Abstract
Calprotectin (CP) is a transition metal-chelating antimicrobial protein of the calcium-binding S100 family that is produced and released by neutrophils. It inhibits the growth of various pathogenic microorganisms by sequestering the transition metal ions manganese and zinc. In this work, we investigate the manganese-binding properties of calprotectin. We demonstrate that the unusual His4 motif (site 2) formed at the S100A8/S100A9 dimer interface is the site of high-affinity Mn(II) coordination. We identify a low-temperature Mn(II) spectroscopic signal for this site consistent with an octahedral Mn(II) coordination sphere with simulated zero-field splitting parameters D = 270 MHz and E/D = 0.33 (E = 81 MHz). This analysis, combined with studies of mutant proteins, suggests that four histidine residues (H17 and H27 of S100A8; H91 and H95 of S100A9) coordinate Mn(II) in addition to two as-yet unidentified ligands. The His3Asp motif (site 1), which is also formed at the S100A8/S100A9 dimer interface, does not provide a high-affinity Mn(II) binding site. Calcium binding to the EF-hand domains of CP increases the Mn(II) affinity of the His4 site from the low-micromolar to the mid-nanomolar range. Metal-ion selectivity studies demonstrate that CP prefers to coordinate Zn(II) over Mn(II). Nevertheless, the specificity of Mn(II) for the His4 site provides CP with the propensity to form mixed Zn:Mn:CP complexes where one Zn(II) ion occupies site 1 and one Mn(II) ion occupies site 2. These studies support the notion that CP responds to physiological calcium ion gradients to become a high-affinity transition metal ion chelator in the extracellular space where it inhibits microbial growth.
Introduction
Transition metal ions are ubiquitous in biology. The bioavailability and distribution of these essential nutrients are key factors in the host/pathogen interaction that contribute to whether the establishment and progression of microbial infection occurs.1–4 Colonizing pathogenic microorganisms employ a variety of strategies to acquire metal ions in the vertebrate host, which include the biosynthesis and utilization of siderophores,5,6 and the expression of high-affinity metal-ion transporters.7–9 Although less celebrated than iron in the context of pathogenesis, manganese and zinc are essential nutrients, and the competition between host and pathogen for these two metal ions influences the pathophysiology of disease.10 Of particular relevance to the work described herein is manganese, which is a co-factor for prokaryotic metalloenzymes that contribute to signal transduction (e.g. phosphatases),11 DNA biosynthesis (e.g. class Ib ribonucleotide reductase),12 metabolism (e.g. adenylyl cyclase, 3-phosphoglycerate mutase),13 and the oxidative stress response (e.g. manganese-containing superoxide dismutase and manganese catalase).14,15 Indeed, manganese acquisition and utilization are often associated with microbial virulence.7,11,16–23 In Salmonella enterica serovar Typhimurium, the genes mntH and sitABCD encode Mn(II) transporters that are necessary for replication in macrophages and virulence in mice.19 Streptococcus pneumoniae expresses PsaA, which is a manganese and zinc transporter important for colonization.24–26 Neisseria gonorrhoeae accumulates intracellular Mn(II), and this phenomenon is associated with increased resistance to oxidative killing by neutrophils.27 Borrelia burgdorferi, the causative agent of Lyme disease, has no metabolic iron requirement28 and its manganese transporter BmtA is required for the establishment of an infectious lifestyle in ticks and for virulence in mammals.29
One general mechanism of the mammalian innate immune response, which serves to inhibit colonization by microbial invaders on a short timescale, involves metal-ion withholding at sites of infection.1,10,30 Calprotectin (CP) was recently identified as a manganese scavenger in the context of Staphylococcus aureus infection.31 CP is reported to reach low-millimolar concentrations in the cytoplasm of neutrophils,32 a type of white blood cell that is recruited to sites of infection to combat invading microbes by releasing a mixture of antimicrobial peptides/proteins (e.g. defensins, lactoferrin, siderocalin, CP) and deleterious reactive oxygen species.33 The antibacterial and -fungal properties of CP have been recognized for decades and were first attributed to its ability to sequester zinc,34–39 which is an essential nutrient for all organisms.40 Recent studies of S. aureus infection in a murine tissue abscess model revealed a calprotectin-dependent depletion of manganese at sites of infection.31 Further in vitro investigations demonstrated that recombinant human CP exhibited antimicrobial activity against S. aureus Newman, and provided protection against Mn(II) toxicity for mutant S. aureus strains lacking the manganese transport regulator MntR (ΔmntR).31 Moreover, human CP exhibited enhanced antimicrobial activity against S. aureus strains deficient in the manganese-uptake proteins MntAB (ΔmntAΔmntB). CP exposure was associated with reduced manganese superoxide dismutase activity, which enhanced the susceptibility of S. aureus to oxidative stress.41 Taken together, these in vitro studies demonstrate that CP interferes with Mn(II) uptake by S. aureus and establish a previously unappreciated role for CP in manganese homeostasis and the host response to infection. Elucidating the manganese-binding properties of CP is therefore necessary to further decipher its biological mechanisms and contributions to transition metal homeostasis.
Human calprotectin is a heterodimer (αβ) or –tetramer (α2β2) of the S100 proteins S100A8 (10.8 kDa, α subunit) and S100A9 (13.1 kDa, β subunit) (Figure 1).42–44 S100 proteins comprise a family of EF-hand domain calcium-binding proteins, and each S100 exhibits two EF-hands. 45, 46 The C-terminal EF-hands of S100A8/S100A9 are similar to those of calmodulin and provide seven-coordinate, “canonical” Ca(II)-binding sites. In contrast, the N-terminal EF-hands provide five-coordinate, “non-canonical” Ca(II)-binding motifs. Crystallographic characterization of the calcium-bound CP α2β2 heterotetramer (human form) revealed two transition metal-binding sites at the S100A8/S100A9 interface (Figure 1A).47 A His3Asp motif (site 1) is formed by A8(H83), A8(H87), A9(H20), and A9(D30), and an unusual His4 motif (site 2) is formed by A8(H17), A8(H27), A9(H91), and A9(H95) (Figure 1A). S100A9 contains an extended C-terminal tail with a number of potential metal-chelating residues (Figure 1B). Prior investigations addressing the Zn(II) coordination chemistry of CP demonstrated that both the His3Asp and His4 sites bind Zn(II) and exhibit Ca(II)-dependent Zn(II) affinity.48 Calcium coordination to the EF-hand domains enhances the Zn(II) affinity of CP such that both sites coordinate Zn(II) with dissociation constants (Kd) in the sub-nanomolar range.
Figure 1.
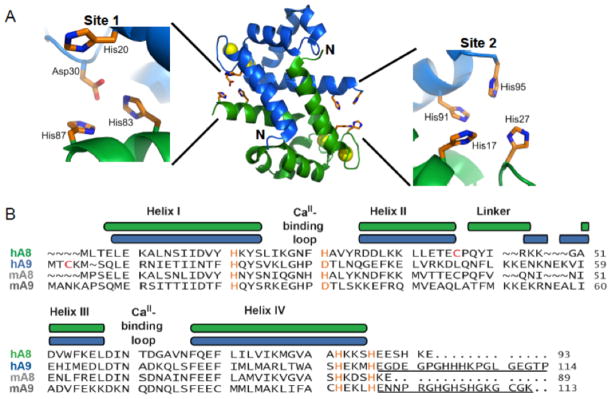
Structure and amino acid sequences of calprotectin. (A) Depiction a heterodimer unit of human CP (PDB: 1XK4). This heterodimer is taken from the crystal structure of the Ca(II)- bound heterotetramer to illustrate the metal-binding sites. No structure of the Ca(II)-free heterodimer is available. S100A8 is colored green, S100A9 is colored blue, and the Ca(II) ions are indicated by yellow spheres. The His3Asp (site 1) and His4 (site 2) metal-binding motifs form at the heterodimer interface. (B) Amino acid sequence alignment for S100A8 and S100A9 from human (indicated by “h”) and mouse (indicated by “m”). The color-coded secondary structure diagrams correspond to the human subunits. The residues of the metal-binding motifs are colored orange. The two Cys residues that were mutated to Ser for metal-binding studies are highlighted in red. The C-terminal extensions of human and mouse A9 are underlined. The numbers indicate amino acid number.
To the best of our knowledge, CP is the only S100 protein with reported Mn(II)-chelating ability, and little is known about this facet of its coordination chemistry.31,41 Whether Mn(II) and Zn(II) share the same coordination sites and calcium-dependence, and whether CP selects for one transition metal ion over the other, is unclear. A recent ITC study supported formation of a high-affinity Mn(II):CP complex.41 In this work, we report extensive spectroscopic and thermodynamic investigations designed to rigorously decipher the Mn(II)-binding properties of human CP and correlate the results to the physiological context. These studies employ an extensive CP mutant family and unambiguously demonstrate that CP employs its unusual His4 motif and two as-yet unidentified ligands to sequester Mn(II) in a calcium-dependent manner. The specificity of Mn(II) for the His4 sites provides CP with the propensity to form mixed Ca(II)/Mn(II)/Zn(II) complexes where the EF-hand domains, the His4 site, and the His3Asp site coordinate Ca(II), Mn(II) and Zn(II), respectively.
Experimental Section
Materials and General Methods
Experimental details for the preparation of buffers for metal-binding studies, including necessary precautions to avoid metal contaminations, are reported elsewhere.48 All aqueous solutions were prepared with Milli-Q water (18.2 MΩ, 0.22 μm filter). For metal-binding experiments, HEPES buffer was prepared by using metal-free Ultrol grade HEPES (free acid, Calbiochem) and TraceSELECT NaCl (Sigma), and metal-free aqueous NaOH (Sigma) was used for pH adjustments. To reduce metal-ion contamination, the buffers were treated with Chelex 100 resin (BioRad; 10 g/1 L) for at least one hour prior to use. The Chelex-treated buffers were filtered through a 0.22 μm filter or centrifuged to remove the Chelex resin, and stored in polypropylene containers. All metal-binding studies were conducted at pH 7.5 in 75 mM HEPES, 100 mM NaCl buffer unless specified otherwise. A Tris buffer (1 mM Tris, pH 7.5) prepared from Tris base (J.T. Baker) was used for circular dichroism spectroscopy. Calcium chloride and 99.999% anhydrous zinc chloride were purchased from Sigma Aldrich, and 99.999% manganese chloride was obtained from Alfa Aesar. Stock solutions of Ca(II) (1 M), Mn(II) (1 M), and Zn(II) (100 mM) were prepared by using Milli-Q water and acid-washed volumetric glassware, and the solutions were immediately transferred to and stored in polypropylene containers. Each working M(II) solution was made fresh daily by diluting the appropriate stock solution to the desired working concentration with either Milli-Q water or buffer (75 mM HEPES, 100 mM NaCl, pH 7.5). Zinpyr-1 (ZP1) was purchased from Strem Chemical Co., and the purity was verified by analytical HPLC analysis, or synthesized from 2′,7′-dichlorofluorescein and di-(2-picolyl)amine as described elsewhere.49 Stock solutions of ZP1 (~2 mM) were prepared in anhydrous DMSO, partitioned into 50-μL aliquots, and stored at −20 °C. Each aliquot was freeze-thawed only once, and the working ZP1 concentration was verified by using the reported extinction coefficient of apo ZP1 (ε515 = 79,500 M−1cm−1).49
Preparation of CP and Mutant CP
Recombinant human CP and mutant proteins were overexpressed, purified, and characterized as reported elsewhere.48 The metal-binding studies presented in this work were conducted by using CP-Ser and mutants thereof. CP-Ser is comprised of the subunits S100A8(C42S) and S100A9(C3S). These Cys-to-Ser mutants were employed to avoid the need for reducing agents in the metal-binding studies. The mutated cysteine residues are not essential for the antibacterial activity of CP.48 CP-Ser, ΔHis3Asp, ΔHis4, ΔΔ, and H27D were prepared multiple times for this work, and experiments conducted with independent batches of protein provided comparable results. The H17A, H27A, H91A, H95A and (H27A)(H91A) mutants were each prepared and purified once. Wild-type CP, which was only employed in analytical size exclusion chromatography, was also prepared once. Following purification, each protein was dialyzed against Chelex resin (10 g/L) for at least 12 h at 4 °C prior to storage. The Chelex-treated protein was stored in aliquots at −80 °C in pH 8.0 buffer (20 mM HEPES, 100 mM NaCl), and each aliquot was freeze-thawed only once. The purification protocol provided wild-type CP and all mutants in the heterodimeric (αβ) form. Protein concentrations were routinely determined by using calculated extinction coefficients (ε280 = 18,450 M−1cm−1 for the CP heterodimer), which are listed in Table S1 (Supporting Information). A BioTek Synergy HT plate reader outfitted with a calibrated Take3 micro-volume plate, or an Agilent 8453 diode array spectrophotometer, was routinely employed to determine protein concentrations. With the exception of CP-Ser(H27D), the site-directed mutagenesis for the mutant proteins utilized in this work was described previously.48 The details for the site-directed mutagenesis and purification CP-Ser(H27D) are provided as Supporting Information, and biochemical characterization of this mutant is summarized in Figure S1. Table S2 summarizes the mutant proteins and protein nomenclature.
Analytical Size Exclusion Chromatography
CP-Ser and the metal-binding site mutants were buffer exchanged into Chelex-treated 75 mM HEPES, 100 mM NaCl buffer adjusted to pH 7.5. The buffer for wild-type CP contained 1 mM β-mercaptoethanol to prevent disulfide formation. The protein samples were adjusted to a concentration of 200 μM and ten equivalents of Mn(II) (14 μL from a 100 mM Mn(II) stock solution added to a final volume of 700 μL) were added. The samples were incubated on ice for 0, 2, or 8–18 h. At each timepoint, a 200-μL aliquot was loaded onto a Superdex 75 10/300 GL (GE Lifesciences) size-exclusion column via a 500-μL sample loop connected to an ÄKTA Purifier FPLC system (GE Lifesciences). The column was pre-equilibrated with 1.5 column volumes of running buffer (Chelex-treated 75 mM HEPES, 100 mM NaCl, pH 7.5), and the sample loop was emptied with 0.5 mL of running buffer. The sample was subsequently eluted over one column volume at a flow rate of 0.5 mL/min at room temperature. The protein-containing fractions, identified by monitoring absorption at 280 nm, were collected and the protein concentration of each fraction was quantified by using the molar extinction coefficient at 280 nm.
Manganese Quantification by Atomic Absorption Spectroscopy
The manganese content of the protein-containing fractions obtained from analytical size exclusion chromatography (SEC) was determined by atomic absorption spectroscopy. A Perkin-Elmer AAnalyst 600 atomic absorption spectrometer was employed for all measurements. The protein-containing fractions were diluted 1:200 by using Milli-Q water. A Mn(II) standard curve (0–5 μg/L) was obtained by using solutions of Mn(II) (1000 μg/mL MnCl2 atomic absorption standard, J. T. Baker) dissolved in 5% nitric acid and diluted into Milli-Q water. The averages obtained from three independent samples are reported and the errors are the standard deviation from the mean.
Optical Absorption and Fluorescence Spectroscopy
Optical absorption spectra were recorded on either a Beckman Coulter (DU800) scanning or an Agilent 8453 diode array UV-visible spectrophotometer thermostated at 25 °C. Fluorescence spectra were collected on a Photon Technologies International QuantaMaster 40 fluorimeter outfitted with a continuous xeon source for excitation, autocalibrated QuadraScopicTM monochromators, a multi-mode PMT detector, and a circulating water bath maintained at 25 °C. The spectrofluorimeter was controlled by the FelixGX software package. Quartz cuvettes (Starna) with a 1-cm pathlength were employed for all optical absorption and fluorescence measurements. To prevent metal-ion contamination, the cuvettes were washed with 20% nitric acid and thoroughly rinsed with Chelex-treated Milli-Q water prior to use. The cuvettes were stored in 20% nitric acid.
Circular Dichroism Spectroscopy
Protein solutions (10 μM, 300 μL) were prepared at pH 7.5 in 1.0 mM Tris-HCl buffer that was treated with Chelex to remove trace metals. The pH was readjusted to 7.5 by addition of hydrochloric acid after the Chelex treatment. For Mn(II)-binding experiments, the CD spectrum of the apo protein was recorded. Subsequently, Mn(II) was added to final concentration of 100 μM, the sample was mixed gently, and the CD spectrum was recorded approximately two minutes after the Mn(II) addition. This experiment was also conducted in the presence of 2 mM Ca(II). The CD spectra were collected from 260–195 nm at 1 nm intervals (3 sec averaging time, three independent scans per wavelength). The data obtained from the three scans were averaged by using Excel and plotted in Kaleidagraph. For thermal denaturation experiments with mutant proteins, a sample of 10 μM ΔHis3Asp (αβ) or ΔHis4 (αβ) was prepared at pH 8.5 in 1 mM Tris buffer containing 0.5 mM EDTA, and the temperature was varied from 25 to 95 °C in 2 °C increments. Samples containing Ca(II) were prepared in the same manner except that Ca(II) was added to a final concentration of 2 mM. These conditions were selected based on prior literature studies that reported the CD spectra of wild-type CP in the absence and presence of Ca(II) under such buffer conditions.42 For thermal denaturation of CP-Ser in the presence of Mn(II), 10 μM CP-Ser was prepared at pH 7.5 in Chelex-treated 1 mM Tris buffer and incubated with 10 equivalents of Mn(II) in the absence and presence of 2 mM Ca(II). For all thermal denaturation experiments, the CD signal at 222 nm was recorded at each temperature after a one-minute equilibration time. A 1-mm path-length quartz CD cell (Hellma) was employed for all CD measurements.
General Methods for Electron Paramagnetic Resonance Spectroscopy
Electron paramagnetic resonance (EPR) spectra (X-band, 9 GHz) were recorded on a Bruker EMX spectrometer outfitted with an ER 4199HS cavity. A flat quartz cell positioned in an E-field null plane within the cavity was used for all room-temperature measurements. An ESR900 cryostat was used for all low-temperature measurements, and the temperature was monitored with either a Cernox sensor or a thermocouple. A copper-EDTA spin standard was used to account for all relevant intensity factors. All spectral analysis and simulations were performed with the Windows software package, SpinCount, developed by Professor Michael P. Hendrich at Carnegie Mellon University. The simulations of the bound-Mn(II) were determined using the standard spin Hamiltonian described by Equation 1 with second-order perturbation theory.
| (Eq. 1) |
D and E are the zero-field splitting (zfs) parameters, g is the g-tensor, and all other parameters have their standard meanings. SpinCount treats the nuclear hyperfine interactions (A) with second-order perturbation theory.
Room-temperature EPR Spectroscopy
In a typical room-temperature EPR titration, a 300-μL sample of CP (approximately 25 or 150 μM for titrations in the presence and absence of Ca(II), respectively) was prepared at pH 7.5 (75 mM HEPES, 100 mM NaCl) and titrated with a freshly-prepared Mn(II) solution prepared in the buffer. The sample was incubated for at least ten minutes at room temperature after each Mn(II) addition and placed in a flat quartz cell (Bruker BioSpin). The EPR spectrum was recorded using the following conditions: microwaves, 9.83 GHz, 20 mW; modulation, 1.0 mT. The concentration of free Mn(II) in each sample was determined by scaling the inner four lines of the EPR spectrum to those of a standard Mn(II) solution prepared from an atomic absorption standard (J.T. Baker) diluted in the buffer and recorded under identical conditions. The Mn(II) standard was also used to verify the concentration of the Mn(II) stock solution employed for the metal-binding titration. Room temperature titrations for the heterodimeric single point mutants H17A, H27A, H27D, H91A, H95A, the double point mutant (H27A)(H91A), and CP-Ser ΔΔ in the absence and presence of Ca(II) were each performed twice. Titrations of CP-Ser (αβ), CP-Ser ΔHis3Asp (αβ), and CP-Ser ΔHis4 (αβ) were conducted at least in triplicate. Titrations of CP-Ser in the presence of Ca(II) were performed in duplicate. The Mn(II) dissociation constant values obtained from room-temperature EPR titrations of CP-Ser were obtained from global fits of the data obtained from the multiple titrations. The equations employed to fit the binding curves are provided as Supporting Information.
Low-temperature EPR Spectroscopy
Low-temperature EPR samples were housed in 4 mm (OD) quartz EPR tubes and frozen in liquid nitrogen prior to analysis. Low-temperature EPR titrations were performed with 200-μL samples of CP at the indicated concentrations. Aliquots of aqueous Mn(II) were added directly into the EPR tube containing a solution of the protein by using a 100-μL Hamilton syringe. The sample was then mixed with a second 250-μL Hamilton syringe and incubated for a minimum of ten minutes at room temperature before being frozen in liquid nitrogen. The EPR spectra of CP samples containing Mn(II) were unperturbed by repeated freeze-thaw cycling. Low-temperature spectra of Mn(II):CP mixtures in the presence or absence of 2 mM Ca(II) or 600 mM NaCl were also recorded. For these spectral acquisitions, the samples containing protein and Mn(II) were thawed and either NaCl or CaCl2 was added to the indicated concentration from a concentrated stock solution (typically >10% dilution of the original sample). The sample was allowed to incubate for a minimum of fifteen minutes at room temperature before being frozen in liquid nitrogen.
Low-temperature Mn(II)-binding EPR titrations were performed in the absence and presence of Ca(II). For the titrations conducted in the absence of Ca(II), the CP-Ser concentration was ~100 μM and 0–1.1 equivalents of Mn(II) were added. For the titrations in the presence of Ca(II), CP-Ser (~100 μM) was incubated with 1 mM of CaCl2 for fifteen minutes or more prior to titrating the sample with Mn(II). The resulting spectra were analyzed for the formation and growth of distinct Mn(II)-bound signals with respect to the concentration of Mn(II) added. For this analysis, the intensity of the highest field hyperfine resonance, defined as the change in y-axis from the positive feature at g = 1.88 to the negative feature at g = 1.873, was determined for each titration point. The intensity of this feature was correlated to concentration of CP-bound Mn(II) by defining the intensity from a sample of CP-Ser with 0.1 equivalent of Mn(II) in the presence of Ca(II) as being equal to the concentration of metal ion added (~10 μM, assuming that all added metal becomes bound metal under these conditions). The concentration of CP-bound Mn(II) for all titrations was determining using this ratio. Each titration was repeated twice and the Mn(II) dissociation constants were determined from a global fit of the combined data.
Mn(II) Competition Experiments with ZP1
For competition experiments between CP (αβ) and ZP1, a 2-mL solution of ZP1 (1–4 μM) was prepared in a quartz cuvette (75 mM HEPES, 100 mM NaCl, pH 7.5). The optical absorption and fluorescence spectra of ZP1 were recorded. An aliquot of CP was introduced to the cuvette to afford the desired protein concentration (1–4 μM) and the optical and fluorescence spectra were recorded again. If metal ion contamination was detected, indicated by a change in ZP1 emission relative to the no protein spectrum, the solution was discarded. For manganese competition experiments, aliquots of aqueous Mn(II) from a freshly-prepared 0.5 or 1 mM working solution were added to the ZP1/CP mixture. Following each Mn(II) addition, the solution was incubated in the dark for ten minutes at room temperature, and the optical absorption and emission spectra were subsequently recorded. This procedure was employed for Mn(II) competitions performed in the presence of varying concentrations of Ca(II) except that the protein was pre-incubated with Ca(II) at the indicated concentration. The same procedure was also utilized for experiments where Ca(II) was added to mixtures containing CP-Ser/ZP1/Mn(II) except that the sample was incubated for fifteen minutes at room temperature after each Ca(II) addition. The emission spectra were recorded from 500 to 650 nm (λex = 490 nm, 0.4 mm excitation and emission slit widths) and integrated over this range by using the FelixGX software package.
Mn(II) and Zn(II) Selectivity Experiments with ZP1
A 1:1 mixture of 4 μM ZP1 and CP-Ser was prepared at pH 7.5 (75 mM HEPES, 100 mM NaCl) and the optical absorption and emission spectra recorded as described above. Aliquots of Mn(II), Zn(II) or a mixed metal solution containing a 1:1 ratio of Mn(II) and Zn(II) were added and ZP1 emission was monitored periodically over the course of several hours.
Results and Discussion
Analytical SEC Reveals that the His4 Site Formed at the CP (αβ) Dimer Interface is Required for Mn(II) Coordination
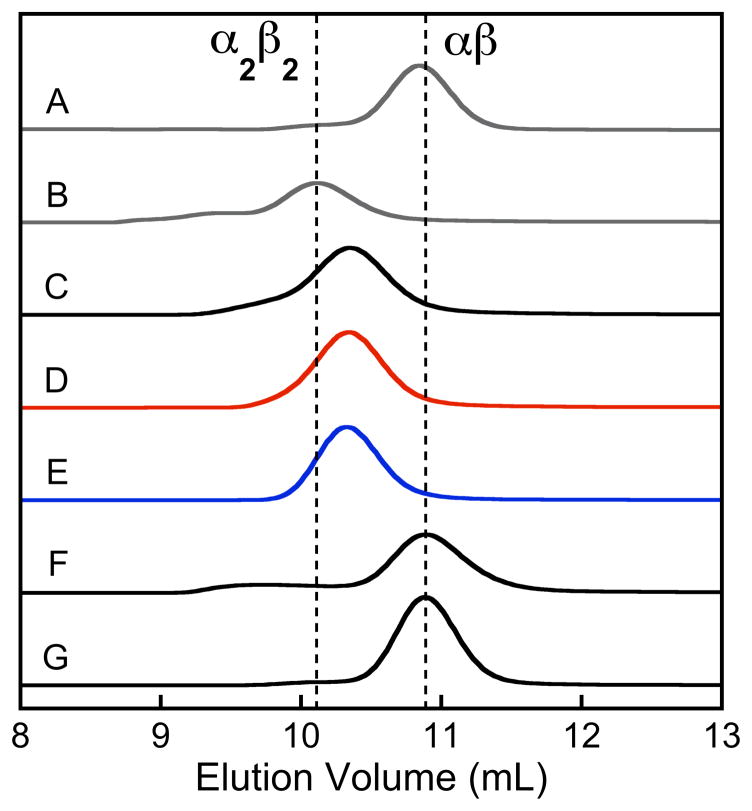
Analytical SEC was performed to determine whether the presence of Mn(II) causes a shift in the elution volume of apo CP, and ascertain whether the protein retained Mn(II) after chromatography. In the absence of divalent cations, analytical SEC of wild-type and mutant CP provided retention volumes of approximately 10.8 mL, which correspond to a molecular weight of 38 kDa. This retention volume was previously assigned to the αβ heterodimer (24 kDa).48 A retention volume of approximately 10.2 mL was observed for the wild-type and mutant α2β2 heterotetramers, which form in the presence of excess Ca(II). This retention volume is in good agreement with the heterotetramer molecular weight of 48 kDa. The retention volumes of CP-Ser and the metal-binding site mutants pre-incubated with ten equivalents of Mn(II) were compared to the CP-Ser αβ and α2β2 standards (Figure 2). Both CP and CP-Ser exhibited a new, Mn(II)-dependent peak with an intermediate retention time of approximately 10.3 mL. A Mn(II)-dependent peak with the same retention time was also observed for ΔHis3Asp, but not for ΔHis4 or ΔΔ. No shift in the peak corresponding to the αβ heterodimer was observed for the latter two mutants as a result of incubation with Mn(II). These comparisons demonstrate that formation of the new 10.3-mL peak requires an intact His4 site.
Figure 2.
Analytical SEC of CP and mutant proteins in the absence and presence of 10 equivalents of Mn(II) (75 mM HEPES, 100 mM NaCl, pH 7.5). (A) CP-Ser. (B) CP-Ser with 2 mM Ca(II) in the SEC running buffer. (C) Wild-type CP pre-incubated with 10 equiv of Mn(II). (D) CPSer pre-incubated with 10 equiv of Mn(II). (E) CP-Ser ΔHis3Asp pre-incubated with 10 equiv of Mn(II). (F) CP-Ser ΔHis4 pre-incubated with 10 equiv of Mn(II). (G) CP-Ser ΔΔ pre-incubated with 10 equiv of Mn(II). The protein concentrations were 100 μM in A and B and 200 μM in C-G. The chromatograms in A and B were scaled 2x. The vertical dashed lines indicate the retention volumes for the CP-Ser αβ and α2β2 forms.
Mn(II) analysis of the protein-containing fractions by atomic absorption spectroscopy revealed that Mn(II) was retained only in the CP, CP-Ser, and ΔHis3Asp samples (Table 1). The Mn(II):CP (αβ) ratios for CP, CP-Ser, and ΔHis3Asp spanned the 0.4–0.7 range. In contrast, only negligible (<0.01 equivalents per CP heterodimer) Mn(II) was detected in the protein-containing fractions obtained for ΔHis4 and ΔΔ, indicating that these mutants did not retain bound Mn(II) following SEC. Taken together, these experiments suggest that the His4 site of CP coordinates Mn(II) and with sufficient affinity that a portion of the metal ion remains bound over the course of the SEC run. Moreover, the Mn(II)-induced shift in elution volume observed in the chromatographs for CP, CP-Ser and ΔHis3Asp indicate that Mn(II) coordination to the His4 site results in a conformational change.
Table 1.
Mn(II) Quantification of Protein-Containing SEC Samples.
| Protein | Elution Volume (mL) | Mn/CPa |
|---|---|---|
| CP | 10.3 | 0.49 ± 0.03 |
| CP-Ser | 10.3 | 0.51 ± 0.03 |
| ΔHis3Asp | 10.3 | 0.70 ± 0.14 |
| ΔHis4 | 10.9 | 0.012 ± 0.003 |
| ΔΔ | 10.9 | 0.005 ± 0.002 |
Protein concentrations were determined by absorbance at 280 nm, and the Mn(II) concentrations were quantified by atomic absorption spectroscopy. The Mn/CP ratios are per CP heterodimer.
The CD spectra of CP-Ser (±Ca) in the absence and presence of ten equivalents of Mn(II) were indistinguishable and exhibit minima at 222 and 208 nm, confirming marked α-helical content of the Mn(II)-bound form (Figure S2). This behavior is in agreement with the CD spectra observed following addition of Co(II) or Zn(II) to CP-Ser.48 Chelation of first-row transition metal ions at the interfacial binding sites has negligible impact on overall secondary structure. We therefore conclude that the conformational change suggested by the SEC experiments does not involve a global perturbation to the α-helical structure of CP-Ser.
To address whether mutations at site 1 or 2 impacts the thermal stability of the protein fold, we determined the Tm values of ΔHis3Asp, and ΔHis4 in the presence and absence of 2 mM Ca(II). We previously reported that CP-Ser has a Tm of approximately 59 °C and that Ca(II) binding causes a shift in the Tm to approximately 79 °C.48 The thermal denaturation curves of ΔHis3Asp and ΔHis4 are comparable to those of CP-Ser in the absence and presence of Ca(II) (Figure S3). The ΔHis3Asp and ΔHis4 mutants exhibit Tm values in the 55–59 (−Ca) and 75–79 (+Ca) °C ranges. Mutation of the His3Asp or the His4 motif residues to alanine moieties has negligible impact on the thermal stability of CP-Ser. Moreover, addition of ten equivalents of Mn(II) to CP-Ser increased the Tm value to approximately 88 °C, and addition of both Mn(II) and Ca(II) precluded unfolding up to 95 °C (Figure S3). Manganese coordination therefore increases the stability of the protein fold to thermal denaturation in both the absence and presence of Ca(II).
Room-Temperature Mn(II) EPR Titrations Confirm the Requirement of the His4 Site
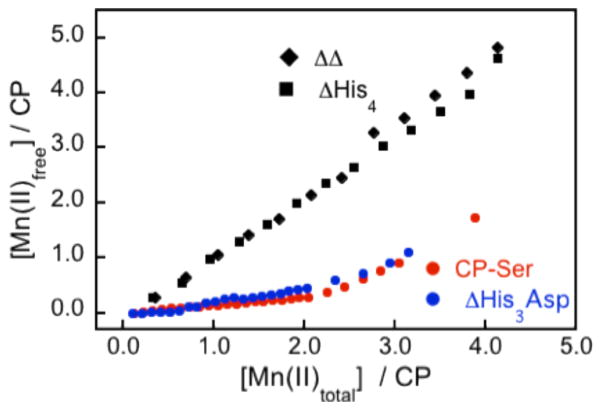
Room-temperature Mn(II)-binding EPR titrations were subsequently performed to further interrogate the Mn(II)-chelating properties of CP. At room temperature, [Mn(H2O)]6 2+ exhibits a six-line pattern centered at g = 2 that results from hyperfine splitting (I = 5/2) of the allowed EPR transition (Δms = 1, ΔmI = 0) in high-spin S = 5/2 55 Mn(II) systems.50,51 In contrast, protein-bound Mn(II) is EPR silent at room temperature because the zero-field splittings broaden the signal beyond detection.52–54 To determine whether CP-Ser or the metal-binding site mutants coordinate Mn(II), each protein (~100 μM, αβ) was titrated with up to four equivalents of Mn(II). All Mn(II) EPR spectra recorded throughout each titration exhibited a six-line hyperfine pattern with a splitting of a = 8.9 mT centered at g = 2. Throughout each titration, the spectra varied only in terms of intensity. The amount of free Mn(II) in each sample was quantified by comparison to a spectrum of a Mn(II) standard recorded under identical conditions as described in the Experimental Section. Plots of [Mn(II)free]/CP versus [Mn(II)total]/CP revealed that attenuation of the free Mn(II) signal was observed for CP-Ser and ΔHis3Asp (Figure 3). In both cases, the concentration of free Mn(II) was less than the concentration of total Mn(II) added, supporting formation of protein-bound Mn(II) species. In contrast, linear relationships with slopes of approximately one were observed for ΔHis4 or ΔΔ (Figure 3, Table S3), which demonstrated that these mutants do not coordinate Mn(II) under the experimental conditions. Moreover, only negligible attenuation of the free Mn(II) signal was observed for the double mutant (H27A)(H91A) and for the single point mutants H17A, H27A, H91A, and H95A (Figure S4). In these cases, plots of [Mn(II)free]/CP versus [Mn(II)total]/CP provided nearly linear relationships. With the exception of the H17A titrations, which afforded slopes ≥1, linear fits of the single-mutant data afforded slopes ranging from 0.74–0.92 (Table S3). The slopes in this range suggest that the single mutants exhibit lower binding affinity for Mn(II) as compared to CP-Ser and ΔHis3Asp, and greater Mn(II) affinity than ΔHis4 and ΔΔ. Because mutation of any one of the four histidine residues significantly perturbs Mn(II) binding, the data indicate that all four histidine residues of site 2 contribute to Mn(II) coordination.
Figure 3.
Plots of [Mn(II)]free versus [Mn(II)]total obtained from room-temperature Mn(II) EPR titrations of 149 μM CP-Ser (red circles), 132 μM ΔHis3Asp (blue circles), 127 μM ΔHis4 (black squares), and 165 μM ΔΔ (black diamonds) at pH 7.5 (75 mM HEPES, 100 mM NaCl). The data from room-temperature EPR titrations of the single- and double-point mutants are provided in Figure S4 and Table S3.
To further evaluate the requirement of the His4 motif comprising the manganese-binding site, a H27D mutant was prepared by site-directed mutagenesis, overexpressed, and purified. We selected H27D because amino acid sequence alignments demonstrate that other S100 proteins, including S100A7, S100A9 and S100A12, house aspartate residues in this position. Room-temperature titration of the H27D mutant provided a similar relationship between free and total Mn(II) as H27A and the other single point mutants (Figure S4, Table S3). This result demonstrates that the Mn(II) coordination is attenuated as a result of replacing His27 with an aspartate residue.
Figure 4 displays the binding curves and fits obtained from Mn(II)-binding titrations monitored by room-temperature EPR spectroscopy. In the absence of Ca(II), a Kd1 value of 4.9 ±1.0 μM was obtained and assigned to the His4 site. Fitting of this titration curve required the incorporation of two low-affinity Mn(II) binding events, which were fixed at Kd values of 1 mM. We contend that these millimolar-affinity binding events are not of physiological relevance and may originate from weak Mn(II) association with the EF-hand domains, which has been observed in other systems,55,56 or other acidic residues.
Figure 4.
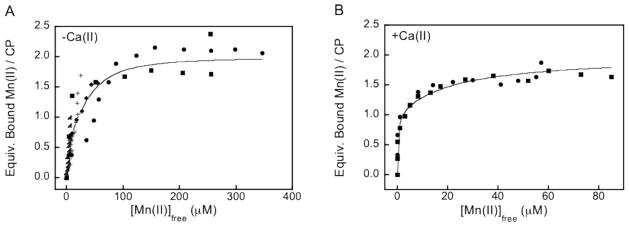
Dissociation constant plots obtained from room-temperature Mn(II) EPR titrations at pH 7.5 (75 mM HEPES, 100 mM NaCl). (A) Titration of approximately 100 μM CP-Ser (αβ) with Mn(II) afforded Kd1 = 4.9 ± 1 μM and Kd2 = 1.0 mM (n = 2; fixed). (B) Titration of 25 μM CP-Ser in the presence of 1 mM Ca(II) afforded Kd1 = 194 ± 203 nM and Kd2 = 21 ± 5 μM. The circles, squares, triangles, diamonds, and plus signs indicate independent titrations. The lines represent the global fits of all data. The equations used in the data analysis are provided as Supporting Information.
The Mn(II) Affinity of the His4 Site is Calcium-Dependent
In prior work, we discovered that the Zn(II) affinity of CP is calcium-dependent.48 We therefore sought to examine the effect of calcium on Mn(II) binding by CP, and room-temperature EPR titrations were performed in the presence of Ca(II). These titrations revealed almost no detectable growth of free Mn(II) until greater than one equivalent of Mn(II) was added to CP-Ser (data not shown). Fits to the resulting binding curves afforded a lower Kd1 value of 194 ± 203 nM (Figure 4). Based on the SEC and RT-EPR experiments, we assign this value to the His4 site. One additional binding event was required to fit this data set, which was allowed to float and provided a Kd2 value of 21 ± 5 μM. We tentatively assign the Kd2 value to weak Mn(II) coordination at the His3Asp site, and further spectroscopic studies are required to rigorously evaluate this possibility. This analysis indicates that the calcium-bound from of CP exhibits higher Mn(II) affinity at the His4 site (and possibility at the His3Asp site) than the αβ heterodimer.
Robust room-temperature EPR analyses require an adequate free Mn(II) signal for quantification. High-affinity Mn(II) binding sites provide relatively low concentrations of observable free Mn(II) at substoichiometric equivalents of the metal ion, which complicates Kd analysis. Because tight Mn(II) binding was observed for CP-Ser by RT-EPR when Ca(II) was added to the buffer, we verified the calcium-dependence and Mn(II) Kd values through a series of Mn(II) competition assays. We previously employed small-molecule Zn(II) sensors in competition assays to delineate the Zn(II)-binding properties of CP-Ser.48 We sought to apply this technique to Mn(II). Most commercially available small molecules that provide an optical response to Mn(II) utilize oxygen-rich chelating moieties and are calcium-sensitive (e.g. MagFura-2, Fura-2). We therefore selected ZP1, a Ca(II)-insensitive turn-on fluorescent Zn(II) sensor with relatively high background fluorescence (Φfree = 0.38 at pH 7.0),49 for Mn(II) competition assays. Paramagnetic first-row transition metal ions quench the background fluorescence of apo ZP1, and this fluorescence turn-off may be employed to monitor the paramagnetic metal ion of interest. Prior investigations reported that ZP1 gives ca. 8-fold fluorescence turn-off in the presence of excess Mn(II), and forms 1:1 and 2:1 Mn(II):ZP1 complexes with Kd1 = 550 nM and Kd2 = 2.2 μM (50 mM PIPES, 100 mM KCl, pH 7.0).57
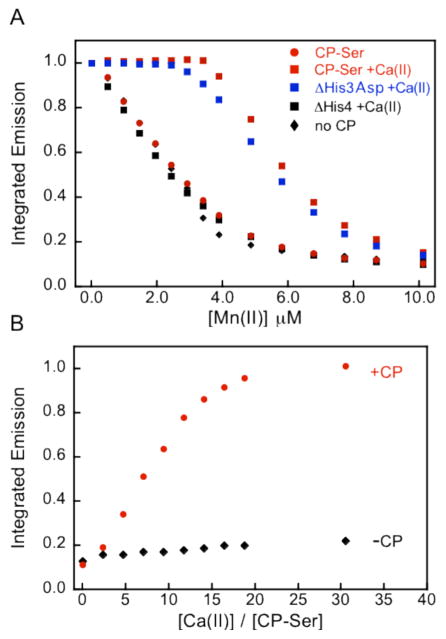
Titration of a 1:1 mixture of CP-Ser (αβ) and ZP1 with Mn(II) resulted in a titration curve comparable to that of the ZP1-only control (Figure 5), which demonstrated that the CP-Ser heterodimer cannot compete with ZP1 for Mn(II) under these conditions. This result suggested that the Kd value of CP-Ser for Mn(II) is > 550 nM, in agreement with the values determined by room-temperature EPR. As expected, ZP1 also outcompeted ΔHis3Asp and ΔHis4 for Mn(II). In contrast, titration of a 1:1 mixture of CP-Ser or ΔHis3Asp and ZP1 with Mn(II) in the presence of fifty-fold excess Ca(II) resulted in marked attenuation of the ZP1 response (Figure 5). Quenching of ZP1 emission was only observed after approximately one equivalent of Mn(II) per CP heterodimer was introduced into the cuvette, revealing that ZP1 cannot compete with the calcium-bound α2β2 form of CP-Ser or ΔHis3Asp for Mn(II). Calcium addition had no effect on the ΔHis4 titration curve. Taken together, these data provide convincing evidence that the Mn(II) affinity of the His4 site is calcium dependent. Calcium binding to the EF-hand domains of CP triggers enhanced Mn(II)-binding affinity as previously observed for Zn(II) chelation. The ZP1 data obtained in both the absence and presence of Ca(II) confirm the results obtained from RT-EPR titrations and support a Mn(II) Kd value for the His4 site that switches from > 550 nM to < 550 nM with calcium binding to the EF-hand domains. These experiments also demonstrate that Mn(II) does not coordinate to the EF-hand domains of CP with an appreciable affinity.
Figure 5.
Competition between ZP1 and CP for Mn(II) in the absence and presence of Ca(II) at pH 7.5 (75 mM HEPES, 100 mM NaCl) and 25 °C. (A) Titration of a 1:1 ratio of approximately 4 μM ZP1 and 4 μM CP with Mn(II) in the absence or presence of 200 μM Ca(II). Black diamonds, ZP1 only; black squares, CP-Ser ΔHis4 +Ca(II); red circles, CP-Ser; blue squares, ΔHis3Asp +Ca(II); red squares, CP-Ser +Ca(II). (B) Titration of a mixture containing 4 μM CP-Ser, 4 μM Mn(II), and 1 μM ZP1 with Ca(II). Excitation was provided at 490 nm and the emission spectra were integrated from 500–650 nm and normalized with respect to apo ZP1 emission (A) or maximum ZP1 emission (B).
To evaluate how many equivalents of Ca(II) are required for CP-Ser to fully sequester Mn(II) from Mn:ZP1, mixtures containing a 4:4:1 ratio of CP-Ser, Mn(II), and ZP1 were titrated with Ca(II) (Figure 5B). Maximum ZP1 fluorescence enhancement, indicating full dissociation of Mn(II) from ZP1, was observed following addition of approximately twenty equivalents of Ca(II) to the mixture.
To further evaluate the consequence of mutating a single His residue of the His4 site on Mn(II) coordination, ZP1 competition assays with the single point mutants CP-Ser H17A, H27A, H91A, H95A, and H27D using a 1:1 ZP1/CP mixture were performed (Figure S5). Variable degrees of competition between ZP1 and the mutant proteins were observed when 500 μM Ca(II) was added to the buffer. Some competition between each mutant and ZP1 was observed; however, no single mutant outcompeted ZP1 for Mn(II) as was observed for CP-Ser and ΔHis3Asp in the presence of Ca(II). A comparison of the titration curves indicated that H91A and H95A provided more competition with ZP1 for Mn(II) than did H17A and H27A. The H27D mutant exhibited similar behavior to the former two proteins. The single mutants retain the ability to coordinate Mn(II) at the mutated His4 site when Ca(II) is present, but do so with lower affinity than CP-Ser. The double mutant (H27A)(H91A) did not compete with ZP1 for Mn(II).
Low-Temperature Mn(II) EPR Spectroscopy of the His4 Site Supports Octahedral Coordination
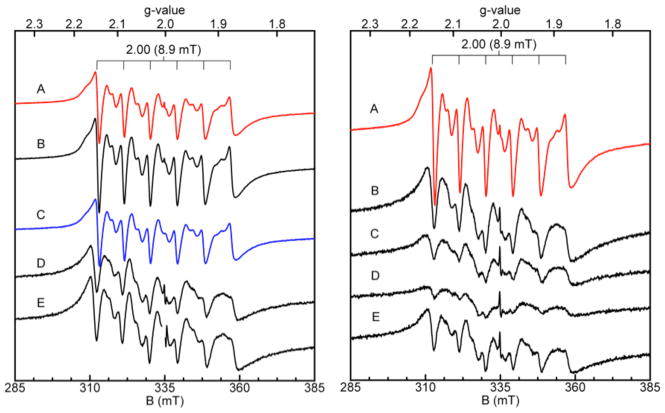
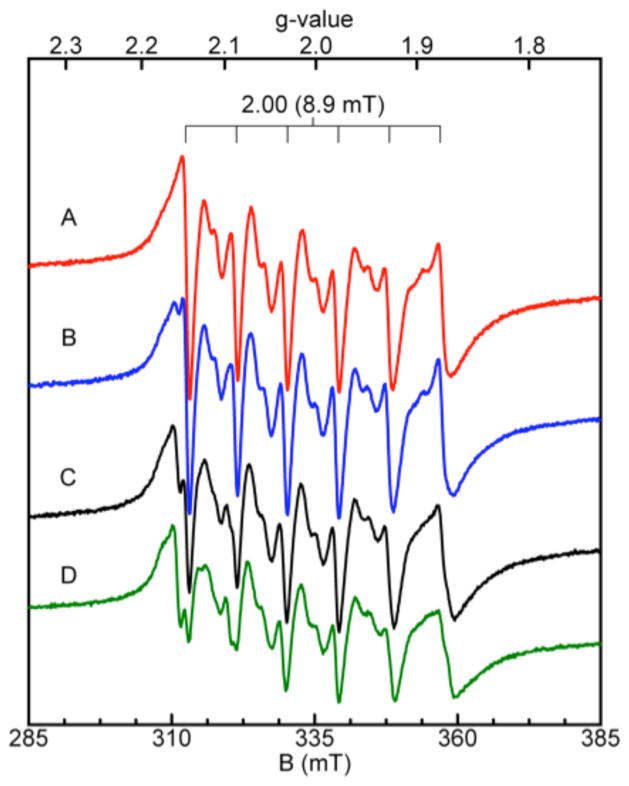
With the analytical SEC, room-temperature EPR, and ZP1 competition assays all demonstrating that the unusual interfacial His4 site of CP is the Mn(II) binding site, we sought to identify low-temperature EPR spectroscopic signals for the Mn(II):CP complex and thereby ascertain further details about the Mn(II) coordination sphere (i.e. coordination number, geometry). Samples containing approximately 100 μM CP-Ser or mutant were incubated with 0.3 equiv of Mn(II) for fifteen minutes and flash frozen in liquid N2 prior to the low-temperature EPR spectroscopic analysis (Figure 6). All spectra are dominated by a six-line pattern with a = 8.9 mT centered at g = 2, as expected for 55 Mn(II) systems (vide supra), and all observed resonances displayed Curie Law-dependence (intensity ~ 1/T). No significant signal intensity away from g = 2, which has been observed for some Mn(II)-containing proteins (e.g. Mn-HPCD,58 MnSOD,59 FosA60), was observed for any of the samples. The intensities and spectral line widths of the six allowed transitions for CP-Ser and ΔHis3Asp were indistinguishable. In contrast, the spectra for the ΔHis4 and ΔΔ samples exhibited significantly broader line widths and decreased signal intensities. These spectra closely resembled the spectrum obtained for Mn(II) in buffer (not shown). The sharper, more intense lines observed upon Mn(II) addition to CP-Ser and ΔHis3Asp may be explained by a more defined ligand field about the Mn(II) center upon coordination to the His4 site. Similar sharpening of the allowed transitions was observed previously for Mn(II) complexation to the hammerhead ribozyme.61 Moreover, a slight shift in the position of the allowed transitions is apparent for CP-Ser and ΔHis3Asp relative to ΔHis4 and ΔΔ. This phenomenon is most apparent in the first allowed transition, which is shifted by approximately 1.5 mT, and is not the result of instrumental artifacts (vide infra). Indeed, the first allowed transition of the CP-Ser sample exhibits a slight shoulder at g = 2.17, which corresponds to the positions of the first allowed transitions in ΔHis4 and ΔΔ.
Figure 6.
Low-temperature Mn(II) EPR spectroscopy of CP-Ser and mutant proteins (75 mM HEPES, 100 mM NaCl, pH 7.5). Left panel: EPR spectra of CP-Ser (A), CP-Ser with 0.6 M NaCl (B), ΔHis3Asp (C), ΔHis4 (D), and ΔΔ (E) each in the presence of 0.3 equiv of Mn(II). A radical signal (S = ½) was masked in E. The protein concentration was approximately 100 μM for each sample. The spectra obtained for the ΔHis4 and ΔΔ are scaled 4x. Right panel: EPR spectra of CP-Ser (A) and the single-point mutants H17A (B), H27A (C), H91A (D), and H95A (E) each in the presence of 0.3 equiv of Mn(II). The CP-Ser spectrum was recorded on a 100 μM sample incubated with 1 mM Ca(II) prior to addition of 20 μM Mn(II). Instrument conditions: temperature, 20 K; microwaves, 0.2 mW at 9.38 GHz; modulation amplitude, 0.5 mT.
Significant substructure, manifest as sets of doublets between each set of allowed transitions, is apparent in all spectra (Figure 6, Figure S6). These inter-doublets arise from formally forbidden transitions (Δms ± 1, ΔmI ± 1) that become “semi-forbidden.”50 The semi-forbidden transitions observed for CP-Ser and ΔHis3Asp are markedly similar and are unique from those observed for ΔHis4, ΔΔ, and Mn(II) in buffer. These differences are most apparent in the forbidden transitions F5 (1) and F5 (2) located between the fifth and six allowed transitions. Any single His→Ala mutation in the His4 site, in addition to the H27D mutation, resulted in a loss of these features (Figures 6 and S7). Moreover, these features were retained under conditions of high salt (Figures 6 and S6), and became more clearly resolved when CP-Ser was pre-incubated with excess Ca(II).
Figure 7 exhibits select spectra from low-temperature EPR titrations of CP-Ser with Mn(II) in the absence and presence of Ca(II), and a side-by-side comparison of these spectra clearly establishes the increased resolution and sharpening of the allowed and semi-forbidden transitions as a result of Ca(II) addition. In the absence of Ca(II), titration of CP-Ser with Mn(II) resulted in a progressive increase in the EPR signal intensity and broadening of the line widths. This trend is especially visible in the lowest field transition. When the spectra are scaled for the concentration of added Mn(II), the EPR intensity decreases with increasing Mn(II) concentration after 0.1 equivalent of Mn(II) is added (Figure S8). Curie Law-dependence was observed throughout the titration, which indicated that the decrease in signal intensity was not from formation of coupled Mn(II). This phenomenon may be explained by the increased metal ion concentration populating an additional and less intense species at higher Mn(II) equivalents. Guided by the ZP1 competition experiments and room-temperature EPR titrations (vide supra), we interpret the additional and less intense species as free or weakly associated Mn(II). In contrast, the Mn(II)-binding titration performed in the presence of Ca(II) results in population of a single, intense species from 0.1 to ~1 equivalent of Mn(II). The corresponding spectra overlay well with one another when scaled for the concentration of added metal (Figure S8), and no progressive line broadening was observed in this titration. The growth in intensity of the highest field resonance used to scale for the concentration of bound metal (see Experimental Section) saturated after one equivalent of added Mn(II), which supported a single tight binding site (Figure 7). A plot of bound Mn(II) ascertained by this approach versus equivalents of added Mn(II) could be fit with Kd values in the nanomolar range (Figure S9), which further supports nanomolar affinity Mn(II) coordination by the His4 site of calcium-bound CP.
Figure 7.
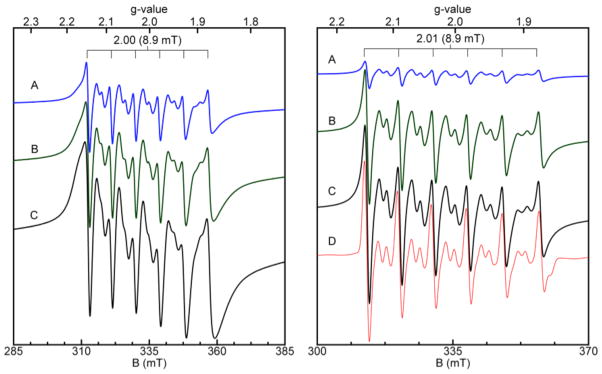
Low-temperature EPR titrations of CP-Ser with Mn(II) at pH 7.5 (75 mM HEPES, 100 mM NaCl) and in the absence (left panel) and presence (right panel) of 1 mM Ca(II). Left panel: Titration of 100 μM CP-Ser with 0.1 (A), 0.5 (B), and 1.0 (C) equivalents of Mn(II). Right panel: Titration of 115 μM CP-Ser with 0.1 (A), 0.5 (B), and 1.0 (C) equivalents of Mn(II) in the presence of Ca(II). Simulation (D) of X-band (9.3 GHz) EPR spectrum (black) of 110 μM CP-Ser with 60 μM Mn(II) in the presence of 1 mM Ca(II) (75 mM HEPES, 100 mM NaCl, pH 7.5). Simulation parameters: S = 5/2, I = 5/2, a = 8.9 mT, g = 2, D = 0.009, E/D = 0.33. The simulation could be fit with no distribution in the zero-field (D, E/D) parameters and was quantitative for the amount of Mn(II) added. Instrument conditions: temperature, 10 K; microwaves, 0.2 mW at 9.38 GHz; modulation amplitude, 0.5 mT.
To the best of our knowledge, this work provides the first low-temperature EPR spectroscopic signals of Mn(II) coordinated to a protein site based on a His4 motif. The EPR signals observed for Mn(II):CP-Ser and ΔHis3Asp are similar to those documented for structurally-characterized mononuclear Mn(II) complexes including [Mn(BPZG)(H2O)]NO3 (BPZG = N,N-bis(3,5-dimethylpyrazolylmethly)glycinate; octahedral N3O3 coordination)62 and Mn(bipy)(H2O)(TBD)] (bipy = 2,2′-bipyridine; heptacoordinate N2O5),63 in addition to Mn(II) in a single crystalline environment.64 The signals are also similar to those observed for the Mn(II)-bound hammerhead ribozyme (octahedral N1O5 coordination).61
Simulation of the X-band EPR spectrum of 110 μM CP-Ser with 60 μM Mn(II) and 1 mM Ca(II) afforded D = 270 MHz and E = 81 MHz (Figure 7, Table 2). This simulation is quantitative for the amount of Mn(II) added. Variations in the zero-field splittings of Mn(II) may be associated with different coordination environments; however, in some instances these associations are not robust,65,66 and qualitative statements about Mn(II) coordination geometry based on Mn(II) zerofield changes are complicated by the complex interactions, which are not fully understood.66–69 A tetragonal or trigonal distortion from cubic symmetry introduces a non-zero value for D whereas rhombic distortion increases E. The simulated zero-field splitting parameter values (Table 2) indicate that CP-Ser coordinates Mn(II) in a nearly idealized, slightly rhombically-distorted octahedral coordination sphere. The small zero-field splitting parameter D suggests a six coordinate rather than five coordinate environment around the Mn(II) center. Five-coordinate Mn(II) complexes typically display significantly higher D (e. g. D ~ 9000 MHz for five-coordinate and ~1500 MHz for six-coordinate). The origin of the very low zero-field splitting values obtained from the simulation of Mn(II):CP-Ser is currently unclear. Guided by the experiments conducted with the site 2 mutants, we propose that the Mn(II) coordination sphere consists of the four histidine residues of the His4 site (H17 and H27 of A8; H91 and H95 of A9) and two additional ligands. These unidentified ligands may be water molecules, buffer components, or derived from protein sidechains.
Table 2.
Reported Zero-Field Splitting Parameters for Mononuclear Mn(II) Determined by EPR.a
| Ligand | D (MHz) | E (MHz) | Ref |
|---|---|---|---|
| CP-Ser | 270 | 81 | This work |
| GMP | 420 | 100 | 50 |
| HHRz | 550–600 | 130 | 50 |
| ATP | 1050 | 300 | 50 |
| EDTA | 3000 | 300 | 50 |
| FosA | 3150 | 600 | 65 |
| FosA + fosfomycin | 7050 | 660 | 65 |
Additional zero-field splitting parameters are given in ref. 50.
CP Prefers to Coordinate Zn(II) at the His4 Site
In prior work, we rigorously established that CP utilizes both the His3Asp and His4 sites to coordinate Zn(II) with high affinity.70 The Zn(II) Kd values are calcium-dependent and span the nanomolar to picomolar range. A comparison of the Zn(II) Kd values to the Mn(II) Kd values presented in this work indicates that CP will select Zn(II) over Mn(II). This evaluation is in agreement with expectations based on the Irving-William series.70 To probe whether Zn(II) displaces Mn(II) from the coordination sphere provided by the His4 site, we monitored Mn(II) release from ΔHis3Asp by low-temperature EPR (Figure 8). A gradual disappearance of the semi-forbidden transitions located between the fifth and sixth lines, attributed to the Mn(II):ΔHis3Asp complex, was observed over the course of ca. 1 h. Moreover, the intensity of the first allowed transition diminished and a new feature at g = 2.17 formed. This observation is consistent with Zn(II)- induced loss of Mn(II) from the His4 coordination sphere, and growth of free Mn(II) at g = 2.17.
Figure 8.
Time course for the displacement of Mn(II) from the His4 site by Zn(II) addition at pH 7.5 (75 mM HEPES, 100 mM NaCl) monitored by low-temperature EPR spectroscopy. (A) 120 μM CP-Ser ΔHis3Asp upon addition of 0.33 equiv of Mn(II). (B) one equiv of Zn(II) was added to A and the sample was frozen right away. This sample was then thawed and allowed to incubate at room temperature for an additional 30 min (C) and 60 min(D). All spectra are scaled for the concentration of added Mn(II). Instrument conditions: temperature, 20 K; microwaves, 0.2 mW at 9.38 GHz; modulation amplitude, 0.5 mT.
We next evaluated the metal-ion selectivity of CP-Ser by using ZP1 as a reporter of free metal (Figure 9). The strategy was to add a total of three equivalents of metal to a CP-Ser/ZP1 mixture and monitor the fluorescence response of ZP1, which affords fluorescence turn-on when complexed with to Zn(II) and turn-off emission when coordinated to Mn(II), to determine the identity of the unbound metal ion (Figure 9). Addition of a mixture containing 1.5 equivalents of Mn(II) and 1.5 equivalents of Zn(II) to CP-Ser/ZP1 resulted in an immediate and slight enhancement of ZP1 emission, indicative of Zn(II) binding to ZP1. Over 100 min, however, the emission from ZP1 was reduced to below that of the apo probe attributed to Mn(II) complexation to ZP1. These data demonstrate that, after sufficient equilibration, CP-Ser sequesters all 1.5 equivalents of Zn(II), leaving only Mn(II) available to modulate the ZP1 response. Moreover, the results suggest that CP has the capacity coordinate both Zn(II) and Mn(I I) simultaneously.
Figure 9.

Metal-ion selectivity of CP-Ser. (A) Addition of 1 equivalent of Zn(II) or Mn(II) to 4 μM ZP1 at pH 7.5 (75 mM HEPES, 100 mM NaCl). Black line: emission from apo ZP1; blue line, emission enhancement observed after addition of Zn(II) to apo ZP1. Red line: quenching observed after addition of Mn(II) to apo ZP1. (B) Addition of a mixed Mn(II):Zn(II) solution to a solution containing 4 μM CP-Ser and 4 μM ZP1. Black line: emission from apo ZP1. Blue line: emission spectrum recorded immediately after addition of a mixture containing 1.5 equiv of Zn(II) and 1.5 equiv of Mn(II). Red line: emission spectrum of the same solution after 100 min. (C) Addition of a mixed Mn(II):Zn(II) solution to a solution containing 4 μM CP-Ser and 4 μM ZP1. In this case, one equivalent of Mn(II) and one equivalent of Zn(II) were added to apo ZP1. Black line: apo ZP1. Red line: addition of one equivalent of Zn(II) to ZP1. Blue line: addition of one equivalent of Mn(II) to ZP1. Green line: addition of one equivalent of Mn(II) and one equivalent of Zn(II) to ZP1. Excitation was provided at 490 nm and T = 25 °C. All samples contained 200 μM Ca(II).
CP Forms Mixed Zn(II):Mn(II) Complexes
We next aimed to delineate whether CP forms mixed Mn/Zn complexes. Based on the specificity of Mn(II) binding to the His4 site, we reasoned that formation of a complex where one Mn(II) ion is housed in the His4 site and one Zn(II) ion is bound to the His3Asp site can exist. Again, we took advantage of the turn-on and turn-off responses of ZP1 for Zn(II) and Mn(II), respectively, to probe this notion. Addition of one equivalent of Zn(II) and one equivalent of Mn(II) to a mixture of CP-Ser (α2β2) and ZP1 resulted in negligible change in ZP1 emission (Figure 9). In contrast, the no CP-Ser control provided ~3- fold fluorescence enhancement following Zn(II) addition and a ~4-fold fluorescence decrease following Mn(II) addition. The lack of ZP1 response to Mn(II) or Zn(II) in the presence of CP-Ser provides convincing evidence for formation of Zn:Mn:CP-Ser where Zn(II) is housed in the His3Asp and Mn(II) is bound in the His4 site.
Summary and Perspectives
Many investigations addressing calprotectin in infectious disease and other human pathologies have highlighted its metal-chelating ability.8,31,39,41,71–73 Nevertheless, the coordination chemistry of calprotectin, in addition to the molecular mechanisms by which calprotectin contributes to the homeostasis of transition metal ions, are largely unexplored. We have therefore initiated a research program designed to evaluate CP from the bioinorganic perspective. This work, motivated by the recently identified role of CP in the homeostasis of manganese,31,41 addresses the question of how CP coordinates Mn(II) and relates this propensity to its function in the host/pathogen interaction.
In particular, thermodynamic and spectroscopic methods allow us to establish that the unusual His4 site of human CP is essential for high-affinity Mn(II) coordination. Moreover, the techniques employed allow us to demonstrate that the Mn(II) affinity of the His4 site is calcium-dependent. This phenomenon is in agreement with recent studies of Zn(II) sequestration by CP.48 Here we reported that calcium coordination lowers the Mn(II) dissociation constant of the His4 site from the micromolar to the nanomolar range. The dissociation constants determined by room-temperature EPR and supported by the ZP1 competition and low-temperature EPR (vide infra) experiments may be compared to those reported and ascertained from a recent ITC analysis (Kd1 = 1.3 nM; Kd2 = 3.7 μM).41 Based on the isotherm obtained for the mutant protein lacking the metal-chelating residues of sites 1 and 2, which revealed no enthalpy change following Mn(II) addition, these Kd values were attributed to Mn(II) binding to each of the interfacial binding sites. The ITC-derived Kd values are each of similar magnitude (e.g. nanomolar, micromolar) to the Kd values determined in this work by room-temperature EPR and in the presence of Ca(II); however, the ITC and EPR/ZP1 titrations were conducted under markedly different conditions, which include buffer composition and calcium ion concentration.
The nanomolar Kd value for the His4 site in the presence of calcium is remarkable given that many characterized manganese-binding proteins, including enzymes and metal sensors, exhibit Mn(II) affinities in the micromolar range.74,75 MntR of Bacillus subtilis has an apparent Mn(II) Kd value of 160 μM,76 Likewise, the B. anthracis transcriptional regulator AntR coordinates Mn(II) with apparent Kd values of 210 and 16.6 μM.77 E. coli Fur chelates Mn(II) with a Kd value of 24 μM,78 and the manganese-dependent phosphatases PrpA and PrpB of Salmonella coordinates Mn(II) with dissociation constants of 65 and 1.3 μM, respectively.79 Moreover, the Mn(II) affinity of calcium-bound CP is within the range of Mn(II) affinities determined for metal ion transporter proteins expressed by human pathogens that include PsaA of Streptococcus pneumoniae (Kd = 3.3 nM)25 TroA of Treponema palladium (Kd = 7.1 nM)80 and YfeA of Yersinia pestis (Kd = 17.8 nM).81
The calcium-dependent Mn(II) affinity of CP has important physiological ramifications. Extracellular Ca(II) concentrations are several orders of magnitude greater than those in the intracellular space. We recently proposed that CP morphs into a potent Zn(II) chelator in the extracellular space as a result of calcium binding.48 The studies presented in this work demonstrate that this calcium-triggered enhancement of binding affinity applies to Mn(II) sequestration. The nanomolar Mn(II) affinity of the His4 site achieved in the presence of Ca(II) suggests that CP can compete with bacterial metal ion transporters for Mn(II), in agreement with the working model of CP-mediated host defense by transition metal deprivation. Indeed, reduction of MnSOD activity was observed following treatment of S. aureus with CP in vitro and attributed to extracellular Mn(II) sequestration.41 This working model is further supported by the high concentrations of CP at sites of infection, are reported to reach >500 μg/mL (ca. 20 μM).32
It is noteworthy that the His4 site of CP is unusual amongst S100 family members.45,82 To the best of our knowledge, CP the only S100 protein thus far reported to coordinate Mn(II). Moreover, characterized metalloproteins employing His4 motifs for coordinating transition metal ions are rare. Two structurally-characterized proteins that each exhibit mononuclear Mn(II) coordinated by a His4 motif are the photochemical reaction center from Rhodobacter spaeroides (PDB: 1YST)83 and a cupin from Thermotoga martima (PDB: 1VJ2)84 Both of these proteins exhibit six-coordinate Mn(II) where the coordination sphere is completed by two oxygen donor atoms from either a bidentate carboxylate sidechain or water molecules. The low-temperature EPR spectroscopic investigations described in this work support an octahedral coordination geometry for Mn(II) bound to the His4 site. This coordination number agrees with expectations based on chemical intuition. Moreover, recent studies of the Co(II)-bound CP suggested an octahedral geometry for Co(II) complex to the His4 site.48 At present, the identities of the additional ligands that complete the Mn(II) coordination sphere and the structural basis for the Ca(II)-triggered enhancement of Mn(II) affinity are unknown. Potential candidates for the unidentified ligands include water molecules and metal-chelating residues in the vicinity of the His4 site such as those found in the C-terminal tail of A9 (Figure 1B). Identification of the ligands completing the Mn(II) coordination sphere, in addition to the precise coordination geometry and further details of electronic structure, will require advanced EPR spectroscopic (e.g. ESEEM) and structural initiatives. Such investigations will also address additional unresolved and fascinating issues, which include understanding the molecular basis for why CP utilizes the His4 site instead of the His3Asp site for Mn(II) coordination, the details of the conformational change that occurs upon Mn(II) binding to the heterodimer, and the origins of the calcium-dependent Mn(II) affinity of the His4 site. We previously suggested that Ca(II) coordination by the EF-hand domains organizes the M(II) coordination spheres by causing conformational changes in the Ca(II) binding loops, because A8(H27) and A9(D30) are components of these loops. Calcium coordination also causes CP to oligomerize from a heterodimeric to heterotetrameric, and whether the heterotetrameric form is ultimately responsible for the enhanced Mn(II) affinity is currently unclear.
Investigations of S. aureus infection that employed a murine model provided the first in vivo evidence for a role of CP in Mn(II) homeostasis. The amino acid sequence alignment of S100A8 and S100A9 from human and mouse reveals that the metal-binding residues comprising sites 1 and 2 are conserved in both species (Figure 1B). Little is known about the structure and metal-binding properties of murine CP.85 It will be important to evaluate whether mouse CP exclusively utilizes a His4 site for high-affinity Mn(II) coordination or employs other tactics for sequestering this metal ion.
CP coordinates both Zn(II) and Mn(II) with high affinity. As a result, the question of the relevant metal ion arises. This issue is very complex and requires further investigation; however, we contend that the relevant metal ion is context specific. Metal ion availability and relative concentrations at the site of infection, and possibly in other biological circumstances, will likely dictate whether CP sequester Zn(II), Mn(II), or both. A similar concept of metal ion availability dictating protein (mis)metalation states is an emerging theme in metal ion homeostasis.74,86
The currently accepted model for CP-mediated growth inhibition focuses on metal-ion capture in the extracellular space. This work provides molecular-level insight into how CP coordinates Mn(II) and provides a basis for further chemical and biological investigations of the manganese-containing protein. The fate of metal-bound CP, and whether these forms have unidentified physiological functions related to the host/pathogen interaction or other phenomena, is unclear and also requires examination.
Supplementary Material
Acknowledgments
Financial support for this work was provided by the Searle Scholars Program (Kinship Foundation), the MIT Center for Environmental Health Sciences (NIH P30-ES002109), and the Department of Chemistry at MIT. We thank Professor Stephen J. Lippard for use of his atomic absorption spectrometer, Mr. Justin J. Wilson for assistance with the AA measurements, Dr. Jeff Simpson for assistance with the EPR spectrometer, Ms. Sumin Kim for assistance with the protein purification, Dr. Andrew J. Wommack for synthesizing ZP1, and Ms. Aleth Galliard de St. Germain for conducting the Mn(II) thermal denaturation experiments. We thank Professor Michael Hendrich for helpful discussions about the EPR spectroscopic data, and for generously providing the SpinCount simulation software. EPR instrumentation is housed in the Department of Chemistry Instrumentation Facility. Instrumentation for circular dichroism spectroscopy is provided by the MIT Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems, which is supported by grants NSF-0070319 and NIH GM68762.
References
- 1.Weinberg ED. JAMA. 1975;231:39–41. doi: 10.1001/jama.231.1.39. [DOI] [PubMed] [Google Scholar]
- 2.Wakeman CA, Skaar EP. Curr Opin Microbiol. 2012;15:169–174. doi: 10.1016/j.mib.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cassat JE, Skaar EP. Semin Immunopathol. 2012;34:215–235. doi: 10.1007/s00281-011-0294-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fischbach MA, Lin H, Liu DR, Walsh CT. Nat Chem Biol. 2006;2:132–138. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- 5.Miethke M, Marahiel MA. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crouch M-LV, Castor M, Karlinsey JE, Kalhorn T, Fang FC. Mol Microbiol. 2008;67:971–983. doi: 10.1111/j.1365-2958.2007.06089.x. [DOI] [PubMed] [Google Scholar]
- 7.Papp-Wallace KM, Maguire ME. Annu Rev Microbiol. 2006;60:187–209. doi: 10.1146/annurev.micro.60.080805.142149. [DOI] [PubMed] [Google Scholar]
- 8.Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, Restrepo NA, Hosking MP, Edwards RA, Battistoni A, Pasquali P, Lane TE, Chazin WJ, Vogl T, Roth J, Skaar EP, Raffatellu M. Cell Host Microbe. 2012;11:227–239. doi: 10.1016/j.chom.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ammendola S, Pasquali P, Pistoia C, Petrucci P, Petrarca P, Rotilio G, Battistoni A. Infect Immun. 2007;75:5867–5876. doi: 10.1128/IAI.00559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kehl-Fie TE, Skaar EP. Curr Opin Chem Biol. 2009;14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Débarbouillé M, Dramsi S, Dussurget O, Nahori M-A, Vaganay E, Jouvion G, Cozzone A, Msadek T, Duclos B. J Bacteriol. 2009;191:4070–4081. doi: 10.1128/JB.01813-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boal AK, Cotruvo JA, Jr, Stubbe J, Rosenzweig AC. Science. 2010;329:1526–1530. doi: 10.1126/science.1190187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chander M, Setlow B, Setlow P. Can J Microbiol. 1998;44:759–767. doi: 10.1139/cjm-44-8-759. [DOI] [PubMed] [Google Scholar]
- 14.Miller A-F. Curr Opin Chem Biol. 2004;8:162–168. doi: 10.1016/j.cbpa.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 15.Robbe-Saule V, Coynault C, Ibanez-Ruiz M, Hermant D, Norel F. Mol Microbiol. 2001;39:1533–1545. doi: 10.1046/j.1365-2958.2001.02340.x. [DOI] [PubMed] [Google Scholar]
- 16.Kehres DG, Maguire ME. FEMS Microbiol Rev. 2003;27:263–290. doi: 10.1016/S0168-6445(03)00052-4. [DOI] [PubMed] [Google Scholar]
- 17.Zaharik ML, Finlay BB. Frontiers Biosci. 2004;9:1035–1042. doi: 10.2741/1317. [DOI] [PubMed] [Google Scholar]
- 18.Wu H-J, Seib KL, Srikhanta YN, Edwards J, Kidd SP, Maguire TL, Hamilton A, Pan K-T, Hsiao H-H, Yao C-W, Grimmond SM, Apicella MA, McEwan AG, Wang AH-J, Jennings MP. J Proteomics. 2010;73:899–916. doi: 10.1016/j.jprot.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyer E, Bergevin I, Malo D, Gros P, Cellier MFM. Infect Immun. 2002;70:6032–6042. doi: 10.1128/IAI.70.11.6032-6042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston JW, Briles DE, Myers LE, Hollingshead SK. Infect Immun. 2006;74:1171–1180. doi: 10.1128/IAI.74.2.1171-1180.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng B, Zhang Q, Gao J, Han H, Li M, Zhang J, Qi J, Yan J, Gao GF. PLoS One. 2011;6:e19510. doi: 10.1371/journal.pone.0019510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schreur PJW, Rebel JMJ, Smits MA, van Putten JPM, Smith HE. J Bacteriol. 2011;193:5073–5080. doi: 10.1128/JB.05305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakubovics NS, Jenkinson HF. Microbiology. 2001;147:1709–1718. doi: 10.1099/00221287-147-7-1709. [DOI] [PubMed] [Google Scholar]
- 24.Ogunniyi AD, Mahdi LK, Jennings MP, McEwan AG, McDevitt CA, Van der Hoek MB, Bagley CJ, Hoffmann P, Gould KA, Paton JC. J Bacteriol. 2010;192:4489–4497. doi: 10.1128/JB.00064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDevitt CA, Ogunniyi AD, Valkov E, Lawrence MC, Kobe B, McEwan AG, Paton JC. PloS Pathog. 2011;7:e1002357. doi: 10.1371/journal.ppat.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston JW, Myers LE, Ochs MM, Benjamin WH, Jr, Briles DE, Hollingshead SK. Infect Immun. 2004;72:5858–5867. doi: 10.1128/IAI.72.10.5858-5867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tseng H-J, Srikhanta Y, McEwan AG, Jennings MP. Mol Microbiol. 2001;40:1175–1186. doi: 10.1046/j.1365-2958.2001.02460.x. [DOI] [PubMed] [Google Scholar]
- 28.Posey JE, Gherardini FC. Science. 2000;288:1651–1653. doi: 10.1126/science.288.5471.1651. [DOI] [PubMed] [Google Scholar]
- 29.Ouyang Z, He M, Oman T, Yang XF, Norgard MV. Proc Natl Acad Sci U S A. 2009;106:3449–3454. doi: 10.1073/pnas.0812999106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hood MI, Skaar EP. Nat Rev Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, Caprioli RM, Nacken W, Chazin WJ, Skaar EP. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 32.Johne B, Fagerhol MK, Lyberg T, Prydz H, Brandtzaeg P, Naess-Andresen CF, Dale I. J Clin Pathol: Mol Pathol. 1997;50:113–123. doi: 10.1136/mp.50.3.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nathan C. Nat Rev Immunol. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 34.Steinbakk M, Naess-Andresen CF, Lingaas E, Dale I, Brandtzaeg P, Fagerhol MK. Lancet. 1990;336:763–765. doi: 10.1016/0140-6736(90)93237-j. [DOI] [PubMed] [Google Scholar]
- 35.Sohnle PG, Collins-Lech C, Wiessner JH. J Infect Dis. 1991;164:137–142. doi: 10.1093/infdis/164.1.137. [DOI] [PubMed] [Google Scholar]
- 36.Clohessy PA, Golden BE. Scand J Immunol. 1995;42:551–556. doi: 10.1111/j.1365-3083.1995.tb03695.x. [DOI] [PubMed] [Google Scholar]
- 37.Loomans HJ, Hahn BL, Li Q-Q, Phadnis SH, Sohnle PG. J Infect Dis. 1998;177:812–814. doi: 10.1086/517816. [DOI] [PubMed] [Google Scholar]
- 38.Sohnle PG, Hunter MJ, Hahn B, Chazin WJ. J Infect Dis. 2000;182:1272–1275. doi: 10.1086/315810. [DOI] [PubMed] [Google Scholar]
- 39.Lulloff SJ, Hahn BL, Sohnle PG. J Lab Clin Med. 2004;144:208–214. doi: 10.1016/j.lab.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Vallee BL, Falchuk KH. Physiol Rev. 1993;73:79–118. doi: 10.1152/physrev.1993.73.1.79. [DOI] [PubMed] [Google Scholar]
- 41.Kehl-Fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, Garcia C, Munro KA, Chazin WJ, Skaar EP. Cell Host Microbe. 2011;10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunter MJ, Chazin WJ. J Biol Chem. 1998;273:12427–12435. doi: 10.1074/jbc.273.20.12427. [DOI] [PubMed] [Google Scholar]
- 43.Leukert N, Sorg C, Roth J. Biol Chem. 2005;386:429–434. doi: 10.1515/BC.2005.051. [DOI] [PubMed] [Google Scholar]
- 44.Leukert N, Vogl T, Strupat K, Reichelt R, Sorg C, Roth J. J Mol Biol. 2006;359:961–972. doi: 10.1016/j.jmb.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 45.Gifford JL, Walsh MP, Vogel HJ. Biochem J. 2007;405:199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- 46.Chazin WJ. Acc Chem Res. 2011;44:171–179. doi: 10.1021/ar100110d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korndörfer IP, Brueckner F, Skerra A. J Mol Biol. 2007;370(5):887–898. doi: 10.1016/j.jmb.2007.04.065. [DOI] [PubMed] [Google Scholar]
- 48.Brophy MB, Hayden JA, Nolan EM. J Am Chem Soc. 2012;134:18089–18100. doi: 10.1021/ja307974e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walkup GK, Burdette SC, Lippard SJ, Tsien RY. J Am Chem Soc. 2000;122:5644–5645. doi: 10.1021/ja010059l. [DOI] [PubMed] [Google Scholar]
- 50.Stich TA, Lahiri S, Yeagle G, Dicus M, Brynda M, Gunn A, Aznar C, DeRose VJ, Britt RD. App Magn Reson. 2007;31:321–341. doi: 10.1007/BF03166263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reed GH, Poyner RR. Metal Ions Biol Syst. 2000;37:183–207. [PubMed] [Google Scholar]
- 52.Reed GH, Cohn M. J Biol Chem. 1970;245:662–664. [PubMed] [Google Scholar]
- 53.Horton TE, Clardy DR, DeRose VJ. Biochemistry. 1998;37:18094–18101. doi: 10.1021/bi981425p. [DOI] [PubMed] [Google Scholar]
- 54.Hunsicker-Wang L, Vogt M, DeRose VJ. Method Enzymol. 2009;468:335–367. doi: 10.1016/S0076-6879(09)68016-2. [DOI] [PubMed] [Google Scholar]
- 55.Atanasijevic T, Zhang X-a, Lippard SJ, Jasanoff A. Inorg Chem. 2010;49:2589–2591. doi: 10.1021/ic100150e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Senguen FT, Grabarek Z. Biochemistry. 2012;51:6182–6194. doi: 10.1021/bi300698h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.You YM, Tomat E, Hwang K, Atanasijevic T, Nam W, Jasanoff AP, Lippard SJ. Chem Commun. 2010;46:4139–4141. doi: 10.1039/c0cc00179a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emerson JP, Kovaleva EG, Farquhar ER, Lipscomb JD, Que L., Jr Proc Natl Acad Sci U S A. 2008;105:7347–7352. doi: 10.1073/pnas.0711179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whittaker JW, Whittaker MM. J Am Chem Soc. 1991;113:5528–5540. [Google Scholar]
- 60.Smoukov SK, Telser J, Bernat BA, Rife CL, Armstrong RN, Hoffman BM. J Am Chem Soc. 2002;124:2318–2326. doi: 10.1021/ja012480f. [DOI] [PubMed] [Google Scholar]
- 61.Vogt M, Lahiri S, Hoogstraten CG, Britt RD, DeRose VJ. J Am Chem Soc. 2006;128:16764–16770. doi: 10.1021/ja057035p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scarpellini M, Gätjens J, Martin OJ, Kampf JW, Sherman SE, Pecoraro VL. Inorg Chem. 2008;47:3584–3593. doi: 10.1021/ic701953g. [DOI] [PubMed] [Google Scholar]
- 63.Tan XS, Xiang DF, Tang WX, Sun J. Polyhedron. 1997;16:689–694. [Google Scholar]
- 64.Misra SK. Physica B-Condensed Matter. 1994;203:193–200. [Google Scholar]
- 65.Walsby CJ, Telser J, Rigsby RE, Armstrong RN, Hoffman BM. J Am Chem Soc. 2005;127:8310–8319. doi: 10.1021/ja044094e. [DOI] [PubMed] [Google Scholar]
- 66.Duboc C, Collomb M-N, Neese F. Appl Magn Reson. 2010;37:229–245. [Google Scholar]
- 67.Duboc C, Ganyushin D, Sivalingam K, Collomb M-N, Neese F. J Phys Chem A. 2010;114:10750–10758. doi: 10.1021/jp107823s. [DOI] [PubMed] [Google Scholar]
- 68.Zein S, Neese F. J Phys Chem A. 2008;112:7976–7983. doi: 10.1021/jp804062a. [DOI] [PubMed] [Google Scholar]
- 69.Zein S, Duboc C, Lubitz W, Neese F. Inorg Chem. 2008;47:134–142. doi: 10.1021/ic701293n. [DOI] [PubMed] [Google Scholar]
- 70.Irving I, Williams RJP. Nature. 1948;162:746–747. [Google Scholar]
- 71.Nakatani Y, Yamazaki M, Chazin WJ, Yui S. Mediators Inflamm. 2005:280–292. doi: 10.1155/MI.2005.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sampson B, Fagerhol MK, Sunderkötter C, Golden BE, Richmond P, Klein N, Kovar IZ, Beattie JH, Wolska-Kusnierz B, Saito Y, Roth J. Lancet. 2002;360:1742–1745. doi: 10.1016/S0140-6736(02)11683-7. [DOI] [PubMed] [Google Scholar]
- 73.Yanamandra K, Alexeyev O, Zamotin V, Srivastava V, Shchukarev A, Brorsson A-C, Tartaglia GG, Vogl T, Kayed R, Wingsle G, Olsson J, Dobson CM, Bergh A, Elgh F, Morozova-Roche LA. PLoS One. 2009;4:e5562. doi: 10.1371/journal.pone.0005562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cotruvo JA, Jr, Stubbe J. Metallomics. 2012;4:1020–1036. doi: 10.1039/c2mt20142a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma Z, Jacobsen FE, Giedroc DP. Chem Rev. 2009;109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Golynskiy MV, Gunderson WA, Hendrich MP, Cohen SM. Biochemistry. 2006;45:15359–15372. doi: 10.1021/bi0607406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sen KI, Sienkiewicz A, Love JF, vander Spek JC, Fajer PG, Logan TM. Biochemistry. 2006;45:4295–4303. doi: 10.1021/bi052288g. [DOI] [PubMed] [Google Scholar]
- 78.Mills SA, Marletta MA. Biochemistry. 2005;44:13553–13559. doi: 10.1021/bi0507579. [DOI] [PubMed] [Google Scholar]
- 79.Shi L, Kehres DG, Maguire ME. J Bacteriol. 2001;183:7053–7057. doi: 10.1128/JB.183.24.7053-7057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Desrosiers DC, Sun YC, Zaidi AA, Eggers CH, Cox DL, Radolf JD. Mol Microbiol. 2007;65:137–152. doi: 10.1111/j.1365-2958.2007.05771.x. [DOI] [PubMed] [Google Scholar]
- 81.Desrosiers DC, Bearden SW, Mier I, Jr, Abney J, Paulley JT, Fetherston JD, Salazar JC, Radolf JD, Perry RD. Infect Immun. 2010;78:5163–5177. doi: 10.1128/IAI.00732-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ostendorp T, Diez J, Heizmann CW, Fritz G. Biochim Biophys Acta. 2011;1813:1083–1091. doi: 10.1016/j.bbamcr.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 83.Arnoux B, Gaucher J-F, Ducruix A, Reiss-Husson F. Acta Crystallogr D Biol Crystallogr. 1995;D51:368–379. doi: 10.1107/S0907444994013120. [DOI] [PubMed] [Google Scholar]
- 84.Jaroszewski L, et al. Proteins. 2004;56:611–614. doi: 10.1002/prot.20130. [DOI] [PubMed] [Google Scholar]
- 85.Vogl T, Gharibyan AL, Morozova-Roche LA. Int J Mol Sci. 2012;13:2893–2917. doi: 10.3390/ijms13032893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Naranuntarat A, Jensen LT, Pazicni S, Penner-Hahn JE, Culotta VC. J Biol Chem. 2009;284:22633–22640. doi: 10.1074/jbc.M109.026773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.