Abstract
Purpose
There are no good genomic markers of survival in patients with advanced colorectal cancer. The CpG island methylator phenotype (CIMP) marks a distinctive pathway in colorectal cancer. We sought to determine the prognostic significance of CIMP in advanced colorectal cancer patients treated with 5-fluorouracil (5-FU) in an Eastern Cooperative Oncology Group clinical trial.
Experimental Design
We studied 188 patients enrolled on protocol E2290, a five-arm trial comparing 5-FU, 5-FU in combination with N-phosphonoacetyl-L-aspartic acid, oral leucovorin, i.v. leucovorin, or IFNα-2a in patients with advanced colorectal cancer. Methylation of MINT1, MINT31, hMLH1, p14ARF, and p16INK4a in DNA extracted from formalin-fixed paraffin-embedded specimens was evaluated by combined bisulfite restriction analysis, and methylation of MINT2 was studied by methylation-specific PCR.
Results
Methylation frequencies were 21% for MINT1, 23% for MINT2, 24% for MINT31, 4% for hMLH1, 11% for p14ARF, and 17% for p16INK4a. Methylation of MINT1, MINT31, p14ARF, and p16INK4a were correlated, as expected. There was no association between methylation and clinicopathologic factors or response to therapy. Methylation of MINT1, MINT31, p14ARF, or p16INK4a was associated individually with shortened overall survival. Hazard ratios were 1.51 (P = 0.05) for MINT1, 1.70 (P = 0.006) for MINT31, 2.22 (P = 0.001) for p14ARF, and 1.51 (P = 0.05) for p16INK4a. Concurrent methylation of two or more genes of the CIMP-associated subset (MINT1, MINT31, p14ARF and p16INK4a) defined a group of cases with markedly reduced overall survival and hazard ratio was 3.22 (P < 0.0001in multivariate analyses).
Conclusions
CIMP is associated with poor survival in advanced colorectal cancer patients.
Colorectal cancer is the second most common cause of cancer-related deaths in Western countries (1). Patients with recurrent or metastatic colorectal cancer have a poor outcome, with usual survival of <2 years (2). Treatment with 5-fluorouracil (5-FU) alone or with combinations of 5-FU and CPT11/irinotecan or oxaliplatin somewhat increases survival, but most patients eventually succumb to the disease (2). Markers to assist in selection of therapy for individual patients are needed, rather than the current empirical decision-making process.
A substantial number of genetic changes have been described in colorectal cancer of various stages (3, 4). Some of these are clearly associated with natural history and survival. For example, microsatellite instability (MSI) is associated with a more favorable outcome, and chromosome 18q deletions are associated with shortened survival in stage II and III colorectal cancer (5, 6). However, there are no known genomic markers of survival in patients with advanced colorectal cancer, where MSI is rare and chromosome 18 deletion is very frequent.
DNA methylation of promoter-associated CpG dinucleotide–rich regions, termed CpG islands, is associated with permanent loss of gene expression (epigenetic regulation) in mammalian cells (7). Aberrant hypermethylation of DNA is common in human cancers and has been associated with silencing of important tumor-suppressor genes (8). Methylation of many genes has now been described in colorectal cancer (9). Significantly, methylation of many of these genes is concurrent in a subset of cancers, a phenomenon that has been termed the CpG island methylator phenotype (CIMP; ref. 10). CIMP cancers seem to have distinct clinical characteristics (more common in proximal tumors, in women, and in older patients), a distinct histology (mucinous and poorly differentiated tumors), and distinctive genetic changes (high rate of MSI and mutations of the KRAS and BRAF genes and low rate of p53 mutations; ref. 11).
DNA methylation/CIMP has been reported to have variable prognostic significance in colorectal cancer in different studies (12–17). This issue is considerably complicated by the described associations between methylation and factors known to affect prognosis in colorectal cancer, such as high levels of microsatellite instability (MSI-H) that occur as a result of hypermethylation and silencing of the hMLH1 mismatch repair gene (18). Recurrent and/or metastatic colorectal cancers represent a potentially more uniform group of cancers to study, with rare MSI-H and very frequent chromosome 18 deletions. The Eastern Cooperative Oncology Group has conducted a clinical trial (E2290) of various 5-FU–based combinations in patients with advanced colorectal cancer (19). Using pathology specimens from individuals enrolled on E2290, we studied DNA methylation in this setting. We now report that CIMP is associated with a very poor prognosis in advanced colorectal cancer treated with 5-FU chemotherapy, findings that have implications for selection of therapy.
Materials and Methods
Patients and samples
Routine formalin-fixed, paraffin-embedded pathology specimens from patients enrolled in E2290 were used. Consistent with the fact that this was a study of metastatic or recurrent disease (19), these patients have a median survival of 14 months, and during more than 5 years of follow-up, most (87%) of the patients died, making the overall survival end point very mature. Eligibility criteria were standard criteria including pathologically verified colon cancer stage IV or recurrent colorectal cancer, adequate performance status (between 0 and 2, as required for patients to be eligible for the E2290 clinical trial), and willingness to participate in a randomized trial. A total of 1,122 patients were enrolled on E2290, and samples from 188 cases were available for the current studies. Patients were selected solely based on tissue availability and all available tissues were evaluated. The studies described here were approved by the M. D. Anderson Cancer Center Institutional Review Board. Microdissection of colon tumor tissue and preparation of DNA were done as previously described (20, 21). The majority of samples corresponded to primary tumor.
DNA methylation analysis
We used bisulfite-based approaches to study the methylation status on MINT1, MINT2, MINT31, hMLH1, P14ARF, and p16INK4a. These loci and genes were selected based on our previous studies showing that they often have concurrent methylation and therefore are excellent markers of CIMP (10, 22). Bisulfite treatment of DNA was done as previously described (23). We used combined bisulfite restriction analysis (COBRA) as a quantitative assay to study methylation of all genes (24). For MINT2, we were unable to develop an assay that was reliable for routine formalin-fixed paraffin-embedded tissues. Consequently, for MINT2 we used methylation-specific PCR (MSP), which is a sensitive, but not quantitative, assay (25). Primers, PCR conditions, and restriction enzymes used for COBRA are listed in Table 1. All PCR products were visualized by 6% PAGE followed by staining with ethidium bromide and imaging and quantitation with a Bio-Rad Geldoc 2000 imager (Bio-Rad).
Table 1.
Summary of primer sequences, PCR conditions, and restriction enzymes used for bisulfite-PCR
| Gene or CpG island | Primer sequence | Annealing temperature, °C (no. cycles) | Restriction enzyme for COBRA |
|---|---|---|---|
| COBRA | |||
| MINT1-F | GGGTTGGAGAGTAGGGGAGTT | 55 (40) | TaqI |
| MINT1-R | CCATCTAAAATTACCTCRATAACTTA | ||
| MINT31-F | GAYGGYGTAGTAGTTATTTTGTT | 58 (3), 56 (4), 54 (5), 52 (28) | BstUI |
| MINT31-R | CATCACCACCCCTCACTTTAC | ||
| P16-F | GGTTTTGGYGAGGGTTGTTT | 58 (3), 56 (4), 54 (5), 52 (28) | EcoRV |
| P16-R | ACCCTATCCCTCAAATCCTCTAAAA | ||
| P14-F | ttagtttgtagttaagggggtaggag | 58 (3), 56 (4), 54(5), 52 (28) | TaqI |
| P14-R | AAAAATCACCAAAAACCTAC | ||
| MLH1-F | TAGTAGTYGTTTTAGGGAGGGA | 60 (5), 57 (5), 54 (5), 51 (25) | RsaI |
| MLH1-R | TCTAAATACTCAACRAAAATACCTT | ||
| MSP | |||
| MINT2-MF | TTGTTAAAGTGTTGAGTTCGTC | 60 (40) | |
| MINT2-MR | AATAACGACGATTCCGTACG | ||
| MINT2-UF | GATTTTGTTAAAGTGTTGAGTTTGTT | 60 (40) | |
| MINT2-UR | CAAAATAATAACAACAATTCCATACA | ||
For COBRA, the identity of the amplified fragment was verified by digestion with multiple restriction enzymes and/or cloning and sequencing. Initial setup of the PCR assays included positive controls (two colon cancer cell lines, SW48 and RKO) and negative controls (normal colon mucosa), mixing experiments to rule out bias, and repeat experiments to assess reproducibility. Because low levels of methylation can be found in normal tissues, and gene silencing requires relatively high levels of methylation, we used a cutoff of 15% to consider a marker methylated. This approach was consistent with our previous studies, and using a cutoff of 10% or 20% instead gave very similar results (10).
Statistical analysis
In addition to usual descriptive statistics of counts and frequencies, standard statistical methods for survival analysis (time to event end points) were used in the analysis. These included log-rank tests to determine univariate differences between groups, Kaplan-Meier analysis for calculation and display of survival curves, and the proportional hazards regression model for time to event regression models with continuous and multiple predictor variables. To analyze the effect of methylation of multiple genes on overall survival, we used a linear model that assumed equal weight for methylation of each gene, built CIMP sum of all genes or a set of genes by calculating the sum of methylated markers among all genes or four genes (MINT1, MINT31, P14ARF, and p16INK4a) for each patient, and included CIMP sums in survival analysis. Statistical significance was determined when P < 0.05 and all statistical tests were two sided.
Results
The clinical results of E2290 have been reported elsewhere (19). Overall, the response rate was 14% and median survival was 14 months. There were no significant differences in response or survival among the different treatment arms. All available samples from 188 patients were evaluated in this study. To test whether availability of tissue samples could introduce potential bias, we compared patient demographic/clinicopathologic characteristics between the cohort of patients with tissue available and those without and found no statistically significant differences of clinical variables in most on-study patients (Supplementary Table S1). The only exception was age at entry in which the cases with tissue available were slightly older (50% of cases >65 years old versus 40% of cases >65 years old; P = 0.02). The laboratory analyses of the 188 patients were done in two separate groups of 93 to 95 patients each. The results were not significantly different in the two groups and were therefore merged for statistical analysis.
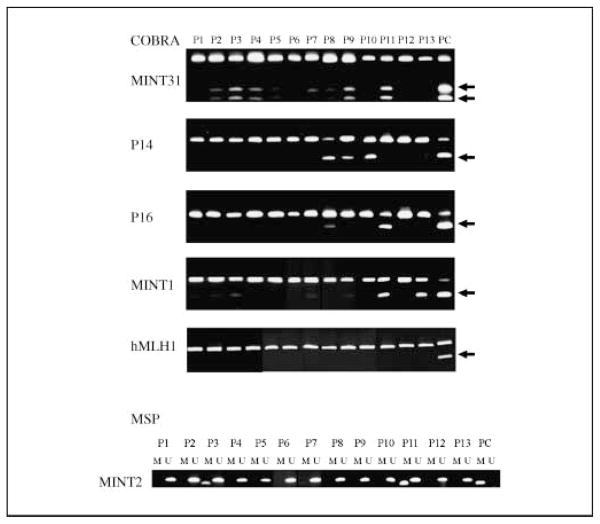
We analyzed methylation of a panel of six genes chosen because they detect CIMP with precision as reported by multiple groups (10, 26, 27). Methylation was studied by the quantitative method COBRA (24) for all genes except MINT2, for which we were unable to design a COBRA assay and used MSP instead. Representative examples of methylation for each gene are shown in Fig. 1. Methylation frequencies in the 188 patients were 21% for MINT1, 23% for MINT2, 24% for MINT31, 4% for hMLH1, 11% for P14ARF, and 17% for p16INK4a. There were significant positive associations among methylation of MINT1, MINT31, p16INK4a, and p14ARF as reported in our previous studies (10, 22), indicating the presence of a hypermethylator phenotype (CIMP) in a subset of cases (see Supplementary Table S2 for correlations between each gene analyzed by Spearman correlation analysis). Methylation of MINT2 was not associated with the other genes, perhaps because it was the only gene studied with a nonquantitative method. hMLH1 methylation was rare (4%), as expected in patients with advanced disease.
Fig. 1.
Representative examples of methylation analysis by COBRA (top) or by MSP (bottom). For COBRA, the arrows point to band shifts after restriction enzyme digestion, which indicate the presence of methylation. For MSP, amplification using M primers represents methylation. P1 to P13, 13 different cancers; PC, positive control (DNA from the RKO or SW48 cell lines).
We used univariate analyses to examine the associations among methylation and clinicopathologic characteristics. No consistent associations were found. In particular, methylation of this selected group of tumor-specific genes was not associated with age, which is distinct from many other genes that have age-related methylation (28, 29). When methylation of multiple genes (MINT1, MINT31, p14ARF, and p16INK4a) was combined as a group and cases divided based on methylation of 0 or 1 gene versus >1 gene as previously reported for the CIMP phenotype, 28 patients were defined as CIMP positive (15%) and 157 patients were defined as CIMP negative. For three patients, we were unable to define CIMP because methylation data were missing for three or more genes. In 182 patients with clinicopathologic characteristics available for analyses, there were no statistically significant differences in age, gender, race, presence of liver and/or nodal metastases, or patient performance status between the two groups (Table 2). However, CIMP-positive cases were more likely to be proximal in location (P = 0.01; see Table 2).
Table 2.
Association between demographic/clinicopathologic characteristics and methylation of multiple genes
| Clinical characteristics | CIMP sum
|
P* | |
|---|---|---|---|
| 0–1
|
2+
|
||
| n (%) | n (%) | ||
| Sex | |||
| Female | 48 (31) | 14 (50) | 0.08 |
| Male | 106 (69) | 14 (50) | |
| Age (y) | |||
| ≤65 | 78 (51) | 13 (46) | 0.84 |
| >65 | 76 (49) | 15 (54) | |
| Race | |||
| White | 135 (88) | 26 (93) | 0.55 |
| Black | 10 (6) | 0 (0) | |
| Hispanic | 9 (6) | 2 (7) | |
| Primary tumor site | |||
| Distal | 107 (69) | 12 (43) | 0.01 |
| Proximal | 47 (31) | 16 (57) | |
| Liver metastases | |||
| No | 34 (22) | 8 (29) | 0.47 |
| Yes | 120 (78) | 20 (71) | |
| Nodal metastases | |||
| No | 100 (65) | 13 (46) | 0.09 |
| Yes | 54 (35) | 15 (54) | |
| Performance status | |||
| 0 | 83 (54) | 9 (32) | 0.08 |
| 1 | 63 (41) | 17 (61) | |
| 2 | 8 (5) | 2 (7) | |
From Fisher’s exact test.
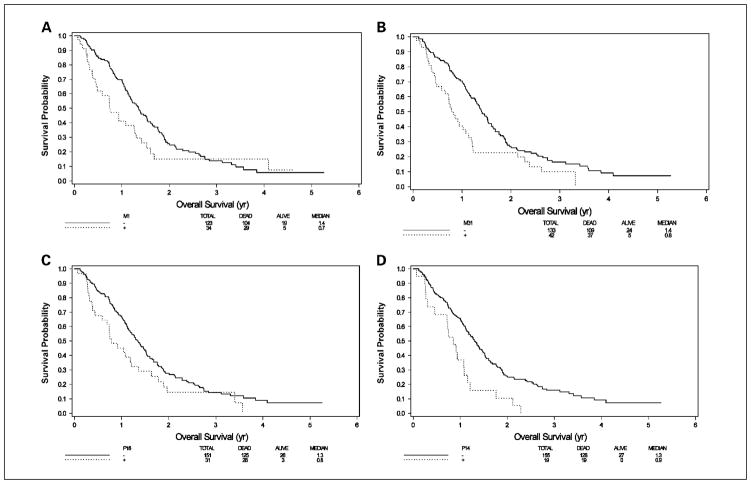
We next analyzed the relationship between methylation and outcome measures. No association was found between methylation of single gene or groups of genes with different schedules of 5-FU therapy. We also did not observe any correlations between methylation and response to therapy. However, MINT1, MINT31, p14ARF, and p16INK4a each showed significant associations with shortened survival in univariate analyses (Table 3). Hazard ratios (HR) for predicting overall survival were 1.51 (P = 0.05) for MINT1, 1.70 (P = 0.006) for MINT31, 2.22 (P = 0.001) for p14ARF, and 1.51 (P = 0.05) for p16INK4a. Figure 2 shows Kaplan-Meier survival curves for each of these four genes. Interestingly, MINT2, which was not studied by quantitative methods, did not show any association with survival. hMLH1 was too rarely methylated for meaningful survival analysis. Multivariate analyses that included known demographic/clinicopathologic characteristics associated with survival (location of tumor, presence of liver and/or nodal metastases, performance status, gender, race, age, and treatment arm) confirmed that MINT1, MINT31, and p14ARF were each associated with overall survival (HR = 1.91, P = 0.005 for MINT1; HR = 1.65, P = 0.02 for MINT31; and HR = 1.97, P = 0.0096 for p14ARF; see Table 3 and Supplementary Table S3 for details on the adjustment covariates used in the models).
Table 3.
HRs/P values for predicting overall survival for advanced colorectal cancer (E2290)
| Gene/marker | Methylation alone (HR/P value) | Methylation adjusted for demographics* (HR/P value) |
|---|---|---|
| MINT1 | 1.51/0.051 | 1.91/0.005 |
| MINT2 | 1.12/0.56 | 1.17/0.46 |
| MINT31 | 1.70/0.006 | 1.65/0.020 |
| P16 | 1.51/0.050 | 1.42/0.13 |
| P14 | 2.22/0.0014 | 1.97/0.0096 |
| MLH1 | 1.26/0.53 | 1.23/0.64 |
| SUM (linear) | 1.32/0.0002 | 1.31/0.0006 |
| MINT31 + P16 + P14 + MINT1 (>1) | 3.22/<0.0001 | 2.92/<0.0001 |
NOTE: SUM indicates the sum of all markers in a linear model. The reduced set of markers is shown analyzed both linearly (0<1<2<3<4) or divided into two groups (0, 1 versus >1), which was our preferred analysis method a priori based on the CIMP concept (10).
The adjustment variables were gender, age, race, primary tumor site, liver metastases, nodal metastases, and performance status (all as in Table 2), as well as treatment arm.
Fig. 2.
Kaplan-Meier curves of survival by methylation of individual genes. A, MINT1; B, MINT31; C, p16INK4a; D, p14ARF; solid line, unmethylated; stippled line, methylated.
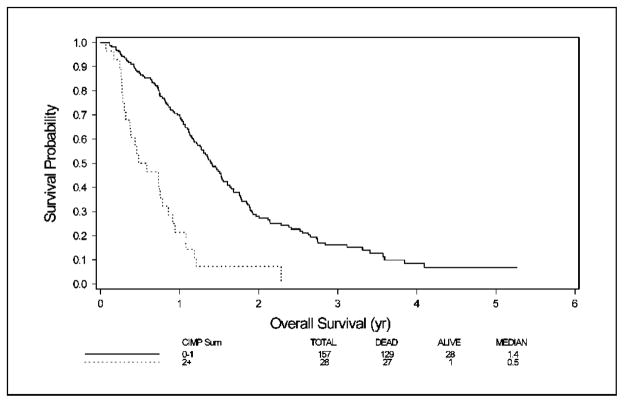
Finally, we analyzed the effect of methylation of multiple genes on overall survival. A linear model that assumed equal weight for methylation of each gene showed a significant association between methylation and survival. This association was similar whether all six genes or a reduced set of four genes (MINT1, MINT31, p14ARF, and p16INK4a) were evaluated (HR = 1.32, P = 0.0002 for all genes and HR = 3.22, P < 0.0001 for reduced set of four genes; see details in Table 3). The original CIMP classification called a tumor CIMP positive if three or more genes of seven studied were methylated. Analysis of the original and extended sets indicated that a reduced set of four markers, with methylation of two or more positive, yields a similar classification. On applying this reduced model to MINT1, MINT31, p14ARF, and p16INK4a, we found that CIMP was present in 28 of 185 (15%) cases. CIMP cases had a significantly shorter survival in multivariate analysis (HR, 2.9; P < 0.0001). As shown in Fig. 3, median survival in the CIMP subset was 6 months versus 17 months in the group without CIMP (P < 0.001). Two-year survival was 8% in the CIMP group versus 28% in the group without CIMP. All but one long-term survivor were in the group with no or only one methylated marker, and no patient with two or more methylated markers survived for 3 years.
Fig. 3.
Kaplan-Meier curve of survival by methylation of multiple genes. Solid line, methylation of 0 to 1 gene; stippled line, methylation of >1 gene.
Discussion
Genomic markers have distinct advantages as prognostic markers in cancer. DNA is more stable than RNA or protein, and DNA changes in tumors are less likely to be labile than other changes. In colorectal cancer, MSI and loss of heterozygosity of the long arm of chromosome 18 provide powerful markers of prognosis in early-stage disease (5), but there are no similar markers of prognosis in metastatic disease. Our data show that DNA methylation defines a group of advanced colorectal cancer patients with a very poor prognosis following 5-FU–based therapy. The results were generated from a multi-institution trial, were consistent across four separate genes, and are likely therefore to be clinically relevant.
The mechanism by which DNA methylation confers a poor prognosis is unclear. The methylation markers studied here have distinct functions. All the MINT markers correspond to the promoters of unique genes except MINT2; MINT1 corresponds to synaptic vesicle glycoprotein 2C gene (SV2C) located on Chr5q13; MINT31 corresponds to a CpG island upstream of the calcium channel CACNA1G gene located on Chr17q21; p14ARF is a upstream regulator of p53 function; p16INK4a is an important cell cycle regulatory gene; and hMLH1 is a mismatch repair gene. Because each of MINT1, MINT31, p14ARF, and p16INK4a had similar effects on prognosis, they may be simply markers for a hypermethylator phenotype associated with a poor prognosis. The association between increasing levels of DNA methylation and poor prognosis is a recurrent theme in oncology, consistent across multiple tumor types that include liver cancers, esophageal cancers, lung cancers, and various leukemias (30). A plausible hypothesis is that tumors with high degrees of methylation are more likely to inactivate genes critical for tumor progression and response to chemotherapy.
CIMP was originally defined in colon cancer by seven cancer-specific MINT makers, which were identified by a genome-wide technique, called methylated CpG island amplification, in combination with representational differential analysis (MCA/RDA), and the CIMP-positive group was defined by high level of methylation at two or more loci simultaneously (10). The existence of CIMP has been confirmed by several independent groups (26, 27, 31). Based on our previous results and studies from other groups, we used six genes as CIMP markers in this study. They are three MINT markers (MINT1, MINT2, and MINT31) from the classic CIMP panel, p14ARF, p16INK4a, and hMLH1. All these genes have been reported to correlate well with CIMP and did well to define CIMP in colon cancer (26, 27) as well as multiple other cancers (32–35). A recent study by Weisenberger et al. (31) suggested that a new panel of genes outperforms the classic panel in defining CIMP. However, by using very strict criteria to define CIMP, that study likely focused on a fraction of the CIMP group (the frequency of CIMP by Weisenberger et al. was 14%, in contrast to 46% in the original work). In fact, a recent study comparing the new panel of genes with original CIMP markers does not support the notion that the new panel performs better than the classic panel (36). Therefore, the most reliable panel of CIMP markers remains to be determined, and future studies are needed perhaps by using prognosis as a landmark to identify these makers.
The proportion of CIMP cases here is lower than previously reported. This is probably because many CIMP cases (those with hMLH1 methylation and MSI) rarely progress to the advanced stage. Consistent with this, in this study, the frequency of hMLH1 methylation that leads to MSI in sporadic colon cancer is only 4%, as opposed to the 15% to 20% usually reported in sporadic colorectal cancers. Several previous studies have found variable prognostic effect for methylation in colorectal cancer (12–17). For instance, one study examined CIMP in stage III colorectal cancers and found that methylation predicts better prognosis (16); however, the CIMP cases in that study were largely from patients with hypermethylation of hMLH1 and MSI. By comparing single marker such as MSI-low, methylation of ID4, methylation of MINT31, or methylation of p16INK4a with clinical outcomes in stage III colorectal cancer patients, other studies have shown that MSI-low or methylation of each individual genes was associated with poor prognosis in these patients (12–15). One study by Ward et al. (17) examined the prognostic significance of DNA methylation in colorectal cancer patients, mainly stages I to III. By analyzing methylation of MINT1, MINT2, MINT12, MINT31, p16INK4a, and hMLH1, they found that none of these makers were independently predictive of prognosis when analyzed with stage and grade. However, when they divided tumors into microsatellite-stable or microsatellite-unstable groups, they found that individuals with heavily methylated but microsatellite-stable tumors had a significantly worse outcome than those with nonmethylated microsatellite-stable tumors. Very recently, in a study of 30 metastatic microsatellite-stable colon cancers, CIMP was also found to be associated with poor survival (14). Here, by focusing on stage IV cancers in a relatively large sample size, we essentially minimized the contribution of MSI to the results and reached conclusions similar to that of Ward et al., with statistical significance for individual markers. In our study, we showed that not only individual gene methylation but also concordant methylation of multiple genes predicts and likely confers a poor prognosis in advanced colorectal cancer. Interestingly, these findings point out a dichotomous situation where CIMP cases with early-stage disease may have a good prognosis (via hypermethylation of hMLH1 and MSI) and those with microsatellite-stable advanced disease may have a poor prognosis. This result also raises the distinct possibility that there could be more than one type of hypermethylator phenotype in colorectal cancer.
To apply our current results to clinical oncology, technical aspects of methylation measurement must be considered. As we discussed earlier, there is no consensus definition for CIMP at present, although the four genes described here (MINT1, MINT31, p14ARF, and p16INK4a) provide an attractive simplified classification method. However, it is important to note that a quantitative approach to methylation measurement is important in defining the phenotype. Thus, of the five commonly methylated genes, the only one studied by a nonquantitative method (MINT2, studied by MSP) was neither well correlated with the others nor associated with prognosis. Among the quantitative methods, COBRA as used here is accurate but somewhat cumbersome because of restriction digestion and gel analysis. Recently described alternate quantitative approaches (37, 38) to methylation analysis may be more appropriate for clinical laboratories.
The clinical implications of our data are clear. Patients with advanced colorectal cancer and hypermethylation of multiple genes have such a poor outcome following standard 5-FU–based chemotherapy that alternate approaches seem to be indicated. Indeed, it may be useful to stratify patients by methylation status in future clinical trials in advanced colorectal cancer, and possibly assign alternate treatments to patients with high levels of methylation. It will also be important to determine whether methylation remains a prognostic factor in patients treated with recently approved drugs for colorectal cancer (2), such as irinotecan, oxaliplatin, cetuximab, and bevacizumab. Ultimately, patients with high levels of methylation may be best treated from the outset with drugs that affect epigenetic processes (39), perhaps to be followed by standard chemotherapy approaches.
Supplementary Material
Acknowledgments
Grant support: NIH grants CA89245, CA105346, and CA098006. J-P.J. Issa is an American Cancer Society Clinical Research Professor supported by a generous gift from the F.M. Kirby Foundation.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
References
- 1.Jemal A, Murray T, Ward E, et al. Cancer statistics. 55. 2005. pp. 10–30. [DOI] [PubMed] [Google Scholar]
- 2.Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476–87. doi: 10.1056/NEJMra040958. [DOI] [PubMed] [Google Scholar]
- 3.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 4.Robbins DH, Itzkowitz SH. The molecular and genetic basis of colon cancer. Med Clin North Am. 2002;86:1467–95. doi: 10.1016/s0025-7125(02)00084-6. [DOI] [PubMed] [Google Scholar]
- 5.Compton CC, Fielding LP, Burgart LJ, et al. Prognostic factors in colorectal cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med. 2000;124:979–94. doi: 10.5858/2000-124-0979-PFICC. [DOI] [PubMed] [Google Scholar]
- 6.Westra JL, Plukker JT, Buys CH, Hofstra RM. Genetic alterations in locally advanced stage II/III colon cancer: a search for prognostic markers. Clin Colorectal Cancer. 2004;4:252–9. doi: 10.3816/ccc.2004.n.024. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–70. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 8.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 9.Kondo Y, Issa JPJ. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004;23:29–39. doi: 10.1023/a:1025806911782. [DOI] [PubMed] [Google Scholar]
- 10.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JPJ. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–6. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Issa JP. Opinion. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–93. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 12.Esteller M, Gonzalez S, Risques RA, et al. K-ras and p16 aberrations confer poor prognosis in human colorectal cancer. J Clin Oncol. 2001;19:299–304. doi: 10.1200/JCO.2001.19.2.299. [DOI] [PubMed] [Google Scholar]
- 13.Kohonen-Corish MR, Daniel JJ, Chan C, et al. Low microsatellite instability is associated with poor prognosis in stage C colon cancer. J Clin Oncol. 2005;23:2318–24. doi: 10.1200/JCO.2005.00.109. [DOI] [PubMed] [Google Scholar]
- 14.Ogino S, Meyerhardt JA, Kawasaki T, et al. CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Arch. 2007;450:529–37. doi: 10.1007/s00428-007-0398-3. [DOI] [PubMed] [Google Scholar]
- 15.Umetani N, Takeuchi H, Fujimoto A, Shinozaki M, Bilchik AJ, Hoon DS. Epigenetic inactivation of ID4 in colorectal carcinomas correlates with poor differentiation and unfavorable prognosis. Clin Cancer Res. 2004;10:7475–83. doi: 10.1158/1078-0432.CCR-04-0689. [DOI] [PubMed] [Google Scholar]
- 16.Van Rijnsoever M, Elsaleh H, Joseph D, McCaul K, Iacopetta B. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res. 2003;9:2898–903. [PubMed] [Google Scholar]
- 17.Ward RL, Cheong K, Ku SL, Meagher A, O’Connor T, Hawkins NJ. Adverse prognostic effect of methylation in colorectal cancer is reversed by microsatellite instability. J Clin Oncol. 2003;21:3729–36. doi: 10.1200/JCO.2003.03.123. [DOI] [PubMed] [Google Scholar]
- 18.Kane MF, Loda M, Gaida GM, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–11. [PubMed] [Google Scholar]
- 19.O’Dwyer PJ, Manola J, Valone FH, et al. Fluorouracil modulation in colorectal cancer: lack of improvement with N-phosphonoacetyl-L-aspartic acid or oral leucovorin or interferon, but enhanced therapeutic index with weekly 24-hour infusion schedule-an Eastern Cooperative Oncology Group/Cancer and Leukemia Group B Study. J Clin Oncol. 2001;19:2413–21. doi: 10.1200/JCO.2001.19.9.2413. [DOI] [PubMed] [Google Scholar]
- 20.Mostoslavsky R, Singh N, Kirillov A, et al. κ chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev. 1998;12:1801–11. doi: 10.1101/gad.12.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rashid A, Shen LL, Morris JS, Issa JPJ, Hamilton SR. CpG island methylation in colorectal adenomas. Am J Pathol. 2001;159:1129–35. doi: 10.1016/S0002-9440(10)61789-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen L, Kondo Y, Hamilton SR, Rashid A, Issa JPJ. p14 methylation in human colon cancer is associated with microsatellite instability and wild-type p53. Gastroenterology. 2003;124:626–33. doi: 10.1053/gast.2003.50102. [DOI] [PubMed] [Google Scholar]
- 23.Clark SJ, Harrison J, Paul CL, Frommer M. High-sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–7. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong ZG, Laird PW. COBRA: A sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–4. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goel A, Nagasaka T, Arnold CN, et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127–38. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Samowitz WS, Albertsen H, Herrick J, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–45. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 28.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JPJ. Aging and DNA methylation in colorectal mucose and cancer. Cancer Res. 1998;58:5489–94. [PubMed] [Google Scholar]
- 29.Rashid A, Issa JPJ. CpG island methylation in gastroenterologic neoplasia: A maturing field. Gastroenterology. 2004;127:1578–88. doi: 10.1053/j.gastro.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 30.Issa JPJ. Methylation and prognosis: Of molecular clocks and hypermethylator phenotypes. Clin Cancer Res. 2003;9:2879–81. [PubMed] [Google Scholar]
- 31.Weisenberger DJ, Siegmund D, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 32.Marsit CJ, Houseman EA, Christensen BC, et al. Examination of a CpG island methylator phenotype and implications of methylation profiles in solid tumors. Cancer Res. 2006;66:10621–9. doi: 10.1158/0008-5472.CAN-06-1687. [DOI] [PubMed] [Google Scholar]
- 33.Shen L, Ahuja N, Shen Y, et al. DNA methylation and environmental exposures in human hepatocellular carcinoma. J Natl Cancer Inst. 2002;94:755–61. doi: 10.1093/jnci/94.10.755. [DOI] [PubMed] [Google Scholar]
- 34.Toyota M, Ahuja N, Suzuki H, et al. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999;59:5438–42. [PubMed] [Google Scholar]
- 35.Ueki T, Toyota M, Sohn T, et al. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res. 2000;60:1835–9. [PubMed] [Google Scholar]
- 36.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–8. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colella S, Shen L, Baggerly KA, Issa JPJ, Krahe R. Sensitive and quantitative universal Pyrosequencing (TM) methylation analysis of CpG sites. Biotechniques. 2003;35:146–50. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 38.Cottrell SE, Laird PW. Sensitive detection of DNA methylation. Ann N Y Acad Sci. 2003;983:120–30. doi: 10.1111/j.1749-6632.2003.tb05967.x. [DOI] [PubMed] [Google Scholar]
- 39.Egger G, Liang GN, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–63. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.