Abstract
A method for highly sensitive and rapid detection of Pseudomonas aeruginosa, based on magnetic enrichment and magnetic separation, is described in this paper. The magnetic nanoparticles (MNPs) were applied to adsorb genome DNA after the sample was lysed. The DNA binding MNPs were directly subjected to polymerase chain reaction (PCR) to amplify gyrB specific sequence of Pseudomonas aeruginosa. The biotin labeled PCR products were detected by chemiluminescence when they were successively incubated with the probes-modified MNPs and alkaline phosphatase (ALP) labeled streptavidin (SA). Agarose gel electrophoresis analyses approved the method of in situ PCR to be highly reliable. The factors which could affect the chemiluminiscence were studied in detail. The results showed that the MNPs of 400 nm in diameter are beneficial to the detection. The sequence length and the binding site of the probe with a target sequence have obvious effects on the detection. The optimal concentration of the probes, hybridization temperature and hybridization time were 10 μM, 60 ºC and 60 mins, respectively. The method of in situ PCR based on MNPs can greatly improve the utilization rate of the DNA template ultimately enhancing the detection sensitivity. Experiment results proved that the primer and probe had high specificity, and Pseudomonas aeruginosa was successfully detected with detection limits as low as 10 cfu/mL by this method, while the detection of a single Pseudomonas aeruginosa can also be achieved.
Keywords: Pseudomonas aeruginosa, Magnetic Enrichment, gyrB, In Situ PCR, Chemiluminescence.
Introduction
Nanotechnology has made rapid development in the past decades, opening new doors of its applications in various fileds. The magnetic nanoparticles (MNPs) possess high surface area and they can easily be controlled under the external magnetic field, which emphasizes their potential use in the detection of biological signals 1-4. For possessing many advantages such as high sensitivity, high specificity, low background, and the ease for quantitative analysis, etc., chemiluminescence is broadly applied in bioanalysis. The combined use of magnetic particles and chemiluminescence technology for the development of efficient biosensors for clinical purposes is an emerging trend 5-7.
Pseudomonas aeruginosa is one of the most extensive and serious problems in nosocomial infection which can infect wounds, fester otitis media and cause pneumonia, cystic fibrosis, sepsis, and other diseases 8-10. Pseudomonas aeruginosa infection is the main reason of septicaemia for patients who receive organ transplants 11. The study by Van der Waaij's showed that 10 to 100 cells of Pseudomonas aeruginosa can lead to gut colonization in patients admitted in intensive care units and immunosuppressed 12. It is also a primary cause of morbidity and mortality in patients with cystic fibrosis (CF), and early antibiotic treatment is crucial 9,13-16. Therefore, the development of rapid and sensitive technique for early detection of Pseudomonas aeruginosa is very important for effective treatment of those diseases.
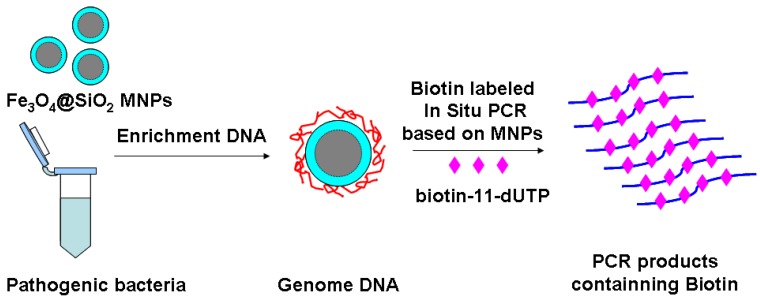
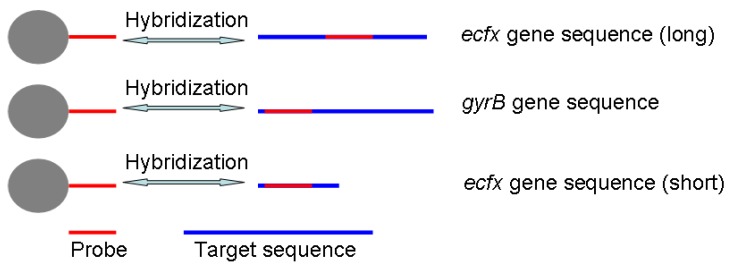
In this paper, we have concocted a simple method of in situ PCR based on MNPs. From the Fig. 1, the DNA binding Fe3O4@SiO2 MNPs were directly added into the polymerase chain reaction (PCR) system to amplify gyrB specific sequence of Pseudomonas aeruginosa, which can improve both the utilization rate of the DNA template and detection sensitivity. dTTP was partly replaced with biotin-labeled-dUTP. The carboxyl-modified MNPs were conjugated with amino-modified probes by incubation, and the probes-modified MNPs were used to capture the specific PCR products. These complexes were bonded with alkaline phosphatase (ALP) through the special coupling function between biotin and streptavidin. Furthermore, the chemiluminescent signal was detected by adding AMPPD which was the substrate reagent of ALP. Different primers and probes, based on ecfx gene and gyrB gene, were devised to detect Pseudomonas aeruginosa in order to study the effects of the product length and probe location on the chemiluminiscent signal intensity (Fig. 2).
Fig 1.
The schematic of magnetic enrichment and in situ PCR.
Fig 2.
The schematic of probe and prime design.
Materials and Methods
Materials
Pseudomonas aeruginosa strain (ATCC27853) was purchased from Huankai Microbial. Sci. & Tech. Co. Ltd. Salmonella enteritidis, Staphylouocus aureus, Shigella dysenteriae, and Escherichia coli were friendly donated by Dr. Ru Zhang of Hunan Institute of Engineering. The oligonucleotides were synthesized and HPLC purified by the Sangon Company (Table 1). 3-(2'-spiroadamantane)-4-methoxy-4-(3'-phosphoryloxy) phenyl-1,2-dioxetane (AMPPD) was purchased from the Biochem-ZX (China). Taq DNA polymerase and other polymerase chain reaction (PCR) reagents were obtained from ShangHai Biocolor BioScience Technology Company (China). Streptavidin-modified alkaline phosphatase (SA-AP) and PEG-4000 were ordered from the Sangon Company (China). The biotin-11-dUTP was purchased from the Fermentas (USA). Other unnamed reagents were domestic analytical reagents. The chemiluminescent intensity was detected using a multi-function micro hole plate reading machine Victor X3 (Perkin Elmer, USA). The PCR amplifier used in this experiment was ABI9700 (Applied Biosystems, USA).
Table 1.
The oligonucleotides used in this study.
| Target | Sequence Name | Sequence (5'-3') | Length | Reference |
|---|---|---|---|---|
| gyrB gene | gyrB-F | CCTGACCATCCGTCGCCACAAC | 222 bp | Qin,2003 Motoshima,2007 |
| gyrB-R | CGCAGCAGGATGCCGACGCC | |||
| gyrB-P | CCGTGGTGGTAGACCTGTTCCCAGACC- (15T)-NH2 | Anuj,2009 | ||
| ecfx gene | ecfx-F | TTCCGTGGTTCCGTCTCGC | This study | |
| ecfx-R-S | TCCGCCGAACTGCCCAGGTGC | 86 bp | This study | |
| ecfx-R-L | CAACAAGTTGCGGGCGATC | 195 bp | This study | |
| ecfx-P | AGCACCTGGGCAGTTCGGCGGA-(15T)-NH2 | This study | ||
| Universal gene | Uni-F | CGGTCCAGACTCCTACGGG | 206bp | Anuj,2009 |
| Uni-R | TTACCGCGGCTGCTGGCA |
Preparation of Fe3O4 MNPs and Fe3O4@SiO2 MNPs
Fe3O4 nanoparticles were prepared by the solvothermal method. 2.03 g of FeCl3·6H2O, 2 g of PEG-4000 and 4 g of NaAc were dissolved in ethylene glycol. The solution was placed in a reaction kettle and allowed to react for 12 h at 180 ºC. The obtained precipitate was washed with deionized water 3 times. The obtained nanoparticles were dispersed in 150 mL solution of ethanol/water (4:1) with the help of an ultrasonic. 3 mL of tetraethoxysilane (TEOS) and 4 mL of ammonia were added into the mixture and then the obtained mixture was stirred for 3 hours. Finally, Fe3O4@SiO2 MNPs were magnetically separated from the reactant mixture by applying an external magnetic field and washed. The amino-modification of Fe3O4@SiO2 MNPs was performed as follows. 150 mg of Fe3O4@SiO2 MNPs were separated by the external magnetic field and dispersed in 30 mL of ethanol/water (199:1) solution by the ultrasonic, and then 60 μL of 3-aminopropyl triethoxysilane (APTES) was added into this mixture. The solution was stirred for 7 h at room temperature. The product was separated from the reactant mixture by the external magnetic field, then washed with N,N-dimethylformamide (DMF) for 5 times. The amino-modified MNPs were dispersed in DMF to a concentration of 5 mg/mL. For carboxyl-modification, 15 mL of amino-modified MNPs solution prepared as above was dropped into equal volume of succinic anhydride solution; the mixture was stirred at 20 °C for 24 h. 7.5 mL of carboxyl-modified MNPs was then acquired by magnetic separation after washing with DI water for 3 times 17-21.
Bacterial Culture
Pseudomonas aeruginosa, Salmonella enteritidis, Staphylococcus aureus, Shigella dysenteriae and Escherichia coli were grown in a LB broth seperately. The bacteria were cultured for 24 h in an incubator at 37 °C, shaking at 180 rpm. The pathogens were inactivated for 48 h by 0.5 % of formaldehyde solution.
The Method of Pseudomonas aeruginosa Genomic DNA Extraction and in situ PCR Based on MNPs
100 µL of Pseudomonas aeruginosa sample, 20 µL of protease K and 100 µL of lysis solution were mixed in a 1.5 mL sterile centrifugal tube; the mixture was incubated for 20 min at 56 °C. 400 µg of Fe3O4@SiO2 MNPs were dispersed in 300 µL of binding buffer after lysis, the sample of Pseudomonas aeruginosa was added into the binding buffer. After 5 minutes, MNPs were separated from the mixture under an external magnetic field and cleaned twice. Taq DNA polymerase buffer, MgCl2, primer, dNTPs, sterile water and Taq DNA polymerase were successively added into the PCR tube and mixed with the Fe3O4@SiO2 MNPs bound with genomic DNA. PCR was conducted under the following thermocycling conditions: 95 °C for 5 min, followed by 35 cycles of 94 °C for 40 sec, 60 °C for 30 sec and 72 °C for 30 sec. A final extension step was continued for another 7 min at 72 °C. A tube containing the reaction mixture and sterile water was included in all reactions as a negative control. The PCR products were analysed by electrophoresis (1.5 % agarose gel) 22-24.
Detection of Pseudomonas aeruginosa Based on Chemiluminescence and Magnetic Separation
The probes was attached onto the surface of carboxyl-modified MNPs according to convalent binding 24. 90 μL of 10 μmol/L amino-modified probes diluted with pH 6.0 25 mmol/L MES and 60 μL of 5 mg/mL 1-Ethyl-3-(3-dimethyllaminopropyl) carbodiie hydrochlide (EDC) were mixed with 4.5 mg carboxyl-modified MNPs, incubated for 4 h at 4 ºC, and then the probes-modified MNPs were washed with pH 7.4 50 mmol/L Tris-HCl buffer for 4 times. At last the probes-modified MNPs were dispersed in 150 μL of PBS buffer. Per mg MNPs could bond to 120 pmol of probes if the reaction is completed. Li's method was followed for the detection of specific gyrB products with some slight modifications 22. 10 μL of probes-modified MNPs (10 mg/mL) were put into a PCR tube, then the supernatant was abandoned after magnetic separation. A 10 μL hybridization solution, 19.5 μL of deionized water and 1 μL gyrB PCR products (10 μL gyrB PCR products were used for sensitivity detection) were added in to the PCR tube. Universal primer PCR products were used as negative control and deionized water in place of the gyrB PCR products as a blank control. The mixture was hybridized at 60 °C for 1 hour, then it was washed for 3 min with 60 μL 2×SSC and cleaning solution (50 mM Tris, 0.15 M NaCl), respectively. The supernatant was abandoned after magnetic separation. The hybrid products were blocked for 30 min by cleaning solution containing 0.25 % of bovine serum albumin (BSA), then were incubated with 10 μL of ALP-SA for 30 min. Furthermore, the MNPs were cleaned twice with cleaning solution. Finally, the chemiluminescent intensity were detected by adding 100 μL of 0.25 mM AMPPD, which was the substrate reagent of AP. Each process was repeated 3 times. The response of chemiluminescence was studied for different concentration of MNPs, different concentration of probes, different hybridization temperatures and different hybrid times.
Specificity and Sensitivity Test
The specificity of gyrB primer and probe to Pseudomonas aeruginosa was tested using Salmonella enteritidis, Staphylouocus aureus, Shigella dysenteriae and Escherichia coli, acting as controls. The Pseudomonas aeruginosa was inactivated for 48 hour in 0.5 % formaldehyde, and bacteria were calculated by plate cultivation which is a classical method, 5 readings were recorded each time. The bacterial concentration was diluted to 10 cfu/mL, 100 cfu/mL and 1000 cfu/mL. 100 μL of the above samples were used in every test. Each experiment or treatment was repeated 5 times.
Data Analysis
All of the data were expressed as mean ± SD (Standard Deviations). Student's t-test was used for comparing the chosen level of statistical significance. The data analysis was made using SPSS 13.0 software.
Results and Discussion
Genomic DNA Extraction and In Situ PCR Based on MNPs
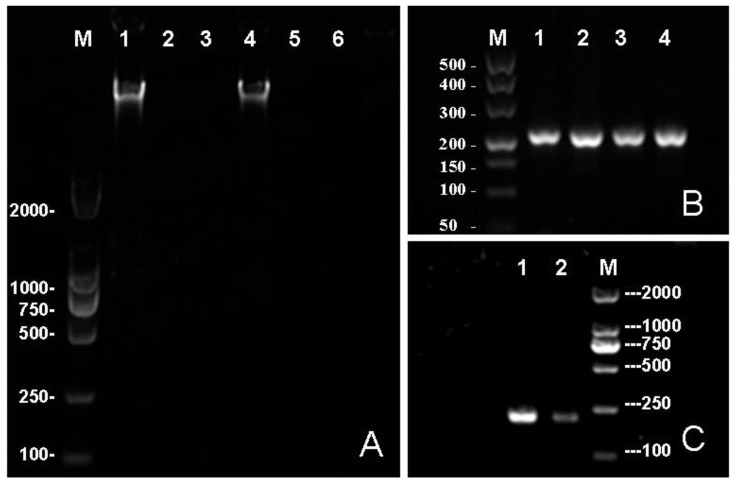
Because of the needs for nucleic acid automated testing and the characteristics of MNPs to help achieving the objective of automation, the genomic DNA extraction technology has rapidly improved in recent years 25. Many researchers have explored the potential use of different magnetic nanoparticles for nucleic acid extraction, as well as the improvement of the extraction process 26-28. However, the studies on the direct use of MNPs combined with genome DNA in in situ PCR are still rare. The automated testing of pathogens will be more convenient and the detection will be more sensitive if an in situ PCR based on the DNA binding MNPs can be developed. In this paper, we have described a method of genomic DNA enrichment based on the Fe3O4@SiO2 MNPs and an in situ PCR which featured the direct use of DNA bound at the surface of MNPs as a DNA template. The in situ PCR is based on the purification of genomic DNA acquired by enrichment of Fe3O4@SiO2 MNPs (Shown in Fig. 3A) and the ability of deionized water to elute the genomic DNA from Fe3O4@SiO2 MNPs at 65 ºC. In clinical samples, the quantity of the pathogenic bacteria is usually very low. The detection results may present a false negative if the utilization of DNA templates is too low. Most DNA templates (Which bonded with Fe3O4@SiO2 MNPs) were used for amplification in in situ PCR, therefore, theoretically, its sensitivity should be better than traditional PCR which usually uses few DNA templates. The results were shown in Fig. 3B-C, demonstrating our method can amplify sufficient PCR products for detection, and the efficiency of amplification is greater than the traditional PCR.
Fig 3.
The electrophoresis results. (A) The effects of different elution buffers to elute the genome DNA from MNPs. The TE buffer was used in the 1st, 2nd and 3rd lane while deionized water was used in the 4th, 5th and 6th lane. primary culture of bacterium was loaded in the 1st and 4th well while the 10 time and 100 time diluted samples were loaded in the 2nd and 5th, 4th and 6th well respectively. (B) An in situ PCR for gyrB based on Fe3O4@SiO2 MNPs: 1st, 2nd, 3rd and 4th lanes all are gyrB PCR products. (C) Lane 1 contains the results of in situ PCR; lane 2 contains the traditional PCR. In this study, the concentration of agrose gel was 1.0% in A and 1.5% in B and C.
The Detection of Pseudomonas aeruginosa Based on Chemiluminescence and Magnetic Separation
AMPPD is a safe and stable chemiluminescence reagent acting as the substrate for ALP 29. In this study, the carboxyl-modified MNPs were conjugated with amino-modified gyrB probes or amino-modified ecfx probes and then applied for the detection of Pseudomonas aeruginosa. The biotin-dUTP-labeled DNA fragments were amplified by in situ PCR. After the probes-modified MNPs were hybridized with gyrB gene fragments, SA-ALP was added. A universal bacterial primer pair was used as negative control. The chemiluminescent intensity was detected by adding AMPPD as the substrate.
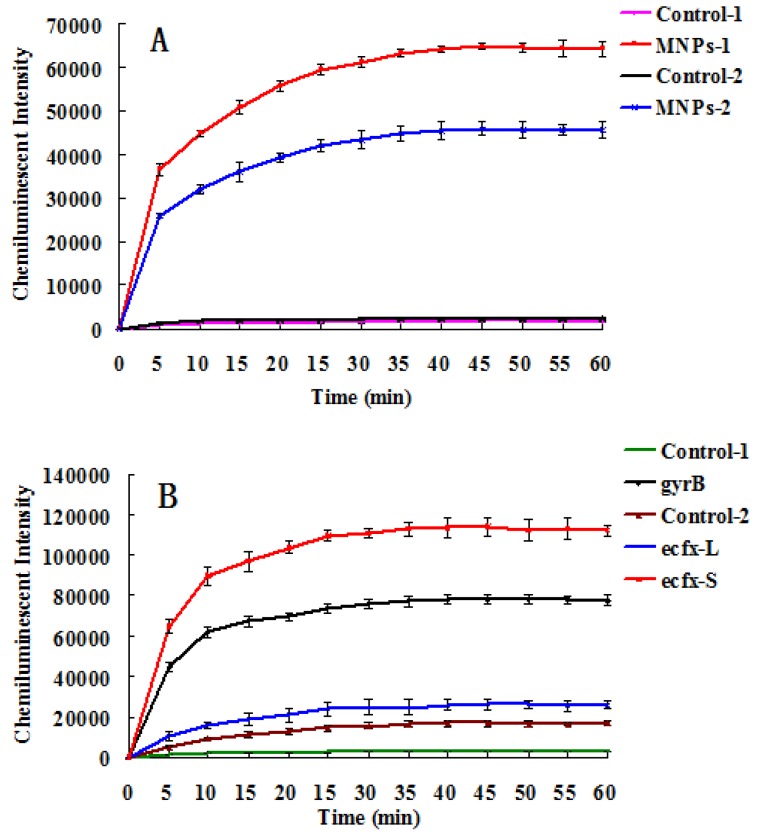
Fig. 4A indicates that the chemiluminescent intensity of the sample was very strong in comparison with the control. The chemiluminescent intensity reached maximum after 30~40 mins, and then became stable. The MNPs with a diameter of 400 nm performed better than the smaller ones for detection of gyrB gene fragments of Pseudomonas aeruginosa. The reason is that bigger MNPs have a higher recovery rate and less steric hindrance. Different primers and probes were devised to detect Pseudomonas aeruginosa based on ecfx gene and gyrB gene. The short ecfx sequences and gyrB probes, complementary with the upstream sequence of targets, gave higher chemiluminescent intensity than the long ecfx sequence probes complementary with the middle sequence of the target (Fig. 2 and Fig. 4B). Owing to its shorter length, the short ecfx sequence had higher efficiency than gyrB sequence. Therefore, the sequence length and the binding site of probe clearly have effects on the detection of Pseudomonas aeruginosa since they are important factors of hybridization. The gyrB sequence was chosen in the following study because the specificity of the short ecfx sequence was very poor.
Fig 4.
The chemiluminescent detection of Pseudomonas aeruginosa based on gyrB gene. (A) Effect of MNPs size on chemiluminescent intensity. MNPs-1: 400 nm sized probes modified MNPs, used to capture PCR products of gyrB gene. Control-1: the probes modified MNPs that captured the PCR products of bacterial universal gene. MNPs-2: 250 nm sized probes modified MNPs were used to capture PCR products of gyrB gene. Control-2: the probes modified MNPs that captured the PCR products of bacterial universal gene. (B) The effects of different binding sites of probes and different lengths of the target sequence on chemiluminescence intensity (Figure 2). 400 nm sized probes modified MNPswere used. gyrB: The gyrB probes were used to capture PCR products of gyrB gene. Control-1: The gyrB probes that captured PCR products of bacterial universal gene. ecfx-S: The ecfx probes were used to capture short PCR products of ecfx gene. ecfx-L: The ecfx probes were used to capture long PCR products of ecfx gene. Control-2: the ecfx probes that captured PCR products of bacterial universal gene.
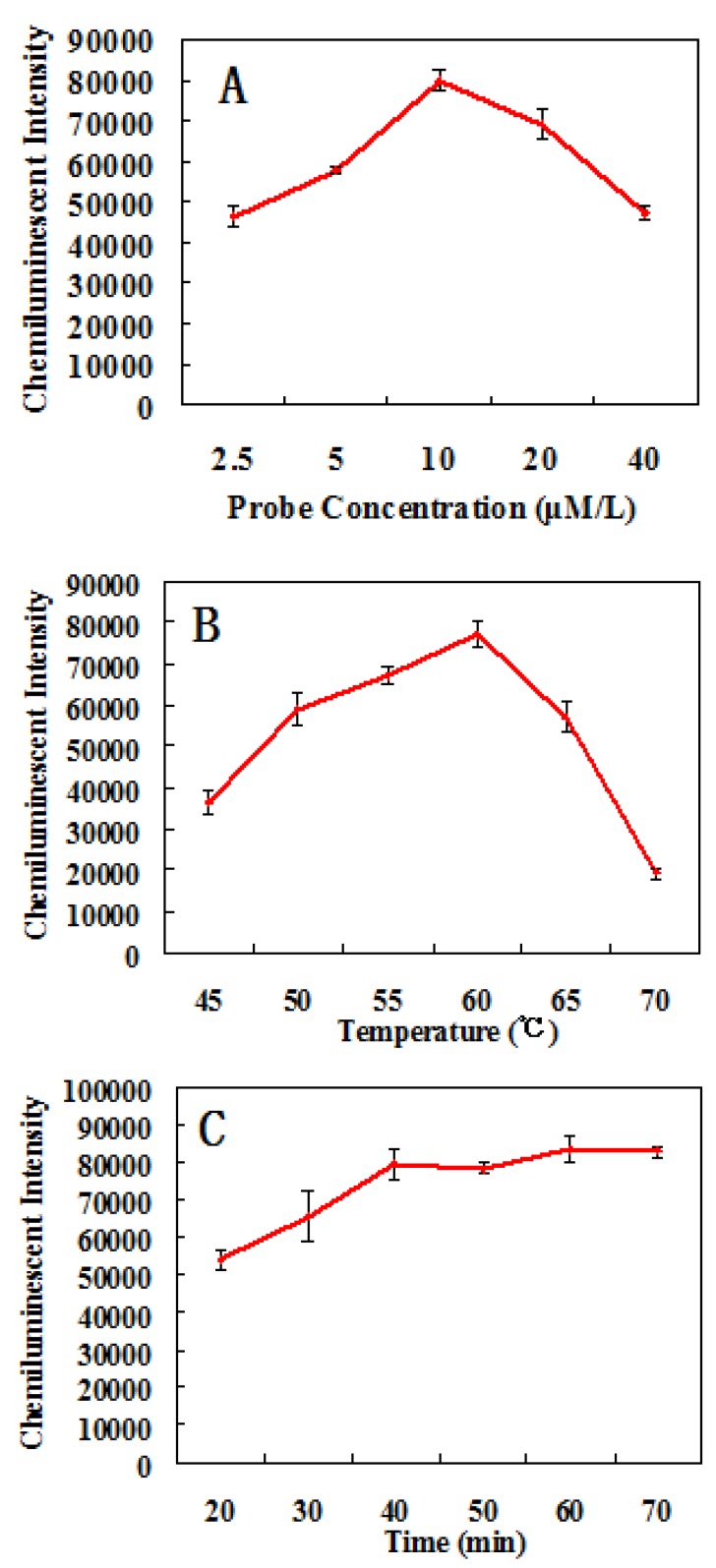
In order to obtain the optimal assay conditions, the effects of different factors on the detection of Pseudomonas aeruginosa were studied. The results shown in Fig. 5 indicates that the optimal concentration of probes is 10 μM/L, the optimal hybridization temperature is 60 ºC, and the optimal hybridization time is 60 mins.
Fig 5.
The effects of different conditions on the chemiluminescent detection of Pseudomonas aeruginosa. (A) Relationship between probes concentration and chemiluminescent intensity. (B) Relationship between hybridization temperature and chemiluminescent intensity. (C) Relationship between hybridization time and chemiluminescent intensity.
The Test of Specificity and Sensitivity
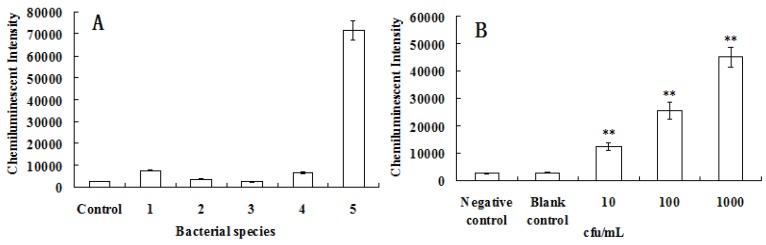
The specificity was largely determined by primer and probe choice. Qin et al studied the specificity of gyrB gene primer and probe used in this study, and no unspecific amplification was found 30-32. The specificity of this method for the detection of Pseudomonas aeruginosa was tested, using Salmonella enteritidis, Staphylouocus aureus, Shigella dysenteriae and Escherichia coli as controls. As shown in Fig. 6A, the primer and probe of gyrB had high specificity.
Fig 6.
The specificity and sensitivity analyses of Pseudomonas aeruginosa detection based on the chemiluminescence. (A) The specificity test of gyrB gene for detection of chemiluminescence. 1. Staphylococcus aureus, 2. Salmonella enteritidis, 3. Shigella dysenteriae, 4. Escherichia coli, 5. Pseudomonas aeruginosa. PCR products of bacterial universal gene were used as the control. (B) The sensitivity was tested after the Pseudomonas aeruginosans were diluted to about 10 cfu/mL, 100 cfu/mL and 1000 cfu/mL. 100 μL of above samples were used in every test. The sterile water was used as the blank control. PCR products of bacterial universal gene were used as the negative control. **, P<0.01 versus control.
For clinical samples, the sensitivity of a test for detecting pathogens is very important 33. Usually, the sensitivity can be influenced by many factors, such as the sampling procedure, the DNA-extraction method or the PCR procedure 13,34. In this paper, in situ PCR based on MNPs was introduced greatly improving detection sensitivity. In addition, the nucleic acid signal was further amplified since the PCR products were labeled with ALP which produced a chemiluminescent signal when reacting with AMPPD. To test the sensitivity of the system for detecting gyrB of Pseudomonas aeruginosa, the Pseudomonas aeruginosa samples were diluted to different concentration, the results are presented in Fig. 6B. It was clearly shown that the chemiluminescent intensity was different than that of the control (P<0.01 versus control) even if the concentration of Pseudomonas aeruginosa is about 10 cfu/mL. Indeed, only 100 μL of Pseudomonas aeruginosa samples were used in every test, so theoretically only single Pseudomonas aeruginosa was checked while the detection results were very positive. Although nanoparticles have many applications in the biomedical field 35-50, we believe the present method has demonstrated the lowest detection limit for Pseudomonas aeruginosa and is a very promising approach.
Conclusion
The development of highly sensitive techniques for early detection of Pseudomonas aeruginosa is an important consideration of medical research 51,52. Herein, we introduced an in situ PCR based on MNPs and a method of detection based on chemiluminiscence and magnetic separation. The parameters for detection were optimized in this method. Finally, a highly sensitive and rapid detection method for Pseudomonas aeruginosa based on magnetic enrichment and magnetic separation has been developed with detection limits as low as about 10 cfu/mL, while the detection of a single Pseudomonas aeruginosa is also recognized. Automation of pathogen detection is a developing trend. The DNA enrichment and in situ PCR based on MNPs can simplify the process of detection, making automation more convenient.
Acknowledgments
We acknowledge the financial supports of the National Key Special Science Program (NO. 2009ZX10004-311), the National Key Program for Developing Basic Research (NO. 2010CB933903), the National Natural Science Foundation of China (NO. 61271056, 60927001 and 61201033), Hunan Science and Technology Projects (2012SK3105 and 2010SK2003), Scientific Research Fund of Hunan Provincial Education Department (11A030), China Postdoctoral Science Foundation (2011M500841, 2012M520980 and 2012M511660), and Natural Science Foundation of Hunan Province (12JJ6060 and 12JJ9038). We are thankful to Jonathan Louis De Wees Foy for revising the English of this manuscript.
References
- 1.Child HW, Del PP, De La Fuente JM. et al. Working together: the combined application of a magnetic field and penetratin for the delivery of magnetic nanoparticles to cells in 3D. ACS Nano. 2011;5:7910–9. doi: 10.1021/nn202163v. [DOI] [PubMed] [Google Scholar]
- 2.Park KS, Kim MI, Cho DY, Park HG. Label-free colorimetric detection of nucleic acids based on target-induced shielding against the peroxidase-mimicking activity of magnetic nanoparticles. Small. 2011;7:1521–5. doi: 10.1002/smll.201001886. [DOI] [PubMed] [Google Scholar]
- 3.Fornara A, Johansson P, Petersson K. et al. Tailored magnetic nanoparticles for direct and sensitive detection of biomolecules in biological samples. Nano Lett. 2008;8:3423–8. doi: 10.1021/nl8022498. [DOI] [PubMed] [Google Scholar]
- 4.Sponarova D, Horak D, Trchova M. et al. The use of oligoperoxide-coated magnetic nanoparticles to label stem cells. J Biomed Nanotechnol. 2011;7:384–94. doi: 10.1166/jbn.2011.1289. [DOI] [PubMed] [Google Scholar]
- 5.Liu H, Li S, Tian L, Liu L, He N. A novel single nucleotide polymorphisms detection sensors based on magnetic nanoparticles array and dual-color single base extension. J Nanosci Nanotechnol. 2010;10:5311–5. doi: 10.1166/jnn.2010.2386. [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Li S, Wang Z. et al. High-throughput SNP genotyping based on solid-phase PCR on magnetic nanoparticles with dual-color hybridization. J Biotechnol. 2007;131:217–22. doi: 10.1016/j.jbiotec.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 7.Liu H, Li S, Ji M. et al. Fabrication and application of single nucleotide polymorphisms library on magnetic nanoparticles using adaptor PCR. J Nanosci Nanotechnol. 2008;8:405–9. [PubMed] [Google Scholar]
- 8.Lee CS, Wetzel K, Buckley T, Wozniak D, Lee J. Rapid and sensitive detection of Pseudomonas aeruginosa in chlorinated water and aerosols targeting gyrB gene using real-time PCR. J Appl Microbiol. 2011;111:893–903. doi: 10.1111/j.1365-2672.2011.05107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cattoir V, Gilibert A, Le Glaunec JM. et al. Rapid detection of Pseudomonas aeruginosa from positive blood cultures by quantitative PCR. Ann Clin Microbiol Antimicrob. 2010;9:21. doi: 10.1186/1476-0711-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Dou H, Chen K, Zhang H, Hu C. Development of a colloidal gold-based immunochromatographic test strip for the rapid, on-site detection of Pseudomonas aeruginosa in clinical samples. Scand J Infect Dis. 2011;43:329–38. doi: 10.3109/00365548.2011.552519. [DOI] [PubMed] [Google Scholar]
- 11.Khan AA, Cerniglia CE. Detection of Pseudomonas aeruginosa from clinical and environmental samples by amplification of the exotoxin A gene using PCR. Appl Environ Microbiol. 1994;60:3739–45. doi: 10.1128/aem.60.10.3739-3745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Waaij D. Colonization resistance of the digestive tract: clinical consequences and implications. J Antimicrob Chemother. 1982;10:263–70. doi: 10.1093/jac/10.4.263. [DOI] [PubMed] [Google Scholar]
- 13.Deschaght P, Van Daele S, De Baets F, Vaneechoutte M. PCR and the detection of Pseudomonas aeruginosa in respiratory samples of CF patients. A literature review. J Cyst Fibros. 2011;10:293–7. doi: 10.1016/j.jcf.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Liu C, Zeng GM, Tang L. et al. Electrochemical detection of Pseudomonas aeruginosa 16S rRNA using a biosensor based on immobilized stem-loop structured probe. Enzyme Microb Technol. 2011;49:266–71. doi: 10.1016/j.enzmictec.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Wang KY, Zeng YL, Yang XY, Li WB, Lan XP. Utility of aptamer-fluorescence in situ hybridization for rapid detection of Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis. 2011;30:273–8. doi: 10.1007/s10096-010-1074-0. [DOI] [PubMed] [Google Scholar]
- 16.Rao AR, Laxova A, Farrell PM, Barbieri JT. Proteomic identification of OprL as a seromarker for initial diagnosis of Pseudomonas aeruginosa infection of patients with cystic fibrosis. J Clin Microbiol. 2009;47:2483–8. doi: 10.1128/JCM.02182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun H, Zeng X, Liu M. et al. Synthesis of size-controlled Fe3O4@SiO2 magnetic nanoparticles for nucleic acid analysis. J Nanosci Nanotechnol. 2012;12:267–73. doi: 10.1166/jnn.2012.5170. [DOI] [PubMed] [Google Scholar]
- 18.Lee SJ, Lee HJ, Moon MJ. et al. Superparamagnetic iron oxide nanoparticles-loaded polymersome-mediated gene delivery guided by enhanced magnetic resonance signal. J Nanosci Nanotechnol. 2011;11:7057–60. doi: 10.1166/jnn.2011.4861. [DOI] [PubMed] [Google Scholar]
- 19.Saiyed ZM, Ramchand CN, Telang SD. Isolation of genomic DNA using magnetic nanoparticles as a solid-phase support. J Phys Condens Matter. 2008;20:1–5. doi: 10.1088/0953-8984/20/20/204153. [DOI] [PubMed] [Google Scholar]
- 20.Ma C, Li C, He L. et al. Preparation and characterization of monodisperse core-shell Fe3O4@SiO2 micropheres and its application for magnetic separation of nucleic acids from E.coil BL21. J Biomed Nanotechnol. 2012;6:1000–5. doi: 10.1166/jbn.2012.1454. [DOI] [PubMed] [Google Scholar]
- 21.Puri S, Mukhopadhyay G. Scattering and absorption properties of multiply coated magnetic nanoparticles. J Nanosci Nanotechnol. 2009;9:5177–82. doi: 10.1166/jnn.2009.1147. [DOI] [PubMed] [Google Scholar]
- 22.Tang Y, Li Z, He N. et al. Preparation of Functional Magnetic Nanoparticles Mediated with PEG-4000 and Application in Pseudomonas Aeruginosa Rapid Detection. J Biomed Nanotechnol. 2013;2:312–7. doi: 10.1166/jbn.2013.1493. [DOI] [PubMed] [Google Scholar]
- 23.Liu H, Li S, Wang Z. et al. PCR amplification on magnetic nanoparticles: application for high-throughput single nucleotide polymorphism genotyping. Biotechnol J. 2007;2:508–11. doi: 10.1002/biot.200600214. [DOI] [PubMed] [Google Scholar]
- 24.Li Z, He L, He N. et al. Polymerase chain reaction coupling with magnetic nanoparticles-based biotin-avidin system for amplification of chemiluminescent detection signals of nucleic acid. J Nanosci Nanotechnol. 2011;11:1074–8. doi: 10.1166/jnn.2011.3061. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Jiang L, Zhang C. et al. A silicon dioxide modified magnetic nanoparticles-labeled lateral flow strips for HBs antigen. J Biomed Nanotechnol. 2011;7:776–81. doi: 10.1166/jbn.2011.1352. [DOI] [PubMed] [Google Scholar]
- 26.Kang K, Choi J, Nam JH. et al. Preparation and characterization of chemically functionalized silica-coated magnetic nanoparticles as a DNA separator. J Phys Chem B. 2009;113:536–43. doi: 10.1021/jp807081b. [DOI] [PubMed] [Google Scholar]
- 27.Intorasoot S, Srirung R, Intorasoot A, Ngamratanapaiboon S. Application of gelatin-coated magnetic particles for isolation of genomic DNA from bacterial cells. Anal Biochem. 2009;386:291–2. doi: 10.1016/j.ab.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 28.Rittich B, Spanova A, Horak D. et al. Isolation of microbial DNA by newly designed magnetic particles. Colloids Surf B Biointerfaces. 2006;52:143–8. doi: 10.1016/j.colsurfb.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 29.Bronstein I, Fortin J, Voyta JC, Kricka LJ. Nitro-Block enhancement of AMPPD chemiluminescent signal in the detection of DNA. Biotechniques. 1992;12:500–2. [PubMed] [Google Scholar]
- 30.Qin X, Emerson J, Stapp J. et al. Use of real-time PCR with multiple targets to identify Pseudomonas aeruginosa and other nonfermenting gram-negative bacilli from patients with cystic fibrosis. J Clin Microbiol. 2003;41:4312–7. doi: 10.1128/JCM.41.9.4312-4317.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motoshima M, Yanagihara K, Fukushima K. et al. Rapid and accurate detection of Pseudomonas aeruginosa by real-time polymerase chain reaction with melting curve analysis targeting gyrB gene. Diagn Microbiol Infect Dis. 2007;58:53–8. doi: 10.1016/j.diagmicrobio.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Anuj SN, Whiley DM, Kidd TJ. et al. Identification of Pseudomonas aeruginosa by a duplex real-time polymerase chain reaction assay targeting the ecfX and the gyrB genes. Diagn Microbiol Infect Dis. 2009;63:127–31. doi: 10.1016/j.diagmicrobio.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 33.West SE, Zeng L, Lee BL. et al. Respiratory infections with Pseudomonas aeruginosa in children with cystic fibrosis: early detection by serology and assessment of risk factors. JAMA. 2002;287:2958–67. doi: 10.1001/jama.287.22.2958. [DOI] [PubMed] [Google Scholar]
- 34.Goto M, Shimada K, Sato A. et al. Rapid detection of Pseudomonas aeruginosa in mouse feces by colorimetric loop-mediated isothermal amplification. J Microbiol Methods. 2010;81:247–52. doi: 10.1016/j.mimet.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Ruan J, Song H, Li C, Bao C, Fu H, Wang K, Ni J, Cui D. DiR-labeled Embryonic Stem Cells for Targeted Imaging of in vivo Gastric Cancer Cells. Theranostics. 2012;2:618–28. doi: 10.7150/thno.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh R, Nalwa HS. Medical Applications of Nanoparticles in Biological Imaging, Cell Labeling, Antimicrobial Agents, and Anticancer Nanodrugs. J Biomed Nanotechnol. 2011;7:489–503. doi: 10.1166/jbn.2011.1324. [DOI] [PubMed] [Google Scholar]
- 37.Chen X. Integrin Targeted Imaging and Therapy. Theranostics. 2011;1:28–9. doi: 10.7150/thno/v01p0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao X, Kan B, Gou M. et al. Preparation of Anti-CD40 Antibody Modified Magnetic PCL-PEG-PCL Microspheres. J Biomed Nanotechnol. 2011;7:285–91. doi: 10.1166/jbn.2011.1280. [DOI] [PubMed] [Google Scholar]
- 39.Zhang F, Zhu L, Liu G. et al. Multimodality Imaging of Tumor Response to Doxil. Theranostics. 2011;1:302–9. doi: 10.7150/thno/v01p0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naresh M, Sharma M, Mittal A. Intracellular Magneto-Spatial Organization of Magnetic Organelles inside Intact Bacterial Cells. J Biomed Nanotechnol. 2011;7:572–7. doi: 10.1166/jbn.2011.1323. [DOI] [PubMed] [Google Scholar]
- 41.Zhen Z, Xie J. Development of Manganese-Based Nanoparticles as Contrast Probes for Magnetic Resonance Imaging. Theranostics. 2012;2:45–54. doi: 10.7150/thno.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren HX, Liu L, Liu C. et al. Physiological Investigation of Magnetic Iron Oxide Nanoparticles Towards Chinese Mung Bean. J Biomed Nanotechnol. 2011;7:677–84. doi: 10.1166/jbn.2011.1338. [DOI] [PubMed] [Google Scholar]
- 43.Xie J, Jon S. Magnetic nanoparticle-based theranostics. Theranostics. 2012;2:122–4. doi: 10.7150/thno.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh SP. Multifunctional Magnetic Quantum Dots for Cancer Theranostics. J Biomed Nanotechnol. 2011;7:95–7. doi: 10.1166/jbn.2011.1219. [DOI] [PubMed] [Google Scholar]
- 45.Niu G, Chen X. Molecular Imaging with Activatable Reporter Systems. Theranostics. 2012;2:413–23. doi: 10.7150/thno.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Llandro J, Palfreyman JJ, Ionescu A. et al. Magnetic biosensor technologies for medical applications: a review. Med Biol Eng Comput. 2010;48:977–98. doi: 10.1007/s11517-010-0649-3. [DOI] [PubMed] [Google Scholar]
- 47.Cui DX, Zhang H, Wang K. et al. Gold Nanoparticles Enhance Efficiency of In Vitro Gene Transcription-Translation System. Nano Biomed Eng. 2011;3:120–5. [Google Scholar]
- 48.Wang F, Ma C, Zeng X. et al. Chemiluminescence Molecular Detection of Sequence-Specific HBV-DNA Using Magnetic Nanoparticles. J Biomed Nanotechnol. 2012;8:786–90. doi: 10.1166/jbn.2012.1446. [DOI] [PubMed] [Google Scholar]
- 49.Wang ZL, Gao JC, Jia DB. et al. Design and Fabrication of Slidable Pedicle Screw Frame System for Surgery Therapy of Patients with Spinal Deformity in Growth Period. Nano Biomed Eng. 2011;3:126–9. [Google Scholar]
- 50.Gao G, Zhang XQ, Wang K. et al. Experimental and Theoretical Studies of 1, 3-diaminopropylene Derivatives as Potential Antioxidative Agents for Copper Nanoparticles in Hydrothermal System. Nano Biomed Eng. 2011;3:130–6. [Google Scholar]
- 51.Logan C, Habington A, Lennon G, Cronin F, O'Sullivan N. Evaluation of the efficacy of n real-time polymerase chain reaction for the routine early detection of Pseudomonas aeruginosa in cystic fibrosis sputum and throat swab specimens. Diagn Microbiol Infect Dis. 2010;68:358–65. doi: 10.1016/j.diagmicrobio.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 52.Kernen Y, Sauty A, Roulet M. Detection and management of early Pseudomonas aeruginosa infection in patients with cystic fibrosis. Rev Med Suisse. 2005;1:637–640. [PubMed] [Google Scholar]