Preclinical class 1 integron with a complete Tn402-like transposition module. Appl Environ Microbiol. 2011;77:335–7. doi: 10.1128/AEM.02142-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liebert CA, Hall RM, Summers AO. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev. 1999;63:507–22. doi: 10.1128/mmbr.63.3.507-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tato M, Coque TM, Baquero F, Cantón R. Dispersal of carbapenemase blaVIM-1 gene associated with different Tn402 variants, mercury transposons and conjugative plasmids in Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2010;54:320–7. doi: 10.1128/AAC.00783-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martínez N, Mendoza MC, Rodríguez I, Soto S, Bances M, Rodicio MR. Detailed structure of integrons and transposons carried by large conjugative plasmids responsible for multidrug resistance in diverse genomic types of Salmonella enterica serovar Brandenburg. J Antimicrob Chemother. 2007;60:1227–34. doi: 10.1093/jac/dkm336. [DOI] [PubMed] [Google Scholar]
- 92.Nógrády N, Pászti J, Pikó H, Nagy B. Class 1 integrons and their conjugal transfer with and without virulence-associated genes in extra-intestinal and intestinal Escherichia coli of poultry. Avian Pathol. 2006;35:349–56. doi: 10.1080/03079450600827007. [DOI] [PubMed] [Google Scholar]
- 93.Reith ME, Singh RK, Curtis B, Boyd JM, Bouevitch A, Kimball J, et al. The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics. 2008;9:427. doi: 10.1186/1471-2164-9-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.de la Cruz F, Grinsted J. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol. 1982;151:222–8. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grinsted J, de la Cruz F, Schmitt R. The Tn21 subgroup of bacterial transposable elements. Plasmid. 1990;24:163–89. doi: 10.1016/0147-619X(90)90001-S. [DOI] [PubMed] [Google Scholar]
- 96.Petrova M, Gorlenko Z, Mindlin S. Tn5045, a novel integron-containing antibiotic and chromate resistance transposon isolated from a permafrost bacterium. Res Microbiol. 2011;162:337–45. doi: 10.1016/j.resmic.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 97.Sundström L, Swedberg G, Sköld O. Characterization of transposon Tn5086, carrying the site-specifically inserted gene dhfrVII mediating trimethoprim resistance. J Bacteriol. 1993;175:1796–805. doi: 10.1128/jb.175.6.1796-1805.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tseng SP, Hsueh PR, Tsai JC, Teng LJ. Tn6001, a transposon-like element containing the blaVIM-3-harboring integron In450. Antimicrob Agents Chemother. 2007;51:4187–90. doi: 10.1128/AAC.00542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Scott JR. Sex and the single circle: conjugative transposition. J Bacteriol. 1992;174:6005–10. doi: 10.1128/jb.174.19.6005-6010.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boyd EF, Almagro-Moreno S, Parent MA. Genomic islands are dynamic, ancient integrative elements in bacterial evolution. Trends Microbiol. 2009;17:47–53. doi: 10.1016/j.tim.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 101.Krizova L, Nemec A. A 63 kb genomic resistance island found in a multidrug-resistant Acinetobacter baumannii isolate of European clone I from 1977. J Antimicrob Chemother. 2010;65:1915–8. doi: 10.1093/jac/dkq223. [DOI] [PubMed] [Google Scholar]
- 102.Waldor MK. Mobilizable genomic islands: going mobile with oriT mimicry. Mol Microbiol. 2010;78:537–40. doi: 10.1111/j.1365-2958.2010.07365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Juhas M, Crook DW, Hood DW. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell Microbiol. 2008;10:2377–86. doi: 10.1111/j.1462-5822.2008.01187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, et al. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2006;2:e7. doi: 10.1371/journal.pgen.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Post V, Hall RM. AbaR5, a large multiple-antibiotic resistance region found in Acinetobacter baumannii. Antimicrob Agents Chemother. 2009;53:2667–71. doi: 10.1128/AAC.01407-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Post V, White PA, Hall RM. Evolution of AbaR-type genomic resistance islands in multiply antibiotic-resistant Acinetobacter baumannii. J Antimicrob Chemother. 2010;65:1162–70. doi: 10.1093/jac/dkq095. [DOI] [PubMed] [Google Scholar]
- 107.Krizova L, Dijkshoorn L, Nemec A. Diversity and evolution of AbaR genomic resistance islands in Acinetobacter baumannii strains of European clone I. Antimicrob Agents Chemother. 2011;55:3201–6. doi: 10.1128/AAC.00221-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hall RM. Salmonella genomic islands and antibiotic resistance in Salmonella enterica. Future Microbiol. 2010;5:1525–38. doi: 10.2217/fmb.10.122. [DOI] [PubMed] [Google Scholar]
- 109.Douard G, Praud K, Cloeckaert A, Doublet B. The Salmonella genomic island 1 is specifically mobilized in trans by the IncA/C multidrug resistance plasmid family. PLoS One. 2010;5:e15302. doi: 10.1371/journal.pone.0015302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ahmed AM, Hussein AI, Shimamoto T. Proteus mirabilis clinical isolate harbouring a new variant of Salmonella genomic island 1 containing the multiple antibiotic resistance region. J Antimicrob Chemother. 2007;59:184–90. doi: 10.1093/jac/dkl471. [DOI] [PubMed] [Google Scholar]
- 111.Rajakumar K, Bulach D, Davies J, Ambrose L, Sasakawa C, Adler B. Identification of a chromosomal Shigella flexneri multi-antibiotic resistance locus which shares sequence and organizational similarity with the resistance region of the plasmid NR1. Plasmid. 1997;37:159–68. doi: 10.1006/plas.1997.1280. [DOI] [PubMed] [Google Scholar]
- 112.Roy Chowdhury P, Merlino J, Labbate M, Cheong EY, Gottlieb T, Stokes HW. Tn6060, a transposon from a genomic island in a Pseudomonas aeruginosa clinical isolate that includes two class 1 integrons. Antimicrob Agents Chemother. 2009;53:5294–6. doi: 10.1128/AAC.00687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Skippington E, Ragan MA. Lateral genetic transfer and the construction of genetic exchange communities. FEMS Microbiol Rev. 2011;35:707–35. doi: 10.1111/j.1574-6976.2010.00261.x. [DOI] [PubMed] [Google Scholar]
- 114.Clark NC, Olsvik O, Swenson JM, Spiegel CA, Tenover FC. Detection of a streptomycin/spectinomycin adenylyltransferase gene (aadA) in Enterococcus faecalis. Antimicrob Agents Chemother. 1999;43:157–60. doi: 10.1128/aac.43.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu SY, Lin JY, Chu C, Su LH, Lin TY, Chiu CH. Integron-associated imipenem resistance in Acinetobacter baumannii isolated from a regional hospital in Taiwan. Int J Antimicrob Agents. 2006;27:81–4. doi: 10.1016/j.ijantimicag.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 116.Soto SM, Lobato MJ, Mendoza MC. Class 1 integron-borne gene cassettes in multidrug-resistant Yersinia enterocolitica strains of different phenotypic and genetic types. Antimicrob Agents Chemother. 2003;47:421–6. doi: 10.1128/AAC.47.1.421-425.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hu Z, Zhao WH. Identification of plasmid- and integron-borne blaIMP-1 and blaIMP-10 in clinical isolates of Serratia marcescens. J Med Microbiol. 2009;58:217–21. doi: 10.1099/jmm.0.006874-0. [DOI] [PubMed] [Google Scholar]
- 118.Moubareck C, Brémont S, Conroy MC, Courvalin P, Lambert T. GES-11, a novel integron-associated GES variant in Acinetobacter baumannii. Antimicrob Agents Chemother. 2009;53:3579–81. doi: 10.1128/AAC.00072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hanlon GW. Bacteriophages: an appraisal of their role in the treatment of bacterial infections. Int J Antimicrob Agents. 2007;30:118–28. doi: 10.1016/j.ijantimicag.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 120.Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–32. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 121.Matic I, Taddei F, Radman M. Genetic barriers among bacteria. Trends Microbiol. 1996;4:69–72. doi: 10.1016/0966-842X(96)81514-9. [DOI] [PubMed] [Google Scholar]
- 122.Davison J. Genetic exchange between bacteria in the environment. Plasmid. 1999;42:73–91. doi: 10.1006/plas.1999.1421. [DOI] [PubMed] [Google Scholar]
- 123.Schmieger H, Schicklmaier P. Transduction of multiple drug resistance of Salmonella enterica serovar typhimurium DT104. FEMS Microbiol Lett. 1999;170:251–6. doi: 10.1111/j.1574-6968.1999.tb13381.x. [DOI] [PubMed] [Google Scholar]
- 124.Johnsborg O, Eldholm V, Håvarstein LS. Natural genetic transformation: prevalence, mechanisms and function. Res Microbiol. 2007;158:767–78. doi: 10.1016/j.resmic.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 125.Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004;2:241–9. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 126.Ramírez MS, Merkier AK, Quiroga MP, Centrón D. Acinetobacter baumannii is able to gain and maintain a plasmid harbouring In35 found in Enterobacteriaceae isolates from Argentina. Curr Microbiol. 2012;64:211–3. doi: 10.1007/s00284-011-0052-9. [DOI] [PubMed] [Google Scholar]
- 127.Gestal AM, Liew EF, Coleman NV. Natural transformation with synthetic gene cassettes: new tools for integron research and biotechnology. Microbiology. 2011;157:3349–60. doi: 10.1099/mic.0.051623-0. [DOI] [PubMed] [Google Scholar]
- 128.Partridge SR, Recchia GD, Stokes HW, Hall RM. Family of class 1 integrons related to In4 from Tn1696. Antimicrob Agents Chemother. 2001;45:3014–20. doi: 10.1128/AAC.45.11.3014-3020.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sinclair MI, Holloway BW. A chromosomally located transposon in Pseudomonas aeruginosa. J Bacteriol. 1982;151:569–79. doi: 10.1128/jb.151.2.569-579.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Partridge SR, Brown HJ, Hall RM. Characterization and movement of the class 1 integron known as Tn2521 and Tn1405. Antimicrob Agents Chemother. 2002;46:1288–94. doi: 10.1128/AAC.46.5.1288-1294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Petrovski S, Stanisich VA. Tn502 and Tn512 are res site hunters that provide evidence of resolvase-independent transposition to random sites. J Bacteriol. 2010;192:1865–74. doi: 10.1128/JB.01322-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mazodier P, Davies J. Gene transfer between distantly related bacteria. Annu Rev Genet. 1991;25:147–71. doi: 10.1146/annurev.ge.25.120191.001051. [DOI] [PubMed] [Google Scholar]
- 133.Pettersen AK, Bøhn T, Primicerio R, Shorten PR, Soboleva TK, Nielsen KM. Modeling suggests frequency estimates are not informative for predicting the long-term effect of horizontal gene transfer in bacteria. Environ Biosafety Res. 2005;4:223–33. doi: 10.1051/ebr:2006008. [DOI] [PubMed] [Google Scholar]
- 134.Townsend JP, Bøhn T, Nielsen KM. Assessing the probability of detection of horizontal gene transfer events in bacterial populations. Front Microbiol. 2012;3:27. doi: 10.3389/fmicb.2012.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Harrison E, Brockhurst MA. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 2012;20:262–7. doi: 10.1016/j.tim.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 136.Dionisio F, Conceição IC, Marques AC, Fernandes L, Gordo I. The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol Lett. 2005;1:250–2. doi: 10.1098/rsbl.2004.0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Johnsen PJ, Townsend JP, Bøhn T, Simonsen GS, Sundsfjord A, Nielsen KM. Retrospective evidence for a biological cost of vancomycin resistance determinants in the absence of glycopeptide selective pressures. J Antimicrob Chemother. 2011;66:608–10. doi: 10.1093/jac/dkq512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Björkman J, Andersson DI. The cost of antibiotic resistance from a bacterial perspective. Drug Resist Updat. 2000;3:237–45. doi: 10.1054/drup.2000.0147. [DOI] [PubMed] [Google Scholar]
- 139.Andersson DI. Persistence of antibiotic resistant bacteria. Curr Opin Microbiol. 2003;6:452–6. doi: 10.1016/j.mib.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 140.Enne VI, Delsol AA, Davis GR, Hayward SL, Roe JM, Bennett PM. Assessment of the fitness impacts on Escherichia coli of acquisition of antibiotic resistance genes encoded by different types of genetic element. J Antimicrob Chemother. 2005;56:544–51. doi: 10.1093/jac/dki255. [DOI] [PubMed] [Google Scholar]
- 141.Bergmiller T, Ackermann M, Silander OK. Patterns of evolutionary conservation of essential genes correlate with their compensability. PLoS Genet. 2012;8:e1002803. doi: 10.1371/journal.pgen.1002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Geissler B, Elraheb D, Margolin W. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci U S A. 2003;100:4197–202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Denamur E, Matic I. Evolution of mutation rates in bacteria. Mol Microbiol. 2006;60:820–7. doi: 10.1111/j.1365-2958.2006.05150.x. [DOI] [PubMed] [Google Scholar]
- 144.Johnsen PJ, Townsend JP, Bøhn T, Simonsen GS, Sundsfjord A, Nielsen KM. Factors affecting the reversal of antimicrobial-drug resistance. Lancet Infect Dis. 2009;9:357–64. doi: 10.1016/S1473-3099(09)70105-7. [DOI] [PubMed] [Google Scholar]
- 145.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–71. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 146.Luo N, Pereira S, Sahin O, Lin J, Huang S, Michel L, et al. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc Natl Acad Sci U S A. 2005;102:541–6. doi: 10.1073/pnas.0408966102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pränting M, Negrea A, Rhen M, Andersson DI. Mechanism and fitness costs of PR-39 resistance in Salmonella enterica serovar Typhimurium LT2. Antimicrob Agents Chemother. 2008;52:2734–41. doi: 10.1128/AAC.00205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Barton NH. Genetic hitchhiking. Philos Trans R Soc Lond B Biol Sci. 2000;355:1553–62. doi: 10.1098/rstb.2000.0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kim Y, Stephan W. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics. 2002;160:765–77. doi: 10.1093/genetics/160.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Levin BR, Bergstrom CT. Bacteria are different: observations, interpretations, speculations and opinions about the mechanisms of adaptive evolution in prokaryotes. Proc Natl Acad Sci U S A. 2000;97:6981–5. doi: 10.1073/pnas.97.13.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hochhut B, Lotfi Y, Mazel D, Faruque SM, Woodgate R, Waldor MK. Molecular analysis of antibiotic resistance gene clusters in vibrio cholerae O139 and O1 SXT constins. Antimicrob Agents Chemother. 2001;45:2991–3000. doi: 10.1128/AAC.45.11.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Starikova I, Harms K, Kaugen P, Lunde TTM, Primicerio R, Samuelsen O, et al. A trade-off between the fitness cost of functional integrases and long-term stability of integrons. Plos Pathog. 2012;8:e1003043. doi: 10.1371/journal.ppat.1003043. [DOI] [PMC free article] [PubMed] [Google Scholar]