Summary
The mechanisms underlying the regenerative abilities of certain model species are of central importance to the basic understanding of pattern formation. Complex organisms such as planaria and salamanders exhibit an exceptional capacity to regenerate complete body regions and organs from amputated pieces. However, despite the outstanding bottom-up efforts of molecular biologists and bioinformatics focused at the level of gene sequence, no comprehensive mechanistic model exists that can account for more than one or two aspects of regeneration. The development of computational approaches that help scientists identify constructive models of pattern regulation is held back by the lack of both flexible morphological representations and a repository for the experimental procedures and their results (altered pattern formation). No formal representation or computational tools exist to efficiently store, search, or mine the available knowledge from regenerative experiments, inhibiting fundamental insights from this huge dataset. To overcome these problems, we present here a new class of ontology to encode formally and unambiguously a very wide range of possible morphologies, manipulations, and experiments. This formalism will pave the way for top-down approaches for the discovery of comprehensive models of regeneration. We chose the planarian regeneration dataset to illustrate a proof-of-principle of this novel bioinformatics of shape; we developed a software tool to facilitate the formalization and mining of the planarian experimental knowledge, and cured a database containing all of the experiments from the principal publications on planarian regeneration. These resources are freely available for the regeneration community and will readily assist researchers in identifying specific functional data in planarian experiments. More importantly, these applications illustrate the presented framework for formalizing knowledge about functional perturbations of morphogenesis, which is widely applicable to numerous model systems beyond regenerating planaria, and can be extended to many aspects of functional developmental, regenerative, and evolutionary biology.
Keywords: Regeneration, Ontology, Database, Planaria, Developmental biology
Introduction
Understanding the regenerative abilities found in many organisms is a major challenge in biology, because it is not only a fundamental aspect of complex systems regulation in biology but also raises the possibility that such pathways can be activated in biomedical contexts to address human injury and disease (regenerative medicine) (Birnbaum and Sánchez Alvarado, 2008; Levin, 2011; Levin, 2012). The development of high-resolution genetic tools and molecular-biological approaches for manipulating cells during regeneration in addition to classical grafting experiments has given rise to an ever-increasing dataset linking experimental perturbations with patterning outcomes. For example, certain genetic or pharmacological perturbations can result in planarian worms with either two or no heads (Reddien and Sánchez Alvarado, 2004; Oviedo et al., 2010; Beane et al., 2011), while cutting and grafting experiments provide many examples of changes of limb morphology in insects and amphibians (Endo et al., 2000; Yakushiji et al., 2009).
However, despite these outstanding accomplishments, no mechanistic model has been proposed that accounts for more than one or two key features of regeneration. A satisfactory model of patterning (as distinct from a model of gene regulation, which by itself does not constrain geometry) must unambiguously explain the information and logical steps needed for a system to perform the observed morphogenetic process – including allometric scaling during remodeling, maintenance of anatomical polarity in fragments, precise regulation of stem cell behavior in creating missing tissue, self-limited growth programs, etc. (Lobo et al., 2012). Casting mechanistic models as constructive algorithms (showing the steps sufficient to produce or repair a given shape, not just the necessary genes/proteins without which a shape malformed) is the only way to determine whether our molecular pathways indeed explain the remarkable self-organization and repair properties of regeneration model organisms. Crucially, such models are required for the insights of developmental biology to ever be translated into rational interventions seeking to control and alter shape for regenerative medicine applications.
The main problem that holds back the development of algorithmic, comprehensive models of regeneration is the amount of raw functional and molecular data in the literature, which has become intractable for a single person. While a larger dataset should permit a more precise picture of the system, the lack of standardization greatly impedes the ability to glean comprehensive insights into shape and the relationship between individual cell regulation pathways and large-scale properties like anatomical polarity and size control (Lazebnik, 2002). The decades of work in regeneration has produced very few constructivist models, and the rapidly-increasing body of functional findings is making the problem worse – it is ever more difficult for scientists to come up with models that explain the increasingly constraining dataset.
A formalization must be developed to allow the unambiguous specification of experimental perturbations (e.g. cuts, gene knockdowns, physiological changes, etc.) and the resulting changes of morphology in the model species. In addition, a database is needed to store the total sum of the field's knowledge in this domain. An appropriate user-interface would then allow the database to be filled with data of the form “(experiment, outcome)”, taken from all available primary literature, and queried and mined by both human scientists and, more importantly, by future computational tools for model-building and discovery.
Ontologies for experiments and phenotypes
Natural language is imprecise and ambiguous (King et al., 2011); ontologies formalize knowledge by representing descriptions of concepts and relations relevant to an application domain or field (Bard, 2003). Their utility has been demonstrated in several biological fields, especially in genetics (Soldatova and King, 2005).
Ontologies have been applied to the formalization of experiments, which not only promotes semantic clarity but is also a technological necessity for the application of computational tools to automate the extraction of knowledge from scientific data (Soldatova and King, 2006). A few experiment ontologies exist, both general (Soldatova and King, 2006) and for specific knowledge domains (Whetzel et al., 2006; Ivchenko et al., 2011; Visser et al., 2011). However, no existent ontology can accommodate the specific characteristics of regenerative experiments. We need a specialized ontology that permits precise description of the outcome of regenerative experiments, including complex cuts and amputations, joining of several grafts, irradiation areas, genetic perturbation, etc.
Several valuable ontologies have been also proposed to formalize phenotypes, such as the Worm Phenotype Ontology for C. elegans (Schindelman et al., 2011), the Mammalian Phenotype Ontology (Smith and Eppig, 2009), and the Human Phenotype Ontology (Robinson and Mundlos, 2010). These ontologies are successfully being applied in several databases that link genetic and phenotypic data, such as the WormBase (Yook et al., 2012), the Mouse Genome Informatics Database (Eppig et al., 2012), and the multi-species PhenomicDB (Groth et al., 2007). These ontologies define a standardized structured vocabulary, where terms are placed hierarchically according to their ‘is-a’ or ‘part-of’ relationships. Specific phenotypes are described as a set of such terms, e.g. a phenotype can be described with the terms “barrel chest”, “distended abdomen”, and “decreased muscle weight” (sample terms extracted from the Mammalian Phenotype ontology). Another approach to use ontologies for describing phenotypes is the ‘EQ’ (Entity + Quality) method (Beck et al., 2009; Washington et al., 2009), where an entity (e.g. “eye”, “head”, “tail”) is described by a quality (e.g. “small”, “round”, “reduced length”). The entities are defined in any anatomical ontology, where the qualities are typically chosen from PATO, the Phenotypic Quality Ontology (Mungall et al., 2010).
These phenotype ontologies and databases are a great advancement over natural language descriptions disseminated in the literature; however, regeneration experiments need also a formalization language that permits describe arbitrary geometric relationships between the parts of a morphology. The most informative experiments are those where specific perturbations radically alter a normal morphology, and the resulting configurations must be describable by any useful ontology. What is needed is a mathematical language to describe a morphology by specifying which parts have been regenerated, their general shapes, and the topological connections among them. In contrast to a textual description, this type of formalism implicitly captures the meaning and allows quantitative comparisons between different morphologies: a four-headed worm differs from a two-headed worm in having two extra lateral heads. These formalisms are essential for the application of computational tools for the discovery of regeneration models that can explain the huge experimental dataset available in the literature.
Thus, a new formalism of shape and anatomical configuration is needed to complement the current phenotype ontologies with a balanced degree of morphological detail to describe what is currently known about the patterning changes that can be induced during regeneration. This new formalism will serve as a foundation for deriving insights that enable biomedically-relevant control of shape.
Phenotype formalizations based on shape
The traditional science of biological shape is called morphometrics (Zelditch et al., 2004), which focuses on the detailed quantitative study of shape by means of landmark coordinates and their statistical variation (Adams et al., 2004). However, morphometric methods are usually not the most appropriate techniques for use in describing the outcomes of regeneration studies. While morphometrics is concerned with exact quantitative differences between shared shape characteristics (landmark coordinates), developmental and regenerative biology studies deal mainly with non-shared characteristics of the morphology at the levels of anatomical qualitative identity (e.g. a region being specified to form a head versus a tail, possibly both having the same overall shape), topology (e.g. one-headed versus two-headed phenotypes – shapes that are not directly comparable by shared landmarks), and frequency (e.g. number of specific organs present in the organism). Thus, standard morphometrics are not directly applicable to such problems because such techniques can become trapped in a focus on many small, often irrelevant quantitative differences between morphologies that obscures their overall anatomical similarity, and because they are not suited to comparison of very different shapes that result from changes in the anatomical identity of body regions.
Previous efforts to encode the shape and pattern of organisms have also used formal generative systems, which are based on the iterative application of simple rules (Hogeweg and Hesper, 1974). Examples of generative systems include L-systems (based on rewriting grammar rules (Lindenmayer, 1968a; Lindenmayer, 1968b; Prusinkiewicz et al., 2007)) and cellular automata (based on a grid of cells that dynamically update their states according to a set of rules (Marée and Hogeweg, 2001)). However, it is in general very difficult to find the specific rules of a generative system that produces a given form (Hornby et al., 2003); hence, their usability for morphology formalization is limited.
Mathematical graphs are widely used for modeling biological phenomena (Mason and Verwoerd, 2007), from gene regulatory networks (de Jong, 2002) to evolutionary dynamics (Lieberman et al., 2005). Furthermore, graphs have been proposed to model the connectivity of the cells in a developing embryo (Nagl, 1979; Doi, 1984), as well as the dynamic process of morphogenesis (Bard, 2011; Lobo et al., 2011). Graphs, apart from having been deeply studied in theory and applications, have a long tradition as a powerful tool for pattern representation, classification, and comparison (Conte et al., 2004).
Here, we propose a new formalism for phenotypes based on mathematical graphs. Graph nodes can represent at a symbolic level both the regions and organs present in an organism, while graph links can represent their topological relations. Moreover, geometrical graphs, which are labeled with the spatial relations of the graph nodes (Pach, 1999), can be employed in order to formalize the geometrical properties of morphologies. Therefore, graphs make an ideal candidate for the formalization of organism morphologies in regenerative experiments, as we illustrate using the planarian worm model system.
Planarian model organism
Among the model organisms in regenerative research, the planarian flatworms are of particular interest due to their outstanding regenerative capacity combined with complex behavior and anatomy. The planarian body is characterized by a central nervous system (including a brain and a diverse set of sensory receptors including eyes), intestine, body-wall musculature, and bilateral symmetry (Reddien and Sánchez Alvarado, 2004); yet, planarians can regenerate any body part lost (including full head, brain, eyes, etc.) after almost any form of amputation. A complete worm can be regenerated from body fragments smaller than 1/200th of the adult size (Morgan, 1898). This enormous plasticity has fueled a spectacular effort by the planarian research community, which has produced a complete genome sequence (Robb et al., 2008) and an extensive literature of cutting experiments (Reddien and Sánchez Alvarado, 2004), gene expression maps (Adell et al., 2010; Reddien, 2011), drug-induced phenotypes (Palakodeti et al., 2008; Oviedo et al., 2010; Beane et al., 2011), and RNAi gene-knockdown experiments (Petersen and Reddien, 2008; Forsthoefel and Newmark, 2009; Petersen and Reddien, 2009; Rink et al., 2009; Pearson and Sánchez Alvarado, 2010; Gaviño and Reddien, 2011; Molina et al., 2011; Petersen and Reddien, 2011; Tasaki et al., 2011a).
Currently, there exist a number of planarian databases, including the hybridoma library of D. tigrina (Bueno et al., 1997), the annotated genomic database of S. mediterranea (Sánchez Alvarado et al., 2002; Robb et al., 2008; Adamidi et al., 2011), and the expressed sequence tag (EST) database of D. ryukyuensis (Ishizuka et al., 2007). However, we are far from an understanding of the links between genetic networks and resulting morphologies. The field needs specialized databases on regeneration, linking experiments to resultant morphologies, which can then be mined to extract relevant models of pattern formation.
The planarian dataset is an ideal candidate to illustrate the power of the new kind of phenotype ontology that we propose here – focused on the mathematical properties of the organism morphologies and the experimental manipulations that produce them. It is imperative that knowledge and efforts in understanding pattern formation receive the incredible benefits that genetics and cell biology reaped after the development of widely-available tools for storing and manipulating primary DNA sequences.
Results
Formalism for phenotype morphologies
We propose a mathematical labeled graph to represent phenotype morphologies. A graph is an abstract representation of a set of objects that can be connected to each other with a set of edges (links between two nodes). We use a graph to represent the organism morphology as follows: vertices denote regions and organs, while edges represent the adjacency between two regions or the location of an organ inside a region. In this way, an organism is divided in a mosaic of regions containing organs. The geometric characteristics of the morphology are stored as labels in the nodes and edges. These characteristics include the region and organ type, the overall shape and size of regions, the location and rotation of organs, etc. To better illustrate this morphological graph encoding, we chose the planarian worm as an application of the presented formalism.
The planarian wild-type anatomy is characterized by a long flat body consisting of three main regions: head, trunk, and tail (Fig. 1A,B), although the precise demarcation of each region's borders is not fully understood. The head region is the most anterior and contains the two eyes and two brain lobes; the trunk region contains the pharynx (a muscular tube used for both food intake and waste expelling); the tail region is the most posterior. Two nerve cords run laterally from the brain to the tail. Accordingly, a planarian morphology can be abstracted as a mosaic of two-dimensional regions (head, trunk, and tail) containing the main organs in the morphology (eyes, brain lobes, pharynx, and ventral nerve cords). These flatworm characteristics make the planarian body suitable for a morphological formalism limited to two dimensions, but the formalism can be readily extended to three-dimensional representations. For initial simplicity of illustrating our approach, we ignore here other major organs (such as the branched gastrovascular tract and excretory system) and numerous miscellaneous internal cell types.
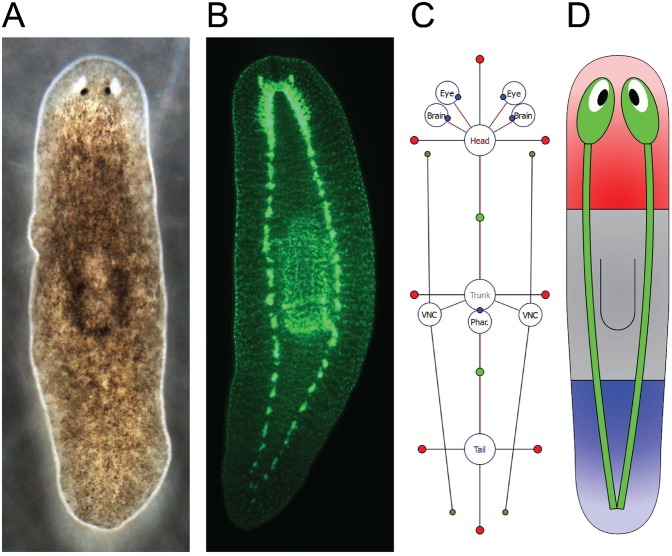
Fig. 1. Wild-type morphology of the planarian Schmidtea mediterranea and its formal representation.
(A) Dorsal side of the body, showing the eyes in the head region and pharynx in the trunk region. (B) Central nervous system (double immunostaining with anti-synapsin and anti-arrestin), highlighting the two brain lobes in the head region, the pharynx in the trunk region, and the two ventral nerve cords running laterally from the brain lobes to the tail region. (C) Diagram of the proposed graph formalism representing the same planarian morphology as in A,B. Regions and organs are represented by vertices labeled with their type; their topological configuration is represented by edges. Parameters store the basic shape (red dots) and adjacency border (green dots) of regions, the rotation of organs (blue dots), and the end points of the ventral nerve cords (gray dots). (D) Cartoon representation of the formalized morphology. Such schematic representations are automatically generated by the software tool to help the visualization of formalized morphologies.
Following the formalism, Fig. 1C shows a schematic representation encoding the phenotype shown in Fig. 1A,B; circles denote vertices and red lines denote edges. Vertices are labeled with the type of region or organ. Region locations are stored as edge labels containing the distance, angle, and location of the border between the two connected regions (represented as green dots in Fig. 1C). Region shapes are abstracted as a list of numerical parameters (included in the vertex label) that represent the distance between the center of the region and its border in a specific direction (red dots connected to region vertices in Fig. 1C). Non-connected regions have four parameters corresponding to the right, anterior, left, and posterior directions; regions connected to one region have three parameters corresponding to +90, +180, and +270 degrees with respect to the direction of the edge (e.g. head and tail regions in Fig. 1C); regions connected to more than one region have a parameter for each bisector of every two consecutive edges (e.g. trunk region in Fig. 1C). Organ locations are stored as vertex labels containing a vector position between the organ center and the center of the region where it is located; in addition, for spot-type organs (eye, brain, and pharynx), the vertex label includes the organ rotation (blue dots) and, in the case of line-type organs (ventral nerve cords), two organ-end vector positions (gray dots). A morphology graph is always connected (there is a path between any two regions or organs), since a region or organ cannot be isolated from the rest of the morphology.
A given morphology can be encoded with the formalism following a simple procedure. For every region present in the morphology, a vertex is added to the graph and labeled with its corresponding type (head, trunk, or tail in planaria). For every two regions adjacent in the morphology, an edge between the corresponding vertices is added and labeled with the distance and angle between the centers of the two regions and the distance between the center of the first region and the border with the other. Next, the shape parameters of the regions are assigned as the distance between the center of the region and its border in the direction of each parameter. Finally, for every organ present in the morphology, a new vertex labeled with its type (eye, brain, pharynx, or ventral nerve cord in the planarian illustration) and a new edge connecting it to the region where it is located are added to the graph; the edge is labeled with the vector position to the center of the region and organ rotation (spot organs) or two vector positions to the ends (line organs). This procedure can be performed manually with the help of the graphical software tool Planform (see below).
To facilitate the visualization of formalized morphologies, we implemented a simple algorithm to draw schematics illustrating the shape of the regions and the position of the organs. Since the graph formalism determines unambiguously morphologies, the algorithm is able to represent automatically a morphology encoded with the formalism as an illustrative cartoon diagram. Fig. 1D shows the worm-like representation generated from the encoded morphology in Fig. 1C. Regions are colored according to its type (head in red, trunk in gray, and tail in blue), and organ placements are sketched. The morphology formalism can also encode the morphological configurations during a regeneration process.
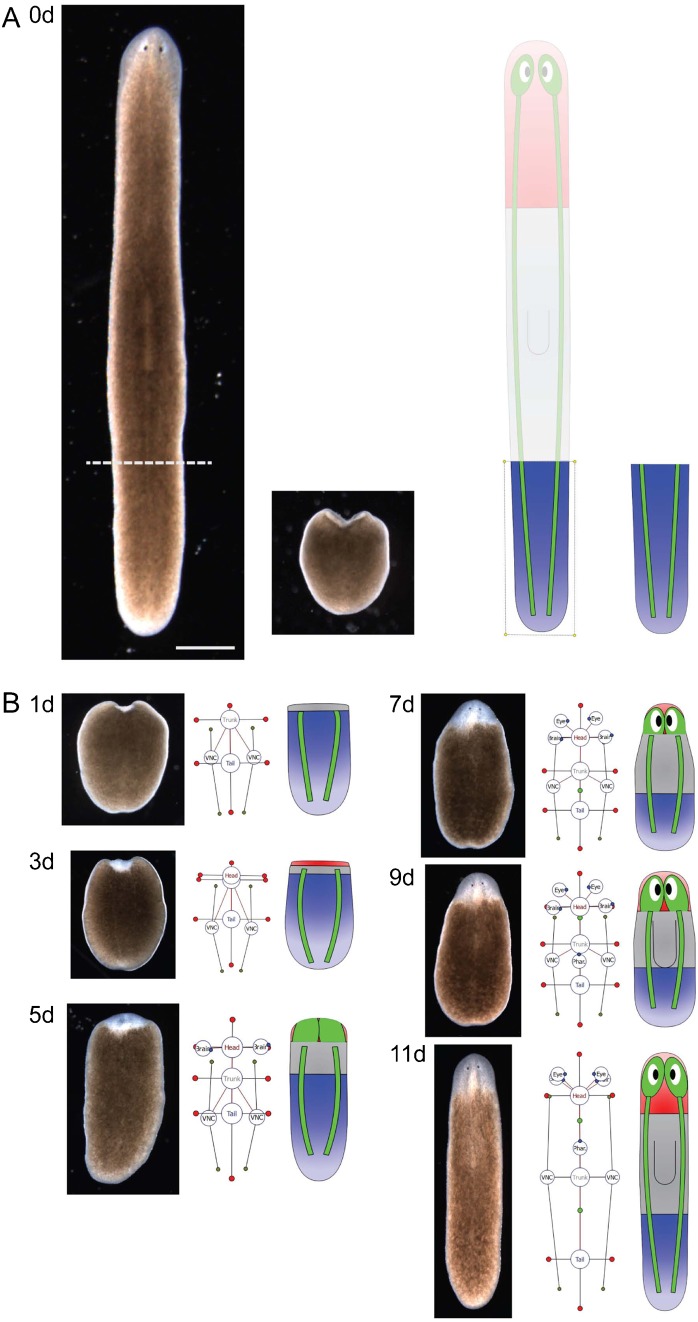
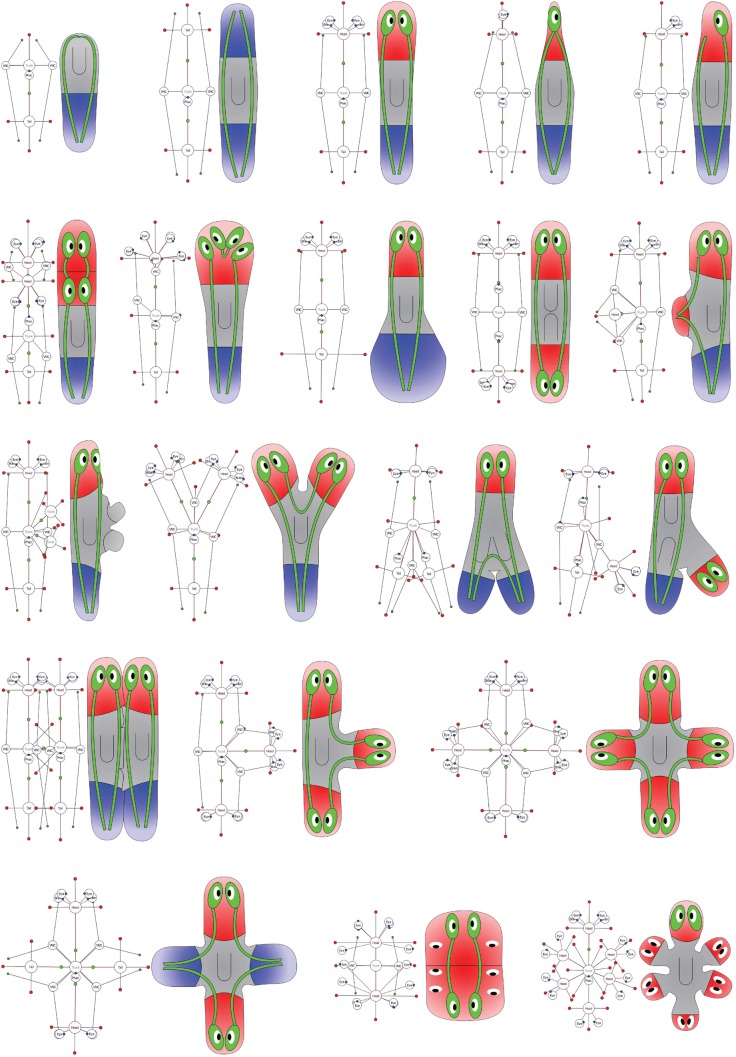
The morphology formalism can represent any possible morphological configuration. This derives from the fact that the formalism can represent both any region topology (a graph can decompose the plane or a volume in any arbitrary configuration) and any organ configuration (their number, position, and orientation are not limited). These characteristics make the formalism complete (universal); that is, all possible morphologies are representable. Fig. 2 illustrates a sequence of encoded two-dimensional morphological configurations during the regeneration of a tail fragment of the planarian S. mediterranea. The in-between morphologies, from a tail piece to a complete worm, can be easily represented and visualized with the formalism. Fig. 3 shows examples of encodings of worm morphologies found in the scientific literature of planarian regeneration. Multiple region types, ectopic organs and their physical characteristics, and the general topology of the worm are clearly and unambiguously specified with the presented formalism.
Fig. 2. A time-lapse of the regeneration process of a tail fragment of the planarian S. mediterranea and the corresponding encoding by the presented formalism.
(A) Amputation of a tail fragment (Day 0). (B) The regeneration sequence of the original regions and organs are unambiguously encoded with the formalism and clearly illustrated by the automatically-generated cartoon diagrams (Days 1–11). Scale bar: 1 mm.
Fig. 3. A selection of formalized morphologies represented by graph and cartoon diagrams and included in the centralized database.
The loss of the original morphological polarity is evident in the cartoon diagram. The formalism allows encoding morphologies practically with any anterior–posterior and medial–lateral patterning.
Formalism for experiment manipulations
During regenerative experiments, surgical, genetic, or pharmacological manipulations are often performed to test regenerative capacities under different conditions. Four basic types of manipulations are included in the current formalism: remove (an area of the organism is cut and discarded), crop (an area of the organism is cut and the rest discarded), join (two pieces are grafted together according to a vector location and a rotation), and irradiate (an area of the organism is exposed to radiation). These basic manipulations can be applied in any combination during the preparation of an experiment.
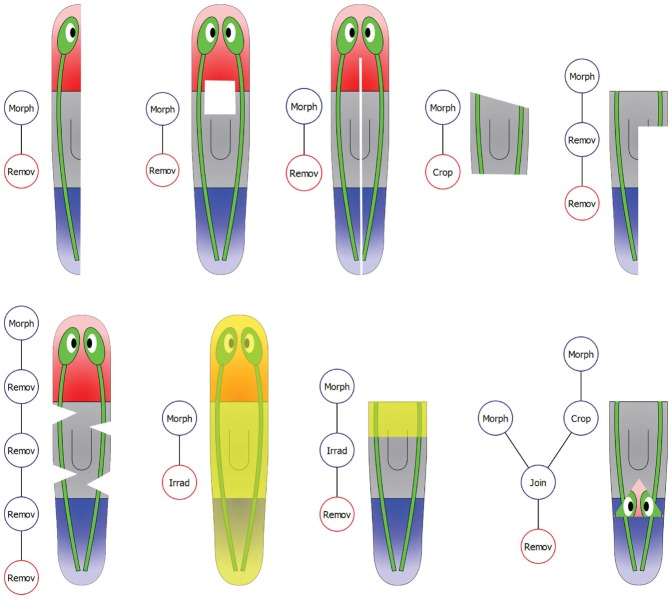
We propose a mathematical labeled tree (a hierarchical graph structure) to abstract the manipulations performed for an experiment: vertices represent basic manipulations or morphologies and edges connect the manipulation outputs and inputs. Fig. 4 shows a diagram of a formalized worm manipulation where one head of a two-headed worm is amputated and replaced by the tail of a wild-type worm. All basic manipulations produce one output, i.e. a piece. Remove, crop, and irradiate manipulations receive one input and are labeled with a list of spatial points defining the removed, cropped, or irradiated piece (yellow dots in the figure). Join manipulations receive two inputs, the two pieces to graft together, and are labeled with the rotation and location of the second piece with respect the first one. The initial morphologies used in the manipulations (the leaves of the tree) are defined according to the formalism for morphologies presented above (top two vertices in the figure). The output of the root of the tree (red vertex in the figure) defines the piece whose regenerative capacity is tested. Fig. 5 shows examples of encoded manipulations from published planarian experiments. All types of complex manipulations present in the literature can be clearly encoded within this formalism.
Fig. 4. An example of a formalized worm manipulation.
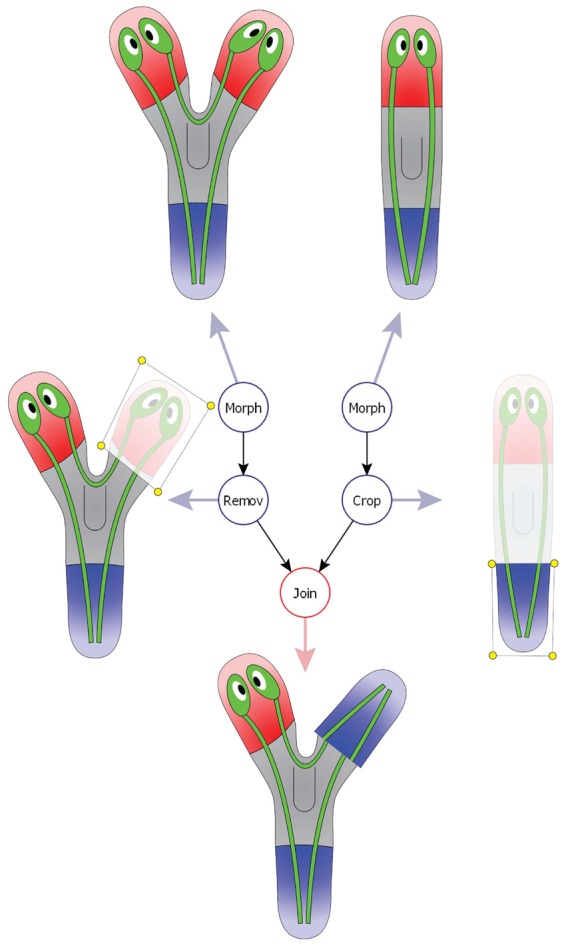
A tree combines morphologies (top two vertices) and basic manipulations, including removing a piece (left vertex), cropping a piece (right vertex), and joining two pieces (bottom vertex). The root of the tree (red) defines the final configuration created in an experiment.
Fig. 5. A selection of formalized manipulations (tree) and final configurations (cartoon) included in the centralized database.
Yellow regions indicate irradiated areas in the worm. Practically, the formalism is able to encode any type of amputation, irradiation, and transplantation (and their combinations).
Formalism for experiment data
Any regenerative experiment can now be described using two formal descriptors: a Manipulation and the resulting Morphologies. In addition, an experiment is encoded with the following information: a descriptive unique name, the publication reporting the results, the species used, any pharmacological compounds in the medium (including the starting and ending time of exposure), and any RNAi injections administered to the organism (which gene(s) have been targeted for knockdown). The results of an experiment (the resultant regenerated morphologies) are grouped by the time at which the morphologies appear. For each documented regeneration period in the experiment, the total number of individuals and the frequency distribution of each resultant morphology are included. Phenotypes with incomplete penetrance in treatments (different resultant morphologies for the same treatment and regeneration period) are supported in the formalism, and any human scientist or automated algorithm that processes the experimental data in this database can utilize this information in modeling endogenous variability and heterogeneity of response among animals. The complete information of an experiment, including where it is published, its setup and results, are unambiguously encoded in the formalism.
Database of regenerative experiments
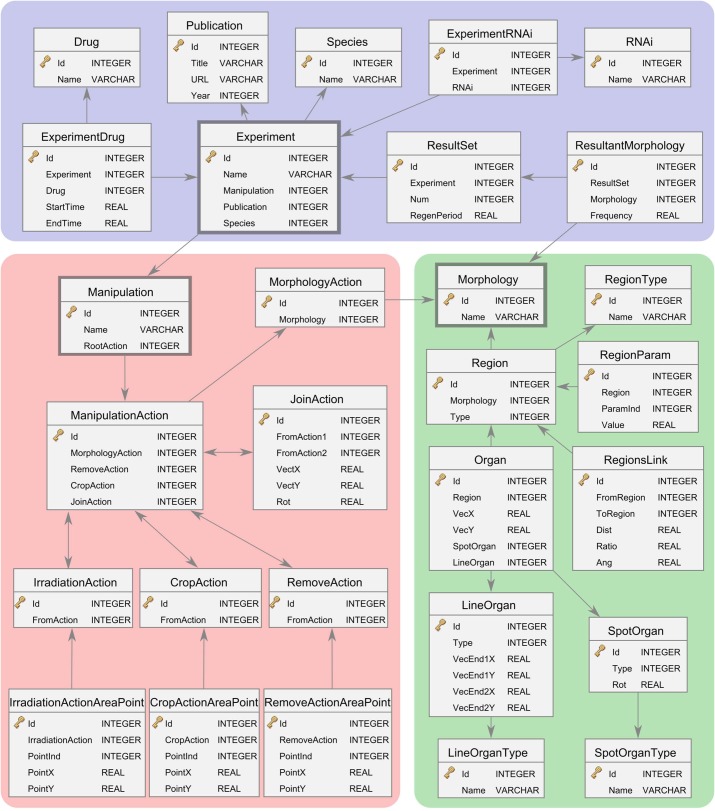
We modeled and implemented the presented formalism for regenerative experiments in a relational database. Fig. 6 shows a diagram representing the database schema – that is, the tables and their logical relations as defined in the database. The schema encodes in an optimal fashion the details of the experiments (tables in the blue area in Fig. 6), manipulations (tables in the red area), morphologies (tables in the green area), and the relations between them (arrows).
Fig. 6. Diagram of the relational database of regeneration experiments, including the tables and their attributes and relations.
The database includes data comprising experiments (blue), manipulations (red), and morphologies (green).
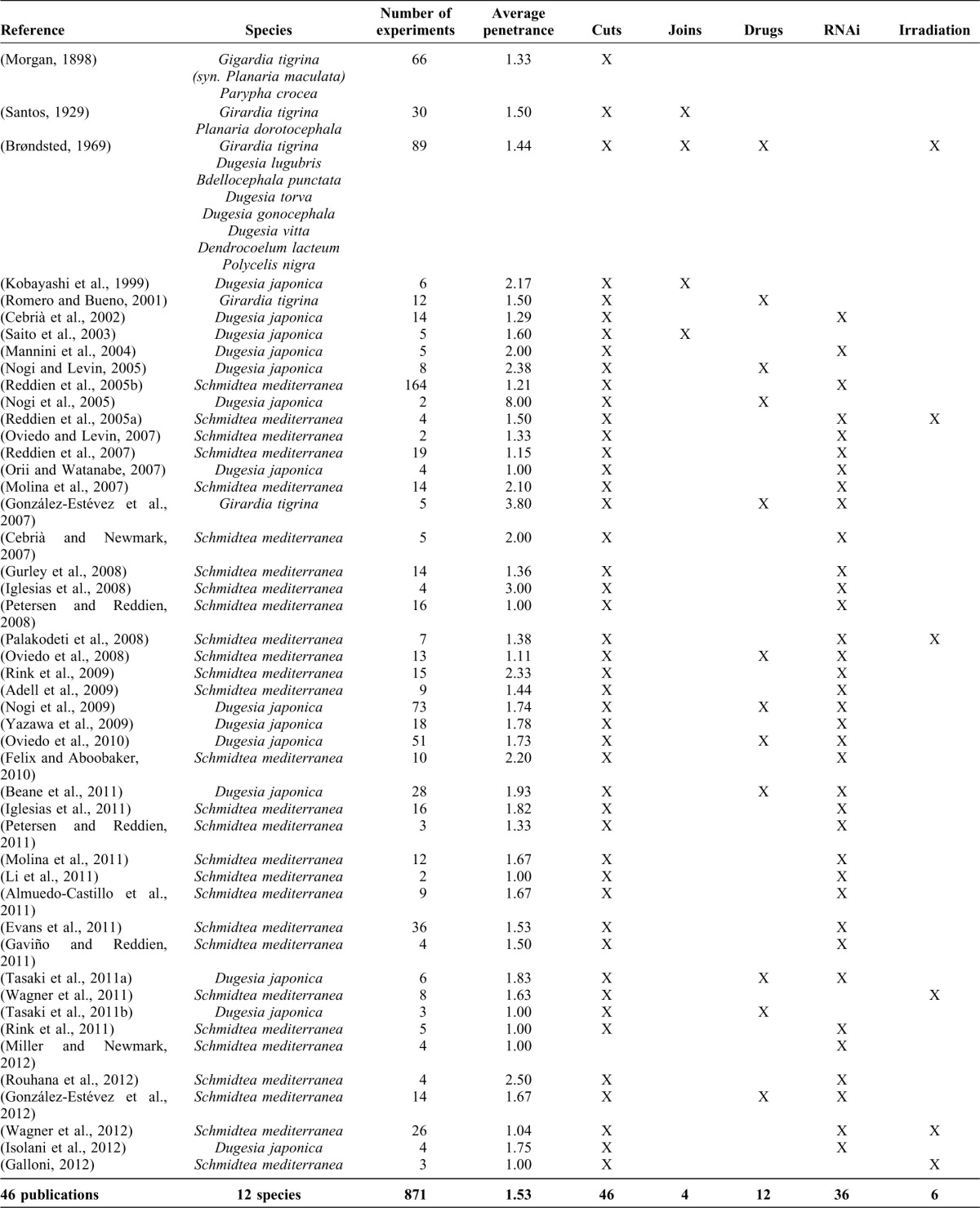
To illustrate the presented formalism, we curated (using a specific software tool, see below) the experiments reported in a selection of primary papers from the planarian literature, creating a centralized database of planarian experiments. Table 1 summarizes the publications and experiments included in the current version of the database, showing the publication reference, the species investigated, total number of experiments in the publication, average penetrance (the average number of different morphologies obtained in a specific experimental setup and time period), and the type of experiments performed (cuts, joins, RNAi injections, irradiation, or drug exposures).
Table 1. Summary of publications currently included in the database.
The database of planarian experiments is freely available on the web (http://planform.daniel-lobo.com). We are continuously expanding the database in our lab as we include more previously published works as well as new results appearing in the literature. Additionally, it is possible for researchers to submit new published data for inclusion in the database. If you would like to submit new data, please use the submission system available in the same website.
Software tool
To facilitate the use of the presented formalism and database, we designed and implemented a software tool. We adapted this tool for planarian experiments, calling it Planform (Planarian formalization). The tool can be used to work with our presented centralized database of planarian experiments published in the literature or with personal databases created by any user. Planform provides a graphical user interface that allows any scientist using a standard desktop computer to query, input, and search for specific planarian experiments.
Planform will be freely available on the web (http://planform.daniel-lobo.com). After downloading the application, a researcher can use Planform for searching specific experiments stored in the centralized database of published planarian experiments, adding new experiments to this database, or creating new personal databases. For example, a researcher studying the effects on pattern generation of a specific drug or gene knockdown can obtain a list of published planarian morphologies (including descriptive names and diagrams) that result from those treatments by typing the specific drug or gene name in the search module. In addition, researchers can unambiguously formalize with Planform the details and results from their own experiments and publications and submit them for inclusion into the centralized database.
Materials and Methods
Database implementation
The database of experiments following the presented formalism was implemented using the database engine SQLite (public domain). SQLite is a very popular embedded relational database management system that implements most of the Structured Query Language (SQL), which facilitates its interoperability with most of the current database applications. In SQLite, a database is contained in a single file, which includes both the schema and the data. In this way, a user can download a database in a single file, which simplifies the access (downloading a single file from the web), extension (the database is completely stored in a single local file that can be extended independently), and sharing of the data (the file containing a database can be copied or sent by e-mail).
Database curation
The centralized database currently contains 871 experiments manually curated from 46 publications from the scientific literature (Table 1), and we are continually expanding it. We have selected for this first version of the database those planarian papers reporting the most fundamental experiments in pattern regeneration, including the regeneration of morphological patterns along the anterior–posterior and medial–lateral axis under specific cuttings, amputations, transplantations, irradiation, drugs, and RNAi treatments. We are currently curating an additional database containing regeneration experiments of vertebrate and insect limbs. Furthermore, future versions of the formalism will be extended to include specific cell types, gene expression, and patterning along the dorsal–ventral axis.
Software implementation
The software tool was implemented and compiled as a native standalone desktop application for the Microsoft Windows, Mac OS X, and Linux platforms. The tool can create, read, and write any database following the presented schema of experiments. Planform write and reads databases stored in a single file, facilitating the organization and sharing of different databases. In the same way, the database of planarian experiments we curated is available as a single file compatible with Planform.
Discussion and conclusion
The field of regenerative biology is producing an ever-increasing amount of information about biological pattern formation following sophisticated surgical manipulations and molecular-genetic or pharmacological perturbations. However, this information is disseminated throughout the scientific literature and encoded in natural language, photographs, and cartoon diagrams of very diverse styles. We believe that the development of constructive models that produce true insight into high-level pattern regulation is inhibited by lack of: (1) a generalized mathematical language for describing experiments based on shape data (formalism), (2) a centralized formal database to store such data, and (3) bioinformatics tools to assist in the search and mining of these huge resources. Moreover, each new publication adds results that further constrain possible models, making it even more difficult for scientists to come up with models that correctly predict available results. Illustrated with the planarian regeneration dataset, we presented here the first steps in a long-term strategy to overcome these problems. Our system is a proof-of-principle platform that extends current ontological efforts and can be applied to numerous existing and future domains of knowledge about functional perturbations of shape in embryogenesis or regeneration. It forms the foundation for the development of automated computational tools for extracting mechanistic model data to enable understanding and control of large-scale patterning of complex structures.
We proposed here a formalism based on the concept of a mathematical graph to unambiguously encode morphologies and manipulations. Current phenotype ontologies store the information in a hierarchical, but textual manner, which is insufficient for a comprehensive database of regenerative experiments and is not understandable by computational applications. In contrast, a graph is a convenient mathematical abstraction to represent the characteristics of morphologies at multiple levels: symbolic (head versus tail), topological (head connected to trunk), and geometrical (general shape of a region). Crucially, this mathematical formalism of morphologies and experiments is not only useful for interactive access by scientists, but makes possible the application of automated artificial-intelligence bioinformatics tools to mine the experimental knowledge on regeneration.
Using this formalism, we implemented a database schema and curated the first database of regenerative experiments based on shape. An experiment in the database unambiguously describes the species, drugs, RNAi injections, surgical manipulations, irradiations, and resultant morphologies after regeneration. We selected the planarian worm to illustrate the presented formalism. All the morphological experiments published in a selection of primary papers from the planarian literature have been curated and introduced in the freely-available database.
Outcomes of planarian regeneration experiments were entered into the database by hand. As the set of results continues to grow, future efforts will be directed to automating the process of adding new results into the database. Automated pattern recognition algorithms have been applied to biological visual data (Shamir et al., 2010). A number of shape representations, key in the segmentation of images in these algorithms (Trinh and Kimia, 2007), have been proposed, including contours (Olson et al., 1980; Kass et al., 1988), deformations from a template shape (Cootes et al., 1995), graph-based representations (Joshi et al., 2002; Pizer et al., 2003), more abstract representations based on topological closed surfaces that model the external shape of an organism (Isaeva et al., 2006), and statistical shape models (Heimann and Meinzer, 2009). These shape formalisms represent great advances in the task of describing precisely (as close as possible to reality) the shapes of biological objects. However, developmental biology studies in general, and the field of regeneration in particular, are often concerned with mechanisms that determine high-level identity of body regions (such as the one-headed versus double-headed phenotypes, or the number of eyes, fingers, etc. in contrast to the exact curve that defines the body). Moreover, the differences (lack of standardization) of microphotography among published studies raises great challenges for the automated extraction of reliable phenotypic morphology data from journal figures. Thus, an interesting line for future research is the application of sophisticated pattern recognition algorithms to microscopy images for automating the addition of new studies' data to the database.
Finally, to facilitate the use of the database, we implemented a software tool (adapted to the planarian dataset) called Planform for the unambiguous specification, centralized storage, and effective search of regenerative experiments. Planform presents a graphical user interface with interactive graphs and user-friendly cartoon diagrams that permit a non-expert user to query and introduce experiments in the database. A search module in the tool allows mining the experimental literature in an easy and effective manner. In summary, storing new data and searching for experiments in the literature containing given characteristics (such as a specific manipulation or regenerated morphology), can be done effortlessly with the help of the software tool Planform and the database of regenerative experiments.
Although this approach is easily extendable, the current version presents some limitations regarding the experimental information that can be formalized. First, a perfectly detailed shape of the organism (useful for morphometric studies) is not the aim of the formalism; instead, a series of parameters approximate to an adequate degree the morphological shape of the regions and ignore contingent, irrelevant features such as body bending, which greatly differs among even normal individuals. This allows automated algorithms processing such data to focus on discovery of models that get the fundamental anatomy correct in predicting the outcomes of functional perturbations. The current dearth of comprehensive, constructive models that correctly predict the major features of the animal after various perturbations suggests that it is necessary to start by facilitating the search for pathway models that explain large-scale bodyplan anatomy, and then move on to fine-scale differences that are so well-suited for morphometric techniques. However, our scheme is compatible with subsequent efforts to incorporate traditional morphometrics for quantitative analysis of subtle deformations, and these can be added as soon as the necessary quantitative data accumulate in the literature.
Cell, cancer, evolutionary, synthetic, and developmental biology have been transformed by bioinformatics: the accessibility of all genetic sequences in one place and in a common format have greatly augmented the ability of scientists to analyze data and plan new experiments. Many of the most important advances in bioengineering and medical technology would be impossible without tools such as those available at NCBI and many other portals. However, this is only the first step, as these tools largely address sequence/structure and transcriptional networks. Current phenotype ontologies have not yet included functional machine-readable morphological data, and our system is an ideal ontological addition towards a new bioinformatics of shape. While the regeneration literature forms a natural and tractable test-bed for these concepts, we anticipate that the same strategy can be applied to numerous areas of developmental biology with the establishment of novel formalisms that complement current ontologies and databases.
The creation of gene ontologies and large gene function databases (Ashburner et al., 2000; Benson et al., 2012) combined with computational methods, including the modeling and simulation of genetic regulatory networks (de Jong, 2002), have allowed for the efficient application of novel bioinformatic algorithms to analyze sequence data, determine protein structures, predict gene functions, and ultimately provide automated discovery of gene regulatory network models consistent with genetic expression data. Similar assistance will be essential if we are to translate the investigation of developmental and regenerative pathways into biomedical strategies that manipulate biological shape. Already the data on just worm regeneration are so complex and plentiful that scientists are finding it very difficult to propose algorithmic, constructive models of pattern regulation that exhibit morphogenetic properties consistent with functional data. This problem will be only exacerbated by increasing numbers of experiments and better high-resolution analyses.
We are currently working on the development of artificial intelligence-based tools to assist scientists in deriving functional models of pattern formation from the presented database of knowledge in this field. The formalism and database we presented here constitute the first step towards a computational system to automate the discovery of models of regeneration from experimental data. The key benefit of the presented formalism is in its mathematical description, which can be interpreted and analyzed by a computer. The automatic comparison of morphologies is an essential element in heuristic search algorithms, which we are implementing for the automation of model discovery based on the mathematical formalism and database presented here. Future modules, built on top of this formalism, will derive candidate mechanistic models, simulate them in silico, and examine their behavior under all of the perturbations in the database to identify models that correctly predict and explain the properties of this remarkable regenerative system.
In summary, we have presented here the first steps towards a bioinformatics of shape that will help us to understand the key properties of pattern formation during regeneration. The development of computational tools to help derive testable, mechanistic models from functional perturbation data in model systems requires a mathematically formalized, deep database of morphological results such as the one presented here. We have illustrated the formalism with planarian experiments; yet, we are currently developing tools and curating experiment databases of other regenerative model organisms, including salamander and insect limbs, and deer antlers.
Acknowledgments
We thank Emma Marshall for her valuable help testing Planform early versions, Junji Morokuma for helpful suggestions and the photographs in Fig. 1, Kiyokazu Agata for the anti-arrestin antibody, and Wendy S. Beane, Tim Andersen, Jeffrey W. Habig, and the Levin Lab members for useful discussions. Grant Sponsors: NIH, G. Harold and Leila Y. Mathers Charitable Foundation, US. Army Medical Research and Materiel Command (USAMRMC), National Science Foundation. Grant Numbers: GM078484, W81XWH-10-2-0058, EF-1124651.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Adamidi C., Wang Y. B., Gruen D., Mastrobuoni G., You X. T., Tolle D., Dodt M., Mackowiak S. D., Gogol-Doering A., Oenal P. et al. (2011). De novo assembly and validation of planaria transcriptome by massive parallel sequencing and shotgun proteomics. Genome Res. 21, 1193–1200 10.1101/gr.113779.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D. C., Rohlf F. J., Slice D. E. (2004). Geometric morphometrics: ten years of progress following the ‘revolution’. Italian Journal of Zoology 71, 5–16 10.1080/11250000409356545 [DOI] [Google Scholar]
- Adell T., Salò E., Boutros M., Bartscherer K. (2009). Smed-Evi/Wntless is required for β-catenin-dependent and -independent processes during planarian regeneration. Development 136, 905–910 10.1242/dev.033761 [DOI] [PubMed] [Google Scholar]
- Adell T., Cebrià F., Saló E. (2010). Gradients in planarian regeneration and homeostasis. Cold Spring Harb. Perspect. Biol. 2, a000505 10.1101/cshperspect.a000505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almuedo-Castillo M., Saló E., Adell T. (2011). Dishevelled is essential for neural connectivity and planar cell polarity in planarians. Proc. Natl. Acad. Sci. USA 108, 2813–2818 10.1073/pnas.1012090108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T. et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard J. (2003). Ontologies: Formalising biological knowledge for bioinformatics. Bioessays 25, 501–506 10.1002/bies.10260 [DOI] [PubMed] [Google Scholar]
- Bard J. (2011). A systems biology representation of developmental anatomy. J. Anat. 218, 591–599 10.1111/j.1469-7580.2011.01371.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beane W. S., Morokuma J., Adams D. S., Levin M. (2011). A chemical genetics approach reveals H,K-ATPase-mediated membrane voltage is required for planarian head regeneration. Chem. Biol. 18, 77–89 10.1016/j.chembiol.2010.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck T., Morgan H., Blake A., Wells S., Hancock J. M., Mallon A. M. (2009). Practical application of ontologies to annotate and analyse large scale raw mouse phenotype data. BMC Bioinformatics 10 Suppl 5, S2 10.1186/1471-2105-10-S5-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D. A., Karsch-Mizrachi I., Clark K., Lipman D. J., Ostell J., Sayers E. W. (2012). GenBank. Nucleic Acids Res. 40, D48–D53 10.1093/nar/gkr1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K. D., Sánchez Alvarado A. (2008). Slicing across kingdoms: regeneration in plants and animals. Cell 132, 697–710 10.1016/j.cell.2008.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brøndsted H. V. (1969). Planarian Regeneration Oxford; New York: Pergamon Press. [Google Scholar]
- Bueno D., Baguñà J., Romero R. (1997). Cell-, tissue-, and position-specific monoclonal antibodies against the planarian Dugesia (Girardia) tigrina. Histochem. Cell Biol. 107, 139–149 10.1007/s004180050098 [DOI] [PubMed] [Google Scholar]
- Cebrià F., Newmark P. A. (2007). Morphogenesis defects are associated with abnormal nervous system regeneration following roboA RNAi in planarians. Development 134, 833–837 10.1242/dev.02794 [DOI] [PubMed] [Google Scholar]
- Cebrià F., Kobayashi C., Umesono Y., Nakazawa M., Mineta K., Ikeo K., Gojobori T., Itoh M., Taira M., Sánchez Alvarado A. et al. (2002). FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature 419, 620–624 10.1038/nature01042 [DOI] [PubMed] [Google Scholar]
- Conte D., Foggia P., Sansone C., Vento M. (2004). Thirty years of graph matching in pattern recognition. Int. J. Patt. Recogn. Artif. Intell. 18, 265–298 10.1142/S0218001404003228 [DOI] [Google Scholar]
- Cootes T. F., Taylor C. J., Cooper D. H., Graham J. (1995). Active shape models – their training and application. Comput. Vis. Image Underst. 61, 38–59 10.1006/cviu.1995.1004 [DOI] [Google Scholar]
- de Jong H. (2002). Modeling and simulation of genetic regulatory systems: a literature review. J. Comput. Biol. 9, 67–103 10.1089/10665270252833208 [DOI] [PubMed] [Google Scholar]
- Doi H. (1984). Graph-theoretical analysis of cleavage pattern: graph developmental system and its application to cleavage pattern of Ascidian egg. Dev. Growth Differ. 26, 49–60 10.1111/j.1440-169X.1984.00049.x [DOI] [PubMed] [Google Scholar]
- Endo T., Tamura K., Ide H. (2000). Analysis of gene expressions during Xenopus forelimb regeneration. Dev. Biol. 220, 296–306 10.1006/dbio.2000.9641 [DOI] [PubMed] [Google Scholar]
- Eppig J. T., Blake J. A., Bult C. J., Kadin J. A., Richardson J. E. (2012). The Mouse Genome Database (MGD): comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Res. 40, D881–D886 10.1093/nar/gkr974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. J., Owlarn S., Tejada Romero B., Chen C., Aboobaker A. A. (2011). Combining classical and molecular approaches elaborates on the complexity of mechanisms underpinning anterior regeneration. PLoS ONE 6, e27927 10.1371/journal.pone.0027927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix D. A., Aboobaker A. A. (2010). The TALE class homeobox gene Smed-prep defines the anterior compartment for head regeneration. PLoS Genet. 6, e1000915 10.1371/journal.pgen.1000915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel D. J., Newmark P. A. (2009). Emerging patterns in planarian regeneration. Curr. Opin. Genet. Dev. 19, 412–420 10.1016/j.gde.2009.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloni M. (2012). Global irradiation effects, stem cell genes and rare transcripts in the planarian transcriptome. Int. J. Dev. Biol. 56, 103–116 10.1387/ijdb.113455mg [DOI] [PubMed] [Google Scholar]
- Gaviño M. A., Reddien P. W. (2011). A Bmp/Admp regulatory circuit controls maintenance and regeneration of dorsal-ventral polarity in planarians. Curr. Biol. 21, 294–299 10.1016/j.cub.2011.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Estévez C., Felix D. A., Aboobaker A. A., Saló E. (2007). Gtdap-1 promotes autophagy and is required for planarian remodeling during regeneration and starvation. Proc. Natl. Acad. Sci. USA 104, 13373–13378 10.1073/pnas.0703588104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Estévez C., Felix D. A., Smith M. D., Paps J., Morley S. J., James V., Sharp T. V., Aboobaker A. A. (2012). SMG-1 and mTORC1 act antagonistically to regulate response to injury and growth in planarians. PLoS Genet. 8, e1002619 10.1371/journal.pgen.1002619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth P., Pavlova N., Kalev I., Tonov S., Georgiev G., Pohlenz H. D., Weiss B. (2007). PhenomicDB: a new cross-species genotype/phenotype resource. Nucleic Acids Res. 35, D696–D699 10.1093/nar/gkl662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley K. A., Rink J. C., Sánchez Alvarado A. (2008). β-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319, 323–327 10.1126/science.1150029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimann T., Meinzer H. P. (2009). Statistical shape models for 3D medical image segmentation: a review. Med. Image Anal. 13, 543–563 10.1016/j.media.2009.05.004 [DOI] [PubMed] [Google Scholar]
- Hogeweg P., Hesper B. (1974). A model study on biomorphological description. Pattern Recognit. 6, 165–179 10.1016/0031-3203(74)90019-3 [DOI] [Google Scholar]
- Hornby G. S., Lipson H., Pollack J. B. (2003). Generative representations for the automated design of modular physical robots. Robotics and Automation, IEEE Transactions on 19, 703–719 10.1109/TRA.2003.814502 [DOI] [Google Scholar]
- Iglesias M., Gomez-Skarmeta J. L., Saló E., Adell T. (2008). Silencing of Smed-βcatenin1 generates radial-like hypercephalized planarians. Development 135, 1215–1221 10.1242/dev.020289 [DOI] [PubMed] [Google Scholar]
- Iglesias M., Almuedo-Castillo M., Aboobaker A. A., Saló E. (2011). Early planarian brain regeneration is independent of blastema polarity mediated by the Wnt/β-catenin pathway. Dev. Biol. 358, 68–78 10.1016/j.ydbio.2011.07.013 [DOI] [PubMed] [Google Scholar]
- Isaeva V., Presnov E., Chernyshev A. (2006). Topological patterns in metazoan evolution and development. Bull. Math. Biol. 68, 2053–2067 10.1007/s11538-006-9063-2 [DOI] [PubMed] [Google Scholar]
- Ishizuka H., Maezawa T., Kawauchi J., Nodono H., Hirao Y., Nishimura O., Nakagawa H., Sekii K., Tasaka K., Tarui H. et al. (2007). The Dugesia ryukyuensis database as a molecular resource for studying switching of the reproductive system. Zoolog. Sci. 24, 31–37 10.2108/zsj.24.31 [DOI] [PubMed] [Google Scholar]
- Isolani M. E., Conte M., Deri P., Batistoni R. (2012). Stem cell protection mechanisms in planarians: the role of some heat shock genes. Int. J. Dev. Biol. 56, 127–133 10.1387/ijdb.113432mi [DOI] [PubMed] [Google Scholar]
- Ivchenko O., Younesi E., Shahid M., Wolf A., Müller B., Hofmann-Apitius M. (2011). PLIO: an ontology for formal description of protein-ligand interactions. Bioinformatics 27, 1684–1690 10.1093/bioinformatics/btr256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S., Pizer S., Fletcher P. T., Yushkevich P., Thall A., Marron J. S. (2002). Multiscale deformable model segmentation and statistical shape analysis using medial descriptions. IEEE Trans. Med. Imaging 21, 538–550 10.1109/TMI.2002.1009389 [DOI] [PubMed] [Google Scholar]
- Kass M., Witkin A., Terzopoulos D. (1988). Snakes: active contour models. Int. J. Comput. Vis. 1, 321–331 10.1007/BF00133570 [DOI] [Google Scholar]
- King R. D., Liakata M., Lu C., Oliver S. G., Soldatova L. N. (2011). On the formalization and reuse of scientific research. J. R. Soc. Interface 8, 1440–1448 10.1098/rsif.2011.0029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi C., Nogi T., Watanabe K., Agata K. (1999). Ectopic pharynxes arise by regional reorganization after anterior/posterior chimera in planarians. Mech. Dev. 89, 25–34 10.1016/S0925-4773(99)00192-6 [DOI] [PubMed] [Google Scholar]
- Lazebnik Y. (2002). Can a biologist fix a radio?—Or, what I learned while studying apoptosis. Cancer Cell 2, 179–182 10.1016/S1535-6108(02)00133-2 [DOI] [PubMed] [Google Scholar]
- Levin M. (2011). The wisdom of the body: future techniques and approaches to morphogenetic fields in regenerative medicine, developmental biology and cancer. Regen. Med. 6, 667–673 10.2217/rme.11.69 [DOI] [PubMed] [Google Scholar]
- Levin M. (2012). Morphogenetic fields in embryogenesis, regeneration, and cancer: non-local control of complex patterning. Biosystems 109, 243–261 10.1016/j.biosystems.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. Q., Zeng A., Han X. S., Wang C., Li G., Zhang Z. C., Wang J. Y., Qin Y. W., Jing Q. (2011). Argonaute-2 regulates the proliferation of adult stem cells in planarian. Cell Res. 21, 1750–1754 10.1038/cr.2011.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman E., Hauert C., Nowak M. A. (2005). Evolutionary dynamics on graphs. Nature 433, 312–316 10.1038/nature03204 [DOI] [PubMed] [Google Scholar]
- Lindenmayer A. (1968a). Mathematical models for cellular interactions in development. I. Filaments with one-sided inputs. J. Theor. Biol. 18, 280–299 10.1016/0022-5193(68)90079-9 [DOI] [PubMed] [Google Scholar]
- Lindenmayer A. (1968b). Mathematical models for cellular interactions in development. II. Simple and branching filaments with two-sided inputs. J. Theor. Biol. 18, 300–315 10.1016/0022-5193(68)90080-5 [DOI] [PubMed] [Google Scholar]
- Lobo D., Vico F. J., Dassow J. (2011). Graph grammars with string-regulated rewriting. Theor. Comput. Sci. 412, 6101–6111 10.1016/j.tcs.2011.07.004 [DOI] [Google Scholar]
- Lobo D., Beane W. S., Levin M. (2012). Modeling planarian regeneration: a primer for reverse-engineering the worm. PLOS Comput. Biol. 8, e1002481 10.1371/journal.pcbi.1002481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannini L., Rossi L., Deri P., Gremigni V., Salvetti A., Saló E., Batistoni R. (2004). Djeyes absent (Djeya) controls prototypic planarian eye regeneration by cooperating with the transcription factor Djsix-1. Dev. Biol. 269, 346–359 10.1016/j.ydbio.2004.01.042 [DOI] [PubMed] [Google Scholar]
- Marée A. F., Hogeweg P. (2001). How amoeboids self-organize into a fruiting body: multicellular coordination in Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 98, 3879–3883 10.1073/pnas.061535198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason O., Verwoerd M. (2007). Graph theory and networks in Biology. IET Syst. Biol. 1, 89–119 10.1049/iet-syb:20060038 [DOI] [PubMed] [Google Scholar]
- Miller C. M., Newmark P. A. (2012). An insulin-like peptide regulates size and adult stem cells in planarians. Int. J. Dev. Biol. 56, 75–82 10.1387/ijdb.113443cm [DOI] [PubMed] [Google Scholar]
- Molina M. D., Saló E., Cebrià F. (2007). The BMP pathway is essential for re-specification and maintenance of the dorsoventral axis in regenerating and intact planarians. Dev. Biol. 311, 79–94 10.1016/j.ydbio.2007.08.019 [DOI] [PubMed] [Google Scholar]
- Molina M. D., Neto A., Maeso I., Gómez-Skarmeta J. L., Saló E., Cebrià F. (2011). Noggin and noggin-like genes control dorsoventral axis regeneration in planarians. Curr. Biol. 21, 300–305 10.1016/j.cub.2011.01.016 [DOI] [PubMed] [Google Scholar]
- Morgan T. H. (1898). Experimental studies of the regeneration of Planaria maculata. Dev. Genes Evol. 7, 364–397 10.1007/BF02161491 [DOI] [Google Scholar]
- Mungall C. J., Gkoutos G. V., Smith C. L., Haendel M. A., Lewis S. E., Ashburner M. (2010). Integrating phenotype ontologies across multiple species. Genome Biol. 11, R2 10.1186/gb-2010-11-1-r2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagl M. (1979). A tutorial and bibliographical survey on graph grammars. Lecture Notes in Computer Science 73, 70–126 10.1007/BFb0025715 [DOI] [Google Scholar]
- Nogi T., Levin M. (2005). Characterization of innexin gene expression and functional roles of gap-junctional communication in planarian regeneration. Dev. Biol. 287, 314–335 10.1016/j.ydbio.2005.09.002 [DOI] [PubMed] [Google Scholar]
- Nogi T., Yuan Y. E., Sorocco D., Perez-Tomas R., Levin M. (2005). Eye regeneration assay reveals an invariant functional left-right asymmetry in the early bilaterian, Dugesia japonica. Laterality 10, 193–205 10.1080/1357650054200001440 [DOI] [PubMed] [Google Scholar]
- Nogi T., Zhang D., Chan J. D., Marchant J. S. (2009). A novel biological activity of praziquantel requiring voltage-operated Ca2+ channel β subunits: subversion of flatworm regenerative polarity. PLoS Negl. Trop. Dis. 3, e464 10.1371/journal.pntd.0000464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson A. C., Larson N. M., Heckman C. A. (1980). Classification of cultured mammalian cells by shape analysis and pattern recognition. Proc. Natl. Acad. Sci. USA 77, 1516–1520 10.1073/pnas.77.3.1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orii H., Watanabe K. (2007). Bone morphogenetic protein is required for dorso-ventral patterning in the planarian Dugesia japonica. Dev. Growth Differ. 49, 345–349 10.1111/j.1440-169X.2007.00931.x [DOI] [PubMed] [Google Scholar]
- Oviedo N. J., Levin M. (2007). smedinx-11 is a planarian stem cell gap junction gene required for regeneration and homeostasis. Development 134, 3121–3131 10.1242/dev.006635 [DOI] [PubMed] [Google Scholar]
- Oviedo N. J., Pearson B. J., Levin M., Sánchez Alvarado A. (2008). Planarian PTEN homologs regulate stem cells and regeneration through TOR signaling. Dis. Model. Mech. 1, 131–143, discussion 141 10.1242/dmm.000117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oviedo N. J., Morokuma J., Walentek P., Kema I. P., Gu M. B., Ahn J.-M., Hwang J. S., Gojobori T., Levin M. (2010). Long-range neural and gap junction protein-mediated cues control polarity during planarian regeneration. Dev. Biol. 339, 188–199 10.1016/j.ydbio.2009.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pach J. (1999). Geometric graph theory. Surveys In Combinatorics (London Mathematical Society Lecture Note Series), Vol. 267 Lamb J D, Preece D A, ed167–200Cambridge; New York: Cambridge University Press. [Google Scholar]
- Palakodeti D., Smielewska M., Lu Y. C., Yeo G. W., Graveley B. R. (2008). The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA 14, 1174–1186 10.1261/rna.1085008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson B. J., Sánchez Alvarado A. (2010). A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development 137, 213–221 10.1242/dev.044297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C. P., Reddien P. W. (2008). Smed-βcatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science 319, 327–330 10.1126/science.1149943 [DOI] [PubMed] [Google Scholar]
- Petersen C. P., Reddien P. W. (2009). Wnt signaling and the polarity of the primary body axis. Cell 139, 1056–1068 10.1016/j.cell.2009.11.035 [DOI] [PubMed] [Google Scholar]
- Petersen C. P., Reddien P. W. (2011). Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science 332, 852–855 10.1126/science.1202143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizer S. M., Fletcher P. T., Joshi S., Thall A., Chen J. Z., Fridman Y., Fritsch D. S., Gash A. G., Glotzer J. M., Jiroutek M. R. et al. (2003). Deformable m-reps for 3D medical image segmentation. Int. J. Comput. Vis. 55, 85–106 10.1023/A:1026313132218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusinkiewicz P., Erasmus Y., Lane B., Harder L. D., Coen E. (2007). Evolution and development of inflorescence architectures. Science 316, 1452–1456 10.1126/science.1140429 [DOI] [PubMed] [Google Scholar]
- Reddien P. W. (2011). Constitutive gene expression and the specification of tissue identity in adult planarian biology. Trends Genet. 27, 277–285 10.1016/j.tig.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W., Sánchez Alvarado A. (2004). Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 20, 725–757 10.1146/annurev.cellbio.20.010403.095114 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Oviedo N. J., Jennings J. R., Jenkin J. C., Sánchez Alvarado A. (2005a). SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310, 1327–1330 10.1126/science.1116110 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Bermange A. L., Murfitt K. J., Jennings J. R., Sánchez Alvarado A. (2005b). Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell 8, 635–649 10.1016/j.devcel.2005.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W., Bermange A. L., Kicza A. M., Sánchez Alvarado A. (2007). BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development 134, 4043–4051 10.1242/dev.007138 [DOI] [PubMed] [Google Scholar]
- Rink J. C., Gurley K. A., Elliott S. A., Sánchez Alvarado A. (2009). Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science 326, 1406–1410 10.1126/science.1178712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J. C., Vu H. T., Sánchez Alvarado A. (2011). The maintenance and regeneration of the planarian excretory system are regulated by EGFR signaling. Development 138, 3769–3780 10.1242/dev.066852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb S. M. C., Ross E., Sánchez Alvarado A. (2008). SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res. 36, D599–D606 10.1093/nar/gkm684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson P. N., Mundlos S. (2010). The human phenotype ontology. Clin. Genet. 77, 525–534 10.1111/j.1399-0004.2010.01436.x [DOI] [PubMed] [Google Scholar]
- Romero R., Bueno D. (2001). Disto-proximal regional determination and intercalary regeneration in planarians, revealed by retinoic acid induced disruption of regeneration. Int. J. Dev. Biol. 45, 669–673. [PubMed] [Google Scholar]
- Rouhana L., Vieira A. P., Roberts-Galbraith R. H., Newmark P. A. (2012). PRMT5 and the role of symmetrical dimethylarginine in chromatoid bodies of planarian stem cells. Development 139, 1083–1094 10.1242/dev.076182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Koinuma S., Watanabe K., Agata K. (2003). Mediolateral intercalation in planarians revealed by grafting experiments. Dev. Dyn. 226, 334–340 10.1002/dvdy.10246 [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A., Newmark P. A., Robb S. M. C., Juste R. (2002). The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development 129, 5659–5665 10.1242/dev.00167 [DOI] [PubMed] [Google Scholar]
- Santos F. V. (1929). Studies on transplantation in Planaria. Biol. Bull. 57, 188–197 10.2307/1536781 [DOI] [Google Scholar]
- Schindelman G., Fernandes J. S., Bastiani C. A., Yook K., Sternberg P. W. (2011). Worm phenotype ontology: integrating phenotype data within and beyond the C. elegans community. BMC Bioinformatics 12, 32 10.1186/1471-2105-12-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir L., Delaney J. D., Orlov N., Eckley D. M., Goldberg I. G. (2010). Pattern recognition software and techniques for biological image analysis. PLOS Comput. Biol. 6, e1000974 10.1371/journal.pcbi.1000974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Eppig J. T. (2009). The mammalian phenotype ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip. Rev. Syst. Biol. Med. 1, 390–399 10.1002/wsbm.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatova L. N., King R. D. (2005). Are the current ontologies in biology good ontologies? Nat. Biotechnol. 23, 1095–1098 10.1038/nbt0905-1095 [DOI] [PubMed] [Google Scholar]
- Soldatova L. N., King R. D. (2006). An ontology of scientific experiments. J. R. Soc. Interface 3, 795–803 10.1098/rsif.2006.0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki J., Shibata N., Nishimura O., Itomi K., Tabata Y., Son F., Suzuki N., Araki R., Abe M., Agata K. et al. (2011a). ERK signaling controls blastema cell differentiation during planarian regeneration. Development 138, 2417–2427 10.1242/dev.060764 [DOI] [PubMed] [Google Scholar]
- Tasaki J., Shibata N., Sakurai T., Agata K., Umesono Y. (2011b). Role of c-Jun N-terminal kinase activation in blastema formation during planarian regeneration. Dev. Growth Differ. 53, 389–400 10.1111/j.1440-169X.2011.01254.x [DOI] [PubMed] [Google Scholar]
- Trinh N. H., Kimia B. B. (2007). A symmetry-based generative model for shape. ICCV 2007: Proceedings Of The Eleventh IEEE International Conference On Computer Vision Rio de Janeiro: IEEE Computer Society. [Google Scholar]
- Visser U., Abeyruwan S., Vempati U., Smith R. P., Lemmon V., Schürer S. C. (2011). BioAssay Ontology (BAO): a semantic description of bioassays and high-throughput screening results. BMC Bioinformatics 12, 257 10.1186/1471-2105-12-257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D. E., Wang I. E., Reddien P. W. (2011). Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811–816 10.1126/science.1203983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D. E., Ho J. J., Reddien P. W. (2012). Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell 10, 299–311 10.1016/j.stem.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington N. L., Haendel M. A., Mungall C. J., Ashburner M., Westerfield M., Lewis S. E. (2009). Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 7, e1000247 10.1371/journal.pbio.1000247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetzel P. L., Parkinson H., Causton H. C., Fan L. J., Fostel J., Fragoso G., Game L., Heiskanen M., Morrison N., Rocca-Serra P. et al. (2006). The MGED Ontology: a resource for semantics-based description of microarray experiments. Bioinformatics 22, 866–873 10.1093/bioinformatics/btl005 [DOI] [PubMed] [Google Scholar]
- Yakushiji N., Yokoyama H., Tamura K. (2009). Repatterning in amphibian limb regeneration: a model for study of genetic and epigenetic control of organ regeneration. Semin. Cell Dev. Biol. 20, 565–574 10.1016/j.semcdb.2008.12.007 [DOI] [PubMed] [Google Scholar]
- Yazawa S., Umesono Y., Hayashi T., Tarui H., Agata K. (2009). Planarian Hedgehog/Patched establishes anterior-posterior polarity by regulating Wnt signaling. Proc. Natl. Acad. Sci. USA 106, 22329–22334 10.1073/pnas.0907464106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook K., Harris T. W., Bieri T., Cabunoc A., Chan J., Chen W. J., Davis P., de la Cruz N., Duong A., Fang R. H. et al. (2012). WormBase 2012: more genomes, more data, new website. Nucleic Acids Res. 40, D735–D741 10.1093/nar/gkr954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelditch M. L., Swiderski D. L., Sheets D. H., Fink W. L. (2004). Geometric Morphometrics For Biologists: A Primer Amsterdam; Boston: Elsevier Academic Press. [Google Scholar]