Abstract
The mode of reproduction has been predicted to affect the proliferation of transposable elements (TEs). A population that switches from sexual to asexual reproduction could either accumulate TEs because purifying selection becomes less efficient, or a decrease in TE load because the opportunity for horizontal transmission is reduced. A third possibility is that the mechanism that induces asexual reproduction affects TE dynamics as a side effect. We propose two such mechanisms that might explain recently described patterns of TE abundance in sexual and asexual lineages of the parasitoid wasp Leptopilina clavipes. Asexual reproduction in this species is induced by endosymbiotic Wolbachia bacteria. In order to achieve parthenogenesis in its host, Wolbachia might remove methylation or interfere with Argonaute proteins. Both methylation and Argonaute proteins are known to control TE activity in other species. By interfering with either, Wolbachia might therefore secondarily hamper the control of specific TEs.
Keywords: transposable elements, sex, asexual reproduction, methylation, Argonaute
The relationship between mode of reproduction and transposable element (TE) dynamics has been the topic of considerable debate (summarized in ref. 1). On the one hand, purifying selection is expected to be less efficient in asexual compared with sexual taxa, leading to an accumulation of TE copies in asexuals. On the other hand, sex allows horizontal transmission of TEs and will facilitate the spread of TEs. Which of these driving factors, if any, will be most important is currently an unresolved question. Recent advances in DNA sequencing technology now allow us to address this question on a genome-wide scale. In a recent paper published in Molecular Ecology, we quantified TE loads in sexual and asexual lineages of the parasitoid wasp Leptopilina clavipes.2 Parthenogenesis in this species is induced by endosymbiotic Wolbachia bacteria, that are thought to have infected L. clavipes several thousand years ago.3 Uninfected lineages reproduce sexually. The results of our study were inconsistent with models that predict increases4 or decreases5 in TE load in asexuals compared with sexuals, regardless of TE type. Instead, we found markedly different patterns between the various types of TEs. Loads of DNA transposons were higher in asexuals, while there was no difference between sexuals and asexuals for LTR and LINE-like TEs, except for one or a few gypsy-like LTR elements. The reasons for these patterns have already been the subject of some speculation.1,2 Here, we elaborate on the possibility that TE dynamics are affected by Wolbachia. More precisely, we suggest that in order to induce parthenogenesis, Wolbachia has to interfere with host cellular processes, which secondarily also interferes with the control of TE activity. While these suggestions are purely speculative at this moment, we discuss them here because we believe that such processes could be of widespread importance.
We suggest two ways in which Wolbachia-induced manipulation of the host reproductive machinery could interfere with the repression of particular TE types. These mechanisms are illustrated in Figures 1 and 2.
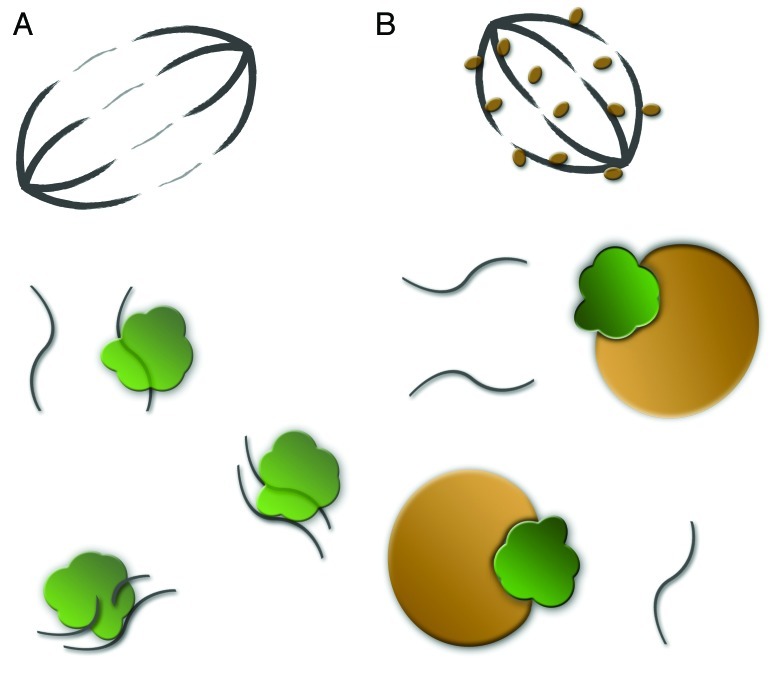
Figure 1. (A) Cartoon of a normal dividing cell with meotic/mitotic spindle at the top. Complexes of Argonaute proteins (green) and antisense TE fragments capture and destroy TE mRNAs. (B) Dividing cell infected with Wolbachia. Wolbachia (brown) associate with microtubuli (top) and capture Argonaute proteins (green). TE derived mRNAs are left to insert back into the host genome.
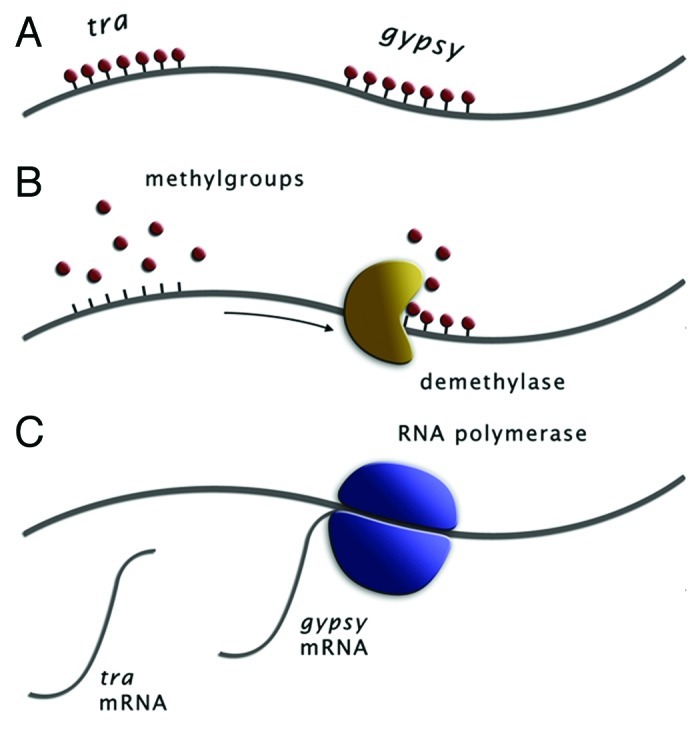
Figure 2. Cartoon of chromosomal region in female germline. (A) The sex determination gene tra is methylated as well as a nearby gypsy element. (B) In order to induce female development, Wolbachia produces a demethylase that removes methyl groups. (C) In the absence of methylation, both tra and gyspsy can be transcribed.
First, Wolbachia might interfere with the normal functioning of proteins from the Argonaute family (Fig. 1). Argonaute-like proteins are involved in many cellular processes, including cell division and gametogenesis.6 In order for Wolbachia to make unfertilized L. clavipes eggs develop as females, it has to ensure that these become diploid. It does so by preventing chromosome segregation at the first mitotic division after meiosis.7 Thus, in gametes infected by Wolbachia, the chromosomes duplicate, condense, but then enter G1 without completing mitosis or cytokinesis. The molecular mechanism through which Wolbachia achieves this effect is currently unknown. However, one way for Wolbachia to prevent the chromosomes from separating after duplication might be to interfere with Argonaute proteins. In mice for example, mutants defective for a protein from the Argonaute family show arrest during early meiosis.8
In addition to their role in cell cycle regulation, Argonaute proteins play an important role in the control of TE activity, through a mechanism known as the ping-pong model.9,10 Briefly, Argonaute proteins form a complex with short anti-sense sequences transcribed from defective TEs. These target full-length TE transcripts (Fig. 1A), which they then degrade, resulting in new short sense TE fragments that can bind to other Argonaute proteins. These in turn target antisense transcripts from the defective TEs, resulting in more antisense bait, and so on. If Wolbachia would interfere with the abundance or functioning of Argonaute proteins as suggested above, it would automatically hamper the Argonaute-driven capturing and degradation of TE mRNA (Fig. 1B). These TE transcripts are then left free to be reverse transcribed into cDNA and pasted back into the genome.
A second way in which Wolbachia-induced manipulation of the host could lead to proliferation of TEs is by disturbing normal patterns of DNA methylation. To make unfertilized eggs develop as females, it is not enough for Wolbachia to cause diplodization of the gametes as described above. Wolbachia also has to prevent diploid zygotes from developing as diploid males. To do so, Wolbachia has to manipulate the host's sex determination mechanism. Several sex determination mechanisms are known in hymenoptera11 and it is currently unknown which of these applies to L. clavipes. However, since strong inbreeding does not result in diploid males in L. clavipes (Kraaijeveld, personal observation), sex determination is unlikely to be based on allelic differences at one or a few genetic loci as in for example the honey bee Apis melifera. We therefore assume that sex determination in L. clavipes is most likely similar to that described for another parasitoid wasp, Nasonia vitripennis. In Nasonia, female development requires at least one active copy of the gene transformer (tra) or a trans-acting factor that regulates tra expression.12 Tra is silenced in the female germline, so simple gamete duplication would result in two silenced copies of tra and hence male development. In the male germline, however, tra is not silenced and males transfer an active copy of tra to their offspring. Fertilized offspring therefore inherit both an active and a silenced copy of tra and develop as females. To achieve female development of diploidized zygotes, Wolbachia has to emulate the male germline and remove silencing of tra. The mechanism through which tra is silenced is not known, but may involve DNA methylation. If so, Wolbachia could either demethylate the host genome completely, or remove methylation only from tra. In the latter case, demethylation could spread to nearby regions of the genome, analogous to the spread of methylation from silenced TEs to nearby genes that has been observed in several species.13-15
Methylation is a common way of silencing TEs. For example, mutant Arabidopsis plants that are defective in their methylation machinery experience bursts of TEs that are normally silent.16 If Wolbachia removes methylation marks in a non-specific manner to induce female development of the zygote, it may also demethylate nearby TEs, thereby reactivating them (Fig. 2).
Whether either of the above mechanisms actually operates in the L. clavipes - Wolbachia system is at this stage unknown. We made a start testing the methylation hypothesis by checking for methylation of gypsy in sexual and asexual L. clavipes. We found that gypsy was not methylated in either,2 suggesting that hypothesis 2 cannot account for the high copy number of gypsy in asexual L. clavipes. We have not tested the first hypothesis. Our reason for elaborating on the ideas here is because mechanisms like these could play an important role in many systems. Molecular mechanisms that control TE proliferation are often closely related to other important processes. As we point out here, methylation controls the transcription of host genes and TEs. Likewise, the ping-pong model for controlling TEs post-transcriptionally contains components that are important in a wide variety of cellular processes. Other mechanisms that control TEs may similarly have other functions in host cells. It follows that interference of methylation, Argonaute proteins or other mechanisms by endosymbiotic bacteria or other environmental factors would disrupt multiple processes at once, including TE control.
Acknowledgments
We thank Bart Pannebakker for helpful comments on an earlier draft of this paper.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/22878
References
- 1.Crespi B, Schwander T. Asexual evolution: do intragenomic parasites maintain sex? Mol Ecol. 2012;21:3893–5. doi: 10.1111/j.1365-294X.2012.05638.x. [DOI] [PubMed] [Google Scholar]
- 2.Kraaijeveld K, Zwanenburg B, Hubert B, Vieira C, De Pater S, Van Alphen JJM, et al. Transposon proliferation in an asexual parasitoid. Mol Ecol. 2012;21:3898–906. doi: 10.1111/j.1365-294X.2012.5582.x. [DOI] [PubMed] [Google Scholar]
- 3.Kraaijeveld K, Franco P, de Knijff P, Stouthamer R, van Alphen JJM. Clonal genetic variation in a Wolbachia-infected asexual wasp: horizontal transmission or historical sex? Mol Ecol. 2011;20:3644–52. doi: 10.1111/j.1365-294X.2011.05150.x. [DOI] [PubMed] [Google Scholar]
- 4.Dolgin ES, Charlesworth B. The fate of transposable elements in asexual populations. Genetics. 2006;174:817–27. doi: 10.1534/genetics.106.060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright S, Finnegan D. Genome evolution: sex and the transposable element. Curr Biol. 2001;11:R296–9. doi: 10.1016/S0960-9822(01)00168-3. [DOI] [PubMed] [Google Scholar]
- 6.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–76. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pannebakker BA, Pijnacker LP, Zwaan BJ, Beukeboom LW. Cytology of Wolbachia-induced parthenogenesis in Leptopilina clavipes (Hymenoptera: Figitidae) Genome. 2004;47:299–303. doi: 10.1139/g03-137. [DOI] [PubMed] [Google Scholar]
- 8.Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–14. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–4. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 10.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 11.Sánchez L. Sex-determining mechanisms in insects. Int J Dev Biol. 2008;52:837–56. doi: 10.1387/ijdb.072396ls. [DOI] [PubMed] [Google Scholar]
- 12.Verhulst EC, Beukeboom LW, van de Zande L. Maternal control of haplodiploid sex determination in the wasp Nasonia. Science. 2010;328:620–3. doi: 10.1126/science.1185805. [DOI] [PubMed] [Google Scholar]
- 13.Kinoshita Y, Saze H, Kinoshita T, Miura A, Soppe WJ, Koornneef M, et al. Control of FWA gene silencing in Arabidopsis thaliana by SINE-related direct repeats. Plant J. 2007;49:38–45. doi: 10.1111/j.1365-313X.2006.02936.x. [DOI] [PubMed] [Google Scholar]
- 14.Martin A, Troadec C, Boualem A, Rajab M, Fernandez R, Morin H, et al. A transposon-induced epigenetic change leads to sex determination in melon. Nature. 2009;461:1135–8. doi: 10.1038/nature08498. [DOI] [PubMed] [Google Scholar]
- 15.Rebollo R, Karimi MM, Bilenky M, Gagnier L, Miceli-Royer K, Zhang Y, et al. Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet. 2011;7:e1002301. doi: 10.1371/journal.pgen.1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukahara S, Kobayashi A, Kawabe A, Mathieu O, Miura A, Kakutani T. Bursts of retrotransposition reproduced in Arabidopsis. Nature. 2009;461:423–6. doi: 10.1038/nature08351. [DOI] [PubMed] [Google Scholar]