SUMMARY
ES cell pluripotency requires bivalent epigenetic modifications of key developmental genes regulated by various transcription factors and chromatin modifying enzymes. How these factors coordinate with one another to maintain the bivalent chromatin state so that ES cells can undergo rapid self-renewal while retaining pluripotency is poorly understood. We report that Utf1, a target of Oct4 and Sox2, is a bivalent chromatin component that buffers poised states of bivalent genes. By limiting PRC2 loading and Histone 3 lysine-27 trimethylation, Utf1 sets proper activation thresholds for bivalent genes. It also promotes nuclear tagging of mRNAs transcribed from insufficiently silenced bivalent genes for cytoplasmic degradation through mRNA de-capping. These opposing functions of Utf1 promote coordinated differentiation. The mRNA degradation function also ensures rapid cell proliferation by blocking the Myc-Arf feedback control. Thus, Utf1 couples the core pluripotency factors with Myc and PRC2 networks to promote the pluripotency and proliferation of ESCs.
Keywords: Utf1, PRC2, Myc, bivalency, pluripotency, self-renewal, ES cells, epigenetics, mRNA degradation, differentiation
INTRODUCTION
Embryonic stem cells (ESCs) undergo rapid self-renewal and can differentiate into any cell type. These features depend on transcription factors including Oct4, Sox2, and Nanog (Boyer et al., 2005; Chen et al., 2008; Kim et al., 2008; Loh et al., 2006) that form the core pluripotency network (Orkin and Hochedlinger, 2011). This ESC-specific network interacts with both the Myc-based transcription network and a network of chromatin-modifying complexes including the polycomb repressive complex 2 (PRC2). Together these three networks occupy and regulate a large number of target genes essential for the self-renewal and differentiation of ESCs (Boyer et al., 2006; Hu et al., 2009; Kim et al., 2010; Ku et al., 2008; Lee et al., 2006). Although a few factors have been shown to coordinate some aspects of these networks in ESCs (Orkin and Hochedlinger, 2011), how all three networks are functionally integrated is unknown.
The Myc-centered network promotes proliferation while regulating lineage-specific differentiation (Lin et al., 2009; Smith et al., 2010; Varlakhanova et al., 2010). However, Myc also activates the Arf tumor suppressor encoded by Cdkn2a (also called Ink4a-Arf), which in turn inhibits cell proliferation. In somatic cells, this feedback mechanism prevents uncontrolled cell proliferation, and its inactivation by mutations leads to tumorigenesis (Cleveland and Sherr, 2004; Eischen et al., 1999). Whereas cancer cells evade the Myc-Arf feedback loop through mutations, how ESCs block this feedback to allow rapid proliferation is unknown. Oct4, Sox2, and Nanog are required for ESCs to maintain a low level of Arf mRNA (Banito et al., 2009; Kawamura et al., 2009; Li et al., 2009; Utikal et al., 2009). As these transcription factors do not directly bind to Cdkn2a, it is unclear how the pluripotency core antagonizes the Myc-Arf feedback loop to ensure rapid ESC self-renewal.
Besides specific transcriptional regulation, two unique chromatin features contribute to the establishment and maintenance of ESC pluripotency. A less compacted chromatin structure compared to differentiated cells endows ESCs with highly dynamic chromatin organization (Meshorer and Misteli, 2006). Another chromatin feature of ESCs is the so-called poised bivalent state of developmentally regulated genes that harbor both transcriptionally active (histone 3-lysine 4 trimethylation, H3K4me3) and transcriptionally repressive (histone 3-lysine 27 trimethylation, H3K27me3) epigenetic marks (Bernstein et al., 2006), which are catalyzed by SET/MLL complexes and PRC2, respectively (Ang et al., 2010; Boyer et al., 2006; Jiang et al., 2010; Landeira et al., 2010; Li et al., 2010; Peng et al., 2009; Shen et al., 2009; Pasini et al., 2010). These chromatin features lead to a global transcriptional activity so that even repressed developmentally-regulated genes are expressed sporadically (Efroni et al., 2008). Since these mRNAs neither accumulate to high levels nor translate into large amounts of proteins in ESCs, active mechanisms must exist to prevent their accumulation and translation. Understanding these mechanisms should shed light on these unique properties of ESCs.
The PRC2-catalyzed H3K27me3 is essential for maintaining both the silenced and poised states of bivalent genes. PRC2 recognizes both CpG islands and chromatin features within the GC-rich regions where most bivalent genes reside (Zhou et al., 2011). However, because both CpG-island densities and chromatin contexts of bivalent genes vary widely, additional regulatory mechanisms are likely required to ensure that appropriate amounts of PRC2 are loaded onto individual bivalent genes in ESCs so that they are neither overly nor insufficiently silenced. Indeed, recent studies of Jarid2, a component of PRC2, have revealed complicated mechanisms of promoting PRC2 loading and activities on bivalent genes. In addition to promoting PRC2 loading, ESCs may actively limit PRC2 binding via a presently unknown mechanism so that the bivalent genes are not overly silenced. The function of PRC2 on bivalent genes must also be coordinated with the core pluripotency factors. Since no physical interaction between the core factors and PRC2 has been detected, the core factors may be coupled to PRC2 through their downstream effector(s) in ESCs.
The Undifferentiated embryonic cell transcription factor 1 (Utf1) is one of the direct downstream target genes of Oct4 and Sox2, and it is highly expressed in mouse and human ESCs (Nishimoto et al., 2005; Okuda et al., 1998a; Tan et al., 2007). The highest amount of Utf1 is found in the inner cell mass of mouse blastocysts. After implantation, Utf1 expression is silenced in most cells with the exception in some embryonic tissues (Okuda et al., 1998b). In ESCs, Utf1 is only found in the nucleus where it tightly associates with chromatin, and during ESC differentiation Utf1 is rapidly down regulated. Although Utf1 has been implicated in regulating ESC proliferation and differentiation, the mechanism remains unclear (Nishimoto et al., 2005; van den Boom et al., 2007). We report here that Utf1 is a component of the bivalent chromatin that regulates gene expression in a context-dependent manner, which connects the pluripotency core to both Myc- and PRC2-networks to ensure rapid proliferation and coordinated differentiation of ESCs.
RESULTS
Utf1 binds to bivalent genes to regulate their expression
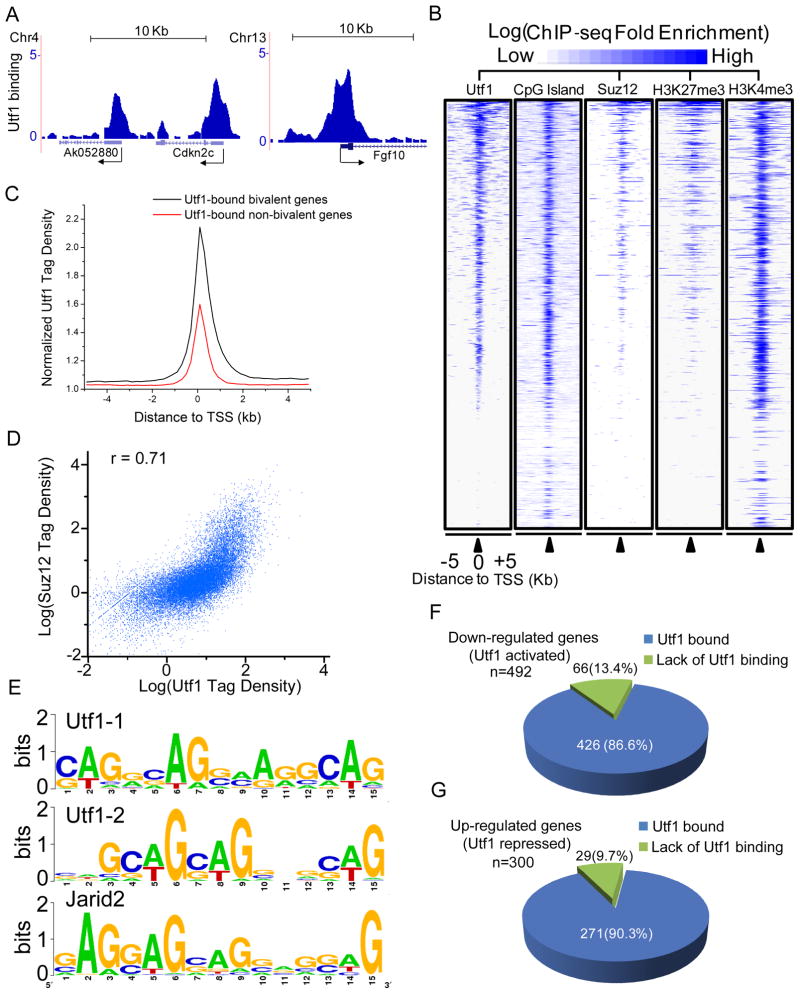
We expressed biotin-Utf1 at less than 5% of the endogenous Utf1 level (Figure S1A) and mapped Utf1 binding sites in ESCs using biotin-mediated and cross-linked ChIP-sequencing (biotin-ChIP-seq) (Kim et al., 2009; Kim et al., 2010; Shen et al., 2009). We identified 75,029 chromatin regions with significant Utf1 enrichments [False Discovery Rate (FDR)<0.001]. Utf1 binding was enriched in gene-rich regions with highest binding near the transcription start sites (TSS) of genes containing dense CpG islands (Figure 1A-C and S1B). Gene ontology (GO) analyses revealed a striking enrichment of Utf1 on genes with functions in organ/system development and cell differentiation (Figure S1C and Table S1).
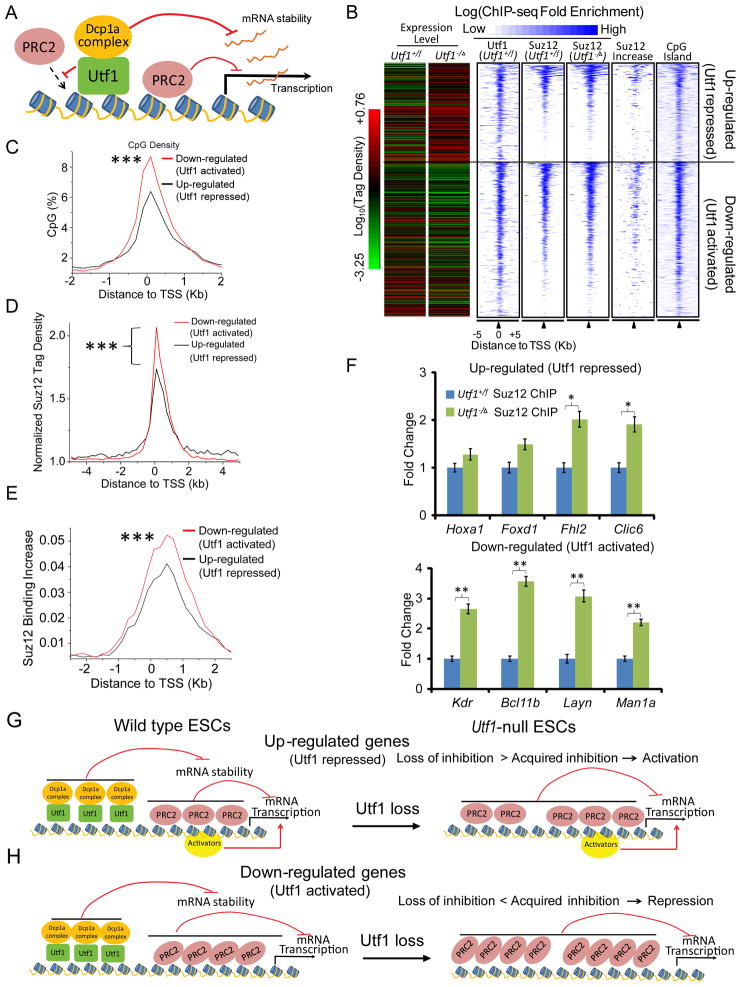
Figure 1. Utf1 binds bivalent genes to regulate their expression.
A. Utf1 binding at bivalent promoters in ESCs. Biotin-ChIP-seq of BirA expressing ESCs showed no significant binding of background biotinylated proteins to chromatin (data not shown). Arrows, the start and direction of transcription. Y-axes, Reads Per Kilo-bases per Million reads (RPKM).
B. Heat-map of annotated mouse-gene promoters enriched for Utf1, CpG, Suz12, H3K27me3 (Mikkelsen et al., 2007), and H3K4me3 in ESCs. ChIP-seq enrichment (identified in this study) for Utf1, Suz12, and H3K4me3 was calculated as the ratio of normalized tag counts of the ChIP-seq and the input sequence in a 1kb window that slides every 200 bp along 10 kb promoter regions. The heat-map is rank-ordered based on the enrichment of Utf1 (blue, enriched; white, not enrichment).
C. Utf1 tag density on bivalent genes is higher than the non-bivalent genes. The normalized Utf1-tag densities, determined as the averaged ratio of normalized tag counts of Utf1-ChIP-seq and the input sample in the 200-bp window, are plotted within the 5 kb up- and down-stream of TSS.
D. Scatter-plot of the correlation between Utf1 and Suz12 binding on promoters. A window of 1kb up- and down-stream of the TSS of all genes was used to calculate the tag enrichment. Spearman’s rank correlation coefficient, r = 0.71.
E. Two AG-rich sequence motifs that are enriched in Utf1-bound loci are highly similar to the predicted Jarid2-bound AG-rich motif.
F and G. Down- (F) or up-regulated (G) genes in Utf1−/Δ ESCs compared to Utf1+/f ESCs as shown by RNA-Seq.
PRC2-ChIP-seq using an antibody to Suz12 (a subunit of PRC2) revealed that Utf1 binding strongly correlated with PRC2 binding (r=0.71; Figure 1B-D and S1B). Our Suz12-ChIP-seq dataset was consistent with previously published datasets (r=0.82) (Ku et al., 2008). Of the total 16380 Utf1-bound genes, ~6116 were bivalent genes (Table S2) and they exhibited significantly stronger Utf1 enrichment within 5kb up- and down-stream of the TSS than those non-bivalent genes (Figure 1C). Utf1 binding were highly correlated (r=0.6) with H3K27me3 catalyzed by PRC2, but poorly correlated with H3K4me3 (r=0.21) present on both bivalent promoters and promoters of strongly expressed genes (Figure 1B, S1B, and S1D). Utf1 binding was not correlated with H3K9me3 (r=−0.17) found on heterochromatin (Figure S1E). Using different motif-find methods (Bailey and Elkan, 1994; Zheng et al., 2011), we predicted two similar AG-rich motifs recognized by Utf1 within CpG islands (Figure 1E) closely resembling one of the two motifs previously predicted to bind to Jarid2 in PRC2 (Peng et al., 2009). Thus, Utf1 is preferentially enriched at the promoters of bivalent genes.
To study how Utf1 regulates gene expression, we generated Utf1-null (Utf1−/Δ) ESCs by gene targeting (Figure S1F and G). Compared to control Utf1+/f ESCs, the Utf1−/Δ ESCs expressed similar amounts of pluripotency proteins and Suz12 (Figure S1H) and maintained similar levels of euploidy under standard ESC culture conditions (Figure S1I). Using RNA-seq, we found 792 genes exhibited changes of expression by at least 1.5-fold (p<10−5) in Utf1−/Δ ESCs compared to Utf1+/f ESCs. Importantly, Utf1 directly bound to 86.6% of the down- and 90.3% of the up-regulated genes (Figure 1F and G, Table S3 and 4). The majority of these are bivalent genes. Thus Utf1 could regulate either repression or activation of bivalent genes in ESCs.
Utf1 limits bivalent gene silencing by preventing excessive PRC2 binding and H3K27me3
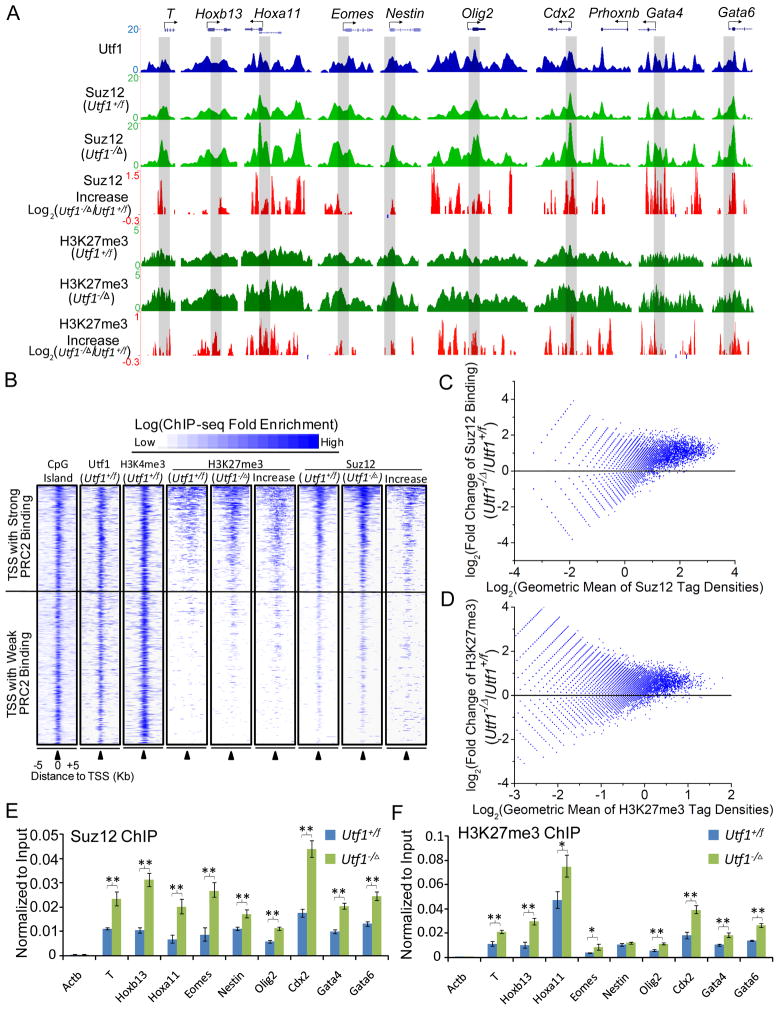
Since Utf1 is strongly enriched on bivalent genes (Figure 1B and C), we focused our study on the bivalent genes. We first studied whether Utf1 could limit gene silencing by preventing excessive PRC2 binding and H3K27me3 on bivalent genes because Utf1 and PRC2 were predicted to bind to similar DNA sequences (Figure 1E). ChIP-seq showed that Utf1−/Δ ESCs had up to 4-fold increase in PRC2 binding to bivalent genes compared to Utf1+/f ESCs, which corresponded to a significant increase in H3K27me3 on these genes (Figure 2A–D and S2A). Further ChIP analyses of Histone H3 and H3K4me3 demonstrated that increased PRC2 binding and H3K27me3 on bivalent genes were not due to some general change of chromatin upon Utf1 deletion (Figure 2E, 2F, S2B and S2C).
Figure 2. Utf1 binds to bivalent genes to limit PRC2 loading and H3K27me3.
A. Plots of ChIP-seq peaks at ten genomic loci. Y-axes, RPKM of ChIP-seq of Utf1, Suz12, and H3K27me3. The increase (red peaks) or decrease (blue peaks below zero, hardly visible due to very little decrease) of Suz12 binding and H3K27me3 in response to Utf1 depletion are plotted as log2 fold changes on Y-axes. Genomic regions that are further analyzed by ChIP–qPCR (see E and F below) are shaded grey.
B. Heat-map of all bivalent genes identified in this study showing the enrichment of CpG, Utf1, H3K4me3, H3K27me3 (in Utf1+/f or Utf1−/Δ ESCs), Suz12 (in Utf1+/f or Utf1−/Δ ESCs), and the relative increase of Suz12 binding or H3K27me3 upon Utf1 depletion. The heat-map is rank-ordered based on the enrichment of Suz12 in control ESCs. The increase in Suz12 binding and H3K27me3 was calculated by the log-ratio of normalized Suz12 and H3K27me3 tag density between Utf1−/Δ and Utf1+/f ESCs.
C and D. MA-plots of Suz12 binding (C) and H3K27me3 (D) on all promoters in Utf1−/Δ and Utf1+/f ESCs. Two biological repeats give similar results. One dataset is shown here. Log2 of the normalized Suz12 or H3K27me3 tag densities within −1kb to 1kb up- and down-stream of the TSS in Utf1+/f and Utf1−/Δ ESCs were averaged and plotted on the X-axes. The Y-axes are the log-ratio of Suz12 or H3K27me3 tag densities between Utf1−/Δ and Utf1+/f ESCs.
E and F. ChIP-qPCR of Suz12 binding (E) and H3K27me3 (F) at selected genes in Utf1+/f and Utf1−/Δ ESCs. The Actb locus was used as controls. Error bars, standard deviations (SD) of triplicates. Student’s T-test, *p<0.05, **p<0.01.
We then used four means to show that Utf1 and PRC2 indeed competitively bound to the same bivalent genes. First, we reduced the PRC2 subunit Jarid2 using RNAi (Figure S2D) and performed Biotin-Utf1-ChIP-qPCR on selected bivalent genes. This resulted in an increased binding of Utf1 to the bivalent genes (graph in Figure S2D). Second, we re-expressed Utf1 in Utf1−/Δ ESCs (Figure S2E) and carried out Suz12-ChIP-qPCR on selected bivalent genes. After re-expressing Utf1, the increase of Suz12 binding to the bivalent genes in Utf1−/Δ ESCs was reduced to similar levels seen in Utf1+/f ESCs (graph in Figure S2E). Third, we performed electrophoretic mobility shift assay (EMSA) to show that Utf1 and PRC2 competitively bound to the DNA oligo containing the predicted binding motifs for Utf1 and PRC2 (Figure S2F). Finally, sequential ChIP-qPCR analyses on selected bivalent genes showed that Utf1 and PRC2 co-occupied the same bivalent genes in wild-type ESCs (Figure S2G). Thus, Utf1 prevents over-silencing of bivalent genes by limiting PRC2 binding.
Utf1 recruits the mRNA decapping protein Dcp1a to bivalent genes
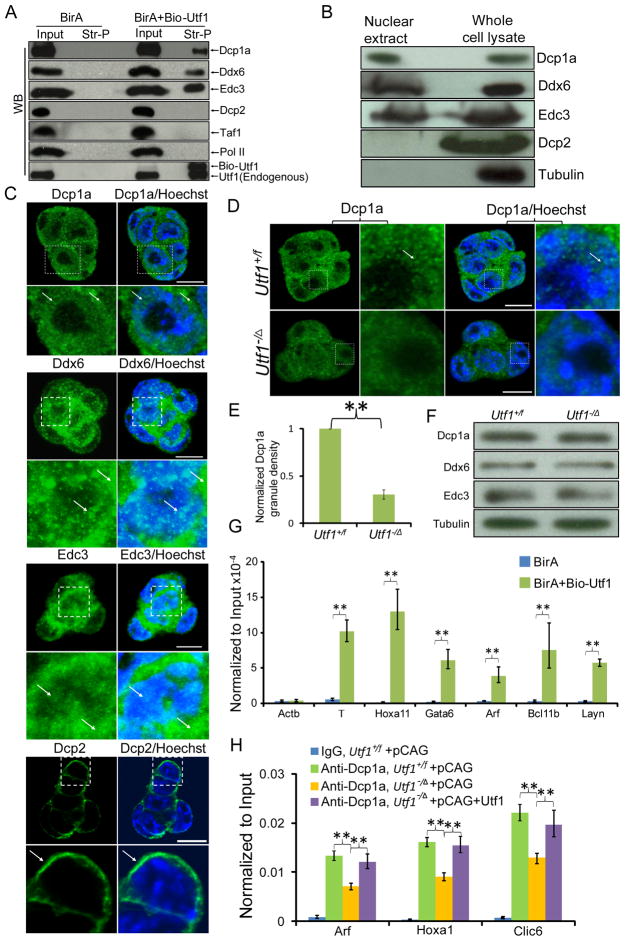
To understand how Utf1 could also repress gene expression, we identified Utf1-interacting proteins using mass-spectrometry. Among the proteins identified by two rounds of mass-spectrometry with a high peptide coverage (Table S5), we did not find either ATF2 or any subunit of TFIID that were shown to interact with Utf1 in somatic cells (Fukushima et al., 1998; Okuda et al., 1998b). However, we identified components of the mRNA decapping complex, Dcp1a, Ddx6, and Edc3, as candidate Utf1-interacting proteins in ESCs (Ling et al., 2011; Tritschler et al., 2009).
By pulling down biotin-Utf1 expressed in ESCs, we showed whereas Dcp1a, Ddx6, and Edc3 co-immunoprecipitated with Utf1, the RNA polymerase II (Pol II) and Taf1, two abundant nuclear proteins, were not detectable in the immunoprecipitate (Figure 3A). Consistent with the absence of the catalytic subunit Dcp2 (cleaves the 5′ methyl cap of mRNAs) in the mass-spec analyses (Table S5), Dcp2 was not detected in Utf1 immunoprecipitates (Figure 3A). We found that Dcp1a, Ddx6, and Edc3 were present in ESC nuclear extracts, whereas Dcp2 was only found in the whole cell lysate (Figure 3B). Similarly, immunofluorescence staining revealed that whereas Dcp1a, Ddx6, and Edc3 were present in both the nucleus and the cytoplasm, Dcp2 was only found in the cytoplasm of ESCs (Figure 3C). Thus Utf1 binds to three non-catalytic subunits of the mRNA decapping complex in the ESC nucleus.
Figure 3. Utf1 recruits the non-catalytic subunit of the mRNA de-capping complex to bivalent genes.
A. Biotin-Utf1 co-precipitates with Dcp1a, Ddx6 and Edc3, but not Dcp2, Taf1, and Pol II in wild-type ESCs. Streptavidin pulled-down (Str-P) of biotin-Utf1 was probed for the indicated antibodies by Western blotting analyses.
B. Western blotting analyses of nuclear extracts and whole cell lysates of wild-type ESCs. Tubulin, cytoplasmic protein control.
C. Immuno-localization of Dcp1a, Ddx6, Edc3, and Dcp2 in wild-type ESCs. Areas in white dashed squares are enlarged at the bottom of each image. Arrows, proteins in the region occupied by chromatin stained by Hoechst (blue) or in the cytoplasm. Scale bar, 10 μm.
D. A reduction of Dcp1a+ granule numbers and intensity in Utf1−/Δ ESCs compared to Utf1+/f ESCs. Areas in dashed squares are enlarged to the right of each image. Arrows, Dcp1a+ granules in the Hoechst-stained chromatin region. Scale bar, 10 μm.
E. Quantifications of Dcp1a+ granules in Utf1−/Δ and Utf1+/f ESCs.
F. Utf1−/Δ and Utf1+/f ESCs express similar amounts of Dcp1a, Ddx6, and Edc3. Loading controls, tubulin.
G. Dcp1a-ChIP samples were re-ChIPed using streptavidin beads followed by qPCR of bivalent genes in Utf1+/f ESCs expressing BirA alone or BirA plus biotin-Utf1.
H. ChIP-qPCR of Dcp1a binding at bivalent genes in Utf1+/f or Utf1−/Δ ESCs transfected with control vector pCAG or pCAG-Utf1. Restoration of Utf1 expression (see Figure S2E for Utf1 Western) rescued Dcp1a binding in Utf1−/Δ ESCs. The IgG antibody was used as controls. Error bars, SD of triplicates. Student’s T-test, **p<0.01.
See also Table S5.
A clear reduction of bright Dcp1a+ granules in both the nucleus and the cytoplasm was seen in ESCs upon Utf1 loss, which was not due to the reduction of the decapping proteins (Figure 3D–F). Instead, as formation of mRNP processing granules requires efficient loading of proteins like Dcp1a onto mRNAs, the reduction of Dcp1a+ granules in Utf1−/Δ ESCs suggests that Utf1 facilitates the recruitment of the non-catalytic subunits of the mRNA decapping complex to bivalent promoters so they can be loaded onto newly transcribed mRNAs in the nucleus. Indeed, sequential ChIP-qPCR of randomly selected bivalent genes showed that Dcp1a and Utf1 co-occupied these genes (Figure 3G). Importantly, the binding of Dcp1a to bivalent genes decreased significantly upon Utf1 loss, which was rescued by re-introducing Utf1 into these ESCs (Figure 3H). Since the reduction of Dcp1a binding in the absence of Utf1 is incomplete, additional factors may mediate Dcp1a recruiting.
Utf1 promotes the binding of Dcp1a to mRNA in the nucleus for cytoplasmic degradation
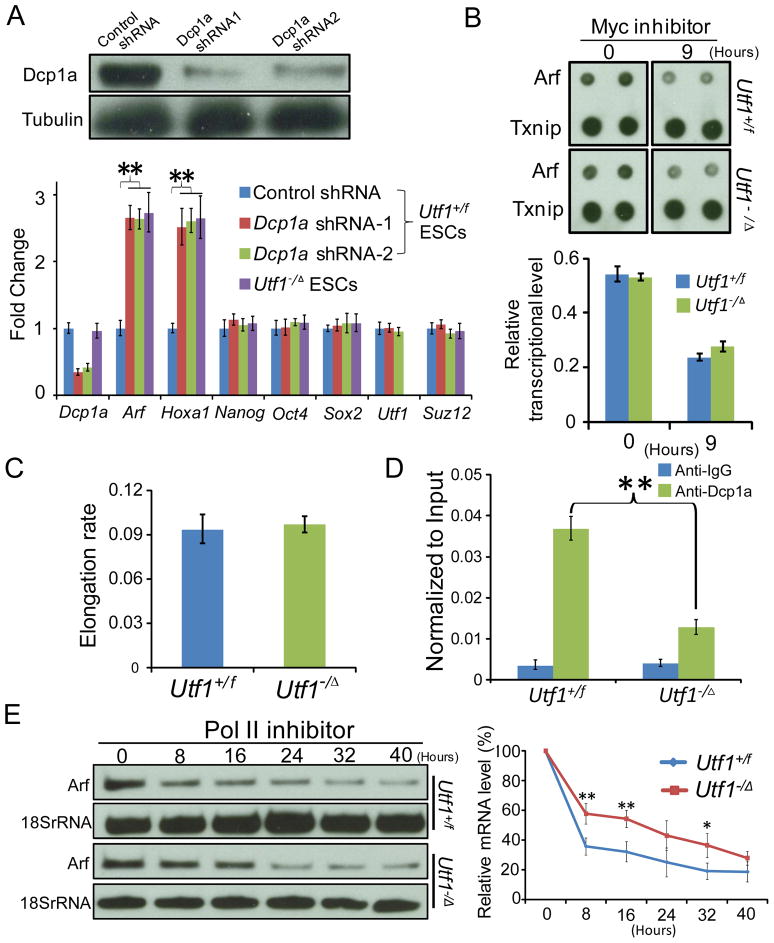
RNA-seq showed that reduction of Dcp1a by two different shRNAs in ESCs resulted in up regulation of many Utf1-bound bivalent genes, including Arf and Hoxa1, without affecting the expression of Nanog, Oct4, Sox2, Utf1, and Suz12 (Figure 4A and Table S6). To show that Utf1 tags the mRNA from leaky bivalent promoters for cytoplasmic degradation by recruiting Dcp1a, we focused on the bivalent gene Arf because it exhibited a low level of expression in Utf1+/f ESCs, which was further up regulated in Utf1−/Δ ESCs (Figure S3A–D). The low level of Arf expression in wild-type ESCs could be due to the binding of the transcriptional activator Myc (Figure S3A). Consistent with the idea that Utf1 represses Arf expression post-transcriptionally, both the nuclear run-on assay and the transcriptional elongation assay showed that Utf1 did not inhibit Arf transcription (Figure 4B and C). Treating ESCs with the Myc inhibitor 10058-F4 (Rahl et al., 2010) caused a similar reduction of Arf transcription in both Utf1+/f and Utf1−/Δ ESCs (Figure 4B), which was consistent with a role of Myc in activating Arf transcription in ESCs. Finally, S1 nuclease protection assays showed that Arf mRNA was properly spliced in the nucleus of Utf1+/f ESCs (Figure S3E). Thus, Utf1 does not regulate Arf transcription or splicing in the nucleus.
Figure 4. Utf1 promotes the binding of Dcp1a to mRNAs in the nucleus for cytoplasmic degradation.
A. Western blotting analyses of Utf1+/f ESCs treated with control or Dcp1a shRNAs. Loading controls, tubulin. The graph shows RT-qPCR of mRNA levels of indicated genes in the control, Dcp1a-shRNA-treated Utf1+/f ESCs, or untreated Utf1−/Δ ESCs.
B. 32P-labeled transcripts from Myc inhibitor treated (9 hours) and untreated (0 hour) ESCs were hybridized to nitrocellulose filters spotted with Arf or Txnip cDNAs (two spots per gene). Txnip is a non-bivalent gene not regulated by Myc (Rahl et al., 2010). The graph shows quantifications of the nuclear run-on assays. The Arf hybridization intensity was normalized to the Txnip intensity and plotted.
C. The elongation rate of Pol II at the Arf locus in Utf1+/f and Utf1−/Δ ESCs measured by the ratio of ChIP-qPCR of Pol II at exon2 and at the promoter region of Arf.
D. RNA immunoprecipitation using Dcp1a antibody in Utf1+/f and Utf1−/Δ ESCs followed by RT-qPCR analyses.
E. Arf mRNA decay was determined by Northern blotting in Utf1+/f and Utf1−/Δ ESCs after transcriptional arrest by flavopiridol. Loading control, 18S rRNA. The Arf mRNA band intensity normalized to the 18S RNA band intensity at time 0 was defined as 100%. The normalized Arf band intensities at other time points were calculated relative to the time 0. Error bars, SD. Student T-test, *p<0.05, **p<0.01.
We then performed RNA immunoprecipitation (RIP) using Dcp1a antibody and nuclear extracts made from Utf1−/Δ and Utf1+/f ESCs. qPCR analyses showed that the binding of Dcp1a to Arf mRNA was significantly reduced upon Utf1 loss (Figure 4D). Next we inhibited Pol II using flavopiridol (Rahl et al., 2010) and probed for Arf mRNA levels using Northern blotting. We found that Utf1−/Δ ESCs were less efficient in degrading Arf mRNA compared to Utf1+/f ESCs (Figure 4E). Thus, Utf1 recruits Dcp1a to bivalent promoters, which facilitates the loading of Dcp1a to mRNAs transcribed from leaky bivalent genes for cytoplasmic degradation.
Utf1 buffers bivalent gene expression in a context-dependent manner
The above findings reveal that whereas Utf1 limits PRC2 binding to prevent excessive H3K27me3 and bivalent gene silencing, it also recruits Dcp1a to bivalent genes to repress gene expression through mRNA pruning (Figure 5A). These dual functions could allow Utf1 to enforce the poised state of bivalent genes residing in different DNA and chromatin contexts. We analyzed whether the down-regulated bivalent genes upon Utf1 loss have different chromatin and DNA contents compared to the up-regulated genes. We found that the up-regulated genes have lower CpG island densities and weaker PRC2 binding than the down-regulated genes (Figure 5B–D). Importantly the up-regulated genes exhibited less PRC2 binding increase upon Utf1 loss than the down-regulated genes (Figure 5B, 5E and F). These analyses suggest the following modes of context-dependent bivalent gene regulation by Utf1. For bivalent genes with low CpG island densities and PRC2 binding or are bound by strong transcriptional activators (see Arf as an example), Utf1 would repress gene expression because the loss of mRNA pruning due to Utf1 loss could not be compensated by the limited gaining of PRC2 (Figure 5G). By contrast, for bivalent genes with high CpG island densities and strong PRC2 binding, Utf1 would prevent gene silencing because upon Utf1 deletion the loss of mRNA pruning would be over compensated by the excessive gaining of PRC2 (Figure 5H).
Figure 5. Utf1 regulates bivalent genes in a context-dependent manner.
A. Illustration of the dual functions of Utf1 on bivalent genes.
B. The Heat-map of gene expression in Utf1+/f and Utf1−/Δ ESCs (red and green, high and low expression, respectively). The profiles of Utf1 binding, Suz12 binding (in Utf1+/f or Utf1−/Δ ESCs), Suz12 binding increase (in Utf1−/Δ ESCs), and CpG densities of these genes are also shown as Heat-maps that are rank-ordered based on Suz12 binding strength in Utf1+/f ESCs. The gene expression heat-map shows that Utf1 buffers gene expression.
C. Quantifications of CpG densities at the TSS of Utf1-activated (down-regulated upon Utf1 loss, red) and Utf1-repressed (up-regulated upon Utf1 loss, black) genes. The CpG density is determined by the percentage of CpG dinucleotides in 200bp windows and plotted within 2 kb up- and down-stream of TSS (Wilcoxon two-sample test, ***p<0.001).
D. Quantifications of Suz12 binding at the TSS of Utf1-activated (down-regulated upon Utf1 loss) and Utf1-repressed (up-regulated upon Utf1 loss) bivalent genes within 5 kb up- and downstream of TSS (Wilcoxon two-sample test, ***p<0.001).
E. Quantifications of Suz12 binding increase upon Utf1 loss at the TSS of Utf1-activated (down-regulated upon Utf1 loss) and Utf1-repressed (up-regulated upon Utf1 loss) genes within 5 kb up- and down-stream of TSS (Wilcoxon two-sample test, ***p<0.001).
F. Suz12-ChIP-qPCR of Utf1-repressed (up-regulated upon Utf1 loss, top plot) and Utf1-activated (down-regulated upon Utf1 loss, bottom plot) genes found by RNA-Seq. Error bars, SD of triplicates. Student’s T-test, *p<0.05, **p<0.01.
G and H. Illustrations for the context-dependent buffering of bivalent genes by Utf1. The dual functions of Utf1 in repressing (by recruiting Dcp1a) and activating genes (by limiting PRC2 binding) are indicated. Upon Utf1 loss, bivalent genes may undergo up regulation (G) or down regulation (H), depending on the degree of PRC2-binding increase, Dcp1a loss, and other transcription activators (such as Myc in the Arf locus) on these genes.
We have shown that Utf1 uses Dcp1a to repress genes such as Hoxa1 and Arf (Figure 4A, Table S4 and 6), which is consistent with the model in Figure 5G. Since our studies suggest that Utf1 recruits Dcp1a to bivalent genes, we analyzed how the loss of Dcp1a could affect the expression of genes that use Utf1 to limit their repression (see Figure 5H). Since these genes exhibit stronger PRC2 binding than the other bivalent genes, Dcp1a loss may or may not affect their expression depending on the strength of PRC2-mediated gene repression. Indeed, we found that 56% (164/295) of them did not show gene expression change upon Dcp1a loss, suggesting that PRC2 was sufficient for their repression. We did find 41% (121 out of 295) of bivalent genes in this group underwent up regulation upon Dcp1a reduction, suggesting that they depended on both PRC2 and Dcp1a for repression. The above analyses predict that whereas Utf1 functions in ESCs to maintain rapid cell proliferation by repressing Arf expression through Dcp1a-mediated mRNA pruning, both the mRNA pruning and the epigenetic functions of Utf1 also ensure proper gene expression during ESC differentiation.
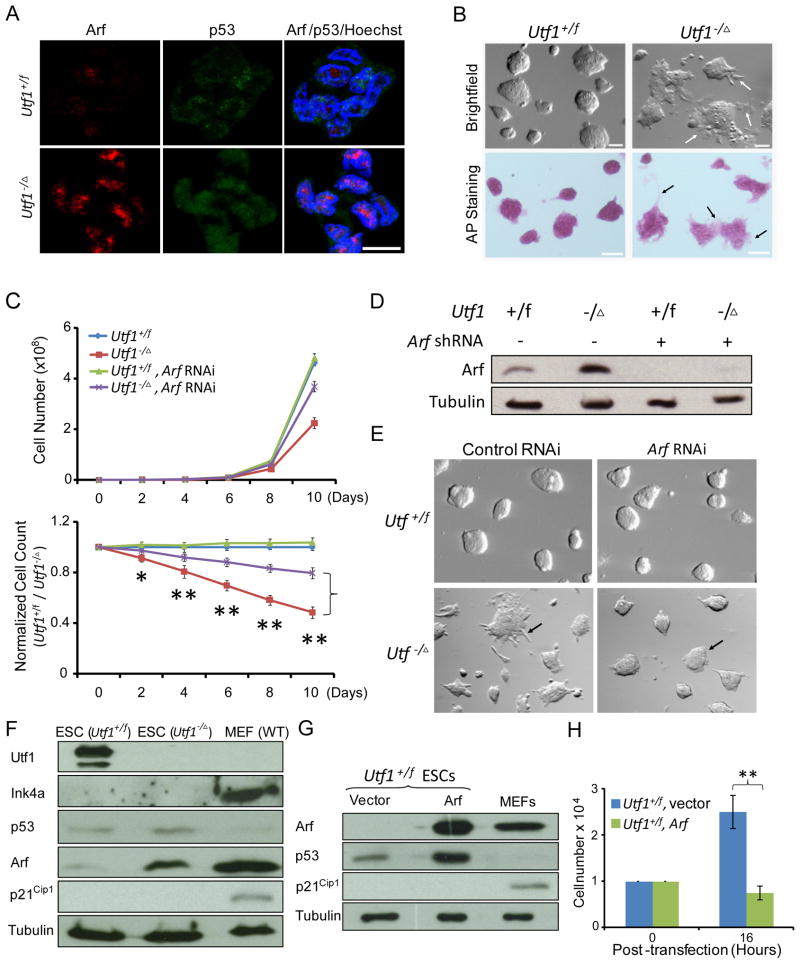
Utf1 antagonizes Myc-mediated activation of Arf to ensure rapid proliferation of ESCs
We found that Arf protein levels in Utf1−/Δ ESCs was reduced by either re-introducing Utf1 or by inhibiting c-Myc (Figure S4A and B), demonstrating that Utf1 functions downstream of Oct4 and Sox2 to antagonize the Myc-Arf feedback loop. Although both Utf1+/f and Utf1−/Δ ESCs expressed similar amounts of pluripotency proteins (Figure S1H and S4C), the Utf1−/Δ ESCs had higher amounts of Arf in the nucleus, flatter colony morphologies, and slower proliferation rates compared to Utf1+/f ESCs (Figure 6A–C). Consistently, teratomas formed by Utf1−/Δ ESCs were much smaller than that of Utf1+/f ESCs (Figure S4D). Inhibiting Arf expression using shRNA (Figure 6D) resulted in a significant increase of cell proliferation rates in Utf1−/Δ ESCs without affecting the rate of Utf1+/f ESCs (Figure 6C). Thus up regulation of Arf upon Utf1 deletion was responsible for the slow ESC proliferation. Reduction of Arf in the Utf1−/Δ ESCs also partially reversed the flattened colony morphology (Figure 6E). The incomplete reversal of the proliferation rate and colony morphology of Arf-RNAi treated Utf1−/Δ ESCs is consistent with our finding that Arf is only one of the targets of Utf1 in ESCs.
Figure 6. Utf1 promotes proliferation of ESCs through inhibiting Arf expression.
A. Representative immunofluorescence images of Arf and p53 proteins in Utf1+/f and Utf1−/Δ ESCs. Scale bar, 10 μm.
B. Utf1−/Δ ESCs appear flatter than Utf1+/f ESCs. Both types of ESCs are positive for the pluripotency marker alkaline phosphatase (AP). Arrows indicate spontaneous differentiation at the edges of Utf1−/Δ ESC colonies.
C. Utf1−/Δ ESCs proliferated slower than Utf1+/f ESCs. Reduction of Arf by shRNA significantly enhanced Utf1−/Δ ESC proliferation without affecting Utf1+/f ESCs. The top graph plots cell numbers. The bottom graph plots the percentages of Utf1−/Δ ESCs normalized against Utf1+/f ESCs. Error bars, SD of triplicates.
D. shRNA reduction of Arf expression in both Utf1+/f and Utf1−/Δ ESCs. Loading controls, tubulin.
E. Inhibiting Arf expression reverted the flattened ESC colonies back to the morphology similar to the Utf1+/f ESCs.
F. Western blotting analyses of Ink4a, p53, Arf, and p21Cip1 in Utf1+/f and Utf1−/Δ ESCs. Controls, wild-type MEFs. Loading controls, tubulin.
G. Western blotting analyses of Arf overexpression 16 hours post transfection of ESCs, which had increased p53 but not p21Cip1. Controls, wild-type MEFs. Loading controls, tubulin.
H. Over-expression of Arf in Utf1+/f ESCs resulted in a significant reduction of cell proliferation 16 hours post-transfection. Student’s T-test in C and H, *p<0.05, **p<0.01.
See also Fig. S4.
The p53-p21Cip1 pathway represents the best-characterized mechanism by which Arf inhibits cell cycle progression (Pomerantz et al., 1998; Zindy et al., 1998). The binding of Arf to Mdm2, an ubiquitin ligase for p53, leads to Mdm2 inhibition and p53 accumulation. The increase in p53 could lead to transcriptional up-regulation of the cell cycle inhibitor p21Cip1, which would slow down cell cycle progression. We found that although Arf up-regulation led to a small increase of p53 levels in Utf1−/Δ ESCs, p21Cip1 was not up regulated (Figure 6A and F). Similarly, although transient overexpression of Arf in Utf1+/f ESCs strongly increased p53 protein levels and inhibited cell cycle progression, p21Cip1 expression was unchanged (Figure 6G, 6H, and S4E–G). Thus, Utf1 functions downstream of Oct4 and Sox2 to ensure rapid cell cycle progression by blocking the Myc-Arf feedback loop independent of p53-p21Cip1 in ESCs.
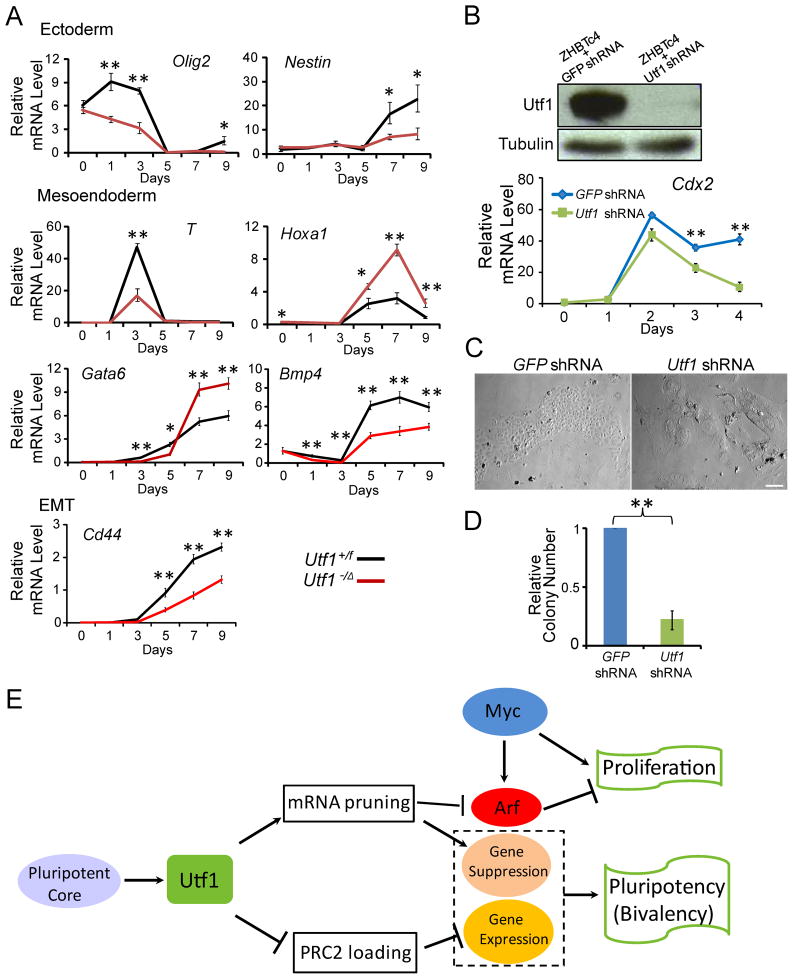
Utf1 coordinates gene expression during ESC differentiation
We next studied whether Utf1 was required for proper bivalent gene expression during ESC differentiation by investigating their expression in either ectoderm (such as Olig2 and Nestin) or mesoendoderm (such as T and Hoxa1). Olig2, Nestin, and T exhibited a significant increase in PRC2 binding and H3K27me3 upon Utf1 loss (see Figure 2A, 2E, and 2F). Hoxa1 was up regulated either upon Utf1 loss or upon Dcp1 RNAi (see Figure 4A, Table S4 and 6), suggesting that the mRNA-pruning mechanism was important for controlling Hoxa1 expression. Using embryoid body (EB)-based differentiation and q-PCR assays, we found whereas Olig2, Nestin, and T were insufficiently up regulated, Hoxa1 underwent excessive up-regulation during EB differentiation of Utf1−/Δ ESCs compared to Utf1+/f ESCs (Figure 7A). Additional qPCR analyses of other bivalent genes expressed in mesoendoderm (Gata6 and Bmp4) or during epithelium to mesenchyme transition (EMT, Cd44) revealed a similar mis-regulation (Figure 7A).
Figure 7. Utf1 regulates proper differentiation by coordinating gene expression in ESCs.
A. Time-course RT-qPCR analyses of developmental regulators normalized to GAPDH during EB differentiation in Utf1+/f and Utf1−/Δ ESCs. Error bars, SD of triplicates.
B. Utf1 shRNA reduced Utf1 protein levels in ZHBTc4 ESCs compared to control GFP shRNA by Western blotting analyses. Time-course RT-qPCR analyses of Cdx2 mRNA during TE differentiation was normalized to GAPDH and plotted. Error bars, SD of triplicates.
C. Images of TS cells or terminally differentiated TE cells derived from GFP-shRNA or Utf1-shRNA-treated ZHBTc4 ESCs, respectively.
D. Quantifications of TS colonies in GFP-shRNA- and Utf1-shRNA-treated cells 6 days after differentiation. Error bars, SD of triplicates. Student’s T-test in A, B, and D, *p<0.05, **p<0.01.
E. A model. Utf1 couples three regulatory networks to ensure proper proliferation and differentiation of ESCs.
To study how Utf1 could affect gene expression and lineage-specific differentiation, we used ZHBTc4 ESCs, in which the endogenous Oct4 was replaced by a tetracyclin (Tc)-regulated Oct4 transgene (Niwa et al., 2000). ZHBTc4 ESCs undergo efficient trophectoderm (TE) differentiation in the presence of Tc. The up regulation of the transcription factor Cdx2 during TE development in vivo is required for establishing progenitor cells for TE (Niwa et al., 2005). In vitro, Cdx2 is not essential for terminal TE differentiation from ESCs but it is required for the establishment and maintenance of trophoblast stem (TS) cells that undergoes further terminal differentiation into TE lineages (Niwa et al., 2005). Upon Utf1 loss, the Cdx2 gene locus exhibited a significant increase of PRC2 binding and H3K27me3 in ESCs (see Figure 2E and F), predicting that Cdx2 would not be sufficiently up regulated when Utf1-knockdown ESCs were induced to differentiate toward TE. Consistently, we found that addition of Tc to ZHBTc4 ESCs treated with Utf1 shRNA led to insufficient Cdx2 up regulation (Figure 7B) and rapid terminal differentiation as judged by the formation of large and flat terminally differentiated TE cells, whereas the control shRNA-treated ZHBTc4 ESCs formed many colonies made of small TS-like cells upon Tc treatment (Figure 7C and D). Thus while Utf1 deletion did not block the differentiation of ESCs to TE, it disrupted proper coupling between proliferation and differentiation.
Discussion
Using a combination of genomic, cell biology, proteomic, and gene-targeting approaches, we uncover a mechanism that integrates the pluripotency core factors with the PRC2-based epigenetic network and Myc regulation. This mechanism has three features. First, the core factors use their direct downstream target Utf1, which itself is a chromatin-associated protein enriched at bivalent genes, to control both self-renewal and proper differentiation. Second, the core factors use Utf1 to block the Myc-Arf feedback loop to ensure rapid cell proliferation. An important function of the core factors in reprograming is to block the expression of Arf and Ink4a. However, since none of the core factors directly binds to Cdkn2a, the mechanism of repression has remained unknown. Our findings provide the critical missing molecular link that allows ESCs to evade the Myc-Arf feedback control. Third, Utf1 integrates all three networks using both epigenetic regulation and a previously unappreciated mRNA pruning mechanism (Figure 7E).
The differential PRC2 loading on bivalent genes invites the question of how the genes with high or low PRC2-binding are neither over-silenced nor de-repressed, respectively. We show that Utf1 and PRC2 bind to bivalent genes competitively because they recognize similar AG-rich motifs, which allows Utf1 to prevent excessive loading of PRC2 in ESCs. How Utf1 or PRC2 binds to chromatin is unclear. Interestingly, H2AZ is one of the Utf1-interacting proteins identified in our mass-spec analyses. Since H2AZ binds to PRC2-bound genes (Creyghton et al., 2008), studying the functional relationship between H2AZ and Utf1 may help to understand how Utf1 is loaded onto bivalent genes. We note that our biotin-Utf1-ChIP-seq data are not consistent with the Native-ChIP-on-chip studies of Utf1 (Kooistra et al., 2010). The native-ChIP procedure involved micrococcal nuclease treatments and additional manipulations for extended time without crosslinking, which could fail to capture the true Utf1-chromatin interactions.
Our data suggest that by binding to Dcp1a, Ddx6, and Edc3, Utf1 ensures that mRNAs transcribed from leaky bivalent genes are tagged for efficient degradation by the mRNA decapping pathway. The mRNA de-capping complex, which has only been studied in fungi and in animal somatic cells, promotes the formation of mRNP processing granules that either prevent mRNA translation or facilitate mRNA degradation (Ling et al., 2011; Tritschler et al., 2009). Our findings have revealed a previously unappreciated function of the mRNA de-capping complex in silencing bivalent genes that are insufficiently repressed in ESCs. As a component of the mRNA decapping complex, Dcp1a could promote both the binding of mRNA to Dcp2 and the decapping activity of Dcp2 to facilitate mRNA degradation in the ESC cytoplasm. Dcp1a could also promote mRNA turnover through microRNAs in ESCs (Ling et al., 2011; Tritschler et al., 2009).
The promoter-mediated mRNA pruning function defined here may represent an important mechanism to ensure ESC maintenance. Interestingly, two recent studies in yeast have shown that promoters can dictate mRNA stability by recruiting proteins that targets mRNAs for cytoplasmic degradation (Bregman et al., 2011; Trcek et al., 2011). Although the proteins involved in the promoter-based marking of mRNAs are different in the yeast studies and in our study, our findings strongly suggest that the promoter-mediated control of mRNA stability is evolutionarily conserved.
Dcp1a, Edc3, and Dcp2 have been found in the same complex with TTF2 (a transcriptional terminator) and they have been implicated in facilitating mRNA decapping during transcription, which inhibits transcriptional elongation in both HeLa and HEK293 cells (Brannan et al., 2012). Dcp1a has also been shown to bind to SMAD4 to activate target genes upon TGFβ signaling in different somatic tissue culture cells (Bai et al., 2002). However, unlike these previous reports, our studies show that the Utf1-mediated recruiting of Dcp1a to bivalent promoters in ESCs does not affect transcription, but it tags mRNA for post-transcriptional degradation in the cytoplasm.
The buffer function of Utf1 we uncovered suggests that while removing Utf1 may not block lineage specification and differentiation, it would lead to inappropriate coupling of developmental processes and cell proliferation. Our preliminary analyses of Utf1-null mice are consistent with this prediction (data not shown). Utf1 is localized on the human chromosome 10q26.3, near the telomere. Humans bearing a heterozygous deletion of 10q26.3 exhibit a number of developmental abnormalities and retarded growth (Kehrer-Sawatzki et al., 2005). Our findings suggest that Utf1 heterozygosity could be responsible for some of the abnormalities in the 10q26.3 patients.
Experimental Procedures
Karyotype analyses of ESCs and differentiation assays
These procedures were performed as described previously (Creyghton et al., 2008; Kim et al., 2011; Niwa et al., 2000; Vong et al., 2009) and in the Supplementary Materials.
Utf1 pull-down, Western blotting, and mass spectrometry
V6.5 ESCs (1–2 ×109) were used to make nuclear extracts. After dialyzing the extract into a low salt buffer, a Utf1 polyclonal antibody was used to immunoprecipitate Utf1 and its associated proteins. ESCs (1×107) expressing either BirA alone or both BirA and biotin-Utf1 were used for streptavidin pull-down of biotin-Utf1. See the Supplementary Materials for more details.
Cell proliferation assay
To perform long-term proliferation assays, 105 Utf1+/f or Utf1−/Δ ESCs were seeded into a single well of a 6-well plate. The cells in each well were counted and re-plated every two days. The Utf1+/f ESCs were diluted so that 105 cells were plated in a fresh well. The same dilution factor was used to dilute the Utf1−/Δ ESCs into a new well. Since Utf1−/Δ ESCs grew slower, fewer Utf1−/Δ ESCs than Utf1+/f ESCs were plated each time. The cumulative total cell number was calculated at each time point and used to plot the growth curve. The cell number ratios of control-treated Utf1+/f ESCs to other experimental groups of ESCs at each time point were used to plot the normalized growth curve.
RNAi-mediated gene silencing in ESCs
Short hairpin (sh) RNAs were purchased from OpenBiosystems to knockdown the mRNA of Arf (TRCN0000077816), Utf1 (TRCN0000081708), Jarid2 (TRCN0000096642) and Dcp1a (shRNA-1:TRCN0000096664, shRNA-2:TRCN0000096665). See the Supplementary Materials for more details.
ChIP assays
The ChIP-qPCR and ChIP-seq assays were performed according to the condition suggested by the manufacturers of ChIP-grade antibodies to H3K27me3 (Abcam, ab6002), H3K4me3 (Cell Signaling, 9751), Suz12 (Cell Signaling, 3737), and Dcp1a (WH0055802) with modifications (Jia et al., 2007). Biotin-ChIP was performed as previously described (Kim et al., 2009). See the Supplementary Materials for detailed procedures, data analyses, and primer information.
Sequential ChIP assay
Sample treatment and immunoprecipitations were performed using standard ChIP assay procedures. After the first immunoprecipitation, chromatin was eluted in a solution of 30 mM DTT, 500 mM NaCl, and 0.1% SDS at 37°C as described (Bernstein et al., 2006). Eluted chromatin was diluted 50-fold and subjected to biotin-Utf1-ChIP as described in the Supplementary Materials.
Whole transcriptome shotgun sequencing (RNA-Seq)
Total RNA was isolated from 107 ESCs with the RNeasy Plus kit (QIAGEN). The poly-A containing mRNAs were purified and libraries were built following Illumina TruSeq RNA protocols. Libraries were sequenced using an Illumina HiSeq 2000. 100 bp-long reads from both ends were obtained. See the Supplementary Materials for more details.
Cell cycle analyses
ESCs were trypsinized and re-suspended in 0.5 ml PBS followed by the addition of 0.5 ml of 100% ice-cold ethanol in a drop-wise manner while vortexing. After incubation for 20 min on ice, cells were harvested and washed by PBS containing 1% FBS. Next, cells were incubated in PBS with 25 μg/ml RNase A at 37°C for 30 min to digest RNA. Finally, cells were stained by 50 μg/ml propidium iodide for 10 min at room temperature, and analyzed by flow-cytometry using BD-FACS Calibur.
Determining mRNA degradation profile
ESCs were cultured as described above. Cells were treated with 1 μM flavopiridol (Sigma cat #F3055) and harvested at different time points. mRNA extraction and Northern blotting analyses are described in the Supplementary Materials.
Nuclear run-on and S1 nuclease protection assays
ESCs were cultured as described above. Cells were treated with 50 μM c-Myc inhibitor 10058-F4 (Sigma cat #F3680) and harvested after 9 hours. Whole ESC and nuclear extract were harvested as described above. S1 nuclease protection assay was performed as previously described (Mendrysa and Perry, 2000). For details see the Supplementary Materials.
Electrophoretic Mobility Shift Assay (EMSA)
A 74 bp mouse genomic sequence containing the predicted Utf1- and Jarid2-binding motifs was labeled by [γ-32P]-ATP and incubated with ESC nuclear extract. The unlabeled competitor probe was used at 100-fold excess to the labeled probe. Super-shift was performed by incubating the reactions with antibodies against Utf1 or Suz12. For details see the Supplementary Materials.
Large Datasets Availability
All ChIP-seq and RNA-seq datasets have been deposited into GEO repository (GSE39513).
Supplementary Material
Highlights.
Utf1 is a component of the bivalent chromatin in ES cells.
Utf1 sets the threshold of transcriptional activation on bivalent genes.
Utf1 promotes the degradation mRNAs from bivalent genes such as Arf.
Utf1 connects core pluripotency factors to the Myc and PRC2 networks.
Acknowledgments
We thank Allison Pinder for deep-seq, Nick Ingolia for advice on data analyses, Chen-ming Fan, Nick Ingolia, Xin Chen, Haiqing Zhao, Dong Yang, Ma Wan, Max Guo, and members of the Zheng lab for comments. JJ and YZ are supported by HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2010;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai RY, Koester C, Ouyang T, Hahn SA, Hammerschmidt M, Peschel C, Duyster J. SMIF, a Smad4-interacting protein that functions as a co-activator in TGFbeta signalling. Nature cell biology. 2002;4:181–190. doi: 10.1038/ncb753. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings/International Conference on Intelligent Systems for Molecular Biology ; ISMB. 1994;2:28–36. [PubMed] [Google Scholar]
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes & development. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, et al. mRNA Decapping Factors and the Exonuclease Xrn2 Function in Widespread Premature Termination of RNA Polymerase II Transcription. Molecular cell. 2012;46:311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman A, Avraham-Kelbert M, Barkai O, Duek L, Guterman A, Choder M. Promoter elements regulate cytoplasmic mRNA decay. Cell. 2011;147:1473–1483. doi: 10.1016/j.cell.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cleveland JL, Sherr CJ. Antagonism of Myc functions by Arf. Cancer cell. 2004;6:309–311. doi: 10.1016/j.ccr.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, et al. Global transcription in pluripotent embryonic stem cells. Cell stem cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes & development. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima A, Okuda A, Nishimoto M, Seki N, Hori TA, Muramatsu M. Characterization of functional domains of an embryonic stem cell coactivator UTF1 which are conserved and essential for potentiation of ATF-2 activity. The Journal of biological chemistry. 1998;273:25840–25849. doi: 10.1074/jbc.273.40.25840. [DOI] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes & development. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Lin M, Zhang L, York JP, Zhang P. The Notch signaling pathway controls the size of the ocular lens by directly suppressing p57Kip2 expression. Molecular and cellular biology. 2007;27:7236–7247. doi: 10.1128/MCB.00780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell. 2010;144:513–525. doi: 10.1016/j.cell.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Daumiller E, Muller-Navia J, Kendziorra H, Rossier E, du Bois G, Barbi G. Interstitial deletion del(10)(q25.2q25.3 approximately 26.11)--case report and review of the literature. Prenatal diagnosis. 2005;25:954–959. doi: 10.1002/pd.1252. [DOI] [PubMed] [Google Scholar]
- Kim J, Cantor AB, Orkin SH, Wang J. Use of in vivo biotinylation to study protein-protein and protein-DNA interactions in mouse embryonic stem cells. Nature protocols. 2009;4:506–517. doi: 10.1038/nprot.2009.23. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan CM, Gaiano N, Ko MS, Zheng Y. Mouse B-Type Lamins Are Required for Proper Organogenesis But Not by Embryonic Stem Cells. Science. 2011;334:1706–1710. doi: 10.1126/science.1211222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra SM, van den Boom V, Thummer RP, Johannes F, Wardenaar R, Tesson BM, Veenhoff LM, Fusetti F, O’Neill LP, Turner BM, et al. Undifferentiated embryonic cell transcription factor 1 regulates ESC chromatin organization and gene expression. Stem cells (Dayton, Ohio) 2010;28:1703–1714. doi: 10.1002/stem.497. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS genetics. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nature cell biology. 2010;12:618–624. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes & development. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. The EMBO journal. 2009;28:3157–3170. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SH, Qamra R, Song H. Structural and functional insights into eukaryotic mRNA decapping. Wiley interdisciplinary reviews. 2011;2:193–208. doi: 10.1002/wrna.44. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nature genetics. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Mendrysa SM, Perry ME. The p53 tumor suppressor protein does not regulate expression of its own inhibitor, MDM2, except under conditions of stress. Molecular and cellular biology. 2000;20:2023–2030. doi: 10.1128/mcb.20.6.2023-2030.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nature reviews. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto M, Miyagi S, Yamagishi T, Sakaguchi T, Niwa H, Muramatsu M, Okuda A. Oct-3/4 maintains the proliferative embryonic stem cell state via specific binding to a variant octamer sequence in the regulatory region of the UTF1 locus. Molecular and cellular biology. 2005;25:5084–5094. doi: 10.1128/MCB.25.12.5084-5094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nature genetics. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–929. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Okuda A, Fukushima A, Nishimoto M, Orimo A, Yamagishi T, Nabeshima Y, Kuro-o M, Boon K, Keaveney M, Stunnenberg HG, et al. UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra-embryonic cells. The EMBO journal. 1998a;17:2019–2032. doi: 10.1093/emboj/17.7.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda A, Fukushima A, Nishimoto M, Orimo A, Yamagishi T, Nabeshima Y, Kuro-o M, Nabeshima Y, Boon K, Keaveney M, et al. UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra-embryonic cells. The EMBO journal. 1998b;17:2019–2032. doi: 10.1093/emboj/17.7.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KN, Singh AM, Dalton S. Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell stem cell. 2010;7:343–354. doi: 10.1016/j.stem.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SM, Wang ST, Hentze H, Droge P. A UTF1-based selection system for stable homogeneously pluripotent human embryonic stem cell cultures. Nucleic acids research. 2007;35:e118. doi: 10.1093/nar/gkm704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Larson DR, Moldon A, Query CC, Singer RH. Single-molecule mRNA decay measurements reveal promoter- regulated mRNA stability in yeast. Cell. 2011;147:1484–1497. doi: 10.1016/j.cell.2011.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F, Braun JE, Motz C, Igreja C, Haas G, Truffault V, Izaurralde E, Weichenrieder O. DCP1 forms asymmetric trimers to assemble into active mRNA decapping complexes in metazoa. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21591–21596. doi: 10.1073/pnas.0909871106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boom V, Kooistra SM, Boesjes M, Geverts B, Houtsmuller AB, Monzen K, Komuro I, Essers J, Drenth-Diephuis LJ, Eggen BJ. UTF1 is a chromatin-associated protein involved in ES cell differentiation. The Journal of cell biology. 2007;178:913–924. doi: 10.1083/jcb.200702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlakhanova NV, Cotterman RF, deVries WN, Morgan J, Donahue LR, Murray S, Knowles BB, Knoepfler PS. myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation; research in biological diversity. 2010;80:9–19. doi: 10.1016/j.diff.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vong QP, Liu Z, Yoo JG, Chen R, Xie W, Sharov AA, Fan CM, Liu C, Ko MS, Zheng Y. A role for borg5 during trophectoderm differentiation. Stem cells (Dayton, Ohio) 2009;28:1030–1038. doi: 10.1002/stem.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Hu GQ, She ZS, Zhu H. Leaderless genes in bacteria: clue to the evolution of translation initiation mechanisms in prokaryotes. BMC genomics. 2011;12:361. doi: 10.1186/1471-2164-12-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes & development. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.