Condensin II initiates structural reorganization of duplicated chromosomes during S phase to prepare for their proper condensation and segregation in mitosis.
Abstract
Condensins I and II are multisubunit complexes that play essential yet distinct functions in chromosome condensation and segregation in mitosis. Unlike condensin I, condensin II localizes to the nucleus during interphase, but it remains poorly understood what functions condensin II might have before mitotic entry. Here, we report that condensin II changes its chromatin-binding property during S phase. Remarkably, advanced premature chromosome condensation (PCC) assays enabled us to visualize condensin II forming “sister axes” in replicated regions of chromosomes in S phase cells. Depletion of condensin II compromised PCC-driven sister chromatid resolution during S phase. Moreover, fluorescence in situ hybridization assays revealed that condensin II, but not condensin I, promotes disjoining duplicated chromosomal loci during S phase. Application of mild replicative stress partially impaired this process and further exacerbated phenotypes arising from condensin II depletion. Our results suggest that condensin II initiates structural reorganization of duplicated chromosomes during S phase to prepare for their proper condensation and segregation in mitosis.
Introduction
Chromosomes undergo drastic conformational changes during the cell division cycle. Since the aesthetic description by Walter Flemming in the late 19th century (Flemming, 1882), the dynamic behavior of chromosomes has attracted countless numbers of cell biologists and geneticists until now. In a typical animal cell, chromosomes can only be visualized in a limited window of the mitotic cell cycle (Morgan, 2007). The first sign of chromosome condensation becomes detectable in early prophase, in which chromatin distributed uniformly throughout the nuclear interior starts to display local compaction and is collapsed toward the nuclear envelope (Kireeva et al., 2004). These structural changes are then followed by the formation of linear chromosomal segments and the appearance of uniformly condensed chromosomes by late prophase. After nuclear envelope breakdown (NEBD) in prometaphase, chromosomes are individualized and sister chromatids within each of the chromosomes are resolved further, eventually leading to the formation of metaphase chromosomes in which rod-like sister chromatids are juxtaposed with each other. This series of structural changes, collectively referred to as chromosome condensation, is thought to be an essential prerequisite for faithful segregation of sister chromatids in subsequent anaphase (Swedlow and Hirano, 2003; Belmont, 2006; Marko, 2008).
Although chromosome condensation is traditionally regarded as an event that starts in mitotic prophase, it is important to note that the template of this process, a duplicated set of chromosomes, is produced in preceding S phase. Then, several fundamental questions arise. For example, how might the two processes of chromosome duplication and condensation mechanistically be linked? Exactly when might chromosomes start to prepare for their condensation and segregation? In retrospect, Mazia (1963) put forth the idea of a continuous “chromosome condensation cycle,” reasoning that some events preparatory to mitosis might take place before the visible process of chromosome condensation begins. Johnson and Rao (1970) then elegantly endorsed this idea by demonstrating that the so-called premature chromosome condensation (PCC) could be induced in interphase nuclei by fusing interphase cells with mitotic ones. Importantly, G1, S, and G2 nuclei were converted into chromosomes displaying different degrees of condensation, providing evidence for progressive changes of chromatin structure during interphase that would otherwise be difficult to visualize. Despite these pioneering studies, the molecular basis of the structural changes of chromosomes throughout the cell cycle has remained elusive.
The discovery and subsequent characterization of the condensin complexes, the major components required for chromosome condensation, now enable us to address this classical question from a molecular point of view (Hudson et al., 2009; Hirano, 2012). It has been established that most if not all eukaryotic cells possess two different condensin complexes, known as condensins I and II (Ono et al., 2003; Yeong et al., 2003). Whereas the two complexes share a pair of structural maintenance of chromosomes (SMC) core subunits (SMC2/CAP-E and SMC4/CAP-C), they have distinct sets of non-SMC regulatory subunits (chromosome-associated polypeptides CAP-D2, -G, and -H in condensin I and CAP-D3, -G2, and -H2 in condensin II). This difference in subunit composition is most likely to confer their differential distributions and actions during the cell cycle (Ono et al., 2004; Hirota et al., 2004; Gerlich et al., 2006; Abe et al., 2011). For instance, condensin I’s action is predominantly mitosis specific: it is sequestered into the cytoplasm during interphase through prophase, and gains access to chromosomes only after NEBD in prometaphase. In striking contrast, the bulk of condensin II localizes to the nucleus during interphase, participates in early stages of chromosome condensation in prophase, and collaborates with condensin I to shape chromosomes after prometaphase. It remains largely unknown, however, what functions condensin II might have within the interphase nucleus. In this respect, a recent intriguing observation is that the nuclear condensin II can be activated prematurely and lead to prophase-like condensation during G2 phase under the condition where MCPH1, a protein responsible for primary microcephaly in humans, is deficient (Trimborn et al., 2006; Yamashita et al., 2011).
In the current study, several lines of evidence are provided to demonstrate that condensin II changes its chromatin-binding property during S phase, long before the entry into mitosis. We then take two complementary approaches to probe potentially cryptic structural changes of chromosomes during interphase. First, advanced, drug-induced PCC assays were developed to differentially visualize unreplicated, replicating, and replicated regions of chromosomes in G1, S, or G2 phase cells and to immunolocalize condensin subunits in those PCC products. We find that condensin II–positive axes become detectable only in replicated regions of S-PCC and G2-PCC products, and that such PCC-driven resolution of sister chromatids is greatly compromised in cells depleted of condensin II. Second, FISH assays are applied to measure the distance between duplicated sister FISH signals in late S phase cells without PCC induction. Our results show that condensin II contributes to increasing the distance between sister FISH signals (i.e., resolving sister chromatids). Furthermore, chromosomal defects observed in condensin II–depleted cells are exacerbated when mild replicative stress is applied in preceding S phase. On the basis of these results, we suggest that condensin II initiates sister chromatid resolution during S phase and prepares for subsequent chromosome condensation and segregation in mitosis.
Results
Condensin II changes its chromatin-binding property during S phase
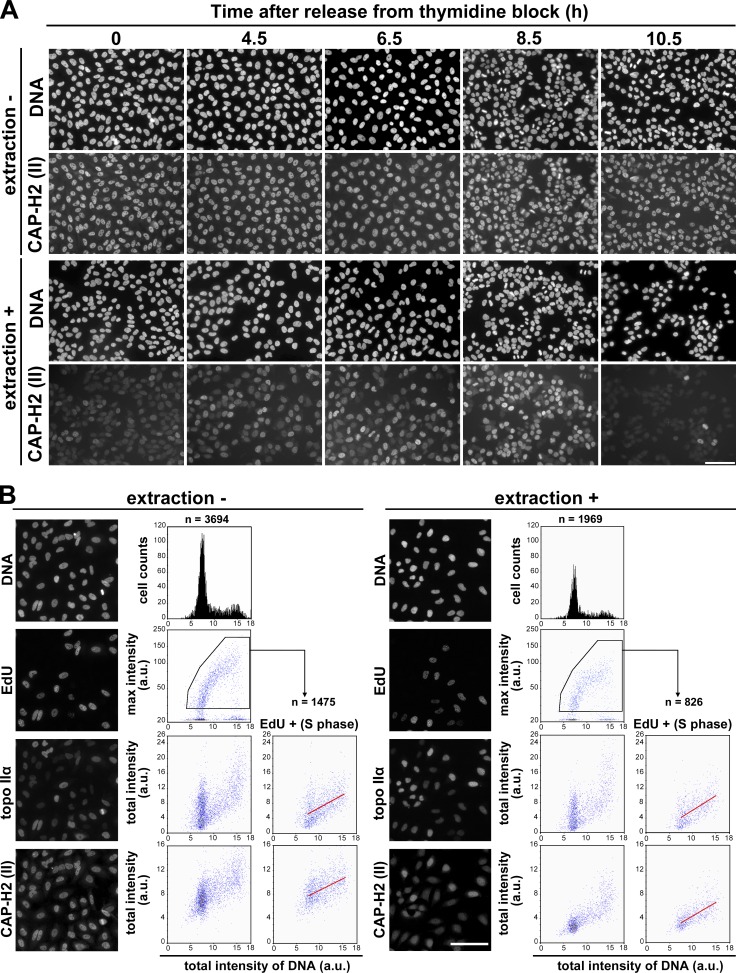
To address the question of what functions condensin II might have within the nucleus before the entry into mitosis, we examined the behavior of condensin II during the cell cycle in great detail. In the first set of experiments, HeLa cells were synchronized and immunolabeled with an antibody against CAP-H2 (a condensin II–specific subunit) at different time points. Under the standard fixation condition, the level of CAP-H2 was relatively constant throughout the cell cycle (Fig. 1 A, extraction −). No obvious change was detectable in the distribution of CAP-H2 from S to G2 phase, which was apparently excluded from the nucleolus and otherwise found evenly within the nucleus. In contrast, we found that, when the synchronized cells were treated with a detergent-containing buffer immediately before fixation, the bulk of the CAP-H2 signal was lost at G1/S phase (Fig. 1 A, extraction +, 0 h). Remarkably, the extraction-resistant population of nuclear CAP-H2 gradually increased in parallel with cell cycle progression from S through G2 phase (Fig. 1 A, extraction +, 4.5, 6.5, and 8.5 h). Upon completion of mitosis, the detergent-resistant population of CAP-H2 largely disappeared (Fig. 1 A, extraction +, 10.5 h). Thus, this extraction protocol allowed us to find that the property of CAP-H2 changes drastically during the cell cycle. Because the CAP-H2 signals in mitotic cells were resistant to the detergent treatment (Fig. 1 A, extraction +, 8.5 h), we considered it reasonable to assume that the detergent-resistant population of CAP-H2 found in interphase cells represents its chromatin-bound form.
Figure 1.
Condensin II changes its chromatin-binding property during S phase. (A) HeLa cells were synchronized by means of double thymidine block and release. Cells were fixed at the time points indicated after release and immunolabeled with anti–CAP-H2 (extraction −). Alternatively, the same set of cells was treated with a detergent-containing buffer before fixation and processed for immunofluorescence labeling (extraction +). Bar, 100 µm. (B) Asynchronously grown HeLa cells were pulse labeled with EdU, fixed, and immunolabeled with anti–topo IIα and anti–CAP-H2 (extraction −). Alternatively, the same set of cells was treated with a detergent-containing buffer before fixation and processed in the same way (extraction +). Quantitative imaging analyses were performed using CELAVIEW RS100. The first row shows DAPI-stained images and profiles of total intensity of DNA per nucleus (x axis) and cells counted (y axis). The second row shows EdU-labeled images and scattergrams of DNA intensity and max intensity of EdU. The third and fourth rows show topo IIα– and CAP-H2–labeled images and scattergrams of DNA intensity and total nuclear intensity of topo IIα and CAP-H2. In the right panels of the third and fourth rows (EdU +), the data of EdU-positive cells were extracted from the pentagonal areas in the second row, and plotted in the same way. In the scattergrams of EdU-positive cells, regression lines were drawn in red. Bar, 100 µm.
In the second set of experiments, asynchronous cultures of HeLa cells were pulse labeled with 5-ethynyl-2’-deoxyuridine (EdU) for detection of nascent DNA, fixed without or with prior extraction, and immunolabeled with antibodies against topoisomerase IIα (topo IIα) and CAP-H2. The resulting samples were subjected to quantitative cell imaging analyses using the high-throughout imaging device CELAVIEW RS100 (see Materials and methods). Consistent with previous studies (Heck et al., 1988; Agostinho et al., 2004), the level of topo IIα increased during S phase progression, peaking at G2 and mitosis. We found that the majority of topo IIα detected here was extraction resistant throughout S phase (Fig. 1 B, topo IIα [S phase]). In contrast, the total level of CAP-H2 increased only modestly during S phase (Fig. 1 B, extraction −, CAP-H2, EdU+ [S phase]). We noticed that more than half of the signals detectable in early S phase cells were sensitive to the detergent treatment whereas the detergent-resistant population of CAP-H2 increased substantially in parallel with S phase progression (Fig. 1 B, extraction +, CAP-H2 and EdU+ [S phase]). Taking these results together, we suggest that a subfraction of condensin II starts to associate with chromatin during S phase, long before chromosome condensation can be visualized in mitosis (for additional evidence, see Fig. S1). Despite this conclusion, it is important to note that siRNA-mediated knockdown of condensin II–specific subunits had little impact on the progression of S phase in HeLa cells (Fig. S2, A–C), as had been implied previously (Ono et al., 2003).
Advanced PCC assays show that condensin II forms sister axes in replicated regions of chromosomes in S phase cells
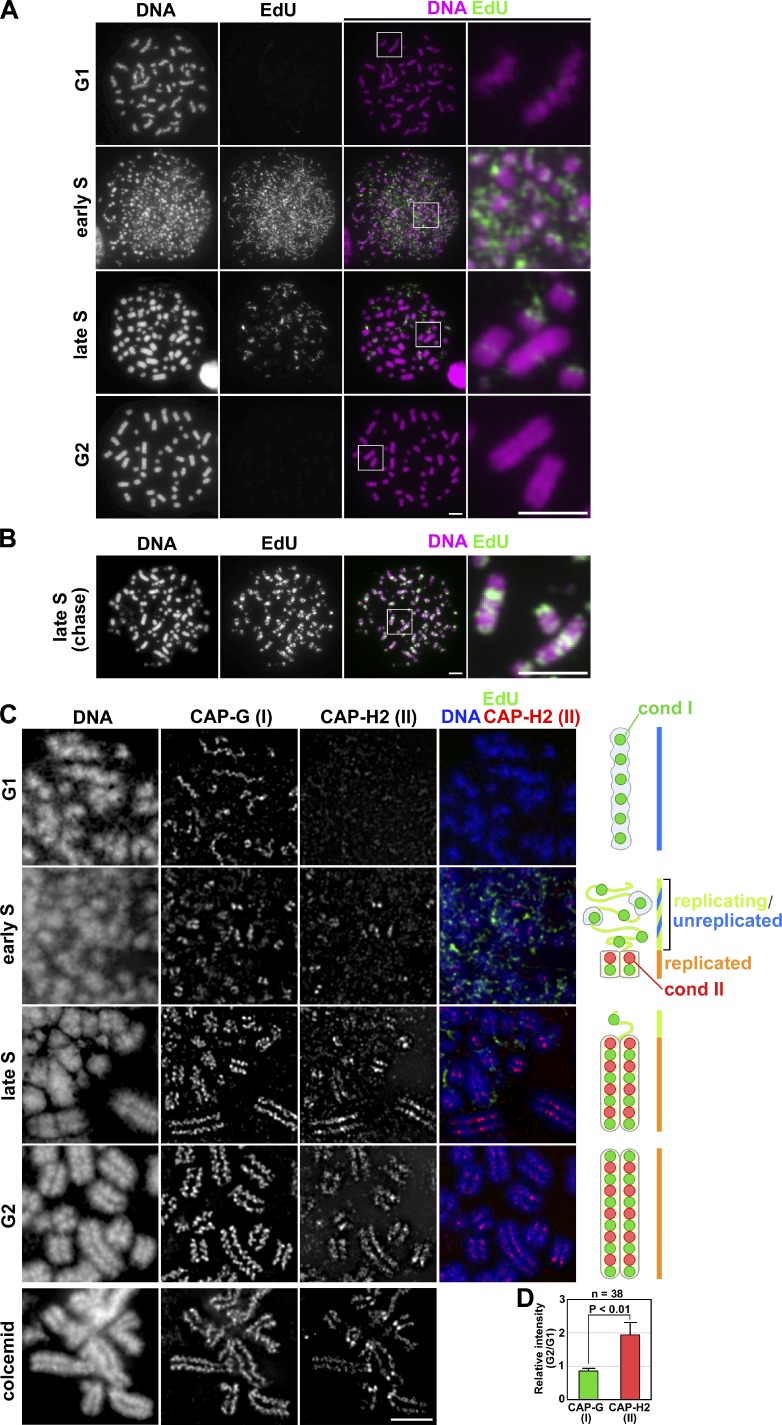
The observations described in the previous section prompted us to hypothesize that condensin II might initiate its role in organizing chromosomes during S phase. To test this hypothesis and to visualize potentially cryptic structural changes of chromosomes outside of mitosis, a drug-induced PCC assay was used in which nuclei at different stages of the cell cycle were converted into characteristic chromosome-like structures. For this purpose, we initially used a lymphoblastoid cell line in combination with calyculin A (a potent inhibitor of type 1 and 2A protein phosphatases), one of the well-characterized protocols for PCC induction (Gotoh and Durante, 2006; Hatzi et al., 2006). In the first set of experiments, asynchronously grown cells were pulse labeled with EdU for 0.5 h and treated with calyculin A for 1 h. The cells were then subjected to a hypotonic treatment, fixed with Carnoy’s solution, and spread on glass slides. EdU-positive cells were classified into early, mid, and late S phase cells on the basis of their characteristic EdU-labeling patterns (El Achkar et al., 2005; Gotoh, 2007). Among EdU-negative cells, G1- and G2-PCC products were judged on the basis of their chromosome morphology: representative G1-PCC cells exhibited a set of single chromatids with a wavy appearance (Fig. 2 A, G1), whereas G2-PCC cells displayed a set of chromosomes in which sister chromatids were clearly distinguishable from each other (Fig. 2 A, G2). Early S nuclei were converted into a scattering of tiny PCC fragments (known as pulverized chromosomes; Fig. 2 A, early S). In contrast, late S-PCC nuclei displayed a mixture of DAPI-dense, chromosome-like bodies and DAPI-poor fibrous structures (Fig. 2 A, late S). Paired sister chromatids were often recognizable in the DAPI-dense bodies, whereas EdU signals (corresponding to late-replicating regions) were primarily detected in the DAPI-poor structures. When cells were pulse labeled with EdU and chased for 3 h (without EdU) before calyculin A treatment, EdU signals overlapped with the DAPI-dense bodies (Fig. 2 B), confirming that they represent replicated regions containing juxtaposed sister chromatids.
Figure 2.
Advanced PCC assays show that condensin II forms sister axes in replicated regions of chromosomes in S phase cells. (A) Asynchronously grown lymphoblastoid cells were pulse labeled with EdU and then treated with calyculin A. The cells were subjected to a hypotonic treatment, fixed with Carnoy’s solution, and spread onto glass slides. The cell cycle stages were judged by EdU-labeling patterns and chromosome morphology. Shown here are representative images of cells at different stages, along with their closeups. Bars, 5 µm. (B) Asynchronously grown lymphoblastoid cells were pulse labeled with EdU and chased for 3 h before treatment with calyculin A. Shown here is a representative image of late S-PCC. Bars, 5 µm. (C) Lymphoblastoid cells were subjected to the same treatment as A and processed for immunolabeling with antibodies against CAP-G and -H2 (see Materials and methods). As a control, metaphase chromosome spreads were prepared in the same way from a culture that had been treated with colcemid (bottom). Cartoons depict the localization of condensins I and II in PCC products at the different stages. Bar, 5 µm. (D) The intensity of fluorescence signals was measured using the Polygon tool of the DeltaVision, and normalized to the intensity of DAPI. Plotted here are the relative intensities of the G2-PCC chromatids to the G1-PCC chromatids (for CAP-G and -H2). The mean and SD were calculated from the data from three independent experiments. The total numbers examined were described in the figure.
In the second set of experiments, we wished to immunolocalize condensin subunits in these PCC products. Because the chromosome spreads prepared with Carnoy’s fixative could not be used for this purpose, a modified fixation/spreading protocol was established to preserve the fragile PCC products for immunofluorescence analysis (see Materials and methods). We found that CAP-G (condensin I) signals were concentrated on the single axis of G1-PCC chromosomes, whereas CAP-H2 (condensin II) signals were barely detectable (Fig. 2 C, G1). In early S-PCC products, CAP-G signals became discontinuous and were often found as pairs of dots or short lines (Fig. 2 C, early S). At this stage, faint signals of CAP-H2 became detectable, overlapping with CAP-G in yet more confined areas. In late S-PCC products, robust enrichments of both CAP-G and -H2 on the sister chromatid axes were observed (Fig. 2 C, late S). Notably, the signals of CAP-H2 and EdU were mutually exclusive, although CAP-G distributed much more broadly than CAP-H2 and partially overlapped with EdU. In G2-PCC chromosomes (Fig. 2 C, G2), the distribution of CAP-G and -H2 were very similar to that found in non-PCC metaphase chromosomes prepared from colcemid-treated cells (Fig. 2 C, colcemid). Notably, although the fluorescent intensity of CAP-G signals was similar between the G1-PCC and G2-PCC chromatids, the intensity of CAP-H2 in the G2-PCC chromatids was approximately twofold higher than that in the G1-PCC chromatids (Fig. 2 D). One plausible interpretation of these observations is that the current PCC assays enable us to visualize the population of condensin II that progressively associates with replicated regions of chromosomes in parallel with S phase progression (also see Discussion).
Condensin II plays a key role in PCC-driven sister chromatid resolution in late S phase cells
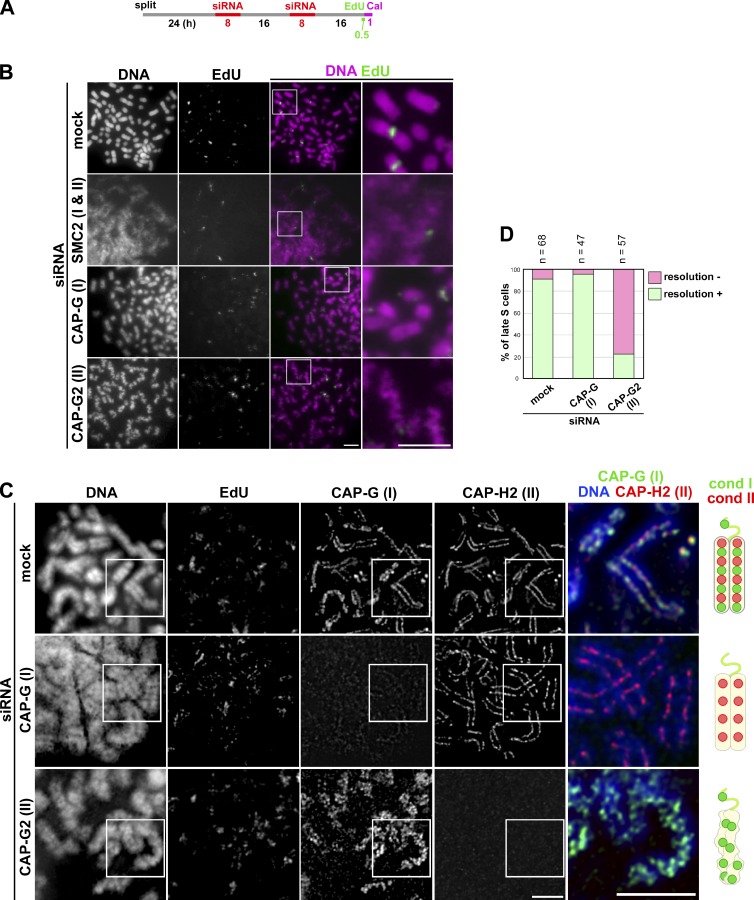
To examine how condensins I and II might contribute to PCC, we set up a protocol in which PCC was induced in cells depleted of different condensin subunits (Fig. 3 A). For this purpose, HeLa cells were used because more efficient gene knockdown with siRNAs was achieved in these cells than in lymphoblastoid cells. Having confirmed that HeLa cells were able to produce a set of PCC products similar to that observed in lymphoblastoid cells (Fig. S3 A), we focused on late S-PCC products in the subsequent analyses. Shown in Fig. 3 B are representative images of late S-PCC products prepared by Carnoy’s fixation. Unlike in mock-depleted cells, cells depleted of SMC2 (a core subunit common to condensins I and II) displayed a very hazy appearance of chromatin in which individual chromosomes were barely observed. In CAP-G–depleted cells, individual chromosomes were discernible, yet they were less structured and had dimmer surfaces compared with control late S-PCC products. The most characteristic morphology of late S-PCC products was observed in cells depleted of CAP-G2: chromosomes were grossly wavy and sister chromatids were hardly recognizable.
Figure 3.
Condensin II plays a key role in PCC-driven sister chromatid resolution in late S phase cells. (A) Experimental protocol for calyculin A–induced PCC from siRNA-treated HeLa cells. After two rounds of siRNA transfection, the cells were pulse labeled with EdU and then treated with calyculin A (Cal). (B) Cells were processed as in Fig. 2 B. Shown here are representative images of late S-PCC cells from populations mock depleted or depleted of SMC2, CAP-G, or CAP-G2. Bars, 5 µm. (C) HeLa cells (mock depleted or depleted of CAP-G or -G2) were pulse labeled with EdU and treated with calyculin A as described in A. The cells were fixed and immunolabled with antibodies against CAP-G and -H2, as described in Fig. 2 C. Shown here are representative images of late S-PCC cells, along with their merged closeups. Bars, 5 µm. Cartoons depict late S-PCC chromosomes observed under the different conditions. (D) Plotted here are the frequencies of resolution defects in late S-PCC products observed under the three conditions. This quantitative evaluation was completed once. For additional information, see Fig. S5 (A–C).
We then examined the localization of condensin subunits in these late S-PCC products prepared by the modified cytospin technique. In mock-depleted cells, both CAP-G and -H2 were concentrated on the axis of sister chromatids in nonoverlapping fashion (Fig. 3 C, mock siRNA), reminiscent of their distributions in metaphase chromosomes described previously (Ono et al., 2003, 2004). In CAP-G–depleted cells, although CAP-G signals were largely diminished, CAP-H2 signals were found along the entire length of sister chromatid axes in a manner similar to that observed in mock-depleted cells (Fig. 3 C, CAP-G siRNA). In striking contrast, in CAP-G2–depleted cells, CAP-G signals exhibited highly disordered, mesh-like structures in the unresolved and wavy chromosomes (Fig. 3 C, CAP-G2 siRNA). Virtually no signal of CAP-H2 was detectable in these late S-PCC products. The resolution defect was observed at a very high frequency in the CAP-G2–depleted cells but not in others (Fig. 3 D). These results clearly demonstrate that condensin II, but not condensin I, plays a crucial role in PCC-driven sister chromatid resolution in late S phase cells.
FISH assays reveal that condensin II promotes disjoining of duplicated chromosomal loci during S phase
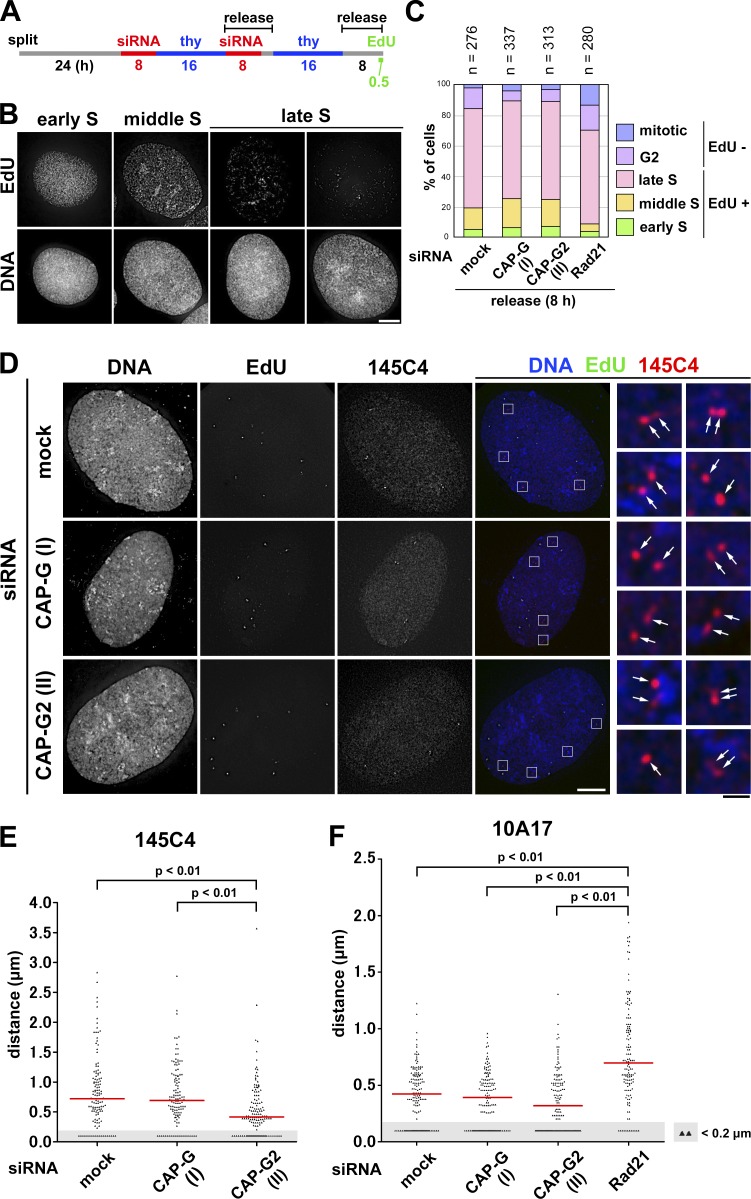
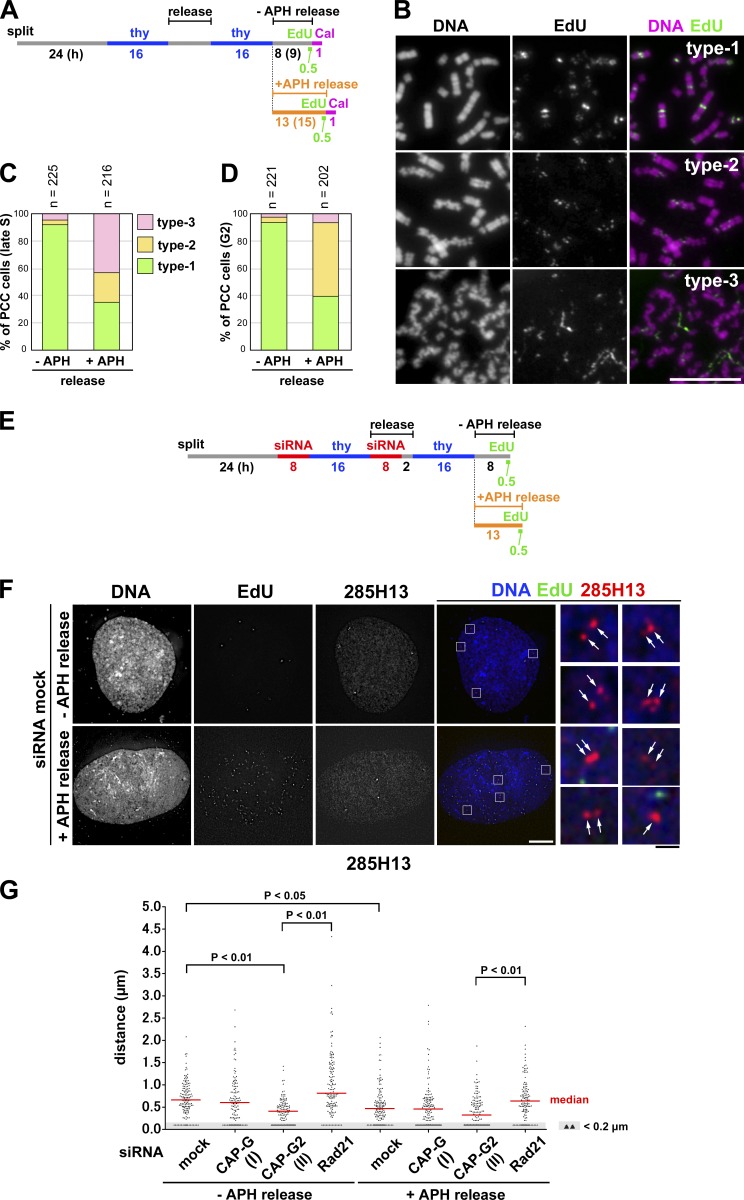
It is well known that FISH assays using single-copy DNA probes detect singlet signals for each of the chromosomal loci before DNA replication and doublet signals after DNA replication (Selig et al., 1992; Kitsberg et al., 1993). We sought to apply FISH assays to obtain independent lines of evidence that condensin II initiates sister chromatid resolution during S phase. If our hypothesis were correct, then the distance between replicated sister loci would decrease in cells depleted of condensin II and would increase instead in cells depleted of cohesin. To this end, we established a protocol to enrich late S phase cells from synchronous cultures depleted of condensin or cohesin subunits (Fig. 4 A; and Fig. S2, E and F, for cohesin depletion). On the basis of the EdU-labeling pattern and the nuclear morphology (Fig. 4 B), the cell cycle stages of more than 200 cells for each condition were determined. According to this protocol, 60–70% of cells for each condition were judged to be in late S phase (Fig. 4 C).
Figure 4.
FISH assays reveal that condensin II promotes disjoining of duplicated chromosomal loci during S phase. (A) Experimental protocol for enriching late S phase cells depleted of condensin subunits (see Fig. S2 E for cohesin depletion). Cells were pulse labeled with EdU before harvest. (B) After a hypotonic treatment, the cells were fixed with Carnoy’s solution and spread onto glass slides. EdU-positive cells were classified into early, middle, or late S on the basis of the EdU-labeling pattern. Representative images of cells at each stage are shown. Bar, 10 µm. (C) Plotted here are cell cycle stages that were judged on the basis of the EdU-labeling pattern and cell morphology. The data shown are from a single representative experiment out of two repeats. (D) FISH assays of a chromosomal locus whose sister distance is susceptible to condensin II depletion. HeLa cells mock depleted or depleted of CAP-G or -G2 were prepared as described in B and hybridized with the BAC clone 145C4. Cells in late S phase were selected as judged by the EdU-labeling pattern, and the distance between sister FISH signals was measured. Shown here are representative images of late S cells from the three different cultures, along with their merged closeups. FISH signals are indicated by the arrows. Bars: (white) 10 µm; (black) 1 µm. (E) Plots of the measured distance between sister FISH signals hybridized with 145C4. Under each condition, >120 pairs of sister FISH signals from late S cells were analyzed. For reasons of convenience, all values judged to be <0.2 µm, the limit of optical resolution, were plotted as 0.1 µm (as indicated by the shadowed areas). The red bar indicates the median value. (F) Plots of the measured distance between sister FISH signals hybridized with 10A17. Under each condition, >130 pairs of sister FISH signals from late S cells were analyzed. Each assessment shown in E and F was completed once. Detailed statistical data for FISH analyses are shown in Tables S1 and S2.
We hybridized these pools of cells with two different DNA probes (each ∼170 kb in length) derived from bacterial artificial chromosomes (BACs), and the distance between sister FISH signals in the late S phase cells was measured. As the HeLa cell line used here was a hypotetraploid in which a modal number of chromosomes was ∼82, most cells therefore displayed four pairs of FISH signals with the BAC probes on chromosome 1 (Fig. 4 D). When the BAC probe 145C4 (on 1p36.13) was used for hybridization, a median distance between sister FISH signals was measured to be ∼0.72 µm in mock-depleted cells (Fig. 4 E and Table S1). Remarkably, the distance significantly decreased in cells depleted of CAP-G2 (∼0.41 µm), but not in cells depleted of CAP-G (∼0.70 µm). Although we noticed that the second BAC probe, 10A17 (on 1p32.1), displayed a short sister distance (∼0.42 µm) even in mock-depleted cells, the value further decreased to ∼0.32 µm in CAP-G2-depleted cells (Fig. 4 F and Table S2). Conversely, when the same probe was applied to cells depleted of the cohesin subunit Rad21, a marked increase in the distance between sister FISH signals was observed (from ∼0.42 to ∼0.70 µm). Thus, the current data from the FISH assays provide independent lines of evidence that condensin II contributes to resolving duplicated chromatids during S phase before the entry into mitosis.
Application of mild replicative stress partially impairs sister chromatid resolution during S phase
To examine how the process of DNA replication might be coupled to subsequent structural reorganization of sister chromatids, we applied mild replicative stress to cells during S phase. When a low concentration (0.1 µg/ml) of aphidicolin was added to cells released from the second thymidine block, the speed of S phase progression slowed down, yet the bulk of replication completed (Fig. S2 D; and Fig. S3, B and C). In the first set of experiments, late S phase cells were enriched from control or aphidicolin-treated cultures, pulse-labeled with EdU, and then treated with calyculin A (Fig. 5 A). In the control culture with no aphidicolin, ∼95% cells displayed a typical product of late S-PCC (Fig. 5, B [type-1] and C). In the culture with aphidicolin, however, significant fractions of cells displayed partially distorted chromosomes with sister chromatids (Fig. 5 B, type-2) or poorly resolved chromosomes with a wavy appearance (Fig. 5, B [type-3] and C). Notably, the morphology of chromosomes classified into type-3 was highly reminiscent of, if not identical to, that observed in the late S-PCC products observed in CAP-G2–depleted cells (Fig. 3 B, CAP-G2 siRNA). When G2-PCC products were prepared after an extended time of incubation (Fig. 5 A), resolution proceeded further even in the presence of aphidicolin (Fig. 5 D). This observation is consistent with the idea that delay in resolution imposed by mild replicative stress can eventually be overcome by metaphase (see Fig. 6).
Figure 5.
Application of mild replicative stress partially impairs sister chromatid resolution during S phase. (A) Experimental protocol for enriching late S and G2 phase cells from synchronized cultures treated with aphidicolin (APH) and/or calyculin A (Cal). HeLa cells were released in a medium with or without 0.1 µg/ml aphidicolin and pulse labeled with EdU before being treated with calyculin A. (B) Cells released for 8 (−APH release) or 13 h (+APH release) were fixed with Carnoy’s solution and spread onto glass slides. Late S cells were selected on the basis of EdU-labeling patterns. The morphology of PCC products was classified into three types, and representative images are shown. Bar, 10 µm. (C) Plotted here are the frequencies of the three types of morphology observed in late S-PCC cells. (D) Cells were incubated for an extended period (9 h, −APH release; 15 h, +APH release; see A) and fixed and treated as in B. G2 cells with no EdU signals were selected, and the frequencies of the three types of morphology observed are plotted. Each experiment shown in C and D was completed once. (E) Experimental protocol for enriching late S phase cells from synchronized cultures treated with siRNAs and aphidicolin. Cells were released in the absence or presence of aphidicolin and pulse-labeled with EdU before harvest. See Fig. S2 E for Rad21 depletion. (F) HeLa cells mock depleted or depleted of CAP-G, CAP-G2, or Rad21 were prepared, hybridized with the BAC clone 285H13, and analyzed as described in Fig. 4 D. Shown here are representative images of late S cells from the mock-depleted culture released in a medium with or without aphidicolin. FISH signals are indicated by the arrows. Bars: (white) 10 µm; (black) 1 µm. (G) Plots of the measured distance between sister FISH signals hybridized with 285H13. In each culture, >110 pairs of sister FISH signals from late S cells were analyzed. This assessment was completed once. Detailed statistical data are shown in Table S3.
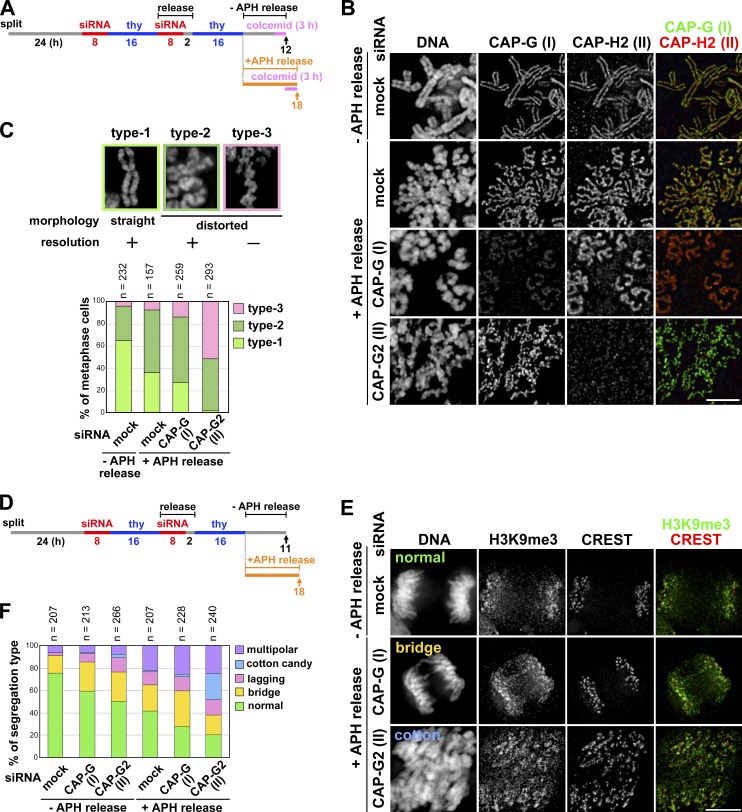
Figure 6.
Application of mild replicative stress to condensin II–depleted cells exacerbates their defects in chromosome architecture and segregation. (A) Experimental protocol for enriching metaphase cells from siRNA-treated synchronized cultures. Cells were harvested at the time points indicated by the arrows after colcemid treatment. (B) Metaphase chromosome spreads were prepared and immunolabeled with antibodies against CAP-G and -H2. Bar, 5 µm. (C) The morphology of metaphase chromosomes observed were classified into three types and plotted for each condition tested. The data shown are from a single representative experiment out of two repeats. For additional information, see Fig. S5 (D and E). (D) Experimental protocol for enriching mitotic cells from siRNA-treated synchronized cultures. No colcemid treatment was applied in this protocol. (E) HeLa cells treated as described in A were fixed on coverslips and immunolabeled with anti-H3K9me3 and a CREST serum. Shown here are representative images of cells displaying anaphase or anaphase-like chromosome morphology: normal segregation, chromosome bridges, and cotton candy chromosomes. Bar, 10 µm. (F) Defects in chromosome segregation were classified into five categories and plotted for each condition. The data shown are from a single representative experiment out of three repeats.
In the second set of experiments, FISH assays were applied to cells exposed to mild replicative stress. We prepared thymidine-blocked cultures depleted of condensin or cohesin subunits, and released them in the presence or absence of the low concentration of aphidicolin (Fig. 5 E). Cell cycle progression of these cultures was indistinguishable from a mock-depleted culture released in the presence of aphidicolin (Fig. S2, C, D, G, and H). We enriched late S cells from these cultures and used them for FISH assays with a third probe (285H13 [on 1p36.11]). With this probe, we first confirmed the effects of depletion of condensin II and cohesin in the absence of aphidicolin (Fig. 5, F and G, −APH release). We then found that, in mock-depleted cells, release into the aphidicolin-containing medium led to a modest decrease in the median sister distance from ∼0.66 to ∼0.47 µm (Fig. 5 G and Table S3). The same tendency toward decreasing distances was found in CAP-G2–depleted cells as well as in Rad21-depleted cells under the condition of aphidicolin release. Thus, both PCC and FISH assays suggest that application of the mild replicative stress partially impairs sister chromatid resolution during S phase.
Application of mild replicative stress to condensin II–depleted cells exacerbates their defects in chromosome architecture and segregation
Next we tested whether mild replicative stress applied to cells during S phase might affect the morphology of metaphase chromosomes in subsequent mitosis, according to the protocol summarized in Fig. 6 A. Metaphase chromosomes prepared from control cells possessed well-resolved sister chromatids in which both condensins I and II were concentrated on their straight axes (Fig. 6 B, −APH release, mock siRNA). In the presence of aphidicolin, sister chromatids were clearly discernible from each other, indicating that chromosomal DNA was fully replicated under this condition. Nonetheless, we noticed that chromosome arms displayed a wavy appearance (Fig. 6 B, +APH release, mock siRNA), a morphology reminiscent of that associated with CAP-G2–depleted chromosomes (Ono et al., 2003). Similarly, the swollen chromosomes in CAP-G–depleted cells acquired an additional, wavy distortion under the condition of aphidicolin release (Fig. 6 B, +APH release, CAP-G siRNA). Remarkably, a far more severe alternation of chromosome morphology was observed when CAP-G2–depleted cells were released in the presence of aphidicolin: continuous axial structures were no longer visible and sister chromatids were barely recognizable in many of the chromosomes (Fig. 6 B, +APH release, CAP-G2 siRNA). A quantitation of these observations is shown in Fig. 6 C.
To test how the combination of replicative stress and condensin II depletion might affect chromosome segregation, a similarly treated set of cells was allowed to enter mitosis without colcemid treatment (Fig. 6 D) and immunolabeled with a CREST serum and an antibody against histone H3 trimethylated on lysine 9 (H3K9me3), a marker for pericentromeric heterochromatin (Guenatri et al., 2004; Sullivan and Karpen, 2004). Anaphase and telophase cells were selected based on their morphology and representative images and quantitation of the observed phenotypes are shown in Fig. 6, E and F, respectively. In the absence of aphidicolin, the frequency of cells with lagging chromosomes and chromosome bridges increased when CAP-G or -G2 was depleted. In the presence of aphidicolin, the occurrence of such anaphase defects including a multipolar phenotype became prominent even in mock-depleted cells and was further elevated in CAP-G–depleted cells. Most remarkably, we found a characteristic defect of chromosome segregation in CAP-G2–depleted cells released in the presence of aphidicolin. In this phenotype, which we refer to as “cotton candy”, chromosomes were swollen and bumpy in appearance; they apparently attempted to segregate yet failed, thereby being scattered widely within the cell (Fig. 6 E, +APH release, CAP-G2 siRNA). Unlike normally segregating chromosomes, the H3K9me3 signals were no longer confined to discrete regions in these cotton candy chromosomes and highly stretched along with similarly distorted CREST signals. These results suggest that, when mild replicative stress is applied to cells depleted of CAP-G2, the architecture of mitotic chromosomes is largely disintegrated, leading to gross defects in their segregation.
Discussion
The chromosome condensation cycle revisited
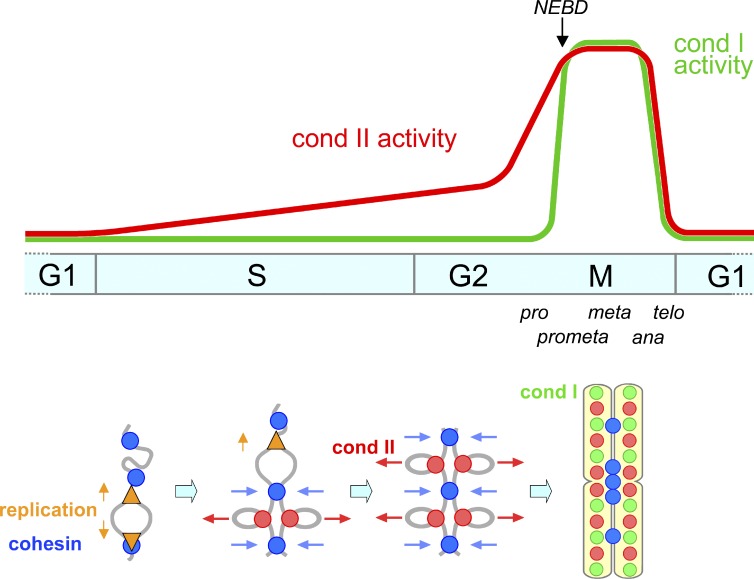
One of the primary motivations behind the current study was to revisit the classical idea of a continuous chromosome condensation cycle (Mazia, 1963) with the modern knowledge, tools, and approaches available today. Unlike in mitosis, large-scale structural changes of chromosomes during interphase are rather cryptic and difficult to probe. We therefore combine several independent approaches to provide evidence that condensin II associates with chromatin during S phase and initiates resolution of sister chromatids long before the entry into mitosis. This process is most likely to be an essential preparatory step for chromosome condensation and segregation in subsequent mitosis. Importantly, there is little sign that condensin I participates in this preparatory step, consistent with its cytoplasmic localization until NEBD in prometaphase. Thus, our results clarify and underscore the division of labor of the two condensin complexes throughout the cell cycle (Fig. 7).
Figure 7.
Models for the action of condensin II from S phase through mitosis. (top) Condensin II associates with chromatin to initiate sister chromatid resolution during S phase. It is assumed here that the activity of condensin II increases gradually over the period of S and G2 phases and rises quickly in prophase. Unlike condensin II, condensin I associates with chromosomes only after NEBD in prometaphase. (bottom) Condensin II (red circles) associates with replicated regions of chromosomes and initiates a “tug-of-war” with cohesin (blue circles) as early as in S phase, which persists until metaphase. Replication forks are indicated by orange triangles.
Advanced PCC and FISH assays uncover a cryptic yet essential role of condensin II in resolving sister chromatids during S phase
In the current study, we have developed and used advanced PCC assays as a potential strategy to uncover the architecture of interphase chromosomes that would otherwise be difficult to visualize. Drug-induced PCC assays had been used primarily in the field of cytogenetics, in combination with a classical chromosome spreading technique using Carnoy’s solution (Gotoh and Durante, 2006; Hatzi et al., 2006). We applied an EdU-labeling technology to the conventional assays so that PCC products derived from different stages of S phase could readily be identified. Furthermore, we modified a fixation/spreading protocol for immunolocalizing condensin subunits in control and siRNA-treated cells so that the differential contributions of condensins I and II to cell cycle–specific PCC could be unveiled. Our results clearly demonstrate that, although both condensin complexes contribute to proper induction of PCC, condensin II is absolutely essential for resolving sister chromatids in late S-PCC cells (Fig. 3). In an excellent agreement with such a role, the localization of condensin II is restricted to replicated regions of chromosomes throughout S phase progression (Fig. 2). In contrast, condensin I is distributed over much broader areas of PCC products, including unreplicated or replicating regions of chromosomes. We also notice that G1-PCC products (with single chromatids) contain condensin I alone and are much more fragile than G2-PCC chromosomes (with double chromatids) containing both condensins. We interpret these results to mean that PCC induction allows us to visualize the fraction of condensin II that has already associated with replicated regions of chromosomes during S phase. In contrast, condensin I, the majority of which is present in the cytoplasm during interphase, quickly associates with virtually all regions of chromosomes upon PCC induction. In fact, when the cells were treated with calyculin A, condensin I was converted into a pseudomitotic form whereas condensin II was not (Fig. S3 D), providing an explanation for the differential behaviors of the two condensin complexes in the PCC assays. Our interpretation is also supported by a previous FRAP analysis, which demonstrated that the interaction of condensin I with chromosomes is much more dynamic than that of condensin II during mitosis (Gerlich et al., 2006).
To complement the observations made with the advanced PCC assays, we used FISH assays. Although several previous studies used this technique to probe cohesin functions (Schmitz et al., 2007; Canudas and Smith, 2009; Remeseiro et al., 2012), the current study represents the first to apply this technique to address condensin functions during interphase in mammalian cells. We tested three different probes to visualize different chromosomal loci, and found consistently that condensin II, but not condensin I, contributes to disjoining of duplicated chromosome loci during S phase (Fig. 4). Our results also imply that the distance between sister chromatids is determined by the balancing action between condensin II–mediated resolving forces and cohesin-mediated cohesive forces along duplicated chromatids during interphase (Fig. 7, bottom), as has been shown in metaphase chromosomes reconstituted in Xenopus laevis egg extracts (Shintomi and Hirano, 2011).
Condensin II is a critical factor that links chromosome duplication to segregation
What might be the physiological relevance of condensin II–mediated sister chromatid resolution during S phase? We argue here that condensin II is a critical factor that links chromosome duplication to segregation. Both PCC and FISH assays show that application of mild replicative stress partially impairs sister chromatid resolution during S phase (Fig. 5). Although exactly how this works remains to be fully understood, one plausible possibility is that slowing down the replication speed triggers the activation of dormant origins and reduces the size of chromatin loops (Ge et al., 2007; Courbet et al., 2008). The small loop size would somehow restrain the distribution and/or action of condensin II during S phase, which is in turn manifested later as structurally distorted axes in metaphase chromosomes. Intriguingly, when condensin II is depleted from cells exposed to the mild replicative stress, far more severe and distinctive defects in chromosome architecture and segregation are observed in subsequent mitosis (Fig. 6): chromatid axes look very fragile and sister chromatids are not discernible from each other. Not surprisingly, they fail to segregate, displaying a highly characteristic cotton candy phenotype. Although these cells appear to have entered anaphase at first glance, they remain in metaphase as judged by the behaviors of cyclin B, aurora B, and Bub1 (Fig. S4, A–C). After a prolonged arrest at the pseudometaphase stage, the cells undergo apoptotic cell death (Fig. S4 D). These observations implicate that, under the condition where dormant origins get activated, the task of organizing and resolving smaller chromatin loops becomes more challenging, thereby helping to unveil the otherwise cryptic requirement for condensin II during S phase. Although we cannot exclude the possibility that the current experimental condition leaves a trace amount of unreplicated regions, the severe architectural defects are observed along the entire length of chromosomes, making this possibility less likely. Taken together, we suggest that condensin II’s action during S phase works as a critical preparatory step for subsequent chromosome condensation and segregation in mitosis.
Implications and perspectives
Emerging and increasing lines of evidence indicate that condensin II participates in several chromosome functions other than chromosome condensation or segregation (Wood et al., 2010; Hirano, 2012). Most notably, Hartl et al. (2008) have demonstrated that condensin II plays a role in antagonizing transvection, a process in which a gene is transcriptionally activated or repressed in trans by regulatory elements located on the homologous chromosome in Drosophila melanogaster. A recent high-throughput genetic screen has also identified condensin II subunits as factors that antagonize somatic homologue pairing in D. melanogaster (Joyce et al., 2012). It is reasonable to speculate that the resolution of sister chromatids within a duplicated chromosome and the restraint of homologue interactions share a common mechanism of action mediated by condensin II. It will be of great interest to test in the future whether analogous mechanisms might operate in other organisms, and to what extent the physical proximity of sister chromatids might affect the programs of gene expression.
The current results also shed new light on our understanding of the evolution of the machinery responsible for chromosome duplication and segregation. Whereas the two processes of duplication and segregation occur at temporally separated phases of the cell cycle in eukaryotic cells, they operate simultaneously in prokaryotic cells. Interestingly, in the gram-positive bacterium Bacillus subtilis, the SMC protein is recruited to the origin region and promotes efficient chromosome segregation (Gruber and Errington, 2009; Sullivan et al., 2009), implicating that SMC might be one of the key components linking duplication to segregation. Our current data show that in human cells condensin II associates with duplicated regions of chromosomes and initiates their resolution during S phase and that this process is most likely to contribute to proper chromosome segregation in mitosis. Thus, we argue that there is a much closer parallel than anticipated before between the eukaryotic and prokaryotic chromosome cycles. Future studies of condensins should further illuminate the mechanistic link between chromosome duplication and segregation.
Materials and methods
Bioethics approval
The use of lymphoblastoid cells in this study was approved by the ethics committees of both RIKEN and the Institute for Developmental Research, Aichi Human Service Center.
Cell culture and EdU labeling
HeLa cells used in this study were obtained from RIKEN BioResource Center (Cell number, RCB0007). A lymphoblastoid cell line was established, at the Institute for Developmental Research, Aichi Human Service Center, through Epstein-Barr virus–mediated transformation of peripheral blood samples collected from a healthy female with her consent (Yamada et al., 2010). HeLa cells were maintained in a modified Eagle’s medium (Sigma-Aldrich) supplemented with 10% FBS (SAFC Biosciences), whereas lymphoblastoid cells were maintained in an RPMI medium (Gibco) supplemented with 20% FBS, both at 37°C in an atmosphere of 5% CO2. For synchronization of HeLa cells, thymidine was added into the culture medium at a final concentration of 2.5 mM for 16 h. In some experiments, aphidicolin (Wako Pure Chemical Industries, Ltd. of Japan) was added at a final concentration of 0.1 µg/ml. To visualize the regions of nascent DNA replication, cells were cultured in the medium supplemented with 10 µM EdU for 0.5 h before fixation. The EdU incorporated was detected with the Click-iT EdU imaging kits (Invitrogen) according to the manufacturer’s instructions. In brief, the coverslips or glass slides were incubated with a reaction solution containing fluorochrome-azide (Alexa Fluor 488 or Alexa Fluor 647) for 30 min at room temperature before they were processed for immunofluorescent labeling.
siRNA treatment
The sequences of the Stealth siRNAs used for depletion experiments were as follows: hSMC2_78, 5′-CUUCAAUGCUAUCACUGGCUUAAAU-3′; hCAP-G_309, 5′-CUUAAAGUCUCAUGAAGCAAACAGC-3′; hCAP-G2_351, 5′-AGCCCUACUGGAAUGUGUUAUUAUA-3′; hCAP-H2_238, 5′-CAGGCCCUUGAUUUCAUCUCUGGAA-3′; hRad21-431, 5′-GGAAGCAGCUUAUAAUGCCAUUACU-3′. HeLa cells were transfected with Stealth siRNA duplexes at a final concentration of 33.3 nM using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. For control experiments, cells were transfected with 33.3 nM Stealth RNAi Negative Control Duplexes (Invitrogen). Medium GC duplex was used as a control for depletion of the condensin subunits, whereas low GC duplex was used for deletion of Rad21.
Antibodies
Rabbit polyclonal antibodies used in this study were anti-human CAP-G (Kimura et al., 2001), anti-human CAP-G2, and anti-human CAP-H2 (Ono et al., 2003). Human CREST serum was provided by Y. Muro (Nagoya University, Nagoya, Japan; Muro et al., 1990). To perform double immunolabeling of condensins I and II (Figs. 2 C, 3 C, 6 B, and S5 D), antibodies against CAP-G and CAP-H2 were labeled with Alexa Fluor 488 and Alexa Fluor 568, respectively, using the Alexa Fluor protein labeling kit (Molecular Probes). An antibody against human cyclin A2 was provided by J. Gannon and T. Hunt (London Research Institute, Cancer Research UK, Herts, UK). The following primary antibodies were purchased from commercial sources: anti–α-tubulin (clone DM1A; Sigma-Aldrich), anti-human topoisomerase IIα (clone 1C5; MBL), and anti-H3K9me3 (EMD Millipore). The following secondary antibodies were purchased from Molecular Probes and used for immunolabeling: Alexa Fluor 568 goat anti–human IgG, Alexa Fluor 488 or 568 goat anti–mouse IgG, and Alexa Fluor 488, 568, or 647 donkey anti–rabbit IgG. For immunoblotting, goat anti–rabbit and goat anti–mouse HRP-conjugated IgG were used as secondary antibodies (Vector Laboratories).
Immunolabeling
HeLa cells grown on poly-l-lysine–coated coverslips were fixed with two different fixation methods according to Ono et al. (2004) with some modifications (Figs. 1 and S1). In the first method, referred to as extraction −, the cells were fixed with 2% PFA in PBS, pH 7.4, at room temperature for 15 min, and then permeabilized in 0.5% Triton X-100 in PBS at room temperature for 5 min. In the second one, referred to as extraction +, the cells were first treated with 0.5% Triton X-100 in CSK buffer (10 mM Hepes, pH 7.5, 100 mM NaCl, 3 mM MgCl2, 300 mM sucrose, 100 nM microcystin, and 0.1 mM PMSF) on ice for 5 min, and then fixed with 2% PFA in CSK at room temperature for 15 min. After fixation, the cells were processed for immunolabeling as described previously (Ono et al., 2003, 2004). In brief, the coverslips were blocked with 3% BSA in PBS for 30 min, and then incubated with primary antibodies in the same solution for 1 h at room temperature, followed by incubation with fluorescent secondary antibodies for 1 h. Metaphase chromosome spreads (Figs. 6 B and S5 D) were prepared as described previously (Ono et al., 2003, 2004) with minor modifications. In brief, colcemid was added to the culture medium at a final concentration of 0.05 µg/ml 3 h before harvesting cells. Mitotic cells were collected by tapping culture dishes, treated with 75 mM KCl at 37°C for 30 min, and then centrifuged onto coverslips at 1,200 rpm for 5 min in a cytocentrifuge (Cytospin 4; Thermo Shandon). The cytospun preparations were fixed with the extraction + method. The coverslips were stained with DAPI, and then mounted with VECTASHIELD (Vector Laboratories). To visualize DNA in the analysis of prolonged cultures (Fig. S4 D), Hoechst 33342 (Invitrogen) was added into culture media at a final concentration of 1 µg/ml and left for 15 min at 37°C before observation.
FACS analysis
For FACS analysis, cells were washed twice with PBS and fixed with cold 70% ethanol. After incubated with 10 µg/ml RNase and 50 µg/ml propidium iodide at 37°C for 30 min, the cells were analyzed using a flow cytometer (LSR; BD). Fluorescence signals were collected at the wavelength of 575 nm for propidium iodide, and the data were analyzed using the FlowJo software (Tree Star).
Advanced PCC assays
To induce PCC, calyculin A (Wako Pure Chemical Industries, Ltd. of Japan) was added into the culture media at a final concentration of 50 nM. After 1 h, cells were treated with a hypotonic solution (75 mM KCl) at 37°C for 15 min, fixed with freshly prepared Carnoy’s solution (methanol/acetic acid, 3:1) through two rounds of centrifugation and resuspension, and spread onto glass slides. After being processed for detection of incorporated EdU as described in the previous section, the preparations were mounted with VECTASHIELD containing 1 µg/ml DAPI (Figs. 2 [A and B], 3 B, 5 B, S3 [A and C], and S5 A). For immunofluorescence labeling of PCC products, a modified fixation/spreading technique was developed. In brief, the cells were fixed with 1% PFA for 10 min, resuspended in PBS containing 0.2% Triton X-100, and centrifuged onto poly-l-lysine–coated coverslips at 800 rpm for 3 min (Cytospin 4; Thermo Shandon). The specimens were refixed with 2% PFA at room temperature for 15 min, treated with PBS containing 0.5% Triton X-100 at room temperature for 5 min, and then processed for EdU detection and immunolabeling (Figs. 2 C, 3 C, and S5 B).
FISH assays
Cells were synchronized into late S phase and pulse labeled with EdU. They were then treated with a hypotonic solution (75 mM KCl) for 8 min at 37°C and fixed with Carnoy’s solution. After spreading onto glass slides, the cells were subjected to hybridization with BAC clones. In this study, we used three different clones: RP11-145C4, RP11-10A17, and RP11-285H13. For annotation of each clone, refer to the University of California Santa Cruz (UCSC) Genome Browser on the UCSC Genome Bioinformatics website (http://genome.cse.ucsc.edu/cgi-bin/hgGateway). The BAC DNAs were labeled with Cy3-dUTP (GE Healthcare) using a Nick Translation kit (Roche), and hybridization was performed as described previously (Ono and Yoshida, 1993) with minor modifications. In brief, 20 µl of a hybridization mixture containing ∼100 ng of labeled DNA was placed on denatured cells on slides and incubated at 37°C for 36–60 h. The cells were then washed with 4× SSC, 4× SSC containing 0.1% Nonidet P-40, and 4× SSC for 10 min each at room temperature. The slides were treated with the procedure to visualize incorporated EdU and mounted with VECTASHIELD containing 1 µg/ml DAPI.
Imaging analyses and microscopy
Fluorescence images were acquired with a microscope (BX51; Olympus) with an UPlanSApo 100×/1.40 NA oil-immersion objective lens or an UPlanSApo 20×/0.75 NA objective lens, equipped with a cooled charge-coupled device camera (DP30; Olympus) and the DP Controller software (Olympus). Grayscale images were pseudocolored and merged with gamma adjustment using Photoshop (Adobe). For the experiment shown in Figs. 1 B and S1 A, immunolabeled cells were analyzed with the high-throughput imaging device CELAVIEW RS100 with a LUCPlanFLN 20×/0.45 NA objective lens (Olympus), as described by Yamashita et al. (2011). In brief, DNA, EdU, topo IIα, and CAP-H2 were detected with DAPI, Alexa Fluor 488 (or 647) azide, Alexa Fluor 568 goat anti–mouse IgG, and Alexa Fluor 647 donkey anti–rabbit IgG (Invitrogen), respectively. Images of >5,000 cells were acquired, and the total nuclear intensities of DNA, topo IIα, and CAP-H2 and the maximum intensity of EdU were evaluated in each cell. The data were plotted as scattergrams. For chromosome spreads (Figs. 2 C, 3 C, 6 B, and S5 [B and D]) and mitotic whole cells (Figs. 6 E and S4 [B and C]), 3D image stacks were acquired with a DeltaVision restoration microscope system (Applied Precision), consisting of an inverted microscope (IX71; Olympus) with an UPlanSApo 100×/1.40 NA oil-immersion objective lens (Olympus). The image stacks of whole cells, calyculin A–induced PCC, and metaphase chromosome spreads were acquired with a Z-step size of 0.5, 0.2, and 0.1 µm, respectively. They were subjected to constrained iterative (10 iterations) deconvolution (Chen et al., 1996), and then projected with five to eight sections using the softWoRx software (Applied Precision). For FISH assays, the 3D image stacks of EdU-labeled late S nuclei and FISH nuclei with a Z-step size of 0.5 µm were subjected to the deconvolution processes and then projected with three sections (Figs. 4 [B and D] and 5 F). Grayscale images were pseudocolored and merged using Photoshop (Adobe). The distance between sister FISH signals was measured on the projected images, and statistical analyses were performed using the GraphPad Prism software (GraphPad Software). Phase-contrast and fluorescence images (Fig. S4 D) were acquired with an inverted microscope (CKX41; Olympus) with a CAchN 10×/0.25 PhP objective lens, equipped with a digital camera (DP21; Olympus) and the DP Controller software (Olympus).
Online supplemental material
Fig. S1 offers an additional control for validating the quantitation of fluorescence signals using CELAVIEW. It also provides an independent line of evidence that condensin II starts to associate with chromatin during S phase. Fig. S2 shows that depletion of condensins or cohesin has little impact on S phase progression even in the presence of mild replicative stress. Fig. S3 shows representative images of calyculin A–induced PCC products in HeLa cells and EdU labeling patterns of late S-PCC products prepared from cells exposed to mild replicative stress. It also provides immunoblotting analysis that revealed differential modifications of condensin subunits under mitotically arrested and PCC-induced conditions. Fig. S4 demonstrates that cells with cotton candy chromosomes are arrested at a pseudometaphase stage and eventually undergo apoptotic cell death. Fig. S5 shows that depletion of CAP-H2 produces a set of defective phenotypes that is indistinguishable from that observed in cells depleted of CAP-G2. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201208008/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201208008.dv.
Supplementary Material
Acknowledgments
We are grateful to Y. Muro for CREST antiserum, J. Gannon and T. Hunt for cyclin A antibody, K. Yoshiura for helpful suggestions concerning FISH assays, and K. Ohtawa for technical support for FACS analysis. We also thank A. Matsuura for excellent technical assistance and members of the Hirano laboratory for critically reading the manuscript.
This work was supported by Grant-in-Aid for Scientific Research C and Grant-in-Aid for Scientific Research Priority Areas (to T. Ono), Grant-in-Aid for Young Scientists B (D. Yamashita), and Grant-in-Aid for Specially Promoted Research (to T. Hirano). D. Yamashita was a RIKEN Special Postdoctoral Researcher.
Footnotes
Abbreviations used in this paper:
- BAC
- bacterial artificial chromosome
- EdU
- 5-ethynyl-2’-deoxyuridine
- NEBD
- nuclear envelope breakdown
- PCC
- premature chromosome condensation
- SMC
- structural maintenance of chromosomes
References
- Abe S., Nagasaka K., Hirayama Y., Kozuka-Hata H., Oyama M., Aoyagi Y., Obuse C., Hirota T. 2011. The initial phase of chromosome condensation requires Cdk1-mediated phosphorylation of the CAP-D3 subunit of condensin II. Genes Dev. 25:863–874 10.1101/gad.2016411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostinho M., Rino J., Braga J., Ferreira F., Steffensen S., Ferreira J. 2004. Human topoisomerase IIalpha: targeting to subchromosomal sites of activity during interphase and mitosis. Mol. Biol. Cell. 15:2388–2400 10.1091/mbc.E03-08-0558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont A.S. 2006. Mitotic chromosome structure and condensation. Curr. Opin. Cell Biol. 18:632–638 10.1016/j.ceb.2006.09.007 [DOI] [PubMed] [Google Scholar]
- Canudas S., Smith S. 2009. Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J. Cell Biol. 187:165–173 10.1083/jcb.200903096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Hughes D.D., Chan T.A., Sedat J.W., Agard D.A. 1996. IVE (Image Visualization Environment): a software platform for all three-dimensional microscopy applications. J. Struct. Biol. 116:56–60 10.1006/jsbi.1996.0010 [DOI] [PubMed] [Google Scholar]
- Courbet S., Gay S., Arnoult N., Wronka G., Anglana M., Brison O., Debatisse M. 2008. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature. 455:557–560 10.1038/nature07233 [DOI] [PubMed] [Google Scholar]
- El Achkar E., Gerbault-Seureau M., Muleris M., Dutrillaux B., Debatisse M. 2005. Premature condensation induces breaks at the interface of early and late replicating chromosome bands bearing common fragile sites. Proc. Natl. Acad. Sci. USA. 102:18069–18074 10.1073/pnas.0506497102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming W. 1882. Zellsubstantz, Kern und Zelltheilung. FCW Vogel, Leipzig, Germany: 424 pp [Google Scholar]
- Ge X.Q., Jackson D.A., Blow J.J. 2007. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 21:3331–3341 10.1101/gad.457807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D., Hirota T., Koch B., Peters J.M., Ellenberg J. 2006. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr. Biol. 16:333–344 10.1016/j.cub.2005.12.040 [DOI] [PubMed] [Google Scholar]
- Gotoh E. 2007. Visualizing the dynamics of chromosome structure formation coupled with DNA replication. Chromosoma. 116:453–462 10.1007/s00412-007-0109-5 [DOI] [PubMed] [Google Scholar]
- Gotoh E., Durante M. 2006. Chromosome condensation outside of mitosis: mechanisms and new tools. J. Cell. Physiol. 209:297–304 10.1002/jcp.20720 [DOI] [PubMed] [Google Scholar]
- Gruber S., Errington J. 2009. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 137:685–696 10.1016/j.cell.2009.02.035 [DOI] [PubMed] [Google Scholar]
- Guenatri M., Bailly D., Maison C., Almouzni G. 2004. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166:493–505 10.1083/jcb.200403109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl T.A., Smith H.F., Bosco G. 2008. Chromosome alignment and transvection are antagonized by condensin II. Science. 322:1384–1387 10.1126/science.1164216 [DOI] [PubMed] [Google Scholar]
- Hatzi V.I., Terzoudi G.I., Paraskevopoulou C., Makropoulos V., Matthopoulos D.P., Pantelias G.E. 2006. The use of premature chromosome condensation to study in interphase cells the influence of environmental factors on human genetic material. ScientificWorldJournal. 6:1174–1190 10.1100/tsw.2006.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck M.M., Hittelman W.N., Earnshaw W.C. 1988. Differential expression of DNA topoisomerases I and II during the eukaryotic cell cycle. Proc. Natl. Acad. Sci. USA. 85:1086–1090 10.1073/pnas.85.4.1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. 2012. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 26:1659–1678 10.1101/gad.194746.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T., Gerlich D., Koch B., Ellenberg J., Peters J.M. 2004. Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell Sci. 117:6435–6445 10.1242/jcs.01604 [DOI] [PubMed] [Google Scholar]
- Hudson D.F., Marshall K.M., Earnshaw W.C. 2009. Condensin: architect of mitotic chromosomes. Chromosome Res. 17:131–144 10.1007/s10577-008-9009-7 [DOI] [PubMed] [Google Scholar]
- Johnson R.T., Rao P.N. 1970. Mammalian cell fusion: induction of premature chromosome condensation in interphase nuclei. Nature. 226:717–722 10.1038/226717a0 [DOI] [PubMed] [Google Scholar]
- Joyce E.F., Williams B.R., Xie T., Wu C.T. 2012. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet. 8:e1002667 10.1371/journal.pgen.1002667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Cuvier O., Hirano T. 2001. Chromosome condensation by a human condensin complex in Xenopus egg extracts. J. Biol. Chem. 276:5417–5420 10.1074/jbc.C000873200 [DOI] [PubMed] [Google Scholar]
- Kireeva N., Lakonishok M., Kireev I., Hirano T., Belmont A.S. 2004. Visualization of early chromosome condensation: a hierarchical folding, axial glue model of chromosome structure. J. Cell Biol. 166:775–785 10.1083/jcb.200406049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitsberg D., Selig S., Brandeis M., Simon I., Keshet I., Driscoll D.J., Nicholls R.D., Cedar H. 1993. Allele-specific replication timing of imprinted gene regions. Nature. 364:459–463 10.1038/364459a0 [DOI] [PubMed] [Google Scholar]
- Marko J.F. 2008. Micromechanical studies of mitotic chromosomes. Chromosome Res. 16:469–497 10.1007/s10577-008-1233-7 [DOI] [PubMed] [Google Scholar]
- Mazia D. 1963. Synthetic activities leading to mitosis. J. Cell. Physiol. 62:123–140 [DOI] [PubMed] [Google Scholar]
- Morgan D.O. 2007. The Cell Cycle: Principles of Control. New Science Press, London: 297 pp [Google Scholar]
- Muro Y., Sugimoto K., Okazaki T., Ohashi M. 1990. The heterogeneity of anticentromere antibodies in immunoblotting analysis. J. Rheumatol. 17:1042–1047 [PubMed] [Google Scholar]
- Ono T., Yoshida M.C. 1993. Chromosomal assignment of retinoblastoma 1 gene (RB1) to mouse 14D3 and rat 15q12 by fluorescence in situ hybridization. Jpn. J. Genet. 68:617–621 10.1266/jjg.68.617 [DOI] [PubMed] [Google Scholar]
- Ono T., Losada A., Hirano M., Myers M.P., Neuwald A.F., Hirano T. 2003. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 115:109–121 10.1016/S0092-8674(03)00724-4 [DOI] [PubMed] [Google Scholar]
- Ono T., Fang Y., Spector D.L., Hirano T. 2004. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell. 15:3296–3308 10.1091/mbc.E04-03-0242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remeseiro S., Cuadrado A., Carretero M., Martínez P., Drosopoulos W.C., Cañamero M., Schildkraut C.L., Blasco M.A., Losada A. 2012. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 31:2076–2089 10.1038/emboj.2012.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J., Watrin E., Lénárt P., Mechtler K., Peters J.M. 2007. Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr. Biol. 17:630–636 10.1016/j.cub.2007.02.029 [DOI] [PubMed] [Google Scholar]
- Selig S., Okumura K., Ward D.C., Cedar H. 1992. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 11:1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K., Hirano T. 2011. The relative ratio of condensin I to II determines chromosome shapes. Genes Dev. 25:1464–1469 10.1101/gad.2060311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B.A., Karpen G.H. 2004. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11:1076–1083 10.1038/nsmb845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N.L., Marquis K.A., Rudner D.Z. 2009. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 137:697–707 10.1016/j.cell.2009.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedlow J.R., Hirano T. 2003. The making of the mitotic chromosome: modern insights into classical questions. Mol. Cell. 11:557–569 10.1016/S1097-2765(03)00103-5 [DOI] [PubMed] [Google Scholar]
- Trimborn M., Schindler D., Neitzel H., Hirano T. 2006. Misregulated chromosome condensation in MCPH1 primary microcephaly is mediated by condensin II. Cell Cycle. 5:322–326 10.4161/cc.5.3.2412 [DOI] [PubMed] [Google Scholar]
- Wood A.J., Severson A.F., Meyer B.J. 2010. Condensin and cohesin complexity: the expanding repertoire of functions. Nat. Rev. Genet. 11:391–404 10.1038/nrg2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Fukushi D., Ono T., Kondo Y., Kimura R., Nomura N., Kosaki K.J., Yamada Y., Mizuno S., Wakamatsu N. 2010. Characterization of a de novo balanced t(4;20)(q33;q12) translocation in a patient with mental retardation. Am. J. Med. Genet. A. 152A:3057–3067 10.1002/ajmg.a.33174 [DOI] [PubMed] [Google Scholar]
- Yamashita D., Shintomi K., Ono T., Gavvovidis I., Schindler D., Neitzel H., Trimborn M., Hirano T. 2011. MCPH1 regulates chromosome condensation and shaping as a composite modulator of condensin II. J. Cell Biol. 194:841–854 10.1083/jcb.201106141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeong F.M., Hombauer H., Wendt K.S., Hirota T., Mudrak I., Mechtler K., Loregger T., Marchler-Bauer A., Tanaka K., Peters J.M., Ogris E. 2003. Identification of a subunit of a novel Kleisin-beta/SMC complex as a potential substrate of protein phosphatase 2A. Curr. Biol. 13:2058–2064 10.1016/j.cub.2003.10.032 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.