Active Rag GTPases are required for recruitment of TFEB to lysosomes and its phosphorylation by mTORC1, inhibiting its function under nutrient-rich conditions.
Abstract
The mTORC1 complex supports cell growth and proliferation in response to energy levels, growth factors, and nutrients. The Rag guanosine triphosphatases (GTPases) activate mTORC1 in response to amino acids by promoting its redistribution to lysosomes. In this paper, we identify a novel role for Rags in controlling activation of transcription factor EB (TFEB), a master regulator of autophagic and lysosomal gene expression. Interaction of TFEB with active Rag heterodimers promoted recruitment of TFEB to lysosomes, leading to mTORC1-dependent phosphorylation and inhibition of TFEB. The interaction of TFEB with Rags required the first 30 residues of TFEB and the switch regions of the Rags G domain. Depletion or inactivation of Rags prevented recruitment of TFEB to lysosomes, whereas expression of active Rags induced association of TFEB with lysosomal membranes. Finally, Rag GTPases bound and regulated activation of microphthalmia-associated transcription factor, suggesting a broader role for Rags in the control of gene expression. Our work provides new insight into the molecular mechanisms that link nutrient availability and TFEB localization and activation.
Introduction
The mammalian target of rapamycin (mTOR) is an evolutionarily conserved serine/threonine kinase that regulates numerous cellular processes, including cell growth, proliferation, cell cycle, and autophagy. mTOR responds to many stresses, and its dysregulation leads to cancer, metabolic disease, and diabetes (Zoncu et al., 2011b). In cells, mTOR exists as two different multiprotein complexes termed mTORC1 (mTOR complex 1) and mTORC2 (mTOR complex 2; Hara et al., 2002; Kim et al., 2002; Sarbassov et al., 2004). Both complexes share the catalytic mTOR subunit, mLST8, DEPTOR, and the Tti1–Tel2 complex. In contrast, raptor and PRAS40 are specific to mTORC1, whereas rictor, mSin1, and protor1/2 are only present in mTORC2. mTORC1 is regulated by multiple upstream factors, including growth factors, glucose, and amino acids, and its function is critical to couple energy and nutrient abundance to cell growth and proliferation (Laplante and Sabatini, 2012). mTORC2 is primarily regulated by growth factor–mediated phosphoinositide-3-kinase signaling and plays important roles in actin reorganization and cell survival (Jacinto et al., 2004).
The activation of mTORC1 by intracellular amino acids is well characterized. In response to amino acid stimulation, mTORC1 is recruited to the lysosomal surface, where it is activated by the small GTPase Rheb (Saucedo et al., 2003; Stocker et al., 2003). The amino acid–dependent translocation of mTOR requires Rag GTPases and Ragulator, a pentameric protein complex that comprises p18, p14, MP1, HBXIP, and C7orf59 and anchors the Rag GTPases to the lysosomes (Sancak et al., 2008, 2010; Bar-Peled et al., 2012). The Rag proteins function as heterodimers in which the active complex consists of GTP-bound RagA or B complexed with GDP-bound RagC or D (Sekiguchi et al., 2001; Gao and Kaiser, 2006). Importantly, amino acids trigger the GTP loading of RagA/B proteins, thus promoting binding to raptor and assembly of an activated mTORC1 complex (Sancak et al., 2008). Active mTORC1 supports synthesis of proteins and cell growth while actively suppressing autophagy. In the absence of amino acids, the Rags turn into an inactive conformation (GDP-bound RagA/B and GTP-bound RagC/D), and mTORC1 is inactivated and shuttled back to the cytosol. Rag GTPases also bind to the lysosomal vacuolar-type H+-ATPase, and this interaction is thought to be a way for the Rags to sense the amino acid content inside lysosomes and, by extension, the nutritional state of the cell (Zoncu et al., 2011a).
One of the most critical functions of the mTORC1 complex is to repress autophagy under conditions in which nutrients are abundant. For this, mTORC1 directly phosphorylates and inhibits Atg proteins involved in autophagy induction, such as Atg13 and Atg1 (ULK1/2; Hosokawa et al., 2009a,b). Recently, we and others have shown that mTORC1 also controls expression of autophagic and lysosomal genes by regulating the localization of the transcription factor EB (TFEB; Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2012). TFEB is a member of the basic helix–loop–helix leucine zipper family of transcription factors that controls lysosomal biogenesis and autophagy by positively regulating genes belonging to the CLEAR (coordinated lysosomal expression and regulation) network (Sardiello and Ballabio, 2009; Sardiello et al., 2009; Palmieri et al., 2011). Activation of TFEB leads to an increased number of autophagosomes and autophagic flux, biogenesis of new lysosomes, and clearance of storage material in several lysosomal storage disorders (Sardiello and Ballabio, 2009; Medina et al., 2011; Settembre et al., 2011). We found that in fully fed cells, active mTORC1 phosphorylates TFEB in several residues, including serine 211 (S211). Phosphorylation in S211 promotes interaction of TFEB with the cytosolic chaperone 14-3-3 and consequent retention of TFEB in the cytosol. Inactivation of mTORC1 by nutrient withdrawal causes dissociation of the TFEB–14-3-3 complex, thus leading to TFEB transport to the nucleus and TFEB-dependent transcription of multiple genes implicated in correcting the nutrient deficiencies of the cell (Martina et al., 2012; Roczniak-Ferguson et al., 2012).
The lysosomal localization of active mTORC1 predicts that TFEB must be recruited to lysosomes to be phosphorylated by mTOR. In fact, the current model suggests that, in fully fed cells, TFEB continuously cycles between lysosomes and cytosol (Martina et al., 2012; Settembre et al., 2012). At any given time, a fraction of TFEB is associated with lysosomes, where it is phosphorylated by mTORC1 and sent back to the cytosol. The recruitment of TFEB to lysosomes is rapid and transient, and at steady state, most TFEB appears freely diffused in the cytosol. However, under conditions in which the formation of the TFEB–14-3-3 complex is blocked, such as mutation of S211 to alanine or inhibition of mTORC1 by Torin-1, TFEB not only accumulates in the nucleus but also shows a stronger and more stable association with lysosomes (Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2012).
The mechanism that controls recruitment of TFEB to lysosomes is critical for the mTORC1-dependent regulation of TFEB and remains to be elucidated. Therefore, in this study, we seek to identify the molecular machinery that regulates association of TFEB with lysosomal membranes as well as understanding the cellular signals that coordinate nutrient availability and TFEB activation.
Here, we show that TFEB interacts with Rag GTPases and is relocated to lysosomes in an amino acid–dependent manner. Changes in the levels of intracellular amino acids alter the nucleotide state of the Rags and, thereby, their ability to bind and recruit TFEB to lysosomes. Binding to active Rags is required for mTORC1-dependent phosphorylation of TFEB and assures retention of TFEB in the cytosol under conditions in which expression of autophagic and lysosomal genes must be repressed. Overall, our work provides new insights into the mechanisms by which the Rag complex provides temporal and spatial coordination of TFEB localization and activation.
Results
Rag GTPases, but not mTORC1, are required for recruitment of TFEB to lysosomes
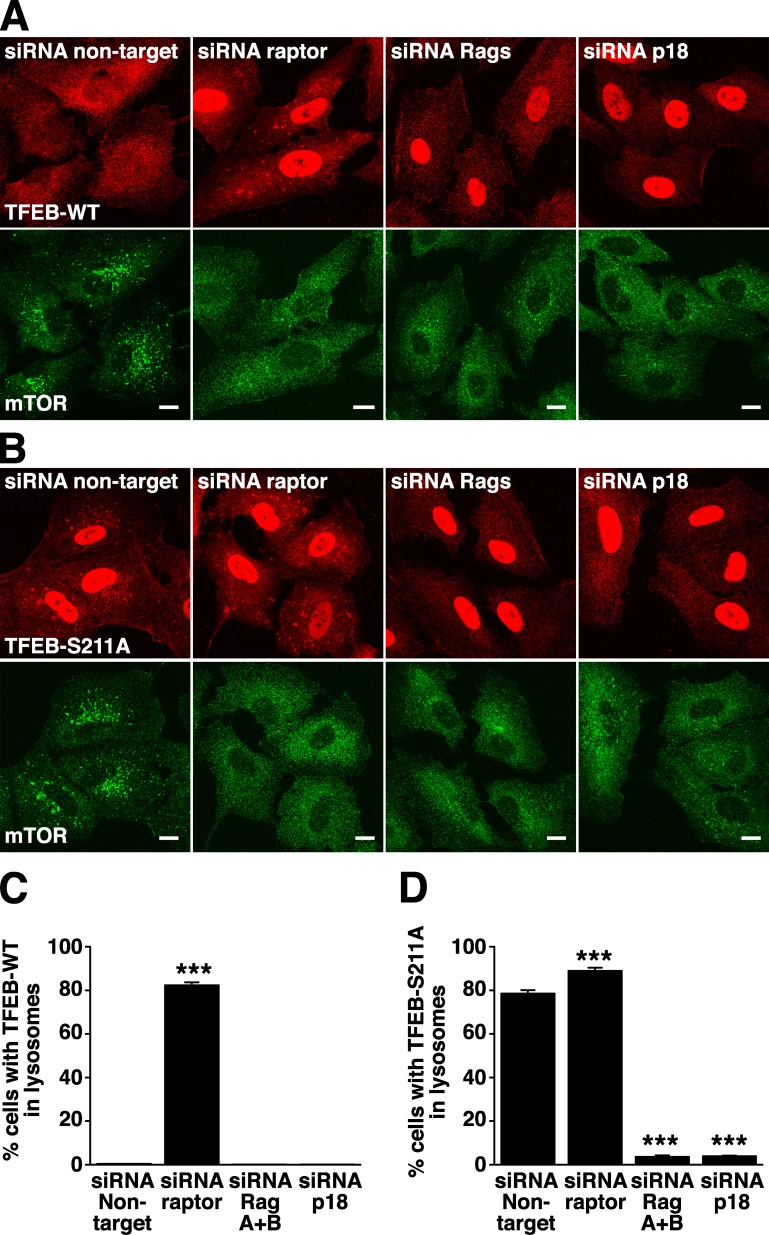
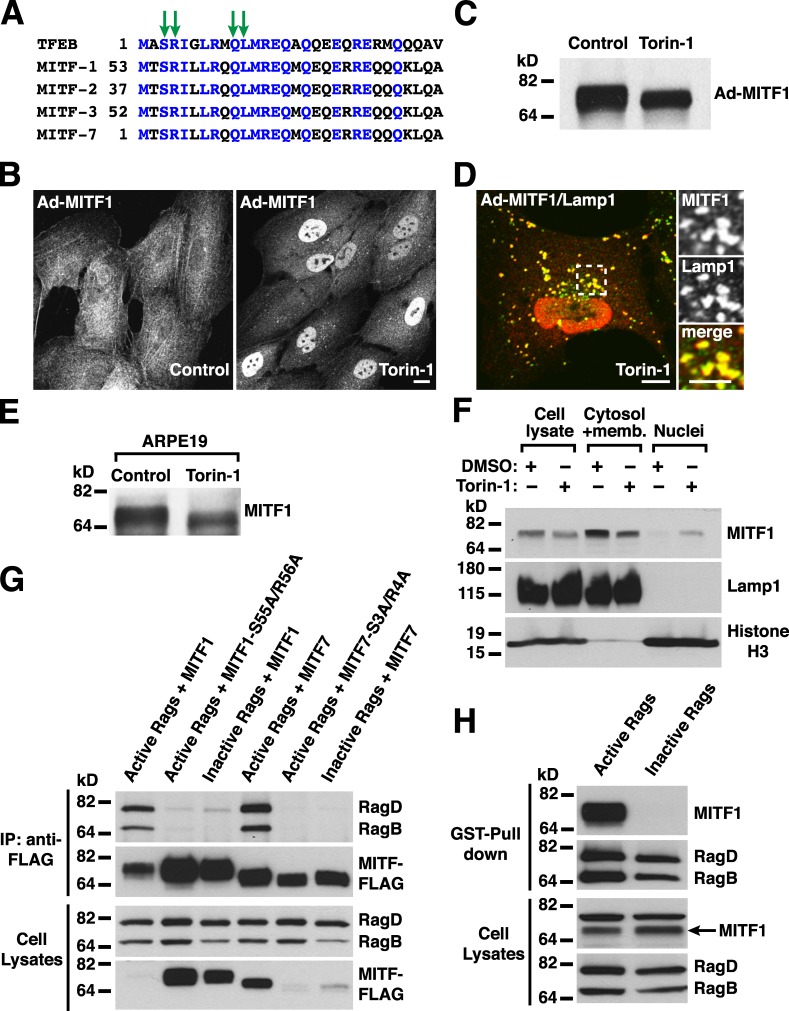
The goal of this study is to characterize the machinery that regulates association of TFEB with lysosomes, a critical step for the proper inhibition of this transcription factor. Based on observations that TFEB is a substrate for mTORC1 (Martina et al., 2012; Settembre et al., 2012) and the fact that other mTORC1 effectors are known to interact with components of the mTORC1 complex (Nojima et al., 2003), our initial hypothesis was that TFEB might directly bind to the mTORC1 complex. To assess this possibility, we treated ARPE-19 cells with specific siRNAs against raptor, one of the components of the mTORC1 complex that mediates recruitment of mTORC1 to lysosomes (a scheme showing changes on the distribution of TFEB as well as alterations on the Ragulator–Rags–mTORC1 complex upon treatment with different siRNAs is depicted in Fig. S1). As previously described, TFEB showed a diffuse cytosolic distribution in cells treated with control nontarget siRNA, whereas mTORC1 localized to lysosomes (Fig. 1 A). Depletion of raptor prevented association of mTORC1 with lysosomes, thus causing inactivation of the kinase (Fig. 1 A; Sancak et al., 2008). Inactivation of mTORC1 in raptor siRNA-treated cells inhibited association of TFEB with 14-3-3, thus leading to accumulation of TFEB in the nucleus (Fig. 1 A and Fig. S2 A; Martina et al., 2012). However, under these conditions, we still observed a significant redistribution of TFEB to lysosomes in >80% of the cells (82.5%, SD ±1.18, n = 697; Fig. 1, A and C). To corroborate that the TFEB-positive vesicles observed in the absence of raptor correspond to lysosomes, we performed double staining with the lysosomal marker Lamp1 (Fig. S2 B). In fact, close to 100% of all the TFEB-positive vesicles analyzed also contained Lamp1 (99.1%, SD ±0.98, n = 422). These results are in agreement with previous studies showing that dissociation of the TFEB–14-3-3 complex (as occurs upon treatment with the mTOR inhibitor Torin-1 or mutation of S211) results in a more stable association of TFEB with lysosomes (Martina et al., 2012; Roczniak-Ferguson et al., 2012). More importantly, the data indicate that neither raptor nor mTORC1 (that is not present in lysosomes under these conditions) are required for association of TFEB with lysosomal membranes.
Figure 1.
Rag GTPases are required for recruitment of TFEB to lysosomes. (A and B) ARPE-19 cells were transfected with siRNA duplexes to raptor, RagA+B, p18, or nontarget. 60 h after transfection, cells were infected with adenovirus expressing either TFEB-FLAG-WT (A) or TFEB-FLAG-S211A (B). 12 h later, cells were fixed, permeabilized with 0.2% Triton X-100, and double stained with antibodies against FLAG (used to detect TFEB) and mTOR. Bars, 10 µm. (C) Quantification of A. (D) Quantification of B. Values are means ± SD of three independent experiments. ***, P < 0.001.
Next, we asked whether Rag GTPases might play a role in the recruitment of TFEB to lysosomes. For this, we used combinations of siRNAs to suppress RagA and RagB or RagC and RagD at the same time. It has been reported that loss of RagA and RagB also led to the loss of RagC and RagD and vice versa, which suggests that, within cells, the Rag proteins are unstable when not in heterodimers (Fig. S2 C; Sancak et al., 2008). As expected, depletion of Rags A and B caused inactivation of mTORC1 (Fig. 1 A, bottom; and Fig. S2 C), inhibition of the interaction between TFEB and 14-3-3 (Fig. S1 C), and accumulation of TFEB in the nucleus (Fig. 1 A and Fig. S2 A). However, under these conditions, no association of TFEB with lysosomes was observed (Fig. 1, A and C). Similar results were obtained upon depletion of RagC and RagD (unpublished data). As expected, depletion of endogenous Rags did not disturb the association of the p18 with lysosomal membranes, thus indicating that the Ragulator complex alone is not capable of recruiting TFEB to lysosomes (Fig. S3 A). Therefore, our data show that Rag GTPases are required for association of TFEB with lysosomes.
Finally, RNAi-mediated suppression of p18, one of the components of the Ragulator that anchors Rags to lysosomal membranes, resulted in dissociation of Rags from lysosomes (Fig. S1 D and Fig. S3 B), inactivation of mTORC1 (Fig. 1 A, bottom; and Fig. S2 C), and accumulation of TFEB in the nucleus (Fig. 1 A and Fig. S2 A). However, the redistribution of TFEB to lysosomes was inhibited, thus confirming a role of Rags in this process (Fig. 1, A and C).
Similar results were observed for the TFEB-S211A mutant. As mentioned in the Introduction, mutation of S211 to Ala blocks the ability of TFEB to bind 14-3-3 and, consequently, inhibits 14-3-3–mediated sequestration of TFEB in the cytosol. At stationary state, TFEB-S211A accumulates in the nucleus but also displays stable association with lysosomes (Fig. 1, B and D; Martina et al., 2012). As expected, depletion of raptor did not affect the ability of TFEB-S211A to associate with lysosomal membranes (Fig. 1, B and D). In contrast, depletion of Rags or p18 strongly inhibited the recruitment of TFEB-S211A to lysosomes (Fig. 1, B and D). Quantification of three independent experiments indicated that ∼80% of the cells treated with control siRNA or siRNA against raptor showed evident association of TFEB-S211A with lysosomes (78.66%, SD ±1.44, n = 768 in siRNA nontarget cells; and 89.04%, SD ±1.35, n = 803 in siRNA raptor-treated cells), whereas <4% (3.7%, SD ±0.54, n = 711) of the Rags-depleted cells retained TFEB-S211A in lysosomes (Fig. 1 D). The cytosolic distribution of TFEB-S211A in cells depleted of Rags or p18 was not caused by alterations in the number or morphology of lysosomes as assessed by normal staining of Lamp1 in these cells (Fig. S3 C). Overall, our data reveal a novel role of Rags in the regulation of TFEB localization.
TFEB interacts with active Rags
Our siRNA experiments indicated that Rags are required for recruitment of TFEB to lysosomes. Therefore, we asked whether TFEB could directly interact with Rags. We used ARPE-19 cells transiently expressing wild-type (WT) TFEB and different combinations of Rag proteins. The Rag proteins function as heterodimers in which the active complex consists of GTP-bound RagA or B complexed with GDP-bound RagC or D. For this reason, we expressed TFEB-FLAG together with Rags mutants predicted to be restricted to the GTP- or GDP-bound conformations. Importantly, we found that TFEB strongly interacted with active heterodimers (RagBGTP/RagDGDP or RagAGTP/RagCGDP) but not with the inactive ones (RagBGDP/RagDGTP or RagAGDP/RagCGTP; Fig. 2 A). In reciprocal experiments, we performed coimmunoprecipitation with anti-HA antibodies in cells expressing Rags-HA and TFEB-GFP and confirmed that TFEB only binds to active Rag heterodimers (Fig. 2 B). In addition, we found that individual Rags (RagBGTP, RagAGTP, RagCGDP, or RagDGDP) failed to interact with TFEB (Fig. 2 C). These results indicate that both the nucleotide binding state and dimerization of Rags are important for TFEB binding. Finally, the stable association of TFEB-S211A with lysosomes implies that this mutant should be capable of binding active Rags. Interaction of TFEB-S211A with active (but not inactive) Rags was indeed detected by coimmunoprecipitation, and it is shown in Fig. 2 D.
Figure 2.
TFEB interacts with active Rag heterodimers. (A) ARPE-19 cells were nucleofected with the indicated Rag-expressing plasmids. 6 h after nucleofection, cells were infected with adenovirus expressing TFEB-FLAG-WT. 12 h later, cells were lysed and subjected to immunoprecipitation with the anti-FLAG antibody. The immunoprecipitates were analyzed by immunoblotting with antibodies against Rag proteins and FLAG (used to detect TFEB-WT). (B) ARPE-19 cells were nucleofected with the indicated Rag- and TFEB-GFP–expressing plasmids. 18 h later, cells were lysed and subjected to immunoprecipitation with the anti-HA antibody (used to immunoprecipitate Rag proteins). The immunoprecipitates were analyzed by immunoblotting with antibodies against GST and GFP (used to detect Rag proteins and TFEB-WT, respectively). (C) ARPE-19 coexpressing TFEB and the indicated Rag-expressing plasmids were lysed and subjected to immunoprecipitation with the anti-FLAG antibody. The immunoprecipitates were analyzed by immunoblotting with antibodies against GST and FLAG (used to detect Rag proteins and TFEB-WT, respectively). (D) Immunoblotting analysis of coimmunoprecipitated TFEB-FLAG-S211A with Rag heterodimers. IP, immunoprecipitation.
Recently, the crystal structure of the Gtr1p–Gtr2p complex, the Rag homologues from Saccharomyces cerevisiae, was solved, leading to the mapping of the interface between Rag GTPases and raptor (Gong et al., 2011). It was shown that depending on the GTP binding status, the Rag heterodimer interacts with raptor mainly via the surface close to switch I and switch II on RagA. To test whether TFEB interacts with Rags via the same surface, we introduced mutations in RagAGTP that abolish binding to raptor. As shown in Fig. S4 A, mutations of residues close to switch I (RagA-R34A/D35A/R37A/R38A) or switch II (RagA-N55A/V57A/N59A/W61A) eliminated the ability of RagAGTP to interact with TFEB. These results explain how nucleotide exchange, which induces alterations on the surface feature of switch I and II, regulate TFEB binding affinity.
Finally, we found that depletion of raptor did not affect the ability of TFEB to interact with endogenous Rags (Fig. S4 B). These results are consistent with our immunofluorescence experiments showing that depletion of raptor did not prevent association of TFEB with lysosomal membranes. All together, our data suggest that Rags bind TFEB and mediate its recruitment to lysosomes.
Endogenous TFEB interacts with active Rags and associates with lysosomes in an amino acid–dependent manner
Most of the published work on TFEB relies on overexpression of the recombinant protein. To corroborate our results, we took advantage of a recently developed commercially available antibody. We were able to detect a band of the correct molecular weight in several cell lines, including ARPE-19, HeLa, HEK-293T, and Raji (Fig. 3 A). More importantly, incubation with Torin-1 changed the electrophoretic mobility of endogenous TFEB, which now appeared as a fast-migrating form (Fig. 3 A). This molecular weight shift was also described for recombinant TFEB, and it is caused by dephosphorylation of TFEB upon mTORC1 inactivation (Settembre et al., 2011; Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2012). As expected, inhibition of mTORC1 resulted in accumulation of endogenous TFEB in the nucleus, as assessed by subcellular fractionation experiments (Fig. 3 B). Immunofluorescence experiments corroborated that, in fully fed cells, endogenous TFEB is excluded from the nucleus and shows a diffuse cytosolic distribution. In contrast, incubation of cells with Torin-1 for 1 h led to a dramatic redistribution of TFEB to lysosomes and the nucleus (Fig. 3 C; an additional example showing colocalization of TFEB-positive vesicles with Lamp1 in the presence of Torin-1 is shown in Fig. 4 C).
Figure 3.
Regulation of endogenous TFEB by mTOR and Rags. (A) The indicated cell lines were incubated in medium containing DMSO or 250 nM Torin-1 for 1 h. Cells were then lysed and subjected to immunoblotting with antibodies against TFEB (used to detect endogenous TFEB) and actin. (B) HeLa cells were treated as indicated in A, and nuclei and membrane plus cytosol fractions were obtained by low speed centrifugation. Proteins from different fractions were subjected to immunoblotting with antibodies against TFEB (used to detect endogenous TFEB), Lamp1, or Histone H3. (C) Immunofluorescence confocal microscopy showing nuclear and lysosomal localization of endogenous TFEB upon treatment of HeLa cells with Torin-1 as indicated in A. (D) HeLa cells were starved in serum- and amino acid–free medium (starvation) for 3 h and analyzed by immunoblotting with antibodies against TFEB. (E) HeLa cells were incubated in normal medium (control) and serum- and amino acid–free medium (Starvation) for 4 h or starved for 4 h followed by restimulation with amino acids (starvation + amino acids) for 30 min and analyzed by immunofluorescence with antibodies against TFEB (endogenous TFEB is shown in green) and Lamp1 (red). The region within the dotted box is magnified in the insets. (F) HeLa cells were nucleofected with the indicated Rag-expressing plasmids. 18 h later, cells were lysed, and RagB/D heterodimers were pulled down using glutathione–Sepharose beads. Proteins bound to the beads were analyzed by immunoblotting with antibodies against TFEB and GST (used to detect endogenous TFEB and Rag proteins, respectively). Bars, 10 µm.
Figure 4.
Inactivation of endogenous Rags by starvation prevents lysosomal localization of TFEB. (A) ARPE-19 cells were infected with adenovirus expressing TFEB-FLAG-WT. 16 h later, cells were incubated with 250 nM Torin-1 for 1 h or starved in serum- and amino acid–free medium for 3 h. Cells were then fixed, permeabilized with 0.2% saponin, and double stained with antibodies against TFEB (used to detect recombinant TFEB) and Lamp1. (B) ARPE-19 cells were infected with adenovirus expressing TFEB-FLAG-S211A. 12 h later, cells were starved in serum- and amino acid–free medium for 4 h (starvation) or kept in complete medium (control). Cells were then fixed, permeabilized with 0.2% Triton X-100, and stained with antibodies against FLAG (used to detect TFEB-S211A). (C) HeLa cells were incubated with 250 nM Torin-1 for 1 h or starved in serum- and amino acid–free medium for 4 h with the addition of Torin-1 during the last hour of starvation. Cells were then washed, fixed, permeabilized with 0.2% Triton X-100, and double stained with antibodies against TFEB (used to detect endogenous TFEB) and Lamp1. Regions within the dotted boxes are magnified in the insets. Bars: (A [main images], B, and C) 10 µm; (A, insets) 5 µm.
To address the regulation TFEB under more physiological conditions, the intracellular localization of endogenous TFEB was analyzed in cells starved or starved and briefly restimulated with amino acids. Endogenous TFEB accumulated into the nucleus when cells were starved in medium without amino acids (Fig. 3 E). Starvation also increased the electrophoretic mobility of endogenous TFEB (Fig. 3 D). In contrast, when cells were restimulated with amino acids for 30 min after starvation, TFEB shuttled back to the cytosol and showed increased association with lysosomes (Fig. 3 E). These results indicate that TFEB associates with lysosomes in an amino acid–dependent manner and corroborate the physiological relevance of the lysosomal localization of TFEB.
Finally, we assessed the ability of endogenous TFEB to interact with Rags. Similar to our experiments with recombinant protein, endogenous TFEB was pulled down by active Rag heterodimers, whereas no interaction was observed with inactive Rags (Fig. 3 F). Therefore, we confirmed that mTORC1 and Rags play an important role in the regulation of the activity and distribution of endogenous TFEB.
The activation state of the Rags determine association of TFEB with lysosomes
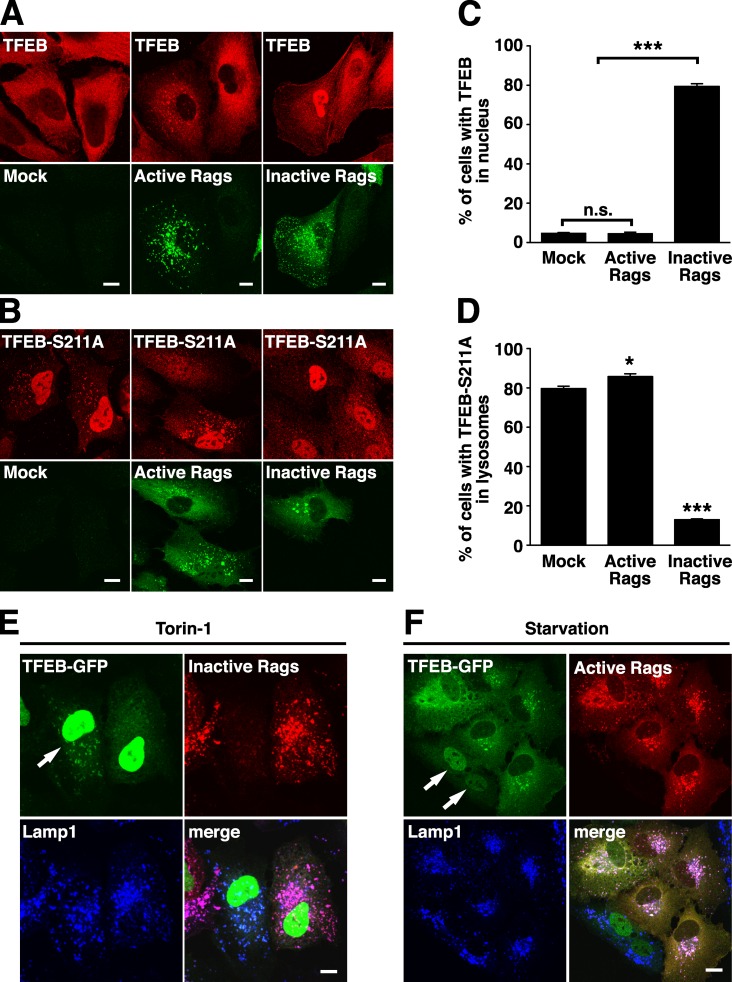
We next tested whether the activation state of the Rag complex might regulate the distribution of TFEB. As previously mentioned, incubation with Torin-1 (that inactivates mTOR but keeps Rags active) caused a dramatic change in the localization of TFEB, which shuttled from the cytosol to the nucleus and lysosomes (Fig. 4 A). In contrast, when cells were starved for 2 h in a medium with no amino acids, a treatment that causes inhibition of both mTORC1 and Rags, TFEB still accumulated in the nucleus, but no association with lysosomes was observed (Fig. 4 A). This suggests that active Rags are required for the redistribution of TFEB to lysosomes upon dissociation of the TFEB–14-3-3 complex. Moreover, the interaction between TFEB and Rags is dependent on the level of amino acids in the cell. Inactivation of endogenous Rags by starvation also caused dissociation of TFEB-S211A from lysosomal membranes (Fig. 4 B) and prevented the recruitment of endogenous TFEB to lysosomes induced by treatment with Torin-1 (Fig. 4 C).
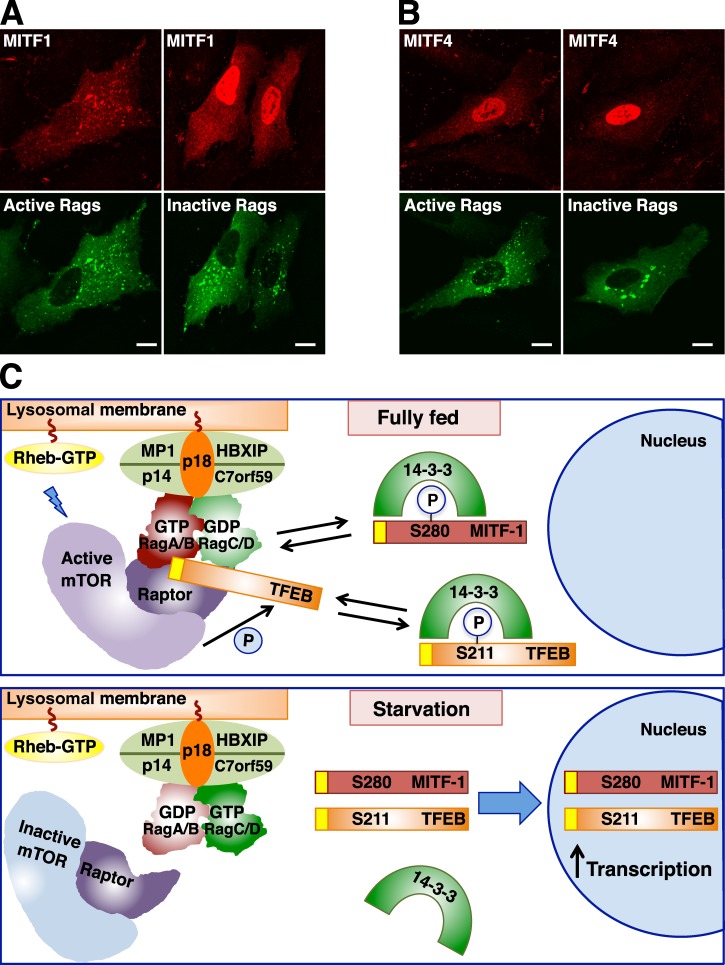
Expression of RagBGTP/RagDGDP active heterodimers was sufficient to induce redistribution of TFEB from the cytosol to lysosomes (Fig. 5 A). Triple staining confirmed colocalization of TFEB and active Rag heterodimers to Lamp1-positive vesicles (Fig. 5 F). Previous studies have shown that expression of inactive Rags is enough to cause inactivation of mTORC1 (Sancak et al., 2008). Consistent with these results, we found significant accumulation of TFEB in the nuclei of cells expressing RagBGDP/RagDGTP (Fig. 5, A and B). However, under these conditions, no association of TFEB with vesicular structures was observed, thus indicating that inactive Rags completely prevented recruitment of the transcription factor to lysosomes (Fig. 5 A). Similar results were observed with the TFEB-S211A mutant. Constitutive association of TFEB-S211A with lysosomes was further enhanced by active Rags but entirely abrogated by expression of inactive Rags (Fig. 5, B and D). Moreover, expression of inactive Rags abolished the recruitment of TFEB to lysosomes induced by Torin-1 (Fig. 5 E), whereas expression of active Rags prevented translocation of TFEB to the nucleus upon starvation (Fig. 5 F).
Figure 5.
The activation state of Rags determines localization and activation of TFEB. (A and B) ARPE-19 cells were transfected with either active or inactive RagB/D heterodimers and infected with adenovirus expressing TFEB-FLAG-WT (A) or TFEB-FLAG-S211A (B). After 12 h, cells were double stained with antibodies against FLAG and GST (used to detect TFEB or Rag proteins, respectively). (C) Quantification of TFEB-WT nuclear localization from A. (D) Quantification of TFEB-S211A lysosomal localization from B. (E and F) ARPE-19 cells were cotransfected with plasmids expressing TFEB-GFP together with inactive (E) or active Rag heterodimers (F). Cells were then incubated with 250 nM Torin-1 for 1 h (E) or starved in serum- and amino acid–free medium for 3 h (F). Anti-GST antibodies were used to detect Rag proteins. Arrows point to cells that do not express Rag heterodimers. Values are means ± SD of three independent experiments. ***, P < 0.001; *, P < 0.05. Bars, 10 µm.
Therefore, consistent with the protein interaction results, these experiments indicate that the activation state of the Rags is sufficient to determine TFEB localization and that active Rags are both sufficient and necessary to promote recruitment of TFEB to lysosomes.
The first 30 amino acids of TFEB are required for binding to Rags and lysosomal distribution
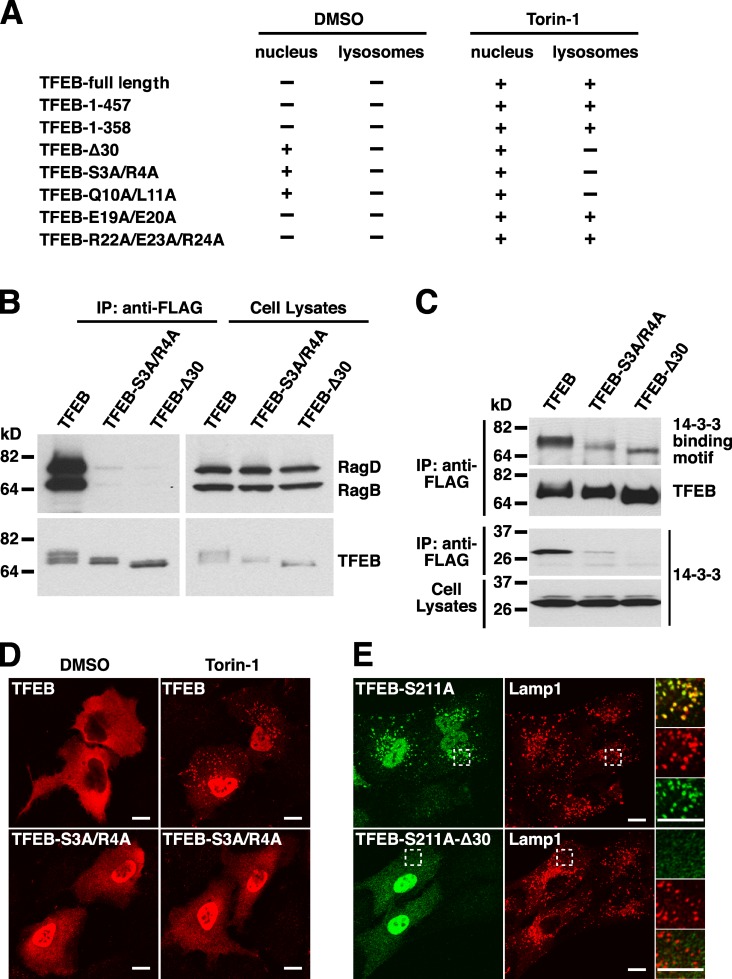
Next, we sought to identify the region of TFEB necessary for association with lysosomes and interaction with Rags. For this, we expressed several FLAG-tagged fragments of TFEB in ARPE-19 cells and assessed their ability to associate with lysosomes in the presence or absence of Torin-1. As previously shown, inhibition of mTORC1 by Torin-1 caused a dramatic redistribution of TFEB to the nucleus and lysosomes (Fig. 4 A; Martina et al., 2012; Settembre et al., 2012). Depletion of various C-terminal fragments did not affect the Torin-1–mediated redistribution of TFEB. In contrast, deletion of just the first 30 amino acids (TFEB-Δ30) completely abolished the ability of TFEB to relocate to lysosomal membranes upon mTORC1 inactivation (Fig. 6 A). Furthermore, point mutation of specific residues within the 30 first residues (TFEB-S3A/R4A and TFEB-Q10A/L11A) was sufficient to prevent association of TFEB with lysosomes (Fig. 6, A and D; and Fig. S5 A). In contrast, mutation of other residues in the same region (TFEB-E19A/E20A and TFEB-R22A/E23A/R24A) had no effect (Fig. S5 A).
Figure 6.
The N-terminal region of TFEB is necessary for interaction with Rag heterodimers and lysosomal localization. (A) Summary of the nuclear and lysosomal distribution of several TFEB amino acid and deletion mutants in ARPE-19 cells treated with either DMSO or Torin-1. (B) ARPE-19 cells were nucleofected with the indicated Rag- and TFEB-expressing plasmids. After 12 h, cell lysates were immunoprecipitated with the anti-FLAG antibody and analyzed by immunoblotting with antibodies against FLAG and GST (used to detect TFEB and Rag proteins, respectively). (C) FLAG-tagged TFEB-WT or TFEB deletion mutants were immunoprecipitated with the anti-FLAG antibody and analyzed by immunoblotting with antibodies against FLAG, 14-3-3 binding motif, or 14-3-3. (D) Immunofluorescence confocal microscopy showing the subcellular distribution of TFEB-WT and TFEB-S3A/R4A mutant upon incubation with DMSO (vehicle) or 250 nM Torin-1 for 1 h. Cells were fixed, permeabilized with 0.2% Triton X-100, and stained with antibodies against FLAG (used to detect TFEB). (E) ARPE-19 cells expressing either TFEB-S211A or TFEB-S211A-Δ30 were double stained with antibodies against TFEB and Lamp1. Regions within the dotted boxes are magnified in the insets. IP, immunoprecipitation. Bars, 10 µm.
To corroborate that binding of TFEB to Rags is necessary for association of TFEB with lysosomes, we assessed the ability to interact with Rags of those mutants that did not relocate to lysosomes in the presence of Torin-1. As predicted, the TFEB-Δ30 and TFEB-S3A/R4A mutants were unable to bind active Rag heterodimers (RagBGTP/RagDGDP) in coimmunoprecipitation experiments, whereas TFEB WT showed a strong interaction (Fig. 6 B). These results confirm that the N-terminal portion of TFEB is required for interaction with Rags and recruitment to lysosomes.
Our model predicts that the association of TFEB with lysosomes is necessary for mTORC1-dependent phosphorylation of TFEB. In agreement with this idea, we found that the levels of phosphorylation at S211 were greatly reduced in TFEB-Δ30 and TFEB-S3A/R4A, and consequently, these mutants were unable to interact with 14-3-3 and accumulated in the nucleus at all times (Fig. 6, C and D). However, because of its incapability to bind Rags, the TFEB-S3A/R4A mutant was unable to associate with lysosomal membranes upon Torin-1 treatment (Fig. 6 D). Mutation of Q10 and L11 to alanines also resulted in constitutive accumulation of TFEB in the nucleus, whereas the TFEB-E19A/E20A and TFEB-R22A/E23A/R24A mutants behaved like TFEB-WT and remained retained in the cytosol (Fig. 6 A and Fig. S5 B). Furthermore, deletion of the first 30 residues also inhibited constitutive association of TFEB-S211A with lysosomes (Fig. 6 E). Finally, expression of active Rags failed to induce redistribution of TFEB-Δ30 and TFEB-S3A/R4A to lysosomes (Fig. S4 C). All together, our data provide, for the first time, causal evidence that TFEB–Rag interaction is required for mTORC1-dependent phosphorylation and thus cytoplasmic sequestration and inhibition of TFEB.
Because TFEB-Δ30 and TFEB-S3A/R4A accumulate in the nucleus, it is possible that the lack of interaction with active Rags is a consequence of the reduced amount of these mutants in the cytosol. To circumvent this, we searched for residues that might potentially function as a nuclear import signal for TFEB and found a stretch of arginines between positions 245 and 248 that are highly conserved among the different member of the TFEB family. In agreement with a recent study (Roczniak-Ferguson et al., 2012), we found that mutation of R245, R246, and R247 to alanines (TFEB-R245-247A) caused retention of TFEB in the cytosol upon mTORC1 inactivation, thus confirming that these amino acids are required for transport of TFEB to the nucleus (Fig. S4 D). Importantly, the TFEB-R245-247A mutant was capable of interaction with active RagBGTP/RagDGDP heterodimers and associated with lysosomal membranes in Torin-1–treated cells (Fig. S4, D and E). However, additional substitution of S3 and R4 by alanines (TFEB-S3A/R4A/R245-247A) resulted in a mutant that is highly expressed in the cytosol but lacks the ability to bind active Rags (Fig. S4 E). These results further confirm that the N-terminal portion of TFEB mediates interaction with Rags.
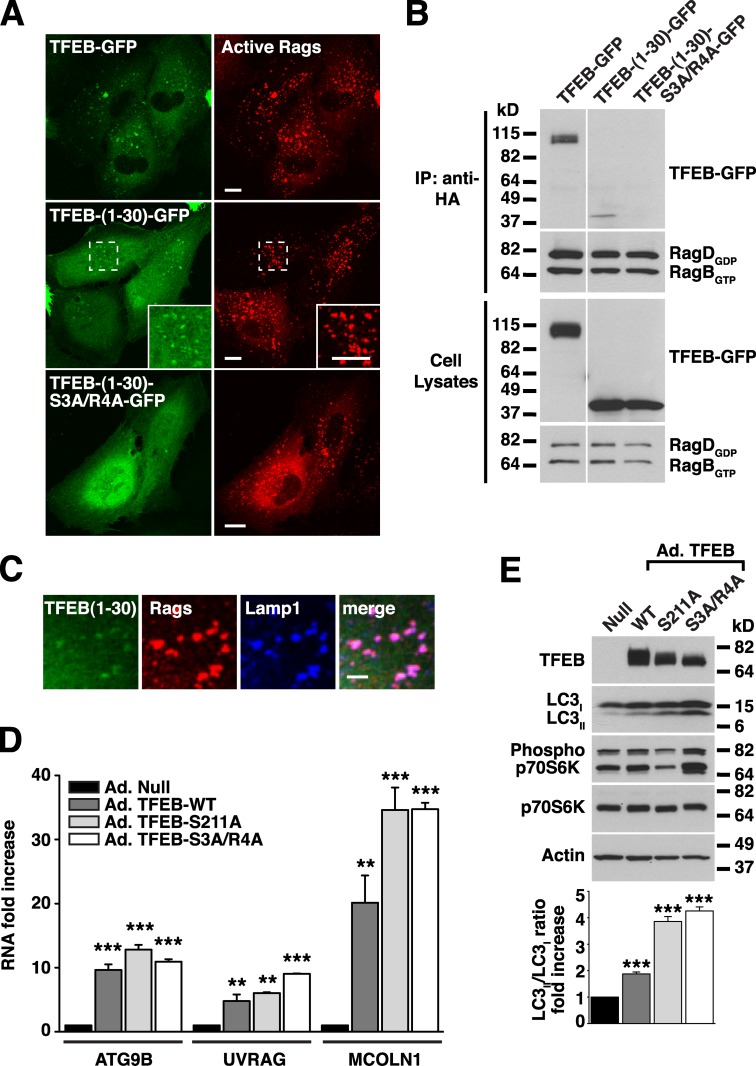
Next, we asked whether the first 30 residues of TFEB are sufficient to promote redistribution of a reporter protein to lysosomes. Our initial attempt to fuse the first 30 amino acids of TFEB to GFP was unsuccessful, and the protein remained cytosolic. However, addition of a leucine zipper to allow dimerization resulted in a chimera (TFEB-(1–30)-GFP) that was efficiently recruited to lysosomes upon expression of active Rags (Fig. 7, A and C). As expected, mutation of S3 and R4 (TFEB-(1–30)-S3A/R4A-GFP) prevented redistribution of the chimera to lysosomal membranes even in the presence of active Rags (Fig. 7 A). Moreover, TFEB-(1–30)-GFP, but not TFEB-(1–30)-S3A/R4A-GFP, interacted with active RagBGTP/RagDGDP heterodimers in coimmunoprecipitation experiments (Fig. 7 B). All together, our results indicate that the first 30 amino acids of TFEB are both necessary and sufficient for interaction with active Rags, recruitment to lysosomes, and mTORC1-dependent phosphorylation of TFEB.
Figure 7.
The first 30 amino acids of TFEB are sufficient for binding to active Rag heterodimers. (A) ARPE-19 cells coexpressing active RagB/D heterodimer and the indicated TFEB plasmids were fixed, permeabilized with 0.2% saponin, and stained with antibodies against GST (used to detect Rag proteins). Regions within the dotted boxes are magnified in the insets. Bars, 10 µm. (B) ARPE-19 cells were cotransfected with active RagB/D heterodimers and the indicated TFEB constructs. After 18 h, cell lysates were immunoprecipitated with the anti-HA antibody (used to immunoprecipitate Rag proteins) and immunoblotted with antibodies against GFP and GST (used to detect TFEB-GFP and Rag proteins, respectively). The white line indicates that intervening lanes have been spliced out. (C) Immunofluorescence confocal microscopy showing the subcellular distribution of TFEB-(1–30)-GFP in ARPE-19 cells coexpressing active RagB/D heterodimers (antibodies against GST were used to detect Rags). Bar, 4 µm. (D) Relative RT-PCR analysis of the mRNA expression of autophagy (ATG9B and UVRAG) and lysosomal (MCOLN1) genes in ARPE-19 cells infected with the indicated adenovirus for 48 h. The values are expressed as a ratio to RNA from cells infected with control adenovirus (Ad.-Null). Values are means ± SD of two independent experiments. ***, P < 0.001; **, P < 0.01. (E) Cells were infected with adenovirus as in D and analyzed by immunoblotting with the indicated antibodies. IP, immunoprecipitation.
Finally, we analyzed the transcriptional activity of the TFEB-S3A/R4A mutant. For this, we infected ARPE-19 cells with adenovirus expressing TFEB-WT, TFEB-S211A, or TFEB-S3A/R4A. 48 h after infections, we used quantitative RT-PCR to assess the expression of several known TFEB targets, including autophagy (ATG9B and UVRAG) and lysosomal (MCOLN1) genes. As seen in Fig. 7 D, TFEB-S3A/R4A induced transcription of ATG9B, UVRAG, and MCOLN1 to similar levels as TFEB-WT or TFEB-S211A. Moreover, overexpression of TFEB-S3A/R4A induced formation of autophagosomes (as assessed by an increased LC3II/LC3I ratio) even when cells were kept in nutrient-rich conditions and mTOR remained active (Fig. 7 E). These results indicate that the TFEB-S3A/R4A is active, and therefore, the lack of interaction with the Rag complex is unlikely to be caused by misfolding of the protein. More importantly, although recruitment to lysosomes is indispensable for the retention of TFEB in the cytosol, it does not seem to play a major role in the regulation of its transcriptional activity.
The Rag complex regulates localization and activation of microphthalmia-associated transcription factor (MITF)
TFEB belongs to a family of basic helix–loop–helix leucine zipper transcription factors that also includes MITF, TFE3, and TFEC. In addition, expression from alternative promoters results in several MITF isoforms differing in their N termini and regulated in a tissue-dependent manner. For example, MITF isoform 4 (also known as MITF-M) is expressed exclusively in melanocytes and melanoma cells, whereas isoforms 1 (MITF-A) and 2 (MITF-H) are expressed in multiple tissues, including RPE cells, cervical cancer, osteoclasts, and mast cells (Yasumoto et al., 1998).
Sequence alignment revealed a high degree of homology between the N-terminal portion of TFEB and some of the MITF isoforms, including human isoforms 1, 2, 3, and 7 (Fig. 8 A). In contrast, no homology was observed between the N terminus of TFEB and MITF isoforms 4, 5, and 6 or TFEC. Importantly, all the residues that were identified as required for the interaction between TFEB and Rag GTPases (S3R4 and Q10L11) were present in the MITF isoforms 1, 2, 3, and 7, thus suggesting that MITF and TFEB might be regulated in similar ways. To test this possibility, we generated an adenovirus expressing MITF-1. Similar to TFEB, MITF-1 was excluded for the nucleus and showed a diffuse cytosolic distribution in fully fed cells, whereas it rapidly accumulated into the nucleus after addition of Torin-1 (Fig. 8 B). Quantification of three independent experiments revealed that, in control conditions, <10% of the cells showed staining of MITF-1 into the nucleus (9.53%, SD ±0.97, n = 448). In contrast, inactivation of mTORC1 induced translocation of MITF-1 to the nucleus in >80% of the cells (80.66%, SD ±3.63, n = 422). Incubation with Torin-1 also caused redistribution of MITF-1 to lysosomes, as confirmed by the presence of Lamp1 (Fig. 8, B and D) and changed its electrophoretic mobility (Fig. 8 C). Quantification of over one thousand vesicles positive for MITF-1 revealed that 99.73% were also positive for Lamp1 (SD ±0.38). Furthermore, endogenous MITF-1 also showed a shift of its molecular weight when cells were treated with Torin-1 (Fig. 8 E), and accumulation of the lower molecular band in the nucleus was observed by subcellular fractionation (Fig. 8 F).
Figure 8.
Rag GTPases and mTORC1 regulate the function of MITF. (A) Multisequence alignment of the first 30 amino acids of TFEB with MITF isoforms 1, 2, 3, and 7. The green arrows indicate the residues identified as essential for the interaction between TFEB and Rag proteins. Blue letters are to indicate conserved amino acid homology between the different proteins. (B–D) ARPE-19 cells were infected with adenovirus expressing MITF1-FLAG. 16 h later, cells were incubated with DMSO or 250 nM Torin-1 for 1 h and analyzed by immunoblotting (C) or immunofluorescence (B and D) with anti-FLAG antibodies. The region within the dotted box is magnified in the insets. Bars: (B and D, main images) 10 µm; (D, insets) 5 µm. (E) ARPE-19 cells were incubated with 250 nM Torin-1 for 1 h. Cells were then lysed and analyzed by immunoblotting with antibodies against MITF. (F) HEK-293T cells were incubated in medium containing DMSO or Torin-1 for 1 h. Cells were lysed, and nuclei and membrane plus cytosol fractions were obtained by low speed centrifugation. Proteins from the different fractions were subjected to immunoblotting with antibodies against MITF, Lamp1, and Histone H3. (G) ARPE-19 cells expressing active (RagBGTP/RagDGDP) or inactive (RagBGDP/RagDGTP) Rag heterodimers were immunoprecipitated with the anti-FLAG antibody and immunoblotted with antibodies against FLAG and GST (used to detect MITF and Rag proteins, respectively). (H) HEK-293T cells expressing active (RagBGTP/RagDGDP) or inactive (RagBGDP/RagDGTP) Rag heterodimers were pulled down using glutathione–Sepharose beads and immunoblotted with antibodies against GST and MITF (used to detect Rag proteins and endogenous MITF, respectively). IP, immunoprecipitation.
Coexpression of GTP- or GDP-bound Rag mutants with various MITF isoforms and subsequent coimmunoprecipitation revealed that, as predicted, MITF-1 and MITF-7 interact with Rags. Similar to TFEB, the binding was only observed upon expression of active Rag heterodimers (RagBGTP/RagDGDP) but not inactive ones (RagBGDP/RagDGTP; Fig. 8 G). As expected, mutation of some of the highly conserved residues to alanines (MITF1-S55A/R56A and MITF7-S3A/R4A) entirely abolished the ability of MITF-1 and MITF-7 to bind Rags (Fig. 8 G). In addition, we detected interaction between endogenous MITF-1 and active Rag heterodimers in GST pull-down experiments (Fig. 8 H). Therefore, our data reveal interesting similarities between the regulation of TFEB and MITF.
Finally, overexpression of active Rags was sufficient to induce relocation of MITF-1 to lysosomal membranes, whereas expression of inactive Rags caused accumulation of MITF-1 into the nucleus (Fig. 9 A). In contrast, MITF-4, which has a different N terminus with no homology to TFEB, accumulated in the nucleus at all times, and its distribution was not affected by expression of active or inactive Rags (Fig. 9 B). All together, these results indicate that Rags and mTORC1 determine the activity and localization of TFEB as well as several MITF isoforms, thus suggesting a more general role for Rags in the regulation of gene expression.
Figure 9.
Rag GTPases regulate recruitment of MITF and TFEB to lysosomes. (A and B) ARPE-19 cells were nucleofected with the indicated Rag- and MITF-expressing plasmids. 12 h later, cells were fixed, permeabilized with 0.2% Triton X-100, and double stained with antibodies against FLAG and GST (used to detect MITF and Rag proteins, respectively). Bars, 10 µm. (C) Model representing the mechanism of TFEB and MITF regulation by Rag GTPases. (top) In nutrient-rich conditions, active Rags promote recruitment of mTORC1 and TFEB to lysosomes, thus facilitating mTORC1-dependent phosphorylation of TFEB. Phosphorylation of TFEB at S211 creates a binding site for 14-3-3 and results in sequestration of TFEB in the cytosol. We suggest that 14-3-3 may mask the Rag-binding domain in TFEB (represented in yellow). (bottom) In the absence of amino acids, Rag GTPases and mTORC1 are inactivated. Dissociation of the TFEB–14-3-3 complex allows transport of TFEB to the nucleus and TFEB-mediated activation of a transcriptional network that promotes autophagy, lysosomal biogenesis, and increased lysosomal degradation. Our model proposes that some MITF isoforms might be regulated in a similar manner. The interaction of MITF-1 with 14-3-3 has been previously described (Bronisz et al., 2006). Please note that mTORC1-dependent phosphorylation of MITF-1 S280 has not been reported and is merely speculative. The representation of the Ragulator–Rag–mTORC1 complex is based on the recent crystal structured described by Gong et al. (2011). P, phosphorylation.
Discussion
One of the most fundamental issues in cell biology is how cells coordinate availability of nutrients and cell growth. Our study provides new insight into the mechanism by which Rag GTPases, mTORC1, and TFEB work together to integrate growth-stimulating and -inhibitory signals and ultimately regulate translation, autophagy, and cell growth. Previously, we showed that mTORC1 mediates phosphorylation-dependent association of TFEB with 14-3-3 proteins that function as cytoplasmic repressors of TFEB. mTORC1 inactivation induces dissociation of the TFEB–14-3-3 complex, thus allowing transport of TFEB to the nucleus (Martina et al., 2012; Roczniak-Ferguson et al., 2012). Here, we characterize the upstream events that regulate recruitment of TFEB to lysosomes, the organelle where active mTORC1 resides.
We found that Rags recruit TFEB to lysosomes in an amino acid–dependent manner and that this redistribution is required for mTORC1-mediated phosphorylation of TFEB. Depletion of endogenous Rags, inactivation of Rags by starvation, or expression of inactive Rags prevented association of TFEB and TFEB-S211A with lysosomal membranes, whereas transfection of cells with dominant-active Rags was sufficient to induce redistribution of TFEB to lysosomes. The Rag complex also regulates association of specific MITF isoforms with lysosomes, thus opening the possibility that amino acids play a role in the expression of MITF target genes.
Our work shows for the first time that TFEB and MITF interact with Rags and that the binding is dependent on the nucleotide binding state of Rags. The interaction of TFEB with Rags shares numerous similarities with the binding between Rags and raptor. Most importantly, both proteins strongly favor the interaction with active heterodimers and bind to the same surface interface containing the switch regions of the G domain of RagA (Gong et al., 2011). Therefore, we suggest that raptor, TFEB, and probably some MITF isoforms function as bona fide effectors of Rag GTPases.
Our study is also the first to identify the region of TFEB and MITF responsible for the binding to Rags. We found that the first 30 residues of TFEB are both necessary and sufficient for the interaction with the Rag complex. The increased ability of the TFEB-S211A mutant to associate with lysosomes when compared with TFEB-WT suggests that the binding of 14-3-3 to TFEB may mask (directly or by causing conformational changes) the N-terminal portion of TFEB, thus favoring retention of the transcription factor in the cytosol (Fig. 9 C). Alternatively, binding to 14-3-3 could inhibit dimerization of TFEB. Dissociation of the TFEB–14-3-3 complex would induce homodimerization of TFEB, thus favoring interaction with Rags. Consistent with this hypothesis, we found that a chimera containing the first 30 amino acids of TFEB was efficiently recruited to lysosomes only when fused to a leucine zipper that promoted its dimerization. In this study, we also identified several arginine residues (R245, R246, and R247) that are indispensable for the transport of TFEB to the nucleus. The proximity of these residues to the 14-3-3 binding site (from L204 to Q218) suggests that 14-3-3 may also mask the TFEB nuclear localization signal.
Overall, we proposed the following model of activation for TFEB depicted in Fig. 9 C. In the presence of amino acids, active Rags promote recruitment of both mTORC1 and TFEB to the lysosomal surface, where mTORC1 is activated and phosphorylates TFEB in several residues, including S211. Phosphorylation of S211 induces interaction of TFEB with 14-3-3 and consequent sequestration of the transcription factor in the cytosol. Upon amino acid deprivation, Rags are inactivated, and TFEB can no longer redistribute to lysosomes or be phosphorylated by mTORC1. As a result, dissociation of the TFEB–14-3-3 complex leads to delivery of TFEB to the nucleus and up-regulation of genes that mediate induction of autophagy and lysosomal biogenesis. Therefore, TFEB follows two different circuits depending on the nutrient status of the cell: it continuously shuttles between lysosomes and cytosol in amino acid–rich conditions, whereas it is transported from the cytosol to the nucleus under starvation. We propose that MITF-1 may share a similar regulatory mechanism. In fact, it has been described that in macrophage/osteoclast precursors, 14-3-3 regulates MITF activity by inducing sequestration of MITF in the cytosol in the absence of signals required for osteoclast differentiation (Bronisz et al., 2006). The interaction between 14-3-3 and MITF is dependent on phosphorylation of S280, a residue that corresponds to TFEB S211 by homology analysis (Bronisz et al., 2006; Martina et al., 2012).
A recent study by Roczniak-Ferguson et al. (2012) suggested that TFEB-GFP and MITF-GFP localize both to the cytoplasm and lysosomes in normal growth conditions. It is important to point out that this study does not disagree with our results. Our present work, as well as previously published studies, suggests that TFEB continuously cycles between cytosol and lysosomes in fully fed cells and that, at any given time, a fraction of TFEB is bound to lysosomal membranes (Martina et al., 2012; Settembre et al., 2012). Therefore, the amount of lysosomal TFEB may vary depending on the level of expression of recombinant TFEB or the method of fixation. However, our analysis of cells expressing low levels of TFEB-FLAG, a stable cell line expressing TFEB-GFP, and more importantly, the examination of the distribution of endogenous TFEB suggest that, in normal growth conditions, the association of TFEB with lysosomes is very transient and the vast majority of the protein resides in the cytosol. However, association of endogenous TFEB with lysosomes was observed after amino acid stimulation, further confirming the physiological relevance of the lysosomal localization of TFEB. Finally, our work also agrees with recently published work showing that expression of inactive Rags or depletion of RagC causes translocation of TFEB to the nucleus (Roczniak-Ferguson et al., 2012; Settembre et al., 2012).
This study also reveals a new and novel role of mTORC1 in the regulation of MITF-1. Two previous studies found that inhibition of mTORC1 leads to melanosome formation and expression of melanogenic genes in MNT-1 melanoma cells, probably through up-regulation of the levels of MITF-4 (Ho et al., 2011; Hah et al., 2012). However, our study is the first one to show that mTORC1 promotes sequestration of this transcription factor in the cytosol. Further studies should address the implications of the regulatory role of amino acids, Rags, and mTORC1 in the expression of genes mediated by specific MITF isoforms. Overall, our work underscores a novel role for lysosomes as signaling centers that synchronize environmental cues, such as amino acid availability, with gene expression, energy production, and cellular homeostasis.
Materials and methods
Cell lines and drug treatment
ARPE-19 cells (American Type Culture Collection) were grown at 37°C in a 1:1 mixture of DMEM and Ham’s F12 media supplemented with 10% fetal bovine serum (Invitrogen), 2 mM GlutaMAX, 100 U/ml penicillin, and 100 µg/ml streptomycin (Gibco) in a humidified 5% CO2 atmosphere. HeLa and HEK-293T cells (American Type Culture Collection) were grown in DMEM media supplemented as indicated for ARPE-19 cell media. Raji cells (a gift from J. Hammer, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD) were grown in Iscove’s modified Dulbecco’s media supplemented as indicated for ARPE-19 cell media. Cells were nucleofected using Cell Line Nucleofector Kit V (Lonza) following the manufacturer’s recommendations. Nucleofected cells were analyzed 12–24 h after nucleofection. For infection experiments, cells were infected with adenoviruses according to the manufacturer’s recommendations. Analyses were performed 16–48 h after infection. Cells were incubated for 1 h at 37°C in medium containing one of the following reagents: DMSO (Thermo Fisher Scientific) or 250 nM Torin-1 (Tocris Bioscience). For starvation experiments, cells were washed three times in Hank’s balanced salt solution (Invitrogen) and incubated for 2–4 h at 37°C in either a 1:1 mixture of DMEM and Ham’s F12 media without amino acids (US Biological) supplemented with 100 U/ml penicillin and 100 µg/ml streptomycin (Gibco) or Earle’s balanced salt solution (Sigma-Aldrich). Recovery after starvation was achieved by the addition of DMEM media supplemented with extra amino acids (MEM amino acids ×50; Invitrogen) and 1% FBS.
Antibodies
The following mouse monoclonal antibodies were used: clone Ab5 to actin (1:10,000; BD), clone 4C5 to FLAG (1:500; Origen), clone H4A3 to LAMP1 (1:3,000; Developmental Studies Hybridoma Bank), clones M2 and M5 (1:2,000) to FLAG (Sigma-Aldrich), clone 16B12 to HA (1:1,000; Covance), and clone C5 to MITF (1:200; EMD Millipore). The following polyclonal antibodies were also used: anti-mTOR (1:200), antiraptor (1:500), anti–14-3-3 (1:1,000), anti–phospho-(Ser) 14-3-3 binding motif, anti-RagC (1:1,000), anti-RagA (1:1,000), anti-p70S6K (1:2,000), anti–phospho-p70S6K (1:1,000), and anti-TFEB (1:1,000) obtained from Cell Signaling Technology, anti-p18 (1:1,000) and anti-LC3 (1:1,000) obtained from Sigma-Aldrich, and anti-LAMP1 (1:800; Abcam), anti-FLAG (1:10,000; Covance), and anti-GFP (1:1,000; MBL International). Alexa Fluor 568–conjugated goat anti–mouse IgG, Alexa Fluor 647–conjugated goat anti–mouse, and Alexa Fluor 488–conjugated goat anti–rabbit IgG were used at a dilution of 1:1,000 (Invitrogen). HRP-conjugated anti–mouse or anti–rabbit IgG were acquired from Cell Signaling Technology and used at a dilution of 1:8,000.
Adenovirus
Adenovirus expressing TFEB-WT, TFEB-S211A, TFEB-S3A/R4A, and MITF1-FLAG were prepared, amplified, and purified by Welgen, Inc.
Plasmids
TFEB-FLAG expression vector was generated by cloning the full-length encoding sequence of human TFEB into p3×FLAG-cytomegalovirus (CMV) with a triple FLAG tag fused to the C termini of TFEB (Sardiello et al., 2009). The p3×FLAG-CMV-TFEB construct was obtained from A. Ballabio (Baylor College of Medicine, Houston, TX; and Telethon Institute of Genetics and Medicine, Napoli, Italy). The cDNA encoding the 30 first amino acids of human TFEB and the leucine zipper motif of S. cerevisiae GCN4p were obtained by PCR amplification followed by in-frame cloning in tandem into XhoI–EcoRI and EcoRI–SalI sites, respectively, of the pmEGFP-N1 vector (Takara Bio Inc.). The full-length cDNA of human TFEB was obtained by PCR amplification followed by in-frame cloning into EcoRI–SalI sites of pmEGFP-N1 vector. The full-length cDNAs of human MITF isoforms 1 and 7 were obtained by RT-PCR amplification of total RNA from ARPE-19 cells followed by in-frame cloning into EcoRI–SalI sites of the pCMV-Tag4A vector (Agilent Technologies). Amino acid substitutions in TFEB, MITF1, MITF7, and RagA and the 30–amino acid deletion in TFEB were made using the site-directed mutagenesis kit (QuikChange Lightning; Agilent Technologies) according to the manufacturer’s instructions. The following constructs were obtained from Addgene: plasmid 19299, pRK5-HA GST RagAGDP; plasmid 19300, pRK5-HA GST RagAGTP; plasmid 19303, pRK5-HA GST RagBGTP; plasmid 19302, pRK5-HA GST RagBGDP; plasmid 19305, pRK5-HA GST RagCGDP; plasmid 19306, pRK5-HA GST RagCGTP; plasmid 19308, pRK5-HA GST RagDGDP; plasmid 19309, pRK5-HA GST RagDGTP (Sancak et al., 2008); and plasmid 31151, pCMV-Tag4A-MITF-M (WT; Cronin et al., 2009). Constructs were confirmed by DNA sequencing.
Immunofluorescence confocal microscopy
Cells were grown on glass coverslips and washed with PBS before fixation with 4% formaldehyde at room temperature for 15 min. Cells were permeabilized with 0.2% Triton X-100 in PBS at room temperature for 10 min. Cells were then incubated with the indicated primary antibodies for 1 h at room temperature in immunofluorescence buffer (PBS containing 10% fetal bovine serum and 0.1% [wt/vol] saponin). Cells were washed three times with PBS and incubated with the corresponding secondary antibodies conjugated to Alexa Fluor 568, Alexa Fluor 488, or Alexa Fluor 647 in immunofluorescence buffer for 30 min at room temperature. After three washes with PBS, the coverslips were mounted onto glass slides with Fluoromount-G (SouthernBiotech). Images were acquired on a confocal system (LSM 510; Carl Zeiss) equipped with a 63×, NA 1.4 oil immersion objective, filter sets for FITC and Rhodamine, 488- and 543-nm laser excitation, a camera (AxioCam; Carl Zeiss), and LSM 510 operating software. Confocal 8-bit RGB images taken with the same acquisition parameters were processed with ImageJ software (National Institutes of Health), and Photoshop CS4 software (Adobe) was used to produce the figures.
RNAi
Knockdown of the indicated genes was achieved by transfection of siRNA duplexes. In brief, cells grown in 6-well plate were transfected with transfection reagent (DharmaFECT; Thermo Fisher Scientific) and 100 nM of ON-TARGETplus nontargeting pool siRNA duplexes or ON-TARGETplus SMARTpool siRNA duplexes targeted against the raptor gene (Thermo Fisher Scientific) or mission siRNA against RagA, RagB, RagC, RagD, and p18 genes (Sigma-Aldrich). Treated cells were analyzed 72 h after transfection.
Coimmunoprecipitation, GST pull-down, electrophoresis, and immunoblotting
Cells were washed with ice-cold PBS, resuspended in lysis buffer (25 mM Hepes-KOH, pH 7.4, 150 mM NaCl, 5 mM EDTA, and 1% Triton X-100) in the presence of phosphatase and protease inhibitors. After a 30-min incubation on ice, cell lysates were passed 10 times through a 25-gauge needle and centrifuged at 16,000 g for 10 min at 4°C, and the collected soluble fractions were subjected to immunoprecipitation. The soluble fractions were incubated with 2 µl anti-FLAG antibody and protein G–Sepharose beads (GE Healthcare) for 2 h at 4°C. For GST pull-down, soluble fractions were incubated with 25 µl glutathione–Sepharose beads at 4°C for 2 h. The immunoprecipitates and pulled down materials were boiled in Laemmli sample buffer, separated on 4–20% SDS-polyacrylamide gels (Invitrogen), and transferred to nitrocellulose membranes. Membranes were incubated with the indicated primary antibodies at 4°C overnight before incubation for 1 h with HRP-conjugated secondary antibodies and detection by Western Lightning Chemiluminescence Reagent Plus (PerkinElmer).
Subcellular fractionation
DMSO- and Torin-1–treated cells were lysed in NP-40 lysis buffer (10 mM Tris, pH 7.9, 140 mM KCI, 5 mM MgCl2, and 0.5% NP-40) supplemented with protease and phosphatase inhibitors. Cell lysates were kept on ice for 15 min. The lysates were then centrifugated at 1,000 g for 5 min. The collected supernatants represent the cytosolic plus the membrane fraction. The pellets corresponding to the nuclear fractions were washed two times with in NP-40 lysis buffer and sonicated in 0.5% SDS and 0.5% Triton X-100 in 100 mM Tris-HCL buffer, pH 7.4.
RNA isolation and relative quantitative real-time PCR
RNA was isolated from cells by using an isolating kit (PureLink RNA Mini; Invitrogen) as recommended by manufacturer. A spectrophotometer (NanoDrop ND-1000; Thermo Fisher Scientific) was used to quantify RNA yields. A reverse transcription kit (SuperScript III First-Strand Synthesis System; Invitrogen) was used to reverse transcribe RNA (2–4 µg) in a 20-µl reaction using oligo(dT)20. Relative quantitative real-time PCR was performed in a total reaction volume of 10 µl using 5 µl SYBR GreenER quantitative PCR SuperMix (Invitrogen), 2 µl cDNA, 1 µl gene-specific primer mix (QuantiTect Primer Assays; QIAGEN), and 2 µl of water. Quantification of gene expression was performed using a real-time PCR system (7900HT Fast; Applied Biosystems) in triplicate. The thermal profile of the reaction was the following: 50°C for 2 min, 95°C for 10 min, and 35 cycles of 95°C for 15 s followed by 60°C for 1 min. Amplification of the sequence of interest was normalized with a reference endogenous gene GAPDH. The value was expressed as a ratio to RNA from cells infected with control adenovirus and then normalized to the mRNA levels of TFEB-WT. For data analysis, the 7900HT Fast Real-Time PCR System Software was used.
Statistical analysis
Obtained data were processed in Excel (Microsoft) and Prism (GraphPad Software) to generate bar charts and carry out statistical analyses. One-way analysis of variance and pairwise post-tests were run for each dependent variable. P < 0.05 was considered statistically significant (*), and P < 0.001 was considered extremely significant (***). P > 0.05 was considered not significant.
Online supplemental material
Fig. S1 shows a schematic representation of changes on the distribution of TFEB upon depletion of different components of the Ragulator–Rags–mTORC1 complex. Fig. S2 further characterizes changes in the localization of TFEB upon mTORC1 inactivation. Fig. S3 provides additional controls for the siRNA-mediated depletion of Rag GTPases and Ragulator complex protein p18. Fig. S4 shows that TFEB binds to the switch regions of the Rags G domain and identifies a stretch of arginines in the central portion of TFEB that functions as a nuclear import signal. Fig. S5 identifies additional residues in the N-terminal portion of TFEB required for association with lysosomes and cytosolic retention. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201209135/DC1.
Supplementary Material
Acknowledgments
This project was supported by the Intramural Research Program of the National Institutes of Health, National Heart, Lung, and Blood Institute.
Footnotes
Abbreviations used in this paper:
- CMV
- cytomegalovirus
- MITF
- microphthalmia-associated transcription factor
- mTOR
- mammalian target of rapamycin
- TFEB
- transcription factor EB
- WT
- wild type
References
- Bar-Peled L., Schweitzer L.D., Zoncu R., Sabatini D.M. 2012. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 150:1196–1208 10.1016/j.cell.2012.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronisz A., Sharma S.M., Hu R., Godlewski J., Tzivion G., Mansky K.C., Ostrowski M.C. 2006. Microphthalmia-associated transcription factor interactions with 14-3-3 modulate differentiation of committed myeloid precursors. Mol. Biol. Cell. 17:3897–3906 10.1091/mbc.E06-05-0470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin J.C., Wunderlich J., Loftus S.K., Prickett T.D., Wei X., Ridd K., Vemula S., Burrell A.S., Agrawal N.S., Lin J.C., et al. 2009. Frequent mutations in the MITF pathway in melanoma. Pigment Cell Melanoma Res. 22:435–444 10.1111/j.1755-148X.2009.00578.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Kaiser C.A. 2006. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat. Cell Biol. 8:657–667 10.1038/ncb1419 [DOI] [PubMed] [Google Scholar]
- Gong R., Li L., Liu Y., Wang P., Yang H., Wang L., Cheng J., Guan K.L., Xu Y. 2011. Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev. 25:1668–1673 10.1101/gad.16968011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah Y.S., Cho H.Y., Lim T.Y., Park D.H., Kim H.M., Yoon J., Kim J.G., Kim C.Y., Yoon T.J. 2012. Induction of melanogenesis by rapamycin in human MNT-1 melanoma cells. Ann. Dermatol. 24:151–157 10.5021/ad.2012.24.2.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 110:177–189 10.1016/S0092-8674(02)00833-4 [DOI] [PubMed] [Google Scholar]
- Ho H., Kapadia R., Al-Tahan S., Ahmad S., Ganesan A.K. 2011. WIPI1 coordinates melanogenic gene transcription and melanosome formation via TORC1 inhibition. J. Biol. Chem. 286:12509–12523 10.1074/jbc.M110.200543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., et al. 2009a. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 20:1981–1991 10.1091/mbc.E08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Sasaki T., Iemura S., Natsume T., Hara T., Mizushima N. 2009b. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 5:973–979 10.4161/auto.5.7.9296 [DOI] [PubMed] [Google Scholar]
- Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M.A., Hall A., Hall M.N. 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6:1122–1128 10.1038/ncb1183 [DOI] [PubMed] [Google Scholar]
- Kim D.H., Sarbassov D.D., Ali S.M., King J.E., Latek R.R., Erdjument-Bromage H., Tempst P., Sabatini D.M. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 110:163–175 10.1016/S0092-8674(02)00808-5 [DOI] [PubMed] [Google Scholar]
- Laplante M., Sabatini D.M. 2012. mTOR signaling in growth control and disease. Cell. 149:274–293 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., Chen Y., Gucek M., Puertollano R. 2012. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 8:903–914 10.4161/auto.19653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina D.L., Fraldi A., Bouche V., Annunziata F., Mansueto G., Spampanato C., Puri C., Pignata A., Martina J.A., Sardiello M., et al. 2011. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell. 21:421–430 10.1016/j.devcel.2011.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima H., Tokunaga C., Eguchi S., Oshiro N., Hidayat S., Yoshino K., Hara K., Tanaka N., Avruch J., Yonezawa K. 2003. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278:15461–15464 10.1074/jbc.C200665200 [DOI] [PubMed] [Google Scholar]
- Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M., Ballabio A. 2011. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20:3852–3866 10.1093/hmg/ddr306 [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson A., Petit C.S., Froehlich F., Qian S., Ky J., Angarola B., Walther T.C., Ferguson S.M. 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5:ra42 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., Sabatini D.M. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 320:1496–1501 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., Sabatini D.M. 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 141:290–303 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov D.D., Ali S.M., Kim D.H., Guertin D.A., Latek R.R., Erdjument-Bromage H., Tempst P., Sabatini D.M. 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14:1296–1302 10.1016/j.cub.2004.06.054 [DOI] [PubMed] [Google Scholar]
- Sardiello M., Ballabio A. 2009. Lysosomal enhancement: a CLEAR answer to cellular degradative needs. Cell Cycle. 8:4021–4022 10.4161/cc.8.24.10263 [DOI] [PubMed] [Google Scholar]
- Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., et al. 2009. A gene network regulating lysosomal biogenesis and function. Science. 325:473–477 [DOI] [PubMed] [Google Scholar]
- Saucedo L.J., Gao X., Chiarelli D.A., Li L., Pan D., Edgar B.A. 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5:566–571 10.1038/ncb996 [DOI] [PubMed] [Google Scholar]
- Sekiguchi T., Hirose E., Nakashima N., Ii M., Nishimoto T. 2001. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J. Biol. Chem. 276:7246–7257 10.1074/jbc.M004389200 [DOI] [PubMed] [Google Scholar]
- Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science. 332:1429–1433 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31:1095–1108 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker H., Radimerski T., Schindelholz B., Wittwer F., Belawat P., Daram P., Breuer S., Thomas G., Hafen E. 2003. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 5:559–565 10.1038/ncb995 [DOI] [PubMed] [Google Scholar]
- Yasumoto K., Amae S., Udono T., Fuse N., Takeda K., Shibahara S. 1998. A big gene linked to small eyes encodes multiple Mitf isoforms: many promoters make light work. Pigment Cell Res. 11:329–336 10.1111/j.1600-0749.1998.tb00491.x [DOI] [PubMed] [Google Scholar]
- Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., Sabatini D.M. 2011a. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 334:678–683 10.1126/science.1207056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R., Efeyan A., Sabatini D.M. 2011b. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12:21–35 10.1038/nrm3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.