Summary
Mutations in rbf1, the Drosophila homologue of the RB tumour suppressor gene, generate defects in cell cycle control, cell death, and differentiation during development. Previous studies have established that EGFR/Ras activity is an important determinant of proliferation and survival in rbf1 mutant cells. Here, we report that Capicua (Cic), an HMG box transcription factor whose activity is regulated by the EGFR/Ras pathway, regulates both proliferation and survival of RB-deficient cells in Drosophila. We demonstrate that cic mutations allow rbf1 mutant cells to bypass developmentally controlled cell cycle arrest and apoptotic pressure. The cooperative effect between Cic and RBF1 in promoting G1 arrest is mediated, at least in part, by limiting Cyclin E expression. Surprisingly, we also found evidence to suggest that cic mutant cells have decreased levels of reactive oxygen species (ROS), and that the survival of rbf1 mutant cells is affected by changes in ROS levels. Collectively, our results elucidate the importance of the crosstalk between EGFR/Ras and RBF1 in coordinating cell cycle progression and survival.
Keywords: rbf1, RB, Capicua, Cell cycle regulation, Eye development
Introduction
The product of the Retinoblastoma tumour suppressor gene RB acts to control the cell cycle, cell death and differentiation during development (Sherr, 1996; Stevaux and Dyson, 2002; van den Heuvel and Dyson, 2008). Thus, loss of RB function, a frequent event in cancer, can lead to defects in various biological processes. A large fraction of these defects are believed to be mediated via the E2F family of transcription factors (Dimova and Dyson, 2005; Dyson, 1998). In quiescent cells, RB family proteins bind to E2F transcription factors, facilitating repression of genes that are necessary for the transition into the S phase of the cell cycle. When RB becomes hyperphosphorylated by Cyclin Dependent Kinases (CDK), interaction with E2Fs is inhibited, and, as a consequence, E2Fs proceed to activate genes involved in the progression of the cell cycle. Accordingly, in cancers, E2F transcription factors are constitutively active resulting in a lack of cell cycle control and uncontrolled proliferation.
While it is clear that E2F transcription factors play a central role, accumulating evidence suggests that there are secondary factors that facilitate development of RB-deficient cancers. Careful analysis of tissue samples from retinoblastoma patients has revealed that an increase in genomic instability correlates with a progression from non-proliferative retinomas to aggressive retinoblastomas (Bowles et al., 2007). Furthermore, inactivation of CDH11, one of the genes that is frequently lost in retinoblastoma, has been shown to promote survival of tumours in a Large T-antigen-induced mouse retinoblastoma model (Marchong et al., 2010). In a more recent study, integrative analysis of epigenomics and gene expression profiles identified that the expression of Spleen Tyrosine Kinase (SYK) is deregulated in retinoblastoma and has proven to be a potential therapeutic target (Zhang et al., 2012). Taken together, these studies indicate that factors other than E2Fs are deregulated in retinoblastoma and contribute to the tumorigenesis of RB-deficient cancer. Evidently, identification of such factors and elucidation of their molecular mechanisms can help us to better understand and target cancer cells.
Drosophila melanogaster provides a context in which the composition of RB/E2F family proteins is simplified, while the biological function is highly conserved. (Stevaux and Dyson, 2002; van den Heuvel and Dyson, 2008). RBF1 is the functional homologue of the RB tumour suppressor protein that regulates the two E2F transcription factors, dE2F1 and dE2F2. dE2F1 behaves as an activator while dE2F2 behaves as a repressor of transcription. This is in contrast to the complex mammalian system, composed of eight E2F proteins and two additional RB family proteins, p107 and p130, which have redundant function to RB. The simplified context of Drosophila RB/E2F provides a convenient genetic system to study their biological function during development.
In the Drosophila eye, RBF1 cooperates with the EGFR/Ras pathway to protect cells entering the morphogenetic furrow (MF) from apoptosis and to maintain cell cycle arrest of differentiating photoreceptors (Firth and Baker, 2005; Moon et al., 2006). At the molecular level, transcription of the pro-apoptotic gene hid is controlled by RBF1 while the activity of the gene product is regulated by the EGFR/Ras pathway via phosphorylation (Bergmann et al., 1998; Kurada and White, 1998; Moon et al., 2006; Moon et al., 2005). Moreover, expression of Dacapo (Dap), the Drosophila homologue of p21/27, accumulates in response to EGFR/Ras signalling (Firth and Baker, 2005). The primary function of Dap is to antagonize CDK2 whose kinase activity requires Cyclin E, a well-known transcriptional target of RBF1. As a consequence, at least in the context of eye development, the EGFR/Ras pathway is an important determinant of proliferation and survival of RB-deficient cells in Drosophila.
In an effort to comprehend the crosstalk that exists between RBF1 and the EGFR/Ras pathway, Capicua (Cic) emerged as a candidate protein that might cooperate with RBF1 to regulate proliferation. Cic is an HMG box transcription factor and was first identified as being essential for the establishment of dorsal–ventral polarity of the Drosophila egg (Jiménez et al., 2012). In this process, the EGFR/Ras pathway post-transcriptionally regulates Cic expression through a MAPK docking site at the C-terminus of Cic (Astigarraga et al., 2007). Notably, in the developing Drosophila eye, Cic is shown to restrict the rate of proliferation in response to EGFR/Ras activity, and the loss of Cic bypasses the requirement for EGFR/Ras activity in proliferation (Tseng et al., 2007). In mammals, it has been demonstrated that CIC forms a stable protein complex with ATXN-1, which is involved in neurodegenerative diseases (Lam et al., 2006). Moreover, CIC is found to form a chimeric protein with Double Homeobox protein 4 (DUX4) in a subset of Ewing-like sarcomas, implicating CIC in cancer (Kawamura-Saito et al., 2006). Despite the clear genetic evidence, molecular mechanisms by which CIC contributes to human disorders remain elusive.
Here, we report that Cic cooperates with RBF1 to restrict proliferation during Drosophila eye development. We demonstrate that Cic, together with RBF1, represses Cyclin E expression and promotes G1 arrest in asynchronously dividing precursor cells as they enter the MF. Moreover, we show that cic mutations can promote the survival of rbf1 mutant cells, which was previously shown to be EGFR/Ras dependent. Strikingly, further investigation of the pro-survival effect of cic mutations revealed that Cic regulates the level of reactive oxygen species (ROS), which are able to modulate the sensitivity of rbf1 mutant cells to undergo cell death. Our results provide evidence to demonstrate the importance of the crosstalk between the EGFR/Ras and RBF1 pathways in coordinating cell cycle progression and survival during Drosophila eye development.
Results
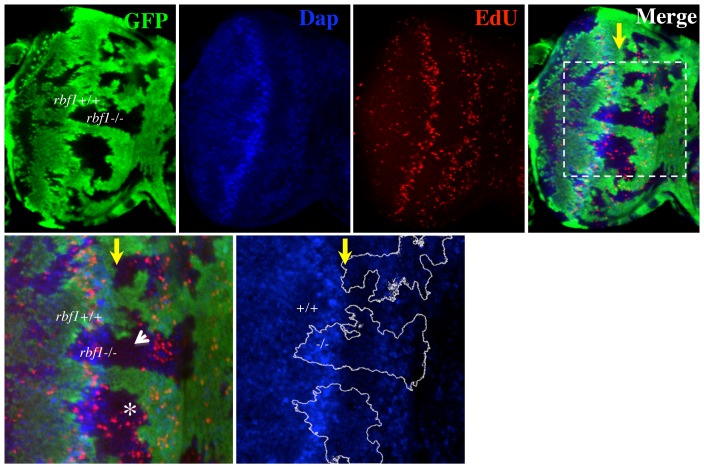
In third instar Drosophila eye imaginal discs, S-phase cells can be observed in two mitotic waves (Baker, 2001). The first mitotic wave is composed of asynchronously dividing cells towards the anterior of the disc. These cells are subsequently arrested in G1 at the morphogenetic furrow (MF) and a fraction of them begin to differentiate. Cells that are not differentiating at the MF enter one more round of synchronous S-phase towards the posterior of the MF, representing the second mitotic wave (SMW). In setting out to further comprehend how RBF1 controls the cell cycle in the eye imaginal disc, we generated mitotic clones of an rbf1 null allele, rbf1Δ14, in an eye-specific manner. Subsequently, third instar larval eye imaginal discs were dissected and fluorescently labelled with EdU, which allows visualisation of S-phase cells. We frequently observed rbf1 null clones with and without ectopic S-phase cells at the MF in a single eye imaginal disc (Fig. 1, asterisk and arrowhead). This result suggests that factors other than RBF1 are at play to promote G1 arrest at the MF. Notably, Dacapo (Dap), a Cyclin Dependent Kinase inhibitor, has been previously shown to cooperate with RBF1 to maintain photoreceptors in G1 phase of the cell cycle (Firth and Baker, 2005). However, immunostaining with anti-Dap revealed that Dap proteins are expressed at a relatively low level in the anterior region of the MF, raising the possibility that factors other than Dap might maintain rbf1 mutant cells in G1 in the anterior region of the MF (Fig. 1).
Fig. 1. rbf1 mutant cells arrest in G1 at the morphogenetic furrow (MF).
Mosaic clones of an rbf1 null allele, rbf1Δ14, were generated in third instar eye imaginal discs using eyFLP. rbf1Δ14 mutant clones are marked by the lack of GFP signal (green). Fluorescently labelled EdU (see Materials and Methods) was used to mark S-phase cells (red) and anti-Dacapo was used to determine the expression pattern of Dacapo (blue) in the eye disc. The position of the MF is indicated by a yellow arrow. A magnified view of the indicated area (white box) is presented below. Note that some rbf1 mutant clones contained S-phase cells at the MF (asterisk) while others do not (arrowhead).
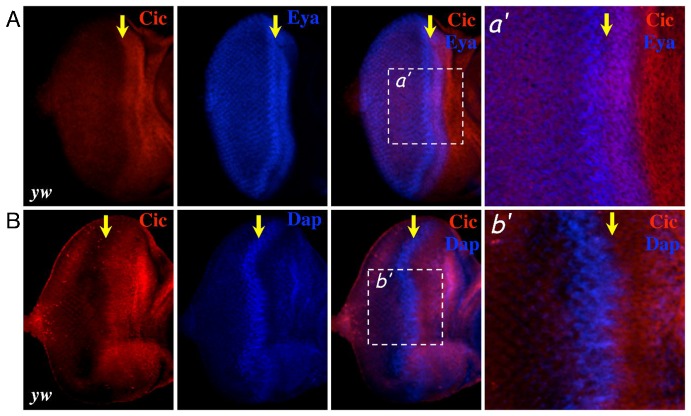
We began to explore factors downstream of the EGFR/Ras pathway that might synergize with RBF1 to promote cell cycle arrest. A candidate upon which we narrowed our focus was an HMG box transcription factor called Capicua (Cic). Similar to Dap, Cic is a negative regulator of proliferation whose expression is controlled by the EGFR/Ras pathway (Astigarraga et al., 2007). We first examined the expression pattern of Cic in eye imaginal discs by co-immunostaining for Cic and a nuclear protein Eyes Absent (Eya), which we used to visualise the nucleus of cells at the MF. As shown in Fig. 2A, Cic is expressed in the progenitor cells anterior to the MF as well as in the cells within the anterior half of the MF. Cic expression sharply decreases in the posterior half of the MF. Interestingly, eye discs co-immunostained for Cic and Dap reveal that Cic expression clearly declines where Dap proteins are highly expressed (Fig. 2B). These expression patterns suggest that the cellular context in which Cic functions as a negative regulator of proliferation might be distinct from that of Dap, and that Cic might be important for controlling proliferation in progenitor cells, including the cells at the anterior region of the MF.
Fig. 2. Expression pattern of Capicua in the third instar eye imaginal disc.
Third instar eye imaginal discs from yw flies are co-immunostained (A) with anti-Capicua (red) and anti-Eye Absence (blue) or (B) with anti-Capicua (red) and anti-Dacapo (blue). The position of the MF is indicated by a yellow arrow. Right panels show magnified views of the indicated areas (white box).
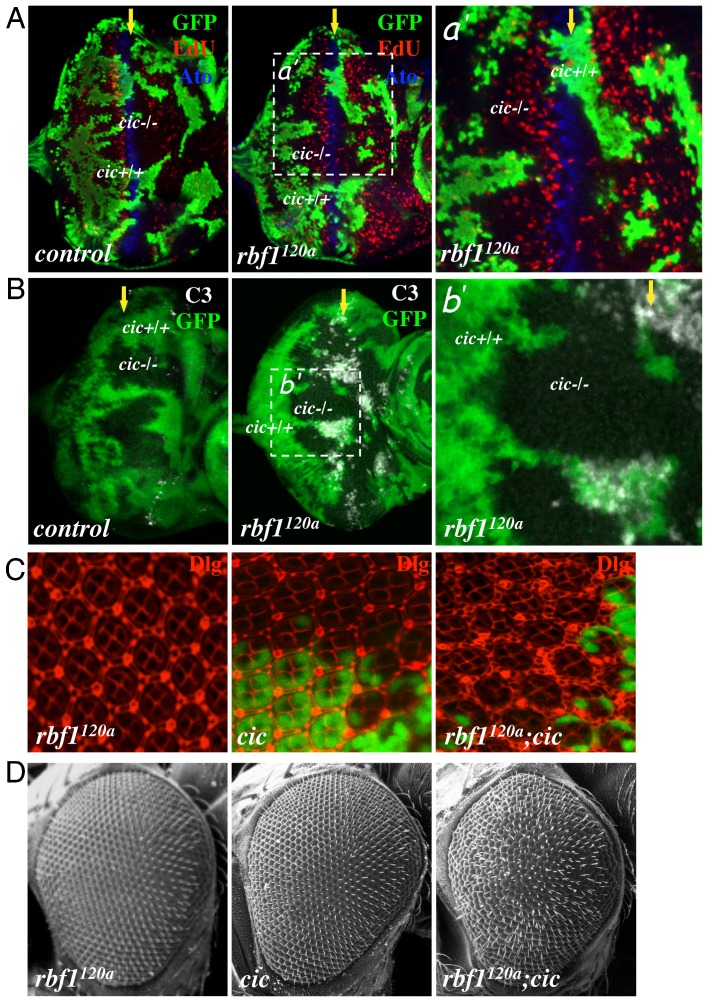
To explore whether Cic cooperates with RBF1 to promote G1 arrest, we examined the consequences of inactivating Cic in an rbf1 mutant background. As homozygous rbf1 null flies die at the first instar larval stage, we took advantage of an rbf1 hypomorphic allele, rbf1120a. Eye discs containing mutant clones of cic were generated in either a control or rbf1120a background. These eye discs were fluorescently labelled with EdU and subsequently immunostained for Atonal (Ato) to visualise S-phase cells and differentiating cells respectively. As previously reported, cic mutation alone cannot overcome the developmentally regulated G1 arrest at the MF (Fig. 3A) (Tseng et al., 2007). In contrast, rbf1 cic double mutant cells are capable of entering S phase in the MF. We also observed that the pattern of S-phase cells in the SMW is disrupted in the double mutant clones, which could be a secondary consequence of failure to arrest at the MF. Of note, we did not observe EdU staining in cells with high levels of Ato, indicating that once the differentiation process has begun, rbf1 cic double mutant cells remain arrested in G1. To better visualise the cell cycle defect, we generated eye discs entirely composed of cic single or rbf1 cic double mutant cells (see Materials and Methods). As expected, we did not observe any ectopic S-phase cells in cic mutant eye discs while ectopic S-phase cells are discernible in rbf1 cic double mutant eye discs ( supplementary material Fig. S1B). We used these rbf1 cic double mutant eye discs to quantify the number of ectopic S-phase cells in the MF and compared it to rbf1 mutant eye discs where we also occasionally observed ectopic S-phase cells (supplementary material Fig. S1C). Because the size of the MF varies between eye discs, we normalised the number of EdU positive cells by the size of the MF. This was achieved by measuring the number of pixels in images taken at the same magnification. We determined that on average 1.6±0.67 ectopic S-phase cells/10,000 pixels are present in rbf1 single mutant discs while 4.55±0.75 ectopic S-phase cells/10,000 pixels are present in rbf1 cic double mutant eye discs, showing a 2.8 fold increase. This result demonstrates that Cic and RBF1 collectively promote G1 arrest at the MF.
Fig. 3. cic mutations cooperate with rbf1 mutation to promote proliferation and survival.
(A) Mosaic clones of cic mutant cells were generated in control or rbf1120a mutant eye discs. Third instar eye imaginal discs were then incubated with EdU (red) to visualise S-phase cells and immunostained with anti-Atonal (blue). cic mutant clones are marked by the lack of GFP signal (green) and the position of the MF is marked by a yellow arrow. (B) Eye imaginal discs of the same genotypes as in A are shown. Anti-cleaved Caspase 3 (C3) was used to detect apoptotic cells (white). (C) Pupal eye discs (42 hours after pupal formation (APF)) of rbf1120a mutant flies, flies with cic mutant clones and rbf1120a mutant flies that contain cic mutant clones were immunostained with anti-Discs Large (red). (D) Scanning Electron Microscopy (SEM) images of Drosophila adult eyes entirely composed of rbf1120a or cic single mutant cells and rbf1120a cic mutant cells are presented.
We also investigated the possibility that Cic regulates survival of rbf1 mutant cells. Normally, rbf1 mutant cells undergo cell death in the anterior region of the MF (Moon et al., 2005; Tanaka-Matakatsu et al., 2009). Intriguingly, immunostaining with anti-cleaved Caspase-3 (C3), which marks apoptotic cells, revealed that the rbf1 cic double mutant cells no longer undergo cell death at the MF (Fig. 3B). In order to better visualise these pro-proliferative and pro-survival effects of the cic mutation on rbf1 mutant cells, we decided to examine the pupal eye. A typical ommatidial cluster at 42 hours after pupal formation (APF) contains 8 photoreceptors that are surrounded by accessory cells: 4 cone cells, 2 primary pigment cells, 6 secondary pigment cells and 3 tertiary pigment cells (Baker, 2001). Anti-Discs Large was used to visualise the cellular outlines in control and rbf1120a pupal eye discs. As shown in Fig. 3C, rbf1 cic double mutant ommatidial clusters display a great surplus of interommatidial cells, while only a few cic single mutant ommatidial clusters contain extra interommatidial cells. As a result, rbf1 cic double mutant adult eyes display a strong rough eye phenotype, which is not apparent in either rbf1 or cic single mutant adult eyes (Fig. 3D). Overall, our results suggest that cic mutations promote ectopic proliferation and survival in rbf1120a mutant eyes.
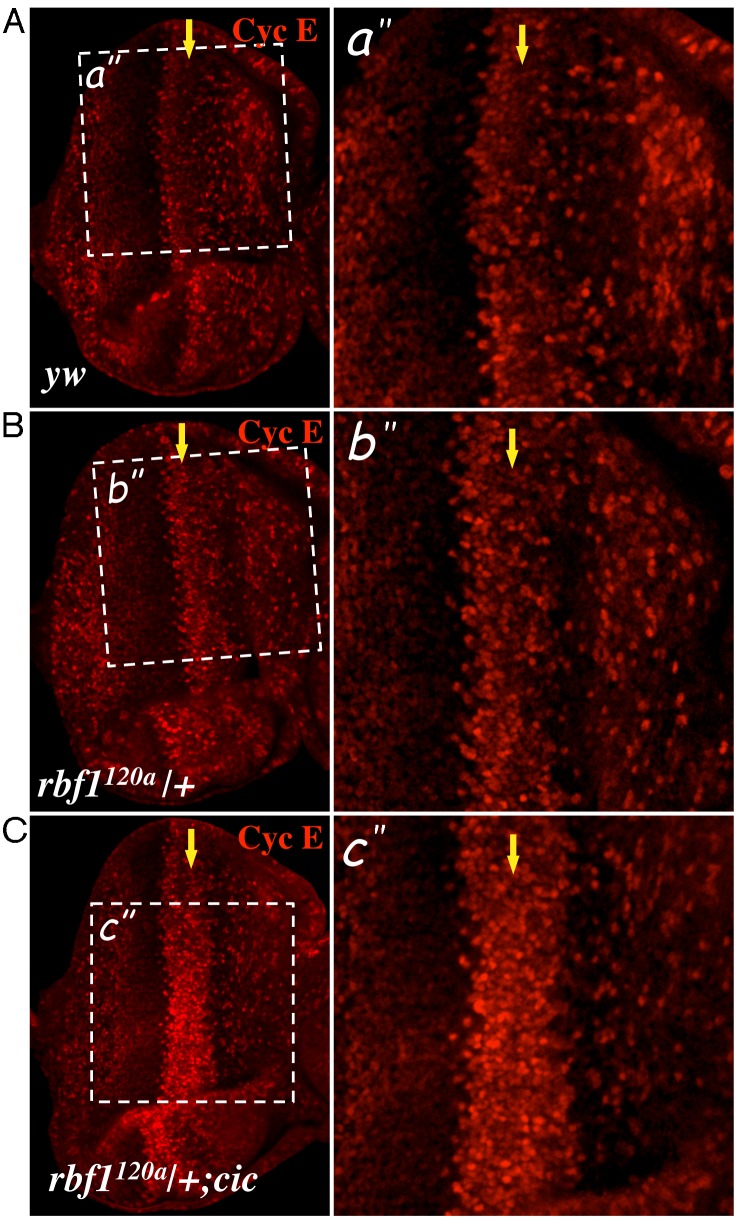
In an effort to understand how cic mutations might cooperate with rbf1 mutations to overcome G1 arrest, we considered the role of Cyclin E, which is a well-known target of RBF1. In developing eye imaginal discs, the expression pattern of Cyclin E largely reflects the global distribution of S-phase cells, exhibiting reduced expression in the MF and high levels of expression at the SMW (Fig. 4A). Surprisingly, we observed that reducing the gene dosage of rbf1 by half was sufficient to derepress Cyclin E expression at the MF (Fig. 4B). In rbf1120a heterozygous eye discs, cells with high levels of Cyclin E expression are more frequently observed compared to control eye discs. However, the cells at the SMW still display the highest level of Cyclin E expression in the rbf1120a heterozygous eye discs. With this is mind, we sought to use rbf1120a heterozygous eye discs as a sensitised genetic background to determine if RBF1 and Cic cooperate to control Cyclin E levels. In order to test this, we generated rbf1120a heterozygous eye discs carrying cic homozygous mutations using the methods described previously (see Materials and Methods). As shown in Fig. 4C, cic mutant cells with rbf1 heterozygous mutations exhibit augmented levels of Cyclin E expression at the MF comparable to the level observed at the SMW. It almost appears as if the SMW is anteriorly expanded. This effect of cic mutations on Cyclin E expression was not evident in the wild-type background, indicating that RBF1 function must be compromised for cic mutations to have an effect on Cyclin E expression (data not shown). Importantly, the increase in Cyclin E expression in the rbf1120a heterozygous background is not simply a consequence of ectopic S-phase cells since cic mutant cells in the rbf1120a heterozygous background properly arrest in G1 at the MF (supplementary material Fig. S1). Since cyclin E is a well-known transcriptional target of RBF1 and the effect of cic mutations on Cyclin E levels could be observed in the rbf1120a heterozygous background, we predicted that Cic would impinge on cyclin E transcription. Therefore, we performed RT-qPCR using eye discs from the genotypes in which we detected a difference in the Cyclin E protein level (supplementary material Fig. S2C). However, we did not observe any appreciable changes in the RNA level, indicating that the effect of cic mutations on Cyclin E expression is likely post-transcriptional. Nevertheless, our result suggests that RBF1 and Cic cooperatively promote G1 arrest at the MF, at least in part, by limiting Cyclin E expression.
Fig. 4. Cyclin E expression is deregulated in cic mutant clones.
Control (yw) and rbf1120a heterozygous (+/rbf1120a) eye discs and rbf1120a heterozygous eye discs also carrying cic homozygous mutations were immunostained with anti-Cyclin E (magenta). The position of the MF is marked by a yellow arrow. Magnified views of the indicated areas (white box) are also presented.
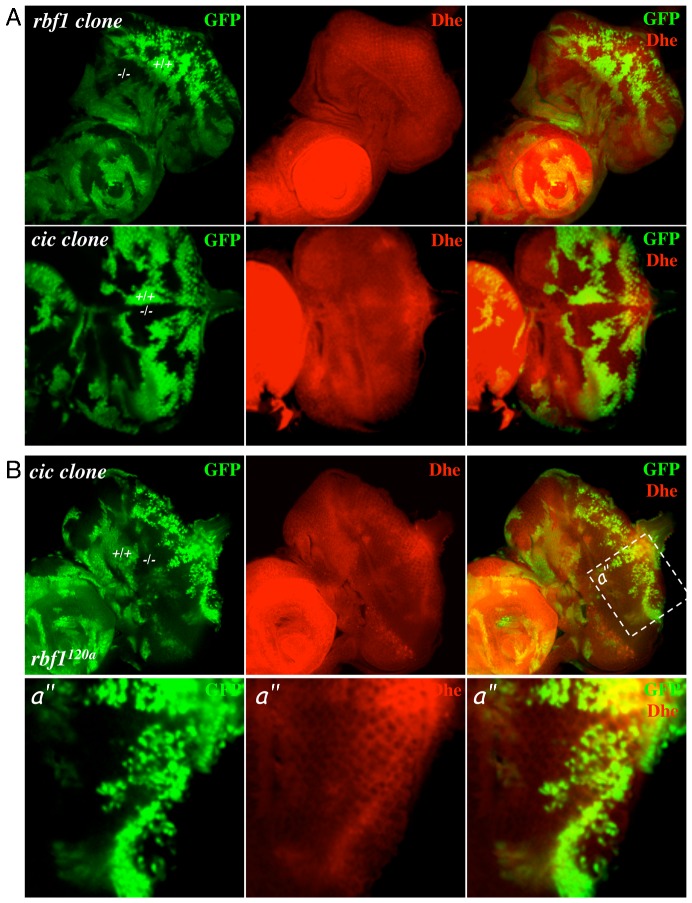
During the course of our study, an article was published demonstrating that Bantam miRNA expression is elevated in cic mutant cells (Herranz et al., 2012). Bantam miRNA has been shown to inhibit Hid expression (Brennecke et al., 2003), lending an explanation as to why cic mutations can inhibit the ectopic cell death in rbf1 mutant eye discs. However, this does not exclude the possibility that Cic possesses other cellular functions that could contribute to its pro-survival effect on rbf1 mutant cells. A handful of recent studies in mammals have made connections between RB and metabolic stress. E2F-1 and RB have proven to be a requirement for the repression of genes involved in oxidative metabolism, and inactivation of the tumour suppressor gene TSC2 induces cell death in RB-deficient cancer cells by promoting oxidative stress (Blanchet et al., 2011; Li et al., 2010). Hence, we set out to test whether cic and/or rbf1 loss had any discernible effect on reactive oxygen species (ROS) production. Eye discs containing homozygous mutant clones of either rbf1 or cic were generated and stained with dihydroethidium (Dhe) to monitor the levels of ROS. rbf1Δ14 clones showed no consistent difference in ROS levels (Fig. 5A). Contrastingly, mutant clones for cic showed a decrease in ROS levels in both wild-type and rbf1120a homozygous mutant backgrounds, suggesting that Cic is required for proper ROS homeostasis (Fig. 5).
Fig. 5. Cic mutant cells have a low level of Reactive Oxygen Species (ROS).
(A) Third instar eye discs containing mosaic clones of either rbf1 null mutant cells or cic mutant cells were stained with Dihydroethidium (Dhe) to monitor the levels of ROS. Note that the intensity of Dhe staining is weaker in cic mutant clones compared to the wild-type clones while it is relatively similar between the rbf1 mutant and control clones. (B) Mosaic clones of cic mutant cells were generated in rbf1120a mutant flies. Third instar eye imaginal discs were stained with Dhe to monitor the level of ROS. Magnified views of the indicated areas (white box) are also presented.
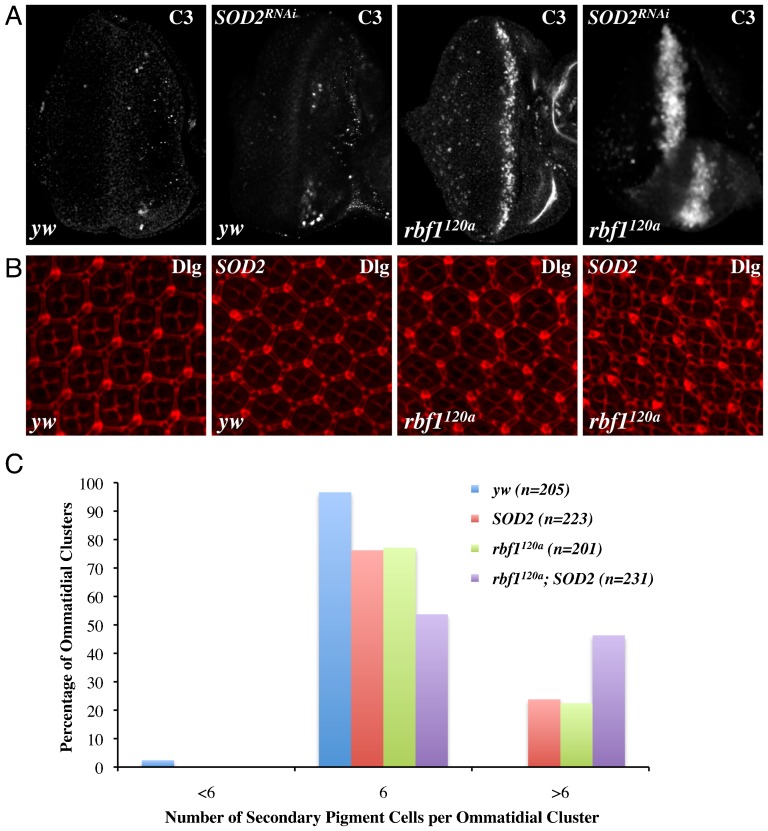
The decrease in ROS levels could simply be a consequence of cic mutations and may not contribute to the cell cycle and cell death phenotypes observed in rbf1 cic double mutant clones. Alternatively, these decreased ROS levels could directly contribute to the phenotypes. In order to distinguish between these two possibilities, we altered the ROS level in rbf1 mutant eye discs by modulating levels of ROS scavenger enzymes. To increase ROS levels, we expressed a Superoxide Dismutase 2 (SOD2) RNAi construct in rbf1120a eye discs. We observed that the stripe of cell death at the MF of rbf1120a eye discs, visualised by anti-C3, becomes wider when SOD2-RNAi is expressed (Fig. 6A). Importantly, there is no apparent increase in cell death when SOD2-RNAi was expressed in a wild-type background, indicating that the effect of SOD2-RNAi is rbf1 specific. Next, in order to test the outcome of reducing ROS, we overexpressed SOD2 in rbf1120a eye discs. We observed inconsistent results with larval eye discs where some eye discs show weak or partial decrease in cell death, while others show no change. Therefore, we decided to study pupal eye discs since the stereotypical organisation of an ommatidial cluster facilitates visualisation of defects in cell number. Pupal eye discs 42 hours APF were immunostained with anti-Discs Large (Fig. 6B). We observed that SOD2 overexpression in rbf1 mutant eyes can result in ommatidial clusters with extra interommatidial cells (Fig. 6B). However, because this effect seems to vary among the discs, and because we did observe that SOD2 overexpression can have an effect in the wild type eye discs, we quantified the ommatidial defect. For each genotype, we counted ommatidia with six secondary pigment cells, which is expected from wild type, and ommatidia with either less or more than six secondary pigment cells. This was done for random regions of ∼10 pupal eye discs for each genotype. As shown in Fig. 6C, SOD2 overexpression produces more ommatidia with extra interommatidial cells in the rbf1 mutant background than in the wild type background (24% in wild type and 46% in rbf1 mutant). Because 22% of the rbf1 mutant ommatidia already have extra interommatidial cells, the effect of SOD2 overexpression in the rbf1 mutant background could be additive. Nevertheless, our results collectively suggest that changes in ROS levels can influence the cell death phenotype in rbf1 mutant eyes and likely contribute to the pro-survival effect of cic mutations.
Fig. 6. SOD2 can promote the survival of rbf1 mutant cells.
(A) Third instar eye discs expressing dsRNA against Superoxide Dismutase 2 (SOD2) in either yw or rbf1120a backgrounds were immunostained with anti-C3. (B) Eye discs (42 hours APF) from flies overexpressing SOD2 in either yw or rbf1120a backgrounds were immunostained with anti-Discs Large. (C) The percentage of ommatidia with abnormal secondary pigment cells was determined for yw, SOD2, rbf1120a and rbf1120a; SOD2 pupal eye discs. In a control background 97% of ommatidial clusters have 6 secondary pigment cells, which is normally expected. However, in an rbf1120a background overexpressing SOD2, 46.3% of ommatidial clusters have greater than 6 secondary pigment cells. SOD2 and rbf1120a pupal eye discs contain 24% and 22% of ommatidial clusters with extra interommatidial cells respectively.
Discussion
We report here that a downstream effector of the EGFR/Ras pathway, Cic, is an important determinant of proliferation and survival of RB-deficient cells in Drosophila. We provide evidence to suggest that Cic synergizes with RBF1 to restrict Cyclin E expression at the MF and that Cic controls ROS levels that can influence the cell death phenotype in rbf1 mutant eyes. Our study strengthens the idea that extensive crosstalk exists between EGFR/Ras and RBF1, and demonstrates the importance of coordinating their functions during development.
Both Dap and Cic are negative regulators of proliferation downstream of the EGFR/Ras pathway. However, EGFR/Ras activity promotes Dap expression while inhibiting Cic expression (Astigarraga et al., 2007; Firth and Baker, 2005). Accordingly, their expression patterns at the MF show that Cic expression drops where Dap expression is most prominent. Immunostaining with anti-phospho-MAPK, which is a marker for EGFR activity and cells initiating differentiation processes, shows a similar expression pattern to that of Dap (Gabay et al., 1997). Perhaps, once cells start to differentiate, Dap plays a predominant role over Cic to maintain differentiating cells in the G1 phase. This would also explain the absence of EdU-positive cells in rbf1 cic double mutant clones that express Atonal (Fig. 3A). We noticed that Dap expression is slightly increased in rbf1 mutant clones. This is likely in response to the Cyclin E activation, a phenomenon which has been previously reported (de Nooij et al., 2000). In contrast to the MF, we could detect co-expression of Dap and Cic proteins in the anterior region of the eye disc (Fig. 2). Perhaps, in this region, EGFR/Ras is activated to a level at which both proteins can coexist. Nevertheless, our results indicate that, at least at the MF, the cellular context in which Dap and Cic act to restrict proliferation is distinct.
In cic homozygous mutant eye discs generated in the rbf1120a heterozygous background, Cyclin E levels are specifically increased at the MF (Fig. 4C). In fact, this is the location where dE2F1 proteins are most highly expressed in the eye disc (Moon et al., 2006). While this observation suggests a strong cooperation between dE2F1 and Cic, we did not observe any change in the dE2F1 protein level nor its activity in cic mutant clones (supplementary material Fig. S2). This result indicates that cic mutations do not affect dE2F1 activity in general. A previous study demonstrated that increases in both Cyclin E and E2F activities are necessary to overcome the cell cycle arrest imposed at the MF (Firth and Baker, 2005). This likely explains why we did not observe ectopic S-phase cells in cic mutant cells generated in rbf1120a heterozygous eye discs despite the elevated level of Cyclin E expression (supplementary material Fig. S1). E2F target genes are consistently expressed at a lower level in cic mutant eye discs generated in the rbf1120a heterozygous background than rbf1120a homozygous mutant eye discs (supplementary material Fig. S2). Nevertheless, we cannot exclude the possibility that dE2F1 is required for the effect of cic mutations on Cyclin E expression. It is interesting to note that both Dap and Cic act on Cyclin E/CDK2 and that their mutations cooperate with rbf1 mutations to cause uncontrolled proliferation. Perhaps, such a context in which either E2F1 or Cyclin E/CDK2 activity is elevated represents a sensitised genetic background where regulators of the other protein can be identified. Indeed, haploinsufficiency of rbf1 is shown to be sufficient to dominantly modify the rough eye phenotype induced by p21 overexpression, the mammalian inhibitor of Cyclin E/CDK2 (Du and Dyson, 1999).
The molecular mechanism by which RBF1 and Cic cooperatively regulate Cyclin E expression remains unclear. RT-qPCR did not reveal that cic mutations produce any discernable changes in cyclin E RNA levels, indicating that the effect of cic mutations on Cyclin E expression is post-transcriptional and likely to be indirect. One interesting observation is that heterozygosity of rbf1 seems to have a general effect on the expression level of RBF1 target genes (supplementary material Fig. S2B). Transcript levels of mcm2 and rnrS are elevated in the rbf1 heterozygous background compared to the wild type. This raises the possibility that increased expression of RBF1 target genes in general provides a specific context that allows the cic mutation to have an effect on Cyclin E expression. We are currently investigating the transcriptional changes that are induced by cic mutations in control and rbf1 mutant backgrounds to determine if Cic regulates different transcriptional programs depending on the status of RBF1.
Whether the alteration of Cyclin E levels is the only molecular mechanism by which cic and rbf1 mutations cooperate to promote ectopic S phase is still unclear. Oddly, we could not detect any discernible change in Cyclin E levels when cic mutant clones were generated in an rbf1120a homozygous background despite the presence of ectopic S-phase cells. One possible explanation for this is that rbf1 homozygous mutations increase Cyclin E expression to the level higher than what is achieved by cic mutations in rbf1120a heterozygous backgrounds. This increase is perhaps near, but not over, the threshold that can overcome the G1 arrest at the MF. In this context, cic mutations would provide the additional Cyclin E expression that is required to actually surpass this threshold. However, once cells enter S phase, Cyclin E is rapidly targeted for degradation, making it difficult to detect the increase in Cyclin E level. The lack of an increase in Cyclin E level in the rbf1120a homozygous background could also indicate that cic mutations can result in additional molecular changes that can cooperate with rbf1 mutations. While it is unclear what these changes might be, we know that Cic's ability to regulate ROS is not likely to contribute to the cell cycle defect. We did not observe any changes in the EdU staining pattern when expression levels of SOD2 were altered in an rbf1 mutant background (data not shown). Presently, we are in the process of comparing transcriptional changes induced by cic mutations in wild-type and rbf1 mutant backgrounds in order to postulate a molecular mechanism.
One of the unforeseen findings from our study is that ROS homeostasis is regulated by Cic. Reduced levels of ROS in cic mutant cells was not simply a secondary consequence of overcoming G1 arrest since cic mutant clones display normal patterns of proliferation in wild-type eye discs (Fig. 3A). Moreover, the changes in ROS levels are most evident at the posterior region of the MF where most cells are arrested in the G1 phase (Fig. 5). We measured the transcript levels of sod1, sod2, and catalase in a cic mutant background to determine if Cic controls ROS levels by regulating transcription of ROS scavenger enzymes. However, we did not observe any changes in their RNA levels, suggesting that Cic regulates ROS levels through an alternative mechanism (supplementary material Fig. S3). While this mechanism is still unclear, it is interesting to note that CIC in mammals is a component of the protein complex that includes Ataxin-1 (Atxn1), which is involved in Spinocerebellar Ataxia (Lam et al., 2006). A recent study demonstrated that CIC mutations can provide improvement of disease phenotypes observed in a polyglutamine-expanded Atxn1 mouse model (Fryer et al., 2011). It will be interesting to investigate if CIC regulates ROS levels in this biological context as well, and whether changes in ROS levels contribute to the effect of CIC mutations on the neurodegenerative phenotype.
Another interesting aspect of CIC in mammals is that CIC likely plays an important role in human cancers. In addition to the CIC–DUX4 chimeric protein mentioned in the introduction, a recent study revealed that CIC is somatically mutated in six out of seven oligodendroglioma patients (Bettegowda et al., 2011). Despite its possible involvement in human cancer, CIC knockout mice were reported to have no obvious tumour phenotype (Lee et al., 2011). It will be particularly interesting to test if CIC mutations can modify the tumour phenotypes of RB-deficient mice. Evidently, CIC is involved in a variety of important biological processes and warrants further investigation.
Materials and Methods
Fly stocks
All fly crosses were performed at 25°C. The rbf1 mutants, rbf1120a and rbf1Δ14 have been previously described (Du and Dyson, 1999). The cic allele used in this study is cicQ474X (Tseng et al., 2007). The SOD2 RNAi allele and the UAS-SOD2 allele were obtained from the Bloomington Stock Center.
Mosaic clones
rbf1Δ14FRT19A/GFPUbiFRT19A; eyFLP/+
rbf1120aeyFLP/Y; FRT82B GFPUbi/FRT82B cicQ474X
yw eyFLP/Y; FRT82B GFPUbi/FRT82B cicQ474X
rbf1120a eyFLP/+; FRT82B GFPUbi/FRT82B cicQ474X
SOD2 flies
rbf1120a eyFLP/Y; Act <CD2 <GAL4, UAS-GFP/SOD2 RNAi
yw eyFLP/Y; Act <CD2 <GAL4, UAS-GFP/SOD2 RNAi
rbf1120aeyFLP/Y; Act <CD2 <GAL4, UAS-GFP/UAS-SOD2
yw eyFLP/Y; Act <CD2 <GAL4,UAS-GFP/UAS-SOD2
Generation of eye discs entirely composed of cic mutant cells
ey-FLP/+ ; FRT 82B P(W+) l(3)cl-R3/FRT82B cicQ474X
rbf1120a ey-FLP/+; FRT 82B P(W+) l(3)cl-R3/FRT82B cicQ474X
rbf1120a ey-FLP/Y ; FRT 82B P(W+) l(3)cl-R3/FRT82B cicQ474X
Immunostaining
The following antibodies were used in this study: anti-C3 (1/200, Cell Signaling), anti-Dacapo (1/100, Developmental Studies Hybridoma Bank, DSHB), anti-Eyes Absent (1/100, DSHB) anti-Discs Large (1/100, DSHB), anti-Capicua (1/1000, a gift from Dr I. Hariharan), anti-Atonal (1/300, a gift from Dr Y.N. Jan), anti-Cyclin-E (1/100. a gift from Dr H. McNeill and Santa Cruz Bio.), anti-dE2F1 (1/1000, a gift from Dr N.J. Dyson).
For immunostaining, third larval instar or pupal eye discs were dissected and fixed with 4% paraformaldehyde for 20 to 30 minutes at room temperature. Next, discs were washed twice with 0.3% PBST (0.3% Triton X-100 in PBS) for 10 minutes. Eye discs were then incubated with primary antibody in 0.1% PBST with 5% normal goat serum (NGS) at room temperature for 2 hours. Eye discs were washed five times with 0.1% PBST and incubated with secondary antibody in 0.3% PBST with 5% NGS for 2 hours at room temperature. The immunostained discs were then washed five times with 0.1% PBST at room temperature and mounted for confocal microscopy imaging (Zeiss LSM).
EdU labeling
To visualise S-phase cells, Ethynyl-2′-Deoxyuridine (EdU) cell proliferation assay from Invitrogen was used with the following modification. Eye discs were dissected at the third larval instar stage and incubated in Schneider's medium containing EdU for 1 hour at room temperature. Eye discs were washed two times with 1× PBS and fixed at room temperature in 4% paraformaldehyde for 20 minutes. After fixation, eye discs were washed with PBS containing 3% BSA two times for 5 minutes. Eye discs were then incubated with the detection cocktail according to the manufacturer's specifications. Eye discs were treated with appropriate antibodies for immunostaining and mounted for confocal microscopy.
Real-time quantitative RT-PCR
Total RNA was isolated from 40 eye-antenna discs with RNeasy Mini Kit (QIAGEN) according to the manufacturer's specifications. RNA was reverse transcribed using DyNAmo cDNA Synthesis Kit (Finnzymes). DyNAmo Flash SYBR Green qPCR kit was subsequently used to perform quantitative PCR reactions. RP49 and β-tubulin were used as normalisation controls. Primers were designed with Primer3 (Whitehead Institute for Biomedical Research primer3 shareware [http://primer3.wi.mit.edu]). Primer pairs used were:
SOD1-Forward (ATTAACGGCGATGCCAAG) and SOD1-Reverse (ATTGGTGTTGTCACCGAACTC), SOD2-Forward (CGTAAAATTTCGCAAACTGC) and SOD2-Reverse (GTAGGTCTGGTGGTGCTTCTG), Catalase-Forward (ATCCCGTTGAGCAAATATCC) and Catalase-Reverse (AGGCATCCTTGATTCCAATG), Cyclin E-Forward (GTTTGTGCAAACCTCACAGT) and Cyclin E-Reverse (AACAGCGTAAAGCCATCTCC), Mcm2-Forward (AGGAACCACAGCTGAAGACC) and Mcm2-Reverse (CGTACATCTTGGCGATCTTG), RnrS Forward (AATGGCGTCCAAGGAAAAC) and RnrS Reverse (ACATCTTGCGAACGTTGTTG), β-tubulin-Forward (ACATCCCGCCCCGTGGTC) and β-tubulin-Reverse (AGAAAGCCTTGCGCCTGAACATAG), Rp49-F (TACAGGCCCAAGATCGTGAAG) and Rp49-R (GACGCACTCTGTTGTCGATACC).
Dhe staining
Dihydroethidium (Dhe) dye was reconstituted in 1 ml Schneider's medium to give a final concentration of 30 µM. Eye discs were subsequently incubated in solution for 5 minutes at room temperature in a dark chamber. Then three 5-minute washes were performed in 1 ml Schneider's medium. Discs were fixed for 5 minutes in 4% paraformaldehyde and washed once with 1× PBS for 5 minutes. Confocal microscopy was used to visualise the Dhe staining.
Supplementary Material
Acknowledgments
We would like to thank Dr Iswar Hariharan for providing us with cic stocks and antibodies. We would also like to thank Dr Helen McNeill and Dr Yuh Nung Jan for sharing their antibodies. Thanks to Bloomington Stock Center for providing the fly stocks and Developmental Studies Hybridoma Banks at the University of Iowa for antibodies. This study was supported by Canada Institute for Health Research grant MOP-93666, Natural Science and Engineering Research Council of Canada grant 355760-2008. N.-S.M. is a recipient of CIHR New Investigators Salary Award and M.-R.B.-G. is a recipient of NSERC Alexander Graham Bell Scholarship.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Astigarraga S., Grossman R., Díaz-Delfín J., Caelles C., Paroush Z., Jiménez G. (2007). A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J. 26, 668–677 10.1038/sj.emboj.7601532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N. E. (2001). Cell proliferation, survival, and death in the Drosophila eye. Semin. Cell Dev. Biol. 12, 499–507 10.1006/scdb.2001.0274 [DOI] [PubMed] [Google Scholar]
- Bergmann A., Agapite J., McCall K., Steller H. (1998). The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell 95, 331–341 10.1016/S0092-8674(00)81765-1 [DOI] [PubMed] [Google Scholar]
- Bettegowda C., Agrawal N., Jiao Y., Sausen M., Wood L. D., Hruban R. H., Rodriguez F. J., Cahill D. P., McLendon R., Riggins G. et al. (2011). Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 333, 1453–1455 10.1126/science.1210557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet E., Annicotte J. S., Lagarrigue S., Aguilar V., Clapé C., Chavey C., Fritz V., Casas F., Apparailly F., Auwerx J. et al. (2011). E2F transcription factor-1 regulates oxidative metabolism. Nat. Cell Biol. 13, 1146–1152 10.1038/ncb2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles E., Corson T. W., Bayani J., Squire J. A., Wong N., Lai P. B., Gallie B. L. (2007). Profiling genomic copy number changes in retinoblastoma beyond loss of RB1. Genes Chromosomes Cancer 46, 118–129 10.1002/gcc.20383 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D. R., Stark A., Russell R. B., Cohen S. M. (2003). bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36 10.1016/S0092-8674(03)00231-9 [DOI] [PubMed] [Google Scholar]
- de Nooij J. C., Graber K. H., Hariharan I. K. (2000). Expression of the cyclin-dependent kinase inhibitor Dacapo is regulated by cyclin E. Mech. Dev. 97, 73–83 10.1016/S0925-4773(00)00435-4 [DOI] [PubMed] [Google Scholar]
- Dimova D. K., Dyson N. J. (2005). The E2F transcriptional network: old acquaintances with new faces. Oncogene 24, 2810–2826 10.1038/sj.onc.1208612 [DOI] [PubMed] [Google Scholar]
- Du W., Dyson N. (1999). The role of RBF in the introduction of G1 regulation during Drosophila embryogenesis. EMBO J. 18, 916–925 10.1093/emboj/18.4.916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson N. (1998). The regulation of E2F by pRB-family proteins. Genes Dev. 12, 2245–2262 10.1101/gad.12.15.2245 [DOI] [PubMed] [Google Scholar]
- Firth L. C., Baker N. E. (2005). Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev. Cell 8, 541–551 10.1016/j.devcel.2005.01.017 [DOI] [PubMed] [Google Scholar]
- Fryer J. D., Yu P., Kang H., Mandel-Brehm C., Carter A. N., Crespo-Barreto J., Gao Y., Flora A., Shaw C., Orr H. T. et al. (2011). Exercise and genetic rescue of SCA1 via the transcriptional repressor Capicua. Science 334, 690–693 10.1126/science.1212673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay L., Seger R., Shilo B. Z. (1997). In situ activation pattern of Drosophila EGF receptor pathway during development. Science 277, 1103–1106 10.1126/science.277.5329.1103 [DOI] [PubMed] [Google Scholar]
- Herranz H., Hong X., Cohen S. M. (2012). Mutual repression by bantam miRNA and Capicua links the EGFR/MAPK and Hippo pathways in growth control. Curr. Biol. 22, 651–657 10.1016/j.cub.2012.02.050 [DOI] [PubMed] [Google Scholar]
- Jiménez G., Shvartsman S. Y., Paroush Z. (2012). The Capicua repressor – a general sensor of RTK signaling in development and disease. J. Cell Sci. 125, 1383–1391 10.1242/jcs.092965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura-Saito M., Yamazaki Y., Kaneko K., Kawaguchi N., Kanda H., Mukai H., Gotoh T., Motoi T., Fukayama M., Aburatani H. et al. (2006). Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum. Mol. Genet. 15, 2125–2137 10.1093/hmg/ddl136 [DOI] [PubMed] [Google Scholar]
- Kurada P., White K. (1998). Ras promotes cell survival in Drosophila by downregulating hid expression. Cell 95, 319–329 10.1016/S0092-8674(00)81764-X [DOI] [PubMed] [Google Scholar]
- Lam Y. C., Bowman A. B., Jafar-Nejad P., Lim J., Richman R., Fryer J. D., Hyun E. D., Duvick L. A., Orr H. T., Botas J. et al. (2006). ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell 127, 1335–1347 10.1016/j.cell.2006.11.038 [DOI] [PubMed] [Google Scholar]
- Lee Y., Fryer J. D., Kang H., Crespo-Barreto J., Bowman A. B., Gao Y., Kahle J. J., Hong J. S., Kheradmand F., Orr H. T. et al. (2011). ATXN1 protein family and CIC regulate extracellular matrix remodeling and lung alveolarization. Dev. Cell 21, 746–757 10.1016/j.devcel.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Gordon G. M., Du C. H., Xu J., Du W. (2010). Specific killing of Rb mutant cancer cells by inactivating TSC2. Cancer Cell 17, 469–480 10.1016/j.ccr.2010.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchong M. N., Yurkowski C., Ma C., Spencer C., Pajovic S., Gallie B. L. (2010). Cdh11 acts as a tumor suppressor in a murine retinoblastoma model by facilitating tumor cell death. PLoS Genet. 6, e1000923 10.1371/journal.pgen.1000923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon N. S., Frolov M. V., Kwon E. J., Di Stefano L., Dimova D. K., Morris E. J., Taylor-Harding B., White K., Dyson N. J. (2005). Drosophila E2F1 has context-specific pro- and antiapoptotic properties during development. Dev. Cell 9, 463–475 10.1016/j.devcel.2005.08.015 [DOI] [PubMed] [Google Scholar]
- Moon N. S., Di Stefano L., Dyson N. (2006). A gradient of epidermal growth factor receptor signaling determines the sensitivity of rbf1 mutant cells to E2F-dependent apoptosis. Mol. Cell. Biol. 26, 7601–7615 10.1128/MCB.00836-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C. J. (1996). Cancer cell cycles. Science 274, 1672–1677 10.1126/science.274.5293.1672 [DOI] [PubMed] [Google Scholar]
- Stevaux O., Dyson N. J. (2002). A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 14, 684–691 10.1016/S0955-0674(02)00388-5 [DOI] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M., Xu J., Cheng L., Du W. (2009). Regulation of apoptosis of rbf mutant cells during Drosophila development. Dev. Biol. 326, 347–356 10.1016/j.ydbio.2008.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng A. S., Tapon N., Kanda H., Cigizoglu S., Edelmann L., Pellock B., White K., Hariharan I. K. (2007). Capicua regulates cell proliferation downstream of the receptor tyrosine kinase/ras signaling pathway. Curr. Biol. 17, 728–733 10.1016/j.cub.2007.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S., Dyson N. J. (2008). Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 9, 713–724 10.1038/nrm2469 [DOI] [PubMed] [Google Scholar]
- Zhang J., Benavente C. A., McEvoy J., Flores-Otero J., Ding L., Chen X., Ulyanov A., Wu G., Wilson M., Wang J. et al. (2012). A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature 481, 329–334 10.1038/nature10733 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.