Abstract
The grain filling rate (GFR) is an important dynamic trait that determines the final grain yield and is controlled by a network of genes and environment factors. To determine the genetic basis of the GFR, a conditional quantitative trait locus (QTL) analysis method was conducted using time-related phenotypic values of the GFR collected from a set of 243 immortalized F2 (IF2) population, which were evaluated at two locations over 2 years. The GFR gradually rose in the 0–15 days after pollination (DAP) and 16–22 DAP, reaching a maximum at 23–29 DAP, and then gradually decreasing. The variation of kernel weight (KW) was mainly decided by the GFR, and not by the grain filling duration (GFD). Thirty-three different unconditional QTLs were identified for the GFR at the six sampling stages over 2 years. Among them, QTLs qGFR7b, qGFR9 and qGFR6d were identified at the same stages at two locations over 2 years. In addition, 14 conditional QTLs for GFR were detected at five stages. The conditional QTL qGFR7c was identified at stage V|IV (37–43 DAP) at two locations over 2 years, and qGFR7b was detected at the sixth stage (44–50 DAP) in all four environments, except at Anyang location in 2009. QTLs qQTL7b and qQTL6f were identified by unconditional and conditional QTL mapping at the same stages, and might represent major QTLs for regulating the GFR in maize in the IF2 population. Moreover, most of the QTLs identified were co-located with QTLs from previous studies that were associated with GFR, enzyme activities of starch synthesis, soluble carbohydrates, and grain filling related genes. These results indicated that the GFR is regulated by many genes, which are specifically expressed at different grain filling stages, and the specific expression of the genes between 16–35 DAP might be very important for deciding the final kernel weight.
Introduction
Grain yield has been a main target in cereal breeding, especially for maize (Zea mays L.), a critical source of food, fuel, feed, and fiber worldwide. [1] In maize, grain yield can be defined as the product of kernel sink capacity and grain filling efficiency, [2] and the GFR is regulated by multi-genes or by QTLs, as well as cultivation conditions, showing complex dynamic changes. To dissect the genetic bases of kernel development, certain genes corresponding to grain size or kernel development in maize, such as rgf1, sh1, sh2, dek1, mn1, and CNR1, have been cloned. [3]–[9] However, because of the difficulty in measuring natural variations in GFR, the molecular roles of genes or QTLs specifically expressed during grain filling have not been fully elucidated.
In cereal crops, grain filling is a critical and dynamic process that determines final grain yield. It depends on carbohydrates derived from two different sources: from photosynthesis in the leaf during the grain filling procedure and from accumulated nonstructural carbohydrates in culms and leaf sheaths. [10] The final kernel weight is mainly determined by the grain filling procedure. [11] In the field, the duration of grain filling is affected by changes in plant density and temperature, whereas the GFR is relative steady. [12] For maize, the final grain weight achieved by maize kernels is largely genetically determined. [13] However, factors such as assimilate availability, [14] the ‘sink capacity’ of an individual kernel, [12] kernel water content, [15], [16] leaf nitrogen dynamics, [10] related enzyme activity, [4] drought, [17] or high temperature [18] affect the GFR or GFD, limiting the achievement of maximum kernel weight.
For convenience, the grain filling procedure has been partitioned into three phases: the lag phase, the effective grain filling period and the maturation drying phase. [15] The lag phase is a period of active cell division, followed by differentiation and DNA endoreduplication, with almost no dry matter accumulation. During this phase, the GFR is low. [19] At the end of the lag phase, the GFR starts to rise and reaching its maximum value in the middle of the effective grain filling period. [15] In the effective grain filling period, the GFR and the duration of the effective grain filling period determine the final weight. [20] After reaching the maximum, the GFR gradually decreases, and the final kernel weight is achieved during the maturation drying phase. [15], [21] Although there are many factors that could affect the GFR during the three phases, the genotype has the most important role in affecting the GFR in cereal crops. [22], [23] Under these circumstances, the GFR shows a logistic curve during the grain filling procedure.
Recently, conditional QTL mapping has been used to dissect the genetic architecture of important quantitative traits in maize, such as plant height [24] and enzyme activity during grain development. [4] Although the GFR is an important developmental trait that directly decides the final grain yield, only three genes related to grain filling in maize and rice have been cloned: rgf1, GS5 and GIF1. [3], [11], [25] The GFR is an important factor that decides grain yield in maize; however, its genetic basis is unclear. In this study, a set of immortalized F2 (IF2) maize plants was used to dissect the genetic basis of the GFR using conditional and unconditional QTL mapping. The goal of this study was to: (1) dissect the genetic basis of the GFR in maize, and (2) identify the unconditional and conditional QTLs that controlled the GFR in the different processes of carbohydrates synthesis.
Results
Climate Conditions in the Two Locations
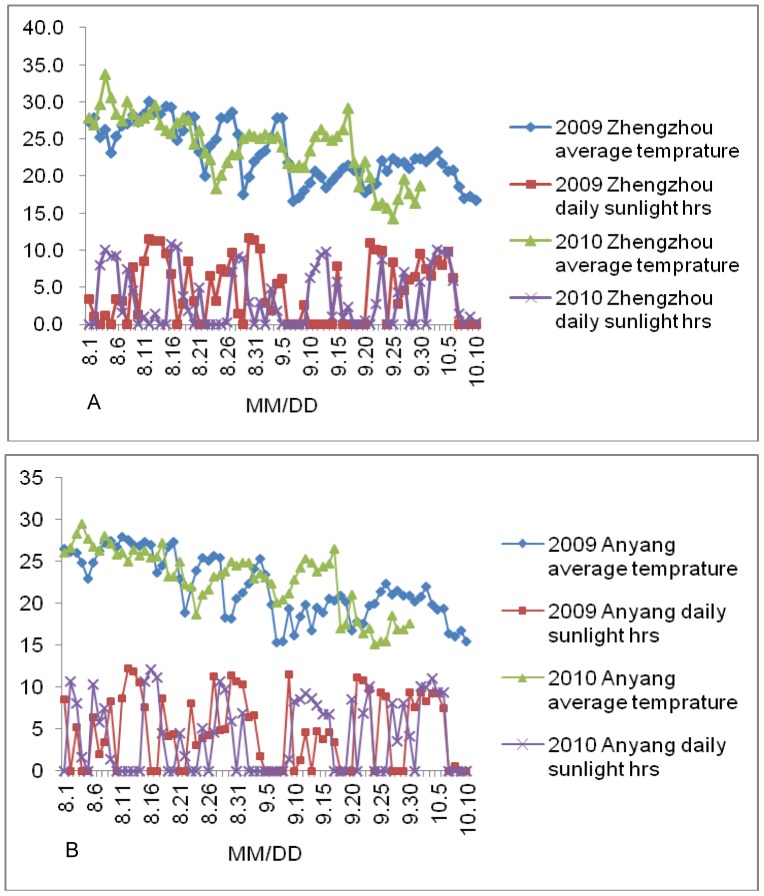
Temperature and sunlight conditions during the maize grain filling duration of the IF2 population across 2 years are shown in Fig. 1a and b. The results of variance analysis showed that the average temperature and sunlight were significant different between the 2 locations (at P = 0.01 significance level), there were large differences at according stages during grain filling duration in related climate factors between the two years at any one location. During grain filling duration at Zhengzhou location, the average of temperature in the IF2 population were 23.1°C and 24.0°C, and the average of daily sunlight were 4.2 hrs and 3.8 hrs in 2009 and 2010, respectively. In Anyang, the average temperature in the IF2 population was 21.9°C and 22.9°C, and the average of daily sunlight was 4.6 hrs and 4.1 hrs in 2009 and 2010, respectively.
Figure 1. Two main climate factors in maize grain filling duration in 2009–2010 at Zhengzhou (a) and Anyang (b).
Variations in GFR
For the two parents (Table 1), the average GFR increased over the initial two or three sampling stages (0–22 DAP in 2009 and 0–29 DAP in 2010), and then decreased over the next one or two sampling stages (30–36 DAP in 2010, 23–36 DAP at Zhengzhou and 23–29 DAP at Anyang in 2009), as did that of the hybrid Nongda 108. Comparing the hybrid and its parents, the maximum GFR of the hybrid in almost all environments was higher than that of the two parents, and the KW of the hybrid was also higher than that of both parents.
Table 1. Performance of grain filling rate, grain filling duration and final kernel weight in the immortalized F2 population at two locations.
| Year | Location | Population | KWa | GFDb | GFRc mg °Cd−1 kernel−1 | ||||||
| g 100−1 kernel−1 | °Cd | I | II | III | IV | V | VI | ||||
| 2009 | Zhengzhou | P1 | Mean | 28.30 | 1169.7 | 0.11 | 0.38 | 0.36 | 0.14 | 0.29 | 0.28 |
| P2 | Mean | 24.98 | 1163.4 | 0.08 | 0.42 | 0.367 | 0.152 | 0.377 | 0.251 | ||
| F1 | Mean | 29.52 | 1192.7 | 0.10 | 0.43 | 0.29 | 0.19 | 0.44 | 0.20 | ||
| IF2 | Mean±SD | 23.87±0.19 | 1178.2±0.8 | 0.06±0.001 | 0.34±0.01 | 0.36±0.01 | 0.28±0.01 | 0.26±0.01 | 0.21±0.01 | ||
| Range | 14.93–35.55 | 1146.8–1207.2 | 0.03–0.13 | 0.11–0.64 | 0.12–0.64 | 0.01–0.56 | 0.01–0.63 | 0.02–0.65 | |||
| Anyang | P1 | Mean | 27.99 | 1006.1 | 0.10 | 0.51 | 0.24 | 0.48 | 0.34 | 0.23 | |
| P2 | Mean | 23.82 | 1026.8 | 0.10 | 0.47 | 0.32 | 0.36 | 0.24 | 0.14 | ||
| F1 | Mean | 29.15 | 1036.4 | 0.11 | 0.53 | 0.19 | 0.44 | 0.42 | 0.24 | ||
| IF2 | Mean±SD | 23.88±0.19 | 1032.8±0.8 | 0.07±0.001 | 0.32±0.01 | 0.39±0.01 | 0.31±0.01 | 0.28±0.01 | 0.22±0.01 | ||
| Range | 16.32–34.36 | 937.7–1051.9 | 0.04–0.15 | 0.09–0.67 | 0.12–0.70 | 0.04–0.71 | 0.01–0.70 | 0.01–0.66 | |||
| 2010 | Zhengzhou | P1 | Mean | 36.38 | 1057.3 | 0.07 | 0.26 | 0.59 | 0.24 | 0.21 | 0.20 |
| P2 | Mean | 24.71 | 1097.4 | 0.08 | 0.36 | 0.43 | 0.07 | 0.39 | 0.22 | ||
| F1 | Mean | 28.32 | 1176.2 | 0.08 | 0.38 | 0.40 | 0.33 | 0.35 | 0.13 | ||
| IF2 | Mean±SD | 24.46±0.22 | 1136.4±3.6 | 0.06±0.001 | 0.31±0.01 | 0.34±0.01 | 0.29±0.01 | 0.27±0.01 | 0.25±0.01 | ||
| Range | 17.64–35.54 | 938.0–1200.9 | 0.03–0.13 | 0.17–0.58 | 0.16–0.57 | 0.06–0.64 | 0.06–0.58 | 0.04–0.76 | |||
| Anyang | P1 | Mean | 36.67 | 987.7 | 0.08 | 0.35 | 0.48 | 0.21 | 0.41 | 0.14 | |
| P2 | Mean | 25.13 | 1060.2 | 0.08 | 0.35 | 0.41 | 0.33 | 0.38 | 0.06 | ||
| F1 | Mean | 28.93 | 1096.5 | 0.09 | 0.25 | 0.52 | 0.34 | 0.42 | 0.26 | ||
| IF2 | Mean±SD | 24.38±0.20 | 1032.1±3.2 | 0.06±0.001 | 0.31±0.01 | 0.35±0.01 | 0.33±0.01 | 0.31±0.01 | 0.26±0.01 | ||
| Range | 17.29–34.41 | 743.6–1108.3 | 0.03–0.12 | 0.15–0.66 | 0.18–0.71 | 0.04–0.65 | 0.01–0.67 | 0.01–0.74 | |||
Note: aThe kernel weight in the IF2 population;
The total thermal time from pollination to the last sampling on average in the IF2 population;
The average grain filling rate in the IF2 population.
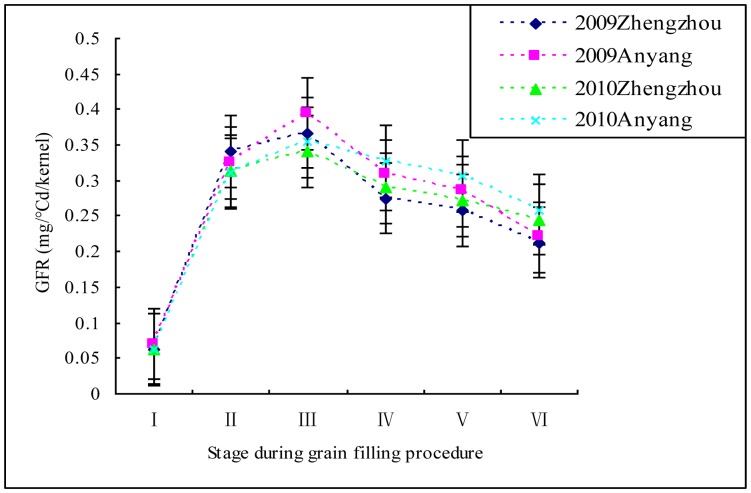
Among the IF2 population (Table 1; Fig. 2), the GFR at the six sampling stages at Anyang were higher than the corresponding sampling stage at Zhengzhou in 2009 and 2010, respectively. However, the GFR during the grain filling process over a year showed a similar tendency at both locations, and the variations in the GFR among the population increased mainly in the middle or later stages (23–50 DAP). Under different environments, there were no significant variations in the KW of the IF2 population; however, the GFD between the 2 years was significantly different at both locations. The dynamic diversification of GFR in all materials shows a tendency for logistic curves: the GFR gradually rose in the first and second sampling stages, reaching a maximum at the third sampling stage for different years or locations.
Figure 2. Dynamic diversity of grain filling rates in maize strains.
In the table 2, the GFR and KW were significantly positively correlated, except at the first sampling stage, which confirmed that the variance of KW is associated with the GFR during the effective grain filling period. Moreover, there were extremely significant positive correlations between sampling stage II (16–22 DAP) and KW, indicating that 16–22 DAP is an important stage for determining the final kernel weight. There was no significant correlation between GFR and GFD.
Table 2. Correlation coefficients between grain filling rate, grain filling duration and final kernel weight.
| Location | I | II | III | IV | V | VI | KW | GFD | |
| Zhengzhou | I | −0.1 | −0.07 | −0.17 | 0.15 | 0.25 | 0.23 | 0.11 | |
| II | 0.02 | −0.29* | 0.08 | 0.01 | 0.03 | 0.40** | 0.23 | ||
| III | −0.21 | −0.32* | −0.05 | 0.04 | 0.09 | 0.30* | −0.10 | ||
| IV | 0.17 | −0.02 | −0.22 | −0.23 | −0.05 | 0.31* | −0.15 | ||
| V | −0.21 | 0.03 | 0.03 | −0.23 | 0.02 | 0.32* | −0.22 | ||
| VI | −0.01 | 0.03 | 0.36* | 0.01 | −0.08 | 0.31* | 0.05 | ||
| KW | −0.01 | 0.42** | 0.48** | 0.31* | 0.1 | 0.39** | 0.08 | ||
| GFD | 0.07 | 0.17 | −0.03 | 0.05 | −0.09 | 0.04 | 0.15 | ||
| Anyang | I | −0.24 | 0.01 | 0.09 | −0.07 | 0.24 | 0.31* | 0.04 | |
| II | −0.26 | −0.37* | 0.05 | 0.2 | 0.06 | 0.39** | −0.02 | ||
| III | −0.07 | −0.35* | −0.04 | −0.21 | −0.17 | 0.02 | 0.09 | ||
| IV | −0.16 | 0.21 | −0.15 | −0.18 | −0.13 | 0.52** | −0.12 | ||
| V | −0.13 | 0.08 | 0.14 | −0.12 | −0.21 | 0.36* | −0.06 | ||
| VI | 0.02 | 0.2 | 0.02 | 0.01 | −0.12 | 0.33* | −0.27 | ||
| KW | 0.04 | 0.52** | 0.55** | 0.31* | 0.36* | 0.51** | −0.24 | ||
| GFD | 0.11 | −0.04 | 0.11 | 0.05 | −0.17 | −0.15 | 0.06 |
Note: *, **Significant effect at probabilities of 0.05 and 0.01, respectively;
The correlation coefficients for 2009 are in the upper triangular area of the table and the correlation coefficients for 2010 are in the lower triangular area of the table.
Unconditional QTLs Detected for Grain Filling Rate
The genetic linkage map for the recombinant inbreed line (RIL) population was constructed using 217 SSR markers, which included 10 linkages, and spanned 2438.2 cM, with an average interval of 11.2 cM. [26] The genotypes of each cross of the IF2 population were deduced from the marker genotypes of their RIL parents, and the molecular linkage map for QTL mapping in the IF2 population was used as the molecular linkage map of the RIL population because it had the same genetic background. [27].
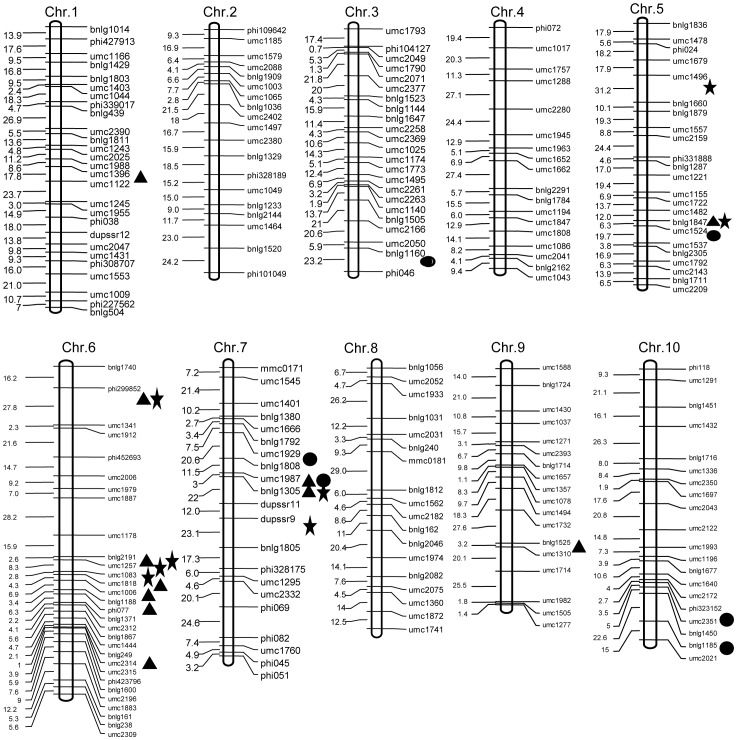
Thirty-three different unconditional QTLs were identified for the GFR at the six sampling stages between 2 years at two locations, and were located on five chromosomes (Table 3; Fig. 3). In the four environments, there were 11, 9, 7 and 7 QTLs detected, respectively. Among them QTLs qGFR7b, qGFR9 and qGFR6d were identified at the same stages at both locations over 2 years. Moreover, QTL qGFR7b was detected at sampling stage II (16–22 DAP) and stage VI (44–50 DAP). QTL qGFR9, derived from the parent Xu 178, showed 7.51%, 6.67%, 6.67% and 7.01% of total variance at the two locations over two years, respectively. Another QTL, qGFR6d, identified at the fifth stage (37–43 DAP), could explain 5.31%, 5.32%, 5.49% and 5.42% of the total phenotypic variance, respectively. However, QTL qGFR6d, coming from the parent Huang-C and detected at stage IV (29–36DAP), could explain 21.14% of the total variance. QTLs qGFR7a, qGFR6a and qGFR6c were detected at the same stages in the four environments, except at Anyang in 2010. QTL qGFR7a, identified at stage II (16–22 DAP), contributed 10.96%, 11.92% and 13.27% of the total variance, respectively. QTL qGFR6a was detected at stage I (0–15DAP) and could explain 33.85%, 23.3% and 19.83% of the total phenotypic variance, with a direct increase of 0.021, 0.016 and 0.013 mg °Cd−1 kernel−1 of GFR, respectively. QTL qGFR6c, detected at stage IV (37–43DAP), contributed 24.37%, 20.75% and 21.67% of the total phenotypic variance, with a direct increase of 0.067, 0.064 and 0.063 mg °C d−1 kernel−1 of GFR, respectively.
Table 3. Unconditional QTLs detected for grain filling rate in the immortalized F2 population.
| Year | Location | Stage/Trait | QTLa | Markers interval | LODb | Ac | Dc | Effectsd | R2e(%) |
| 2009 | Zhengzhou | I | qGFR5 | bnlg1847-umc1524 | 4.67 | 0.012 | −0.006 | PD | 22.05 |
| I | qGFR6a | phi299852-umc1341 | 4.44 | 0.014 | −0.005 | PD | 33.85 | ||
| II | qGFR7a | umc1987-bnlg1305 | 5.12 | 0.043 | 0.005 | A | 10.96 | ||
| II | qGFR7b | bnlg1305-dupssr11 | 3.72 | 0.041 | −0.003 | A | 10.96 | ||
| III | qGFR9 | bnlg1525-umc1310 | 4.29 | −0.029 | 0.058 | OD | 7.51 | ||
| IV | qGFR6b | umc1006-bnlg1188 | 4.49 | 0.084 | −0.042 | PD | 25.04 | ||
| IV | qGFR6c | phi077-bnlg1371 | 4.25 | 0.085 | −0.051 | PD | 24.37 | ||
| IV | qGFR6d | umc1818-umc1006 | 4.16 | 0.077 | −0.030 | PD | 21.14 | ||
| IV | qGFR6e | umc2315-phi423796 | 4.07 | 0.063 | −0.055 | D | 13.52 | ||
| V | qGFR6d | umc1818-umc1006 | 5.27 | −0.035 | 0.077 | OD | 5.31 | ||
| VI | qGFR7b | bnlg1305-dupssr11 | 4.44 | 0.086 | −0.028 | PD | 22.91 | ||
| KW | qKW7a | umc1987-bnlg1305 | 7.46 | 0.020 | 0.003 | A | 10.59 | ||
| KW | qKW10a | bnlg1185-umc2021 | 4.72 | 0.015 | −0.007 | PD | 14.94 | ||
| KW | qKW3a | bnlg1160-phi046 | 3.96 | −0.015 | −0.004 | PD | 12.07 | ||
| KW | qKW7b | umc1929-bnlg1808 | 3.92 | −0.025 | 0.006 | PD | 17.99 | ||
| Anyang | I | qGFR6a | phi299852-umc1341 | 4.07 | 0.012 | −0.006 | PD | 23.3 | |
| II | qGFR7a | umc1987-bnlg1305 | 4.29 | 0.051 | −0.002 | A | 11.92 | ||
| II | qGFR7b | bnlg1305-dupssr11 | 3.92 | 0.056 | −0.016 | PD | 15.73 | ||
| III | qGFR9 | bnlg1525-umc1310 | 4.09 | −0.027 | 0.056 | OD | 6.67 | ||
| IV | qGFR6b | umc1006-bnlg1188 | 4.39 | 0.088 | −0.045 | PD | 25.17 | ||
| IV | qGFR6e | umc2315-phi423796 | 4.08 | 0.063 | −0.059 | D | 12.61 | ||
| IV | qGFR6c | phi077-bnlg1371 | 3.83 | 0.082 | −0.054 | PD | 20.75 | ||
| V | qGFR6d | umc1818-umc1006 | 5.09 | −0.035 | 0.076 | OD | 5.32 | ||
| VI | qGFR7b | bnlg1305-dupssr11 | 3.94 | 0.061 | −0.008 | A | 15.42 | ||
| KW | qKW10a | bnlg1185-umc2021 | 5.49 | 0.016 | −0.004 | PD | 16.64 | ||
| KW | qKW5 | umc1524-umc1537 | 4.91 | −0.017 | −0.001 | A | 17.34 | ||
| 2010 | Zhengzhou | I | qGFR6a | phi299852-umc1341 | 3.78 | 0.010 | −0.009 | D | 19.83 |
| II | qGFR7a | umc1987-bnlg1305 | 4.11 | 0.041 | −0.004 | A | 13.27 | ||
| II | qGFR7b | bnlg1305-dupssr11 | 4.07 | 0.052 | −0.028 | PD | 21.73 | ||
| III | qGFR9 | bnlg1525-umc1310 | 4.07 | −0.027 | 0.056 | OD | 6.67 | ||
| IV | qGFR6c | phi077-bnlg1371 | 3.8 | 0.081 | −0.050 | PD | 21.67 | ||
| V | qGFR6d | umc1818-umc1006 | 4.54 | −0.036 | 0.074 | OD | 5.49 | ||
| VI | qGFR7b | bnlg1305-dupssr11 | 4.7 | 0.085 | −0.021 | PD | 22.08 | ||
| KW | qKW7a | umc1987-bnlg1305 | 6.68 | 0.021 | 0.002 | A | 12.65 | ||
| KW | qKW7b | umc1929-bnlg1808 | 4.98 | −0.021 | 0.003 | A | 18.92 | ||
| KW | qKW10b | phi323152-umc2351 | 4.15 | 0.013 | −0.005 | PD | 10.71 | ||
| Anyang | I | qGFR1 | umc1396-umc1122 | 3.98 | −0.003 | 0.013 | OD | 15.03 | |
| II | qGFR5 | bnlg1847-umc1524 | 5.09 | −0.048 | −0.019 | PD | 16.39 | ||
| II | qGFR7b | bnlg1305-dupssr11 | 4.29 | 0.052 | −0.028 | PD | 22.03 | ||
| III | qGFR9 | bnlg1525-umc1310 | 4.28 | −0.028 | 0.057 | OD | 7.01 | ||
| IV | qGFR6f | bnlg2191-umc1257 | 4.19 | 0.024 | −0.060 | OD | 5.88 | ||
| V | qGFR6d | umc1818-umc1006 | 4.6 | −0.032 | 0.075 | OD | 5.42 | ||
| VI | qGFR7b | bnlg1305-dupssr11 | 4.85 | 0.088 | −0.022 | PD | 23.3 | ||
| KW | qKW7a | umc1987-bnlg1305 | 6.54 | 0.018 | 0.002 | A | 10.45 | ||
| KW | qKW10a | bnlg1185-umc2021 | 4.24 | 0.014 | −0.007 | PD | 13.53 |
Note: aUnconditional QTLs detected for grain filling rate in the IF2 population;
Logarithm of Odds for each QTL.
A, additive values; D, dominant values;
The effect of each QTL; A, additive; PD, partial dominance; D, dominance; OD, overdominance;
R2 contribution rate.
Figure 3. Chromosomal locations of QTLs detected for grain filling rate and kernel weight.
Note: Triangle unconditional QTLs detected for grain filling rate, Star conditional QTLs detected for grain filling rate, Round unconditional QTLs detected for kernel weight.
There were eleven QTLs detected for kernel weight, and were located on four chromosomes at the two locations over 2 years (Table 3; Fig. 3). The QTL qKW10a was detected at both locations in 2009 and at Anyang location in 2010, and contributed 14.94%, 16.64%, and 13.53% of total phenotypic variance, respectively. The QTL qKW7a was detected at Zhengzhou in 2009 and at both locations in 2010 and contributed 10.59%, 12.65% and 10.45% of total variance, respectively. In addition, the qGFR7a was co-located with the QTL qKW7a at Zhengzhou location over 2 years.
Conditional QTL Mapping for Grain Filling Rate
Fourteen conditional QTLs were detected at five stages for the GFR and are distributed on chromosomes 6 and chromosome 7 (Fig. 3; Table 4). These QTLs clustered at chromosome bins 6.01–6.02 and 7.02–7.03, which correlates with the unconditional QTL mapping results. The conditional QTL qGFR7c, identified at stage V|IV (37–43DAP) at two locations over 2 years, contributed 14.77%, 18.70%, 21.40% and 20.08% of the total phenotypic variance in GFR. QTL qGFR7b, detected at the sixth stage (44–50DAP) at Zhengzhou in both years and Anyang in 2010, could explain 14.55%, 10.04% and 10.17% of the total variance, respectively. In addition, QTL qGFR7b, identified at stage II|I (0–15DAP) at Anyang in 2010, could explain 17.81% of the total variance in the GFR. In 2009, QTL qGFR6g was detected at two locations, contributing 11.82% and 14.42% of the total variance, respectively. In 2010, QTL qGFR6h, derived from the parent Huang-C, was detected at two locations, and contribution large proportions of the phenotypic variance: 38.42% and 37.27%, respectively.
Table 4. Conditional QTLs detected for grain filling rate in the immortalized F2 population.
| Year | Location | Stage | QTLa | Markers interval | LODb | Ac | Dc | Effectsd | R2e(%) |
| 2009 | Zhengzhou | I | qGFR5a | bnlg1847-umc1524 | 4.65 | 0.012 | −0.006 | PD | 21.88 |
| I | qGFR6a | phi299852-umc1341 | 4.49 | 0.023 | −0.005 | PD | 33.85 | ||
| IV|III | qGFR6g | umc1257-umc1083 | 3.67 | 0.048 | −0.066 | OD | 11.82 | ||
| V|IV | qGFR7c | dupssr9-bnlg1805 | 3.87 | 0.059 | −0.065 | D | 14.77 | ||
| VI|V | qGFR7b | bnlg1305-dupssr11 | 6.07 | 0.058 | 0.023 | PD | 14.55 | ||
| Anyang | I | qGFR6a | phi299852-umc1341 | 4.03 | 0.012 | −0.006 | PD | 23.18 | |
| IV|III | qGFR6g | umc1257-umc1083 | 3.66 | 0.055 | −0.066 | OD | 14.42 | ||
| V|IV | qGFR7c | dupssr9-bnlg1805 | 4.97 | 0.066 | −0.072 | D | 18.70 | ||
| 2010 | Zhengzhou | I | qGFR6a | phi299852-umc1341 | 3.72 | 0.010 | −0.009 | D | 19.25 |
| IV|III | qGFR6f | bnlg2191-umc1257 | 5.56 | 0.025 | −0.076 | OD | 6.42 | ||
| IV|III | qGFR6h | umc1083-umc1818 | 4.07 | 0.117 | −0.054 | PD | 38.42 | ||
| V|IV | qGFR7c | dupssr9-bnlg1805 | 5.87 | 0.071 | −0.073 | D | 21.40 | ||
| VI|V | qGFR7b | bnlg1305-dupssr11 | 4.59 | 0.049 | 0.032 | PD | 10.04 | ||
| Anyang | I | qGFR1 | umc1396-umc1122 | 3.95 | −0.003 | 0.013 | OD | 15.03 | |
| II|I | qGFR7b | bnlg1305-dupssr11 | 4.38 | 0.062 | −0.014 | PD | 17.81 | ||
| IV|III | qGFR6f | bnlg2191-umc1257 | 4.71 | 0.022 | −0.075 | OD | 5.33 | ||
| IV|III | qGFR6h | umc1083-umc1818 | 4.11 | 0.117 | −0.056 | PD | 37.27 | ||
| V|IV | qGFR7c | dupssr9-bnlg1805 | 5.11 | 0.069 | −0.070 | D | 20.08 | ||
| VI|V | qGFR7b | bnlg1305-dupssr11 | 4.84 | 0.049 | 0.033 | PD | 10.17 |
Note: aConditional QTLs detected for grain filling rate in the IF2 population;
Logarithm of Odds for each QTL.
A, additive values; D, dominant values;
The effect of each QTL; A, additive; PD, partial dominance; D, dominance; OD, overdominance;
R2 contribution rate.
Comparing the results of the unconditional and conditional QTL mapping methods (Table 3; Table 4; Fig. 3), there were five unconditional QTLs detected under conditional mapping in the same environments. At the sixth stage (44–50 DAP), QTL qGFR7b was identified by both QTL mapping methods in all four environments, except at Anyang in 2009 under conditional QTL mapping. qGFR7b showed higher effects (22.91%, 22.08% and 23.30% of the total variance) under unconditional QTL mapping, than under conditional QTL mapping (14.55%, 10.04% and 10.17% of the total variance). At the second stage (16–22 DAP), qGFR7b was identified at Anyang in 2010 using both QTL mapping methods, and contributed 22.03% and 17.81% of the total variance. Additionally, QTL qGFR6f was identified at the fourth sampling stage (30–36 DAP) under both methodologies at Anyang in 2010. Among the new QTLs detected by conditional QTL mapping, qGFR6g and qGFR6h were adjacent to the unconditional qGFR6b and qGFR6d on chromosome 6, and qGFR7c was located at the adjacent locus to the unconditional QTLs qGFR7a and qGFR7b on chromosome 7.
Discussion
In maize, many previous studies on grain filling or kernel development used several inbred lines and hybrids (with different genetic backgrounds), or RIL populations for QTL mapping. [4], [16], [26], [28] The GFR is easily affected by meteorological factors, edaphic conditions, water and fertilizer management levels, as well by plant density. [29]–[32] Comparing with inbred lines and RIL populations, [4], [26] an IF2 population not only has similar heterotic phenotypes to hybrid maize, which are not easily affected by various environmental factors, but also each family of the IF2 population has similar flowering and silking times. Thus, using an IF2 population ensured accurate phenotypic values for the GFR in this study.
As in previous reports, the GFR and GFD were determined by genotype and were influenced by environmental factors. [29]–[32] Stewart et al. reported that when maize is grown under a very broad range of temperatures, plant development in response to temperature is nonlinear during the reproductive period. [29] When grown over a narrower range of temperatures, the response reported by Stewart et al. approximated a linear relationship, with the base temperature near 0°C. [29], [30] In addition, because of the narrower range of temperature encountered in this study, the GFR was evaluated using heat units between sampling times, and daily °C d values for grain filling were measured at the base temperature of 0°C. Using this method, Borrás and Otegui evaluated the effective grain filling rate using two hybrids, [33] and the kernel growth rate was also measured for two hybrids and a set of inbred lines by Borrás et al. [12], [15], [28] In the previous study of Liu et al., a set of RIL population was adopted for identifying GFR related QTL in maize, days between two sampling times were used as grain filling duration for calculating GFR. [26] In this study, thermal time between two sampling times were used for evaluating GFR value, which could benefit of decreasing the affects of temperature.
Kernel development is a complex process with a dynamic character that is regulated by three physiological activities during the reproductive period: (1) cell division and differentiation; (2) the effective grain filling period, and (3) the maturation drying period. [34] The GFR is low speed during the cell division and differentiation phase, during which almost no dry matter accumulates. [19] The effective grain filling period is a process of rapid dry matter accumulation resulting from the deposition of seed reserves. In this period, the GFR rises gradually and reaches its maximum value in the middle of the period. [15] During the maturation drying phase, the GFR decreases gradually, with kernels continuing to lose water. Here, six samplings during effective grain filling period and the maturation drying phase (15–50 DAP) were adapted for GFR evaluation that is because of the dry matter mainly accumulate in the two periods. And, starch synthesis in the kernel begins from 12–15 DAP. [35] In this study, the GFR of the IF2 population gradually rose in the first and second stages, reaching a maximum at the third stage, and then gradually decreased over the last three stages (Fig. 2).
Grain filling determines the final kernel weight, and thus contributes greatly to grain productivity. It is reported that the variation in KW may be achieved through different combinations of kernel growth rates and grain filling durations; however, there was no correlation between kernel growth rate and grain filling duration. [16], [36] In the present study, there was no significant correlation between the correlation between the GFR and GFD. However, in this study, the variation in KW was determined by the GFR during the effective grain filling period and maturation drying stage, and there was no correlation between KW and the GFD.
Although physiologists have directed their attention to the grain filling processes, there have been few genetic studies of grain filling because of its complex and dynamic features. [10] Wang et al. performed a genetic analysis on the GFR and GFD in maize, and their results revealed that general combining ability (GCA) was more important than special combining ability (SCA) for both the GFR and the effective filling duration. [13] QTL mapping for grain filling using a RIL population in maize was reported by Thévenot et al. for enzyme activities and soluble carbohydrates, [4] and by Liu et al. for the GFR. [26] Thévenot et al. reported that a higher density QTLs was detected on chromosome 1 and 2 at 35 DAP, and that QTLs were detected that clustered at bin 5.02–5.03 and 5.04–5.05 at 15 DAP. [4] In this study, a higher density of QTLs was identified on chromosome 6 at 30–36 DAP, clustered at bin 6.01–6.02. However, there is still a number of QTLs for GFR that co-localize to the QTLs at same chromosomal bin for enzyme activities, soluble carbohydrates, and the genes associated with grain filling. [4] For example, qGFR1 is located at the same chromosomal bin as the BT2 gene, a QTL for fresh matter and a QTL for neutral-cytosolic invertase. QTL qGFR5 was identified at stage 0–15 DAP in the same chromosomal fragment as a QTL for glucose, fructose and sucrose content; and QTL qGFR6a was identified in the same chromosomal fragment as the BT1 gene and a QTL for glucose content. In addition, QTL qGFR9 was located in the same chromosomal bin as a QTL for sucrose synthase, glucose content and fructose content. QTLs qGFR7a, qGF7b and qGFR7c co-localize with a QTL for glucose at chromosomal bin 7.02–7.03. At chromosomal bin 6.01–6.02, there were the gene 6PGDH (6-phosphogluconate dehydrogenase) and a QTL for fresh matter co-localize. These results reveal that the grain filling process not only involves starch synthesis, but also other novel activities.
Grain filling represents a process of starch accumulation, [2] and there have been many reports of the starch pathway in cereals. For example, in rice Ohdan et al. analyzed the genes associated with starch synthesis at the level of transcription during the grain filling process. [37] They divided the 27 starch synthesis-associated genes into four groups. Group 1 genes are expressed very early in grain formation and are presumed to be involved in the construction of fundamental cell structure and de novo synthesis of glucan primers. Group 2 genes are highly expressed throughout the grain development process. Group 3 genes are transcribed at a low level at the onset, but rise steeply at the beginning of starch synthesis in the endosperm. Group 4 genes are barely expressed, mainly at the onset of grain development. Group 3 genes are thought to play essential roles in endosperm starch synthesis. Yan et al. compared the starch synthesis genes between maize and rice, and detected thirty starch synthesis genes in the maize genome, which covered all the starch synthesis gene families encoded by 27 genes in rice. [38] Among the unconditional QTLs detected for the GFR in this study, QTL qGFR6a was only identified at the first stage in three out of four environments; this kind of QTL resembles a group 1 and group 4 gene of starch synthesis. [37] However, no QTL was detected for the GFR that was expressed throughout the whole process of grain filling. These results indicated that the GFR is regulated by genes that are selectively expressed at different grain filling stages. Among these QTLs identified for GFR, QTL qGFR7b was identified at different stages in four environments using two QTL mapping methods; therefore, it represents a main QTL for the GFR. In addition, several QTLs, such as qGFR6a, qGFR6d, qGFR9, qGFR6c, qGFR6f and qGFR7c, were detected in different environments, and might represent genes with important effects in regulating grain development. Several QTLs were identified in single environments and stages, which might be caused by the differences in climate factors under the different environments and grain filling stages. Although, thermal time was the main contributor to GFR, the other climate factors also had a certain influence to grain filling rate and grain filling duration. [12], [15], [31], [32] In this study, the average temperature and daily sunlight were significant different (Fig. 1a and 1b) between the two locations. And, there were large differences at according stages in related climate factors between the two years at any one location. So in this study, the thermal time was used as for calculating GFR, and used as input data for QTL mapping However, under the affects of the other different environmental factors, most unconditional and conditional QTLs for GFR expressed selectively.
In recent decades, increases in grain yield in maize were achieved mainly by lengthening the grain filling period and increasing population density, which in turn increased GFR per unit land area. GFD was longer in the newer hybrids; even though harvest maturity remained unchanged. [36] The increase in GFD was the result of delayed physiological maturity rather than a change in flowering date. The GFR is somewhat more stable than GFD, and the latter is easily affected by changes in plant density and temperature, whereas the kernel growth rate is not affected. [13] Additionally, the KW is associated with the GFR during the effective grain filling period, as reported in this study. In many countries or areas of the world, the season for maize growth is very limited, and the tendency for use of mechanical harvesting demands hybrid maize with a relatively short period of dehydration in the field. Thus, commercial hybrids must have a high GFR and an appropriate growth duration to obtain high grain yields.
Materials and Methods
The Development of the Immortalized F2 Population
A population of 166 RILs was constructed by a single-seed descent method from two elite inbred lines, Huang-C and Xu178. The cross was an elite hybrid, Nongda108, which occupied approximately 2.7 million hectares during 2001–2004 in China. One of its parents, Huang-C, was selected from Chinese germplasm, and the other parent, Xu178, was derived from an exotic hybrid. According to the procedure described by Hua et al., [39] the 166 RILs were randomly divided into two groups, each group including 83 RILs. Then, pairs of crosses were made randomly between the lines of the two groups, without repetition, so that 83 different crosses were generated. The procedure was repeated three times. Finally, 249 (83×3) pairs of crosses between the two RILs formed the immortalized F2 population. Six crosses lacked abundant seeds because of a difficulty in mating; thus, 243 crosses were used in this study.
Field Evaluation
The IF2 population, the two parents, and the hybrid were planted in 2009 and 2010 on the Agronomy Farm of Henan Agricultural University (Zhengzhou, 113°42′E, 34°48′N), which is located in the central region of China and has an average daily temperature 14.3°C and an average annual rainfall of 640.9 mm. The maize plants were also planted during the same years at the Anyang Agricultural Institute (Anyang, 114°21′E, 36°6′N), which is located in the center of the north China plain and has an average temperature of 14.1°C and an average of 556.9 mm of rainfall per year. At Zhengzhou, all the plant materials were planted on the 12th and 8th of June in 2009 and 2010, respectively. At Anyang, plant materials were planted on 17th and 12th of June in 2009 and 2010, respectively. The field experimental design followed an incomplete block design approach, with two replications at each location. Each experimental material was applied to two plots of 6 m long×0.67 m wide rows and comprised 50 plants, at a density of 65,250 plants per hectare. The fields were kept free of weeds and nests, and irrigated and fertilized properly to avoid nutritional stress.
Sampling and Measurements of GFR
In each plot, when 50% of the silks spit out of all plants, the pollination date was determined. Samples were hand-collected for five ears at each plot at 15, 22, 29, 36, 43 and 50 days after pollination (DAP) in 2009 and 2010, respectively. The sampling dates were chosen starting at 15 DAP because previous studies have shown that starch synthesis in the kernel begins from 12–15 DAP. [35] Ears with irregular kernel sets along the ear row were discarded to avoid the confounding effect of atypically large kernels adjacent to unpollinated florets. [12] These harvested ears were dried fully under nature condition, and the grains in the center of the ear were threshed. The moisture content of all the grain samples was detected by PM-8188NEW grain moisture determination apparatus. And, the grain moisture values for all the samples were amended to 13%, and then the 100-kernel weight was evaluated. These treatments were used for ensuring all the samples harvested at different grain filling stages in the same moisture. The 100-kernel weight in the center of the ear was then quantified three times, and the average data among the three 100-kernel weights for every sampling time were calculated. The GFR between two sampling stages was calculated as: GFR (mg °Cd−1 kernel−1) = the margin of kernel weight for two sampling times (mg kernel−1)/GFD between two sampling times (°Cd). The GFR of pollination date-15DAP (I), 16–22 DAP (II), 23–29DAP (III), 30–36DAP (IV), 37–43DAP (V) and 44–50DAP (VI) were calculated, respectively. Here, we used thermal time as the GFD, [12], [14], [28] which is calculated using the daily air temperature values between two sampling times. In addition, the daily °Cd value for grain filling was calculated using 0°C as base temperature. [29] The average performance data generated in each replication and location were used as raw data for further analyses. Data analysis was performed using SAS 9.2 statistical software package with the PROC MIXED procedure. [40] The climate data were obtained from the Climate Bureau of Zhengzhou and the Climate Bureau of Anyang, China.
Unconditional and Conditional QTL Mapping
Unconditional QTL mapping was performed using the composite interval mapping method and Model 6 of the Zmapqtl module of QTL Cartographer 2.5. [41] The threshold of a logarithm of Odds (LOD) was calculated using 1,000 permutations at a significance level of P = 0.05, with scanning intervals of 2 cM between markers and a putative QTL, and a 10 cM window. The number of marker cofactors for background control was set by forward-backward stepwise regression with five controlling markers.
For dynamic traits of developmental behavior, the genetic effect (G(t)) at time t is the genetic effect (G(t−1)) at time (t−1) and the extra genetic effect (G(d)). [42]–[44] Thus, it calculates the cumulative gene effects from initial time to t, but not for the independent effects of gene expression in the duration (t−1) to t. To reject the genetic effect of a genetic effect (G(t−1)) at time t, the conditional phenotypic values y (t |t−1) were obtained by the mixed model approach for the conditional analysis of quantitative traits described by Zhu. [42] The conditional phenotypic values were used as input data for conditional QTL mapping, which used the composite interval mapping method.
Funding Statement
This work was supported by a grant from the National Project 973 of China (2012CB723001) and the National High Technology Research and Development Program of China (2012AA10A305). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gore MA, Chia JM, Elshire RJ, Sun Q, Ersaz ES, et al. (2009) A first-generation haplotype map of maize. Science 326: 1115–1117. [DOI] [PubMed] [Google Scholar]
- 2. Yang J, Zhang J (2010) Grain-filling problem in ‘super’ rice. J Exp Bot 61: 1–5. [DOI] [PubMed] [Google Scholar]
- 3. Maitz M, Santandrea G, Zhang Z, Lal S, Hannah C, et al. (2000) rgf1, a mutation reducing grain filling in maize through effects on basal endosperm and pedicel development. Plant J 23: 29–42. [DOI] [PubMed] [Google Scholar]
- 4. Thévenot C, Simond-Côte E, Reyss A, Manicacci D, Trouverie J, et al. (2005) QTLs for enzyme activities and soluble carbohydrates involved in starch accumulation during grain filling in maize. J Exp Bot 56: 945–958. [DOI] [PubMed] [Google Scholar]
- 5. Lid SE, Gruis D, Jung R, Lorentzen JA, Ananiev E, et al. (2002) The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc Natl Acad Sci 99: 5460–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng W, Tallerclo EW, Chourey PS (1996) The miniature seed locus of maize encodes a cell wall invertase required for normal development of endosperm and maternal cells in the pedicel. Plant Cell 8: 971–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carson SJ, Chourey PS (1999) A re-evaluation of the relative roles of two invertases, incw2 and ivr1, in Developing Maize Kernels and other tissues. Plant Physiol 121: 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vilhar B, Kladnik A, Blejec A, Chouery PS, Dermastia M (2002) Cytometrical evidence that the loss of seed weight in the miniature1 seed mutant of maize is associated with reduced mitotic activity in the developing endosperm. Plant Physiol 129: 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo M, Rupe MA, Dieter JA, Zou J, Spielbauer D, et al. (2010) Cell number regulator1 affects plant and organ size in maize: implications for crop yield enhancement and heterosis. Plant Cell 22: 1057–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takai T, Fukuta Y, Shiraiwa T, Horie T (2005) Time-related mapping of quantitative trait loci controlling grain-filling in rice (Oryza sativa L.) J Exp Bot. 56: 2107–2118. [DOI] [PubMed] [Google Scholar]
- 11. Wang E, Wang J, Zhu X, Hao W, Wang L, et al. (2008) Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet 40: 1370–1374. [DOI] [PubMed] [Google Scholar]
- 12. Wang G, Kang MS, Moreno O (1999) Genetic analyses of grain-filling rate and duration in maize. Field Crops Res 61: 211–222. [Google Scholar]
- 13. Borrás L, Westgate ME, Otegui ME (2003) Control of kernel weight and kernel water relations by post-flowering source-sink ratio in maize. Ann Bot 91: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Borrás L, Slafer GA, Otegui ME (2004) Seed dry weight response to source-sink manipulations in wheat, maize and soybean: a quantitative reappraisal. Field Crops Res 86: 131–146. [Google Scholar]
- 15. Borrás L, Westgate ME (2006) Predicting maize kernel sink capacity early in development. Field Crops Res 95: 223–233. [Google Scholar]
- 16. Gambín BL, Borrás L, Otegui ME (2007) Kernel water relations and duration of grain filling in maize temperate hybrids. Field Crops Res 101: 1–9. [Google Scholar]
- 17. Löffler CM, Wei J, Fast T, Gogerty J, Langton S, et al. (2005) Classification of maize environments using crop simulation and geographic information systems. Crop Sci 45: 1708–1716. [Google Scholar]
- 18. Wilhelm EP, Mullen RE, Keeling PL, Singletary GW (1999) Heat stress during grain filling in maize: Effects on kernel growth and metabolism. Crop Sci 39: 1733–1749. [Google Scholar]
- 19. Setter TL, Flannigan BA (2001) Water deficit inhibits cell division and expression of transcripts involved in cell proliferation and endoreduplication in maize endosperm. J Exp Bot 52: 1401–1408. [DOI] [PubMed] [Google Scholar]
- 20. Gupta PK, Rustgi S, Kumar N (2006) Genetic and molecular basis of grain size and grain number and its relevance to grain productivity in higher plants. Genome 49: 565–571. [DOI] [PubMed] [Google Scholar]
- 21.Blewley JD, Black M (1985) Seeds: Physiology of development and germination. Plenum Press New York p: 41–42. [Google Scholar]
- 22. Seka D, Cross HZ (1995) Xenia and maternal effects on maize kernel development. Crop Sci 35: 80–85. [Google Scholar]
- 23. Seka D, Cross HZ, McClean PE (1995) Maize kernel development in vitro: sucrose concentration, xenia, and maternal effects. Crop Sci 35: 74–79. [Google Scholar]
- 24. Liu Z, Tang J, Wang C, Wei X, Tian G, et al. (2007) QTL analysis of plant height under N-stress and N-input at different stages in Maize. Acta Agron Sin 33: 782–789. [Google Scholar]
- 25.Li Y, Fan C, Xing Y, Jiang Y, Luo L, et al.. (2011) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet published online 23 October: 1–5. [DOI] [PubMed] [Google Scholar]
- 26. Liu Z, Ji H, Cui Z, Wu X, Duan L, et al. (2011) QTL detected for grain-filling rate in maize using a RIL population. Mol Breed 27: 25–36. [Google Scholar]
- 27. Tang JH, Yan JB, Ma XQ, Teng WT, Wu WR, et al. (2010) Dissection of the genetic basis of heterosis in an elite maize hybrid by QTL mapping in an immortalized F2 population. Theor Appl Genet (2010) 120: 333–340. [DOI] [PubMed] [Google Scholar]
- 28. Borrás L, Zinselmeier C, Senior ML, Westgate ME, Mnszynski MG (2009) Characterization of grain-filling patterns in diverse maize germplasm. Crop Sci 49: 999–1010. [Google Scholar]
- 29. Stewart DW, Dwyer LM, Carrigan LL (1998) Phenological temperature response of maize. Agron J 90: 73–79. [Google Scholar]
- 30. Muchow RC (1990) Effect of high temperature on grain-growth in field-grown maize. Field Crops Res 23: 145–158. [Google Scholar]
- 31. Zheng H, Dong S, Wang K, Hu C, Guo Y, et al. (2001) Studies on effect of ecological factors on maize kernel growth and corresponding regulative measures. Journal of Maize Science in China 9: 69–73. [Google Scholar]
- 32. Li S, Pai P, Lv X, Liu S, Dong S (2003) Ecological and sowing date effects on maize grain filling. Acta Agron Sin 29: 775–778. [Google Scholar]
- 33. Borrás L, Otegui ME (2001) Maize kernel weight response to post flowering source-sink ratio. Crop Sci 41: 1816–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Borrás L, Gambín BL (2010) Trait dissection of maize kernel weight: Towards integrating hierarchical scales using a plant growth approach. Field Crops Res 118: 1–12. [Google Scholar]
- 35. Prioul J, Jeannette E, Reyss A, Grégory N, Giroux M, et al. (1994) Expression of ADP-glucose pyrophosphorylase in maize (Zea mays L.) grain and source leaf during grain filling. Plant Physiol 104: 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cavalieri AJ, Smith OS (1985) Grain filling and field drying of a set of maize hybrids released from 1930 to 1982. Crop Sci. 25: 856–860. [Google Scholar]
- 37. Ohdan T, Francisco PB, Sawada T, Hirose T, Terao T, et al. (2005) Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot 56: 3229–3244. [DOI] [PubMed] [Google Scholar]
- 38. Yan H, Pan X, Jiang H, Wu G (2009) Comparison of the starch synthesis genes between maize and rice: copies, chromosome location and expression divergence. Theor Appl Genet 119: 815–825. [DOI] [PubMed] [Google Scholar]
- 39. Hua J, Xing Y, Xu C, Sun X, Yu S, et al. (2002) Genetic dissection of an elite rice hybrid revealed that heterozygotes are not always advantageous for performance. Genetics 162: 1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.SAS Institute (2009) SAS/STAT 9.2 user’s guide, second edition, chapter 6th and 56th. SAS Institute Inc. Cary. [Google Scholar]
- 41. Zeng Z (1994) Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhu J (1995) Analysis of conditional genetic effects and variance components in developmental genetic. Genetics 141: 1633–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu W, Zhou Y, Li W, Mao D, Chen Q (2002) Mapping of quantitative trait loci based on growth models. Theor Appl Genet 105: 1043–1049. [DOI] [PubMed] [Google Scholar]
- 44. Wu W, Li W, Tang D, Lu H, Worland AJ (1999) Time-related mapping of quantitative trait loci underlying tiller number in rice. Genetics 151: 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]