Summary
Sequential addition of segments in the posteriorly growing end of the embryo is a developmental mechanism common to many bilaterians. However, posterior growth and patterning in most animals also entails the establishment of a ‘posterior organiser’ that expresses the Caudal and Wnt proteins and has been proposed to be an ancestral feature of animal development. We have studied the functional relationships between the Wnt-driven organiser and the segmentation mechanisms in a basal insect, the cockroach Periplaneta americana. Here, posteriorly-expressed Wnt1 promotes caudal and Delta expression early in development to generate a growth zone from which segments will later bud off. caudal maintains the undifferentiated growth zone by dampening Delta expression, and hence Notch-mediated segmentation occurs just outside the caudal domain. In turn, Delta expression maintains Wnt1, maintaining this posterior gene network until all segments have formed. This feedback between caudal, Wnt and Notch-signalling in regulating growth and segmentation seems conserved in other arthropods, with some aspects found even in vertebrates. Thus our findings not only support an ancestral Wnt posterior organiser, but also impinge on the proposals for a common origin of segmentation in arthropods, annelids and vertebrates.
Key words: Caudal, Wnt, Notch, Posterior organiser, Segmentation, Periplaneta americana
Introduction
Animals display a great variety of morphologies and body plans. Despite this diversity, a common form of development entails the early setting up of a small anteriorized embryo, which then grows from its posterior end until the final body length is achieved (Jacobs et al., 2005). This is the mode of development for most complex bilaterians, except derived ones, such as Echinoderms and Urochordates (Holland, 2002; Mooi and David, 2008). In most posteriorly growing animals this occurs through the sequential addition of serially repeated units called segments or somites (Balavoine and Adoutte, 2003; Couso, 2009; De Robertis, 1997). This is the case of vertebrates, annelids, and most arthropods.
This mode of development requires the establishment of a ‘posterior organiser’ that drives development and growth from the posterior end of the embryo. In vertebrates this involves the expression of genes of the Wnt family (Shimizu et al., 2005; Takada et al., 1994). In fact, Wnts are expressed in the posterior of all bilaterians studied to date and have even been found in non-bilaterian Porifera larvae (Cho et al., 2010; Janssen et al., 2010; Martin and Kimelman, 2009; Niehrs, 2010; Riddiford and Olson, 2011; Ryan et al., 2007). Wnt loss-of-function in vertebrates leads to the absence of posterior body structures highlighting its importance in establishing a posterior organiser (Martin and Kimelman, 2009; Shimizu et al., 2005; Takada et al., 1994). Another gene required for posterior growth is Cdx, the vertebrate homologue of the homeobox transcription factor caudal (cad). Similar to Wnt, Cdx loss-of-function leads to severe posterior truncations (Chawengsaksophak et al., 2004; van de Ven et al., 2011; van den Akker et al., 2002). It has been determined that Cdx expression is controlled by Wnt and, indeed, that the effects of posterior Wnt-signalling are mediated through Cdx (Lohnes, 2003; Shimizu et al., 2005; van de Ven et al., 2011). Thus, the use of a Wnt-cad posterior organiser may be a common mechanism utilised by animals in which posterior growth occurs through the sequential addition of segments (Martin and Kimelman, 2009).
In arthropods this mode of development is the most common, and inferred ancestral, and is referred to as short germ band (Anderson, 1972a; Davis and Patel, 2002; Liu and Kaufman, 2005; Peel and Akam, 2003). It differs from the long germ band mode of development found in higher insects, like Drosophila melanogaster, in which all segments form simultaneously in a syncytial environment (Anderson, 1972b; Nüsslein-Volhard and Wieschaus, 1980). In short germ band arthropods, the most anterior cephalic and thoracic segments of the embryo are determined during the early, syncytial blastoderm stage. However, most segments are subsequently added during germ band elongation from an undifferentiated region of proliferating cells at the posterior end of the embryo called the growth zone (GZ) (Davis and Patel, 2002; Liu and Kaufman, 2005; Peel and Akam, 2003). Comparison of different arthropods has revealed a conserved set of genes involved in early posterior development consisting of caudal and the Wnt-signalling pathway. Similar to vertebrates, knock-down of either cad or Wnt in arthropods results in posterior truncations (Bolognesi et al., 2008; Copf et al., 2004; McGregor et al., 2009; McGregor et al., 2008; Shinmyo et al., 2005).
We have used a basal, short germ band insect, the cockroach Periplaneta americana, to gain insight about the interaction between this posterior organiser and the segmentation mechanisms. Cockroaches are a good system in which to study these mechanisms as they are highly susceptible to RNA interference (RNAi), embryos can be cultured ex ovo, and within a single ootheca there is an age gradient where younger embryos differ from older ones by approximately one half segment (Pueyo et al., 2008). These advantages were instrumental to prove the existence of the cyclic waves of Periplaneta Delta and hairy/Hes that emanate from the posterior and resolve into segmental stripes of expression in the anterior GZ prior to segment formation (Pueyo et al., 2008). Loss of Notch (N) signalling via RNAi resulted in embryos in which the posterior is truncated and unsegmented (Pueyo et al., 2008). This N-mediated segmentation is reminiscent of the ‘clock and wavefront’ mechanism found in vertebrate somitogenesis (Dequéant and Pourquié, 2008; Jiang et al., 2000). Dynamic expression of Dl/N has been found in spiders and determined to be important for proper segmentation and been proposed to be ancestral, though whether these patterns are also oscillatory in the spider remain to be determined (Oda et al., 2007; Stollewerk et al., 2003). Interestingly, a number of studies have indicated that an interplay between Wnt, cad and N signalling during somitogenesis in vertebrates may exist (Aulehla and Herrmann, 2004; Grainger et al., 2012; Savory et al., 2011; Young et al., 2009); however, the nature of such a connection has not yet been established definitively.
Here we show that Pa-cad and Pa-Wnt1 are expressed in the posterior GZ and disrupting their functions using RNAi or chemical inhibitors produce embryos with segmentation defects and revealed two distinct phases of Pa-cad and Pa-Wnt1 function. First, early in development, Pa-Wnt1 is required for Pa-cad expression and together they establish a posterior organiser and a functional GZ. Second, during germ band elongation Pa-Wnt1 regulates axial growth and posterior segmentation by activating Pa-Delta (Dl) and Pa-cad expression in the GZ. Subsequently, Pa-cad maintains the GZ in an unsegmented and proliferative state through which the dynamic waves of Delta (Pueyo et al., 2008) are allowed to progress in order to form segments outside the Pa-cad domain. Reciprocally, Dl-N signalling in the posterior tip is necessary to maintain posterior Pa-Wnt1 expression. Thus, the interplay between cad, Wnt and N signalling pathways regulate posterior growth, elongation and segmentation in Periplaneta. Our two-step model can explain results in other arthropods and shows conserved features in vertebrates, suggesting that Wnt and Notch signalling form an ancestral gene network controlling posterior growth and segmentation.
Materials and Methods
Periplaneta americana rearing
A cockroach colony is kept at 29°C. Freshly laid ootheca were collected and placed in a humidified incubator at 29°C and staged according to Lenoir-Rousseaux and Lender (Lenoir-Rousseaux and Lender, 1970).
Cloning of Pa-caudal and Pa-Wnt1 genes
RNA was extracted from Periplaneta americana embryos of mixed stages (4–9) using the RNAqueous-4PCR kit (Ambion). cDNA was synthesised using the RETROscript kit (Ambion). Degenerate primers for the caudal homeodomain motifs ELEKEF and WFQNRR (supplementary material Fig. S1C) were used to isolate a fragment of 116 bp by RT-PCR. Then Pa-cad specific primers were designed (supplementary material Fig. S1C) to obtain the full length cDNA via 5′ and 3′RACE using the FirstChoice RLM-RACE kit (Ambion). A 1420 bp sequence was assembled from the fragments obtained (AC number KC311251). Pa-Wnt1 was cloned following similar procedures. Degenerate primers (supplementary material Fig. S2C) designed against a Wnt1 conserved region from other arthropods amplified a 535 bp fragment of Pa-Wnt1 by RT-PCR. Specific Pa-Wnt1 primers (supplementary material Fig. S2C) were designed to use in 5′ and 3′RACE. A 2364 bp full length Pa-Wnt1 was assembled from the PCR fragments (AC number KC311252). Sequence and phylogenetic analyses were carried out by DNASTAR Software.
In situ hybridization and immunocytochemistry
Fluorescent and colourimetric in situ hybridisation protocols were carried out according to Pueyo et al. (Pueyo et al., 2008). A 904 bp Pa-cad and a 535 bp Pa-Wnt1 cDNA fragments were used as templates for transcription. Labelled UTP RNA probes (DIG and Biotin – Roche; DNP – Molecular Probes) were fragmented into 100–200 bp sizes by hydrolysis.
Immunostaining was performed as described (Pueyo et al., 2008). Primary monoclonal mouse antibody FP6.87 (α-Ubx/α-AbdA) (1:10) (Rob White, Cambridge University). Apoptosis was detected using a rabbit α-cleaved caspase 3 antibody (1:50) (Cell Signalling Technology) and cells undergoing mitosis were detected using a rabbit α-phosphorylated Histone 3 antibody (1:1000) (Upstate). Nuclei were detected using DAPI (Invitrogen). Secondary antibodies were from Jackson Immunochemicals.
Inhibitor treatment in embryo cultures
For DAPT (Calbiochem) treatment at 100 µM we followed the protocol described (Pueyo et al., 2008). Inhibitor of Wnt Production – compound 3 (IWP-3; Stemgent), which blocks the secretion of Wnt ligands by inhibiting palmitoylation of Wnt by Porcupine (Chen et al., 2009), was diluted in DMSO (DMSO; Sigma) and added to the embryo culture at a final concentration between 20 µM and 40 µM. Exposure response analysis showed various results at different concentrations: <20 µM had no effect; 20–30 µM had hypomorph/weak effects; 30–40 µM had strong effects; >40 µM produced cytotoxic effects. Exposure to IWP-3 for times longer than 18–24 hours also produced cytotoxicity. Before culture, some embryos were fixed to report the developmental stage (Control 0 hours). The remaining embryos were then split into two groups and cultured for 16–24 hours either with or without inhibitor (control). Control cultured embryos were treated with the same amount of DMSO.
RNA interference
Maternal RNAi for Pa-cad and Pa-Wnt1 was carried out according to Pueyo et al. (Pueyo et al., 2008). Specific primers containing the T7 polymerase promoter sequence at the 5′ end were used with Pa-cad cDNA and Pa-Wnt1 RT-PCR templates to amplify a 635 bp and a 500 bp fragment, respectively. In vitro transcription of these fragments was carried out by the T7 RiboMAX kit (Promega). dsRNA was injected into virgin adult female abdomens at a concentration of 2.0 µg/µl (Pa-cad) or 0.5 µg/µl (Pa-Wnt1). Controls were injected with 10 µl H2O.
RT-PCR analysis
RNA isolation and cDNA synthesis from stage 9 wild type, Class ‘T’ Pa-Wnt1RNAi and Pa-cadRNAi embryos were conducted. Equal cDNA concentrations were used in RT-PCR reactions using primers for Pa-Wnt or Pa-cad to determine the amount of each in Pa-cadRNAi or Pa-WntRNAi. Specific primers for Periplaneta 18S ribosomal subunit were used as a positive control: Pa-18S forward 5′GTACCGGCGACGCATCTTTCA3′; Pa-18S reverse 5′CTTTCGGCCAGGCAGGACAC3′.
Results
Isolation and patterns of expression of Pa-caudal and Pa-Wnt1 transcripts
We have cloned the full length transcripts of Pa-cad and Pa-Wnt1 by RT-PCR (supplementary material Figs S1, S2, respectively). A single Pa-cad transcript of 1420 nucleotides that encodes for a 290 aa protein was identified (supplementary material Fig. S1A). Our phylogenetic analysis indicates that Pa-cad is the Periplaneta orthologue as it aligns closely to caudal from related species, such as Gryllus (cricket) and Schistocerca (grasshopper) (supplementary material Fig. S1B). Likewise, we isolated a single 2364 bp Pa-Wnt1 transcript encoding for a 373 aa protein (supplementary material Fig. S2A). Our alignment shows that Pa-Wnt1 is the Periplaneta Wnt1 orthologue closely related to Cryptotermes (termite) and Orthopterans, which follows the predicted insect phylogeny (supplementary material Fig. S2B).
In situ hybridisation using a Pa-cad riboprobe shows that at early, post-blastoderm stages of embryogenesis, Pa-cad is strongly expressed at the posterior end of the embryo (Fig. 1A). This broad domain of expression remains at the onset of germ band elongation (Fig. 1B), but is cleared from the posterior tip at late germ band elongation (Fig. 1C). Pa-cad expression pattern is similar to that observed in other short germ band insects (Copf et al., 2004; Dearden and Akam, 2001; Shinmyo et al., 2005). Likewise, Pa-Wnt1 transcripts are detected at the post-blastoderm stage in the posterior GZ as two symmetrical clusters of cells (Fig. 1D) and anteriorly in the head lobes and the antennal primordia (Fig. 1D). During early germ band elongation anterior stripes of Pa-Wnt1 expression become apparent in the presumptive gnathal and thoracic segments while the two posterior clusters start to fuse (Fig. 1E). By late germ band elongation the two Pa-Wnt1 clusters of cells have joined together forming a wide arc of expression in the posterior end of the GZ, set apart from the posterior tip (Fig. 1F). New stripes of Pa-Wnt1 expression appear one-by-one in the anterior-most GZ and remain in the developing segments and ventral appendages throughout embryogenesis (Fig. 1F). The early, dynamic posterior expression of Pa-Wnt1 lies close to the expression of Pa-cad, and thus would be consistent with a conserved role for these two genes during posterior patterning. The later Pa-Wnt1 segmental stripe pattern is consistent with Wnt1 expression in other insects and arthropods in which its function in parasegmental boundary formation, segmentation and appendage development has been established (Bolognesi et al., 2008; Couso et al., 1993; Grossmann et al., 2009; Martinez-Arias, 1993; Miyawaki et al., 2004).
Fig. 1. Wild type expression patterns of Pa-cad (A–C), Pa-Wnt1 (D–F), and Pa-en (G–I) in Periplaneta americana.

(A,B) Pa-cad is expressed in a broad posterior domain (brackets) in post-blastoderm (A) and early germ band elongation (B) embryos. (C) During late germ band elongation, Pa-cad is restricted to the mid-GZ (bracket) and is lost from the posterior tip (arrow). (D) Pa-Wnt1 post-blastoderm expression in head (arrow) and antennae (arrowhead) and in two posterior clusters of cells (*). (E) By early germ band elongation, Pa-Wnt1 is at the posterior (*) and in segmental stripes reaching T3. (F) During late germ band elongation, Pa-Wnt1 is expressed in an arc-like stripe in the posterior GZ (*) – set apart from the posterior tip (arrow), in segmental stripes in the anterior GZ (open arrowheads), in anterior segments and ventral appendages (black arrowheads). (G) Pa-en segmental expression up to T2 during post-blastoderm. Additional stripes (black arrowheads) are added sequentially from the posterior during early (H) and late (I) germ band elongation and remain throughout development (open arrowheads). T2, T3: second, third thoracic segment; A1, A4: first, fourth abdominal segment.
To establish the spatial and temporal details of Pa-Wnt1 and Pa-cad expression, we used double fluorescence in situ hybridisation (FISH) and compare with the pan-segmental marker engrailed (en) (Patel et al., 1989). Pa-Wnt1 slightly overlaps and is posterior to Pa-cad expression during both post-blastoderm (Fig. 2A–A″) and germ band elongation (Fig. 2B–B″). In comparison, post-blastoderm expression of Pa-en (Marie and Bacon, 2000) appears as several stripes from the head segments up to the T2 segment (Fig. 1G). During early (Fig. 1H) and late (Fig. 1I) germ band elongation additional stripes of Pa-en appear sequentially in the anterior GZ, apparently slightly before the corresponding Pa-Wnt1 segmental stripe expression (compare Fig. 1E,F with Fig. 1H,I). Double FISH experiments confirm the temporal delay of segmental Pa-Wnt1 expression after Pa-en (Fig. 2C–C″), as well as their adjacent expression at the parasegmental boundary as Pa-Wnt1 abuts anterior to Pa-en expression (Farzana and Brown, 2008; Martinez-Arias and Lawrence, 1985; Prud'homme et al., 2003; Martínez-Arias, personal communication).
Fig. 2. FISH reveals spatiotemporal patterns of Pa-Wnt1, Pa-cad and Pa-en.
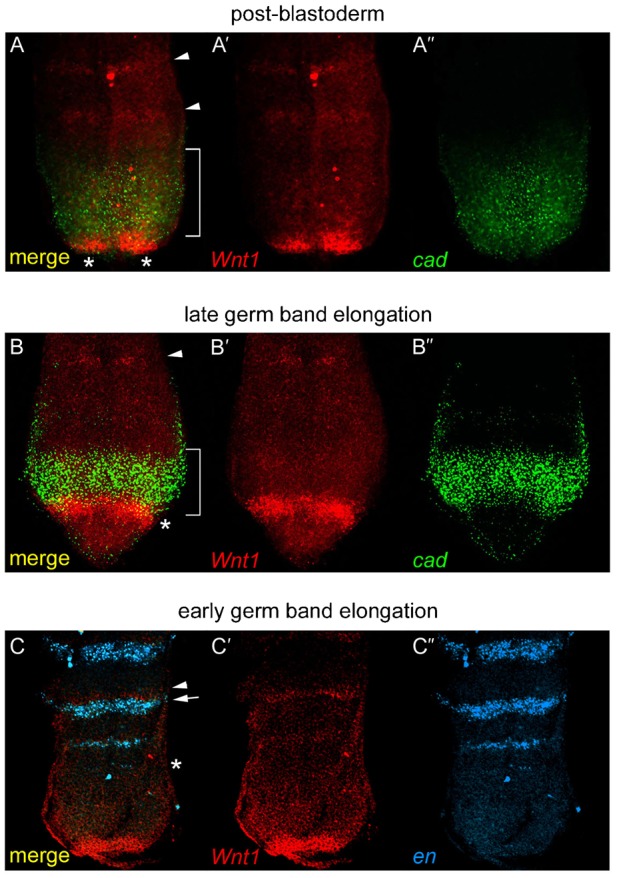
(A–A″) Posterior end of a post-blastoderm embryo (stage 6) showing Pa-Wnt1 (red, A′) and Pa-cad (green, A″) patterns. Pa-Wnt1 expression in segmental stripes (arrowheads), and posterior clusters (*) overlapping with Pa-cad domain (bracket). (B–B″) Posterior end of a germ band elongation embryo (stage 9), showing Pa-Wnt1 expression (B′) in segmental stripes in anterior GZ (arrowhead) and a posterior arc (*) slightly overlapping with Pa-cad (B″, bracket). (C–C″) Pa-Wnt1 (C′) and Pa-en (blue, C″) in the anterior GZ with stripes of Pa-en (*) developing before Wnt1 and defining the parasegment boundary. Pa-Wnt1 (arrowhead) is expressed anterior and adjacent to Pa-en (arrow).
Pa-cad and Pa-Wnt1 act as a posterior organiser
To test the possible conservation of Wnt and Cad function we first used maternal RNAi. In Pa-Wnt1RNAi embryos Pa-cad expression is greatly reduced at post-blastoderm (Fig. 3B, compare with Fig. 3A). By contrast, posterior Pa-Wnt1 expression in Pa-cadRNAi embryos at this stage was similar to wild type, despite a premature fusion of the two Pa-Wnt1 expressing clusters (Fig. 3D, compare with Fig. 3C). Pa-Wnt1RNAi and Pa-cadRNAi embryos developing up to germ band elongation display a truncated or tapering posterior end with growth and development of new segments arrested at the thoracic region (Fig. 3F,H). To ascertain the cellular basis of this reduced and abnormal GZ in Pa-Wnt1RNAi embryos we examined cell death and proliferation (supplementary material Fig. S3). Pa-Wnt1RNAi embryos do not show an increase in apoptosis in the GZ (supplementary material Fig. S3A,B,I), but there is a reduction in cell proliferation (supplementary material Fig. S3C,D,I). In these Pa-Wnt1RNAi embryos, expression of Pa-cad appears completely lost according to both in situ hybridisation (compare Fig. 3F and Fig. 3E) and RT-PCR (supplementary material Fig. S4D). However, in the reciprocal experiment, the number of cells expressing Pa-Wnt1 in Pa-cadRNAi appears similar to that of the wild type, confirmed by RT-PCR (supplementary material Fig. S4D), albeit the region displays an abnormal shape (compare Fig. 3H and Fig. 3G). These results suggest that Pa-Wnt1 regulates Pa-cad expression and that the apparent alteration of the Pa-Wnt1 expression domain in Pa-cadRNAi is a secondary consequence of abnormal posterior tip development (Fig. 3D,H).
Fig. 3. Disruption of Wnt1 and cad function by maternal RNAi and embryo culture.
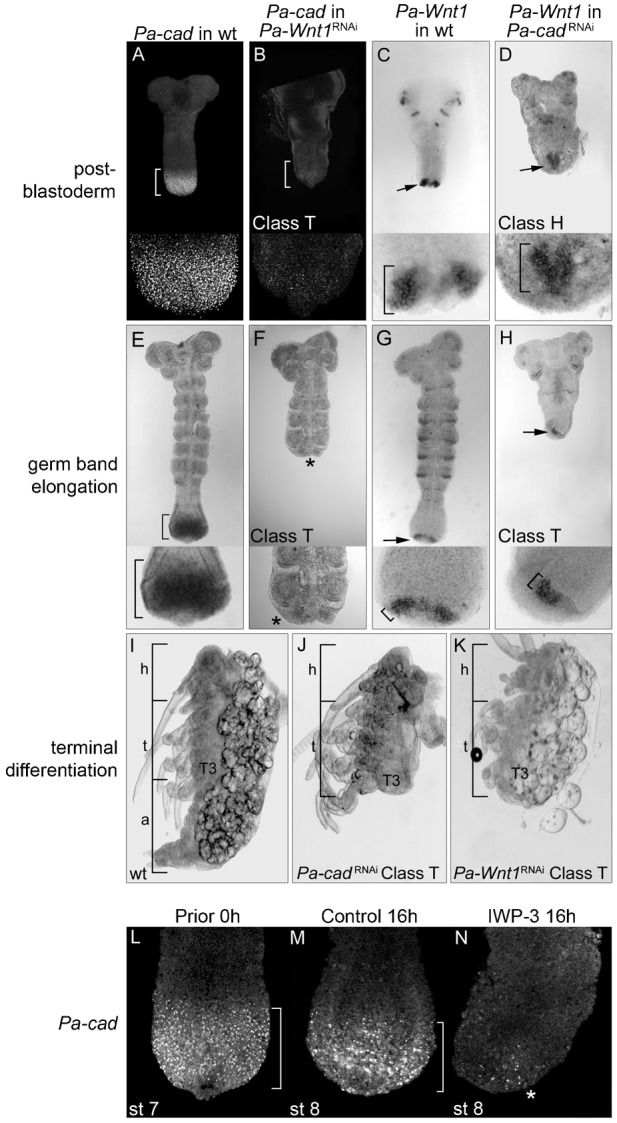
(A–D) RNAi effects on Pa-cad and Pa-Wnt1 expression at post-blastoderm. The Pa-cad broad posterior domain in wild type embryos (bracket, A) is greatly reduced in Pa-Wnt1RNAi (bracket, B). Conversely, the wild type expression of Pa-Wnt1 (arrow, C) is relatively unaltered in Pa-cadRNAi (arrow, D; brackets in C,D insets). (E–H) RNAi affects Pa-cad and Pa-Wnt1 during germ band elongation. The Pa-cad domain (bracket, E) is absent in Pa-Wnt1RNAi embryos (*, F), while the arc of Pa-Wnt1 (arrow, G) is unaffected in location (arrow, H) and width of expressing cells in Pa-cadRNAi embryos (brackets in G,H insets). (I–K) RNAi phenotypes. (I) Stage 22 wild type embryo displaying head, thorax and abdomen. Class ‘T’ Pa-cadRNAi (J) and Pa-Wnt1RNAi (K) embryos show a similar body truncation after T3. (L–N) IWP-3 culture inhibition of Wnt-signalling affects Pa-cad expression. (L) Prior to culture (0-hour control), Pa-cad is in a broad posterior domain (bracket). After 16 hours in DMSO control culture there is no effect on Pa-cad (bracket, M), while IWP-3 cultured embryos show a marked decrease in Pa-cad (*, N). h: head; t: thorax; a: abdomen.
A detailed quantification of the phenotypes indicates a graded variability within a requirement for posterior development. We classified Pa-cadRNAi embryos according to the strength of the phenotype into three different classes (Table 1). In Class ‘H’ embryos only the head segments form properly (supplementary material Fig. S4A) and often followed by undifferentiated tissue, representing a complete or near complete loss of Pa-cad expression. In Class ‘H’ developing embryos, no posterior segmental addition is observed (compare supplementary material Fig. S5A and Fig. S5B). In Class ‘T’ embryos the head and some of the thoracic segments developed normally but with truncations after either T2 or T3 (Fig. 3J and supplementary material Fig. S5E), suggesting that some Pa-cad expression may have remained in the early post-blastoderm, but later expression was lost, leading to the development thoracic segments. This would explain the residual Pa-cad expression detected in RT-PCR using Class ‘T’ Pa-cadRNAi embryos (supplementary material Fig. S4D). Finally, Class ‘A’ embryos show lack of some abdominal segments and a reduced GZ (supplementary material Fig. S5H) resolving into abnormal abdominal development (supplementary material Fig. S4B). Pa-Wnt1RNAi embryos displayed a similar range of posterior segmentation phenotypes (Table 1). Pa-Wnt1RNAi embryos in Class ‘T’ show severe truncations ranging from the first thoracic segment to the first abdominal segment (Fig. 3K). In these embryos the GZ is narrow and posterior segmentation proceeds abnormally (supplementary material Fig. S5F). Class ‘A’ Pa-Wnt1RNAi embryos displayed moderate truncations involving only a few abdominal segments and a slight reduction in the GZ (supplementary material Fig. S4C and supplementary material Fig. S5I). The absence of Class ‘H’ embryos, which correlate with a total loss of Pa-cad, in Pa-Wnt1RNAi may be due to several reasons. First, Pa-Wnt1RNAi may not create a null condition at post-blastoderm or, second, there could be other Wnt genes acting at these early stages. Third, there could be some Wnt-independent, maternally deposited, Pa-cad expression. The absence of Pa-Wnt1 in Pa-Wnt1RNAi RT-PCR may discard the first explanation, but in principle either of the three is compatible with both the residual Pa-cad expression observed in Pa-Wnt1RNAi post-blastoderm embryos (Fig. 3B), but not later, as the Class ‘T’ phenotypes between the two RNAi treatments are similar (Fig. 3J,K).
Table 1. Phenotypic series of Pa-cadRNAi and Pa-Wnt1RNAi embryos.
Range of RNAi phenotypes categorized into three classes. Class ‘H’ embryos displayed head only or head plus posteriorly undifferentiated tissue, only found in Pa-cadRNAi. Class ‘T’ embryos form the head and some thoracic segments properly, but are truncated after either T2 or T3; this was the most common Pa-cadRNAi and Pa-Wnt1RNAi phenotype. Class ‘A’ embryos were observed in both Pa-cadRNAi and Pa-Wnt1RNAi. These embryos developed complete head and thorax with defects in abdominal segmentation, usually tapering at the posterior. Control Pa-H2O embryos were wild type in appearance.

Specificity of Wnt-Cad posterior organiser phenotypes
Altogether these results are compatible with the model where Wnt-signalling activates zygotic cad expression early in development, establishing a posterior organiser (McGregor et al., 2008; Shinmyo et al., 2005). This Periplaneta ‘posterior organiser’ seems required for proper patterning of the posterior end of the embryo, including the GZ, and for subsequent growth and segmentation. However, these RNAi results do not clarify whether this Wnt-Cad organiser is still required during germ band elongation, or whether the phenotypes observed arise secondarily from earlier defects during establishment of the GZ. To clarify this issue we employed the embryo culture technique (Pueyo et al., 2008). We have cultured germ band elongating embryos and exposed them to an inhibitor of secretion of Wnts, IWP-3 (Chen et al., 2009), to decouple the function of Wnt1 and other Wnts during germ band elongation from earlier or indirect perturbations. Embryos exposed to IWP-3 showed a marked decrease in Pa-cad expression and develop a slightly reduced GZ (Fig. 3N, compared to Fig. 3L,M) and segmentation is disrupted (supplementary material Fig. S6), while the expression of other genes, such as Ubx and AbdA, remain unaffected (supplementary material Fig. S3E,F). Compared to control cultured embryos (supplementary material Fig. S3E,G), we observed a slightly increased rate of apoptosis (supplementary material Fig. S3F,J), but a decrease in cell proliferation (supplementary material Fig. S3H,K) in IWP-3 cultured embryos. Staining with the vital dye DAPI confirms that the overall majority of cells are alive throughout the posterior embryo (supplementary material Fig. S3G,H). Overall, these effects are in accordance with Pa-Wnt1RNAi data where Wnt-signalling seems to be required mainly in maintaining cell proliferation in the GZ (supplementary material Fig. S3C,D,I). In addition, dying or proliferating cells are scattered throughout the embryo and not localised to the posterior GZ such that these changes in growth and death cannot account for the proportional reduction in posterior Pa-cad expression. These data confirm that a) Wnt-signalling is required for Pa-cad expression, and b) this requirement extends into the germ band elongation stages.
Regulatory interactions between the posterior organiser and the Notch pathway
As dynamic N-signalling is necessary for posterior segmentation in Periplaneta (Pueyo et al., 2008), we investigated the regulatory interactions between the Wnt-Cad organiser with elements of the N-signalling pathway. First, we compared their GZ expression patterns during post-blastoderm (Fig. 4A,B) and germ band elongation (Fig. 4C–F). At post-blastoderm, five stripes of Pa-Dl expression appear simultaneously in the anterior part of the embryo, yet its expression is noticeably absent from the posterior end (Fig. 4A) where Pa-Wnt1 and Pa-cad are expressed (Fig. 4B). These early Pa-Dl stripes have faded by germ band elongation when a new Pa-Dl pattern emerges at the posterior to include the new sequentially arising segmental stripes (Pueyo et al., 2008). This germ band elongation pattern is composed of a small domain in the posterior tip from which cyclic waves emanate periodically (Pueyo et al., 2008) resulting in 2–3 stripes in the anterior GZ, interestingly placed outside of the Pa-cad expression domain (Fig. 4C–C″). These Pa-Dl stripes regulate Pa-en expression in the anterior GZ (Fig. 4E–E″) leading to segment border formation. When these patterns of Pa-Dl and Pa-en are overlaid with those of Pa-cad and Pa-Wnt1 (Fig. 4C′,D), the GZ can be divided into 4 distinct gene expression regions (Fig. 4F), from posterior to anterior: 1) posterior tip, expressing Pa-Dl; 2) posterior-GZ, expressing an arc of Pa-Wnt1; 3) mid-GZ, expressing a broad Pa-cad domain; and 4) anterior, pre-segmental region, expressing first Pa-Dl, then Pa-en, and finally Pa-Wnt1. Thus, these spatiotemporal patterns indicate that the most significant change in the transition from early post-blastoderm to germ band elongation is the addition of Pa-Dl, and hence N-signalling, to the posterior gene network.
Fig. 4. Co-expression of ‘posterior network’ genes at post-blastoderm (stage 6, A,B) and germ band elongation (stage 9, C–F).
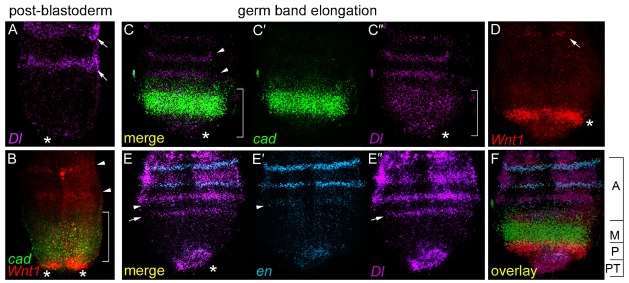
(A) Pa-Dl is expressed in anterior segmental stripes (arrows) but not in the posterior (*). (B) Double FISH from Fig. 2, showing Pa-Wnt1 (red) and Pa-cad (green) expression. (C–C″) Double FISH showing expression of Pa-cad (C′) and Pa-Dl (purple, C″). The wave of Pa-Dl (bracket) emanates from the posterior tip (*), travelling through the Pa-cad domain and resolving into stripes in the anterior GZ (arrowheads). (D) Pa-Wnt1 expression in segmental stripes (arrow) and in a posterior arc (*). (E–E″) Double FISH for Pa-en (blue, E′) and Pa-Dl (E″). Pa-Dl is expressed in the posterior tip (*) and the wave of expression has coalesced into a stripe in the anterior GZ (arrow) preceding the stripes of Pa-en expression (arrowhead). (F) Photomontage of C–E overlaid to show the GZ divided into four distinct posterior gene expression domains; from posterior to anterior – PT: posterior tip expressing Pa-Dl; P: posterior GZ arc of Pa-Wnt1; M: broad Pa-cad domain in mid-GZ; A: anterior GZ region expressing segmental stripes of Pa-Dl, Pa-en, and Pa-Wnt1.
To understand the regulatory relationships between the genes of this posterior gene network we examined their expression patterns in different maternal RNAi conditions. We use Pa-NRNAi instead of Pa-DlRNAi, which has fewer effects on oogenesis, as the latter produces sterile females (R. Lanfear, The evolution of animal body plans, DPhil thesis, University of Sussex, 2007). Pa-NRNAi leads to a loss of Pa-Dl expression in the posterior tip and a failure to form stripes in the anterior GZ (supplementary material Fig. S7A,B), corresponding to the loss of sequential segment addition (Pueyo et al., 2008). A similar effect is observed in embryos cultured in the N-signalling inhibitor DAPT, compared to controls (supplementary material Fig. S7C,D). In both cases, neurogenic phenotypes are also observed (supplementary material Fig. S7A′–D′).
In post-blastoderm Pa-Wnt1RNAi embryos the early segmental stripes of Pa-Dl expression remain (Fig. 5B, compare with Fig. 5A). Conversely, Pa-NRNAi had no effect on the Wnt-Cad organiser at this stage, as Pa-cad expression is normal (Fig. 5E,F). However, during germ band elongation, Pa-Wnt1RNAi embryos show an absence of Pa-Dl expression in the posterior tip and anterior GZ stripes, whereas neural Pa-Dl expression remains (Fig. 5C,D). Similarly, embryos cultured with the Wnt-signalling inhibitor, IWP-3, during germ band elongation experienced an almost total loss of Pa-Dl in the posterior tip (Fig. 5K). Interestingly, the Pa-Dl segmental stripes do not disappear in IWP-3 embryos, however, only one new stripe can form in the anterior GZ (Fig. 5K) compared to the two stripes that form in control culture (Fig. 5J), indicating a temporal component to the stronger phenotypes revealed previously by Pa-Wnt1RNAi.
Fig. 5. Pa-Dl posterior tip expression during germ band elongation requires Pa-Wnt1 and Pa-cad.
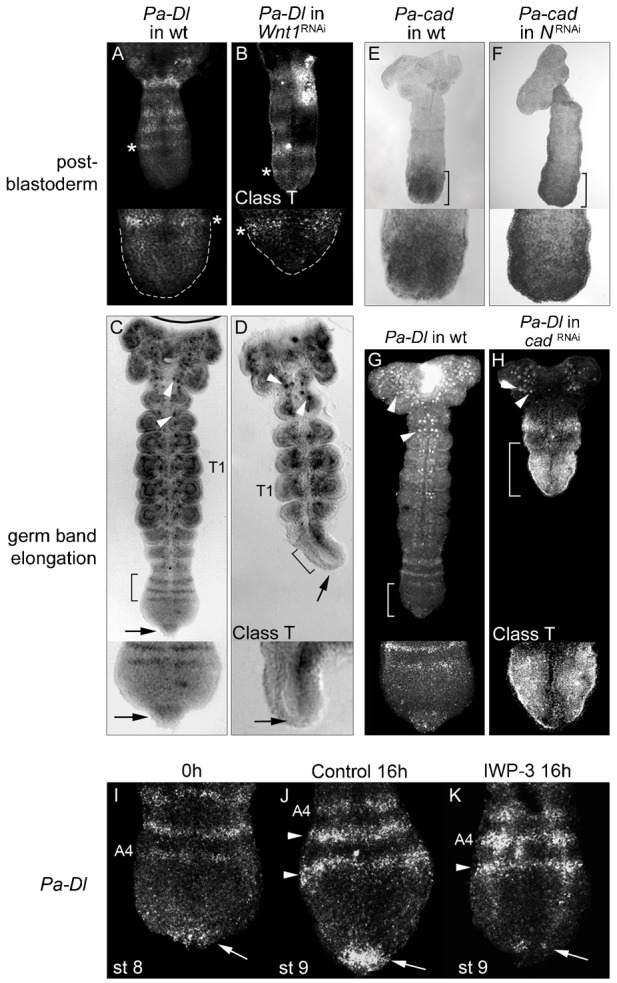
(A,B) Post-blastoderm Pa-Dl segmental expression (*) in wild type (A) is unaffected in Wnt1RNAi (B), despite the GZ being reduced (GZ outlines in A,B insets). (C) Wild type expression of Pa-Dl during germ band elongation. (D) Loss of Pa-Dl at the posterior tip (arrow) and absence of stripes in the anterior GZ (bracket), but not in neuroblasts (arrowheads) in Pa-Wnt1RNAi embryo. (E,F) Pa-cad expression (brackets) is similar in post-blastoderm wild type (E) and NRNAi embryos (F). Wild type expression of Pa-Dl (G) is expanded in Pa-cadRNAi embryos (H) covering the posterior region and does not form stripes (bracket); neural expressions remain unaffected (arrowheads). (I–K) Inhibition of Wnt-signalling in culture affects Pa-Dl expression. (I) Pa-Dl in the posterior tip (arrow) and in segmental stripes (A4) in a 0-hour control embryo. (J) After 16 hours in DMSO control culture Pa-Dl is expressed in the posterior tip (arrow) and two additional stripes have been added in the anterior GZ (arrowheads). (K) Embryos cultured for 16 hours in the IWP-3 Wnt-inhibitor show greatly reduced Pa-Dl expression in the posterior tip (arrow) and only one new stripe (arrowhead) has formed. T1: first thoracic segment; A4: fourth abdominal segment.
This function of Pa-Wnt1 in activating Pa-Dl posterior tip expression does not seem to be mediated by Pa-cad. In Pa-cadRNAi embryos at germ band elongation the tip expression of Pa-Dl remains, but the anterior stripes do not form (Fig. 5H). Instead, there is a strong, expanded expression of Pa-Dl covering the posterior region of the embryo, in what corresponds to the entire GZ of a wild type embryo. The lack of effect of Pa-cad on Pa-Dl posterior tip expression is not unexpected as these expression domains do not overlap. However, our results reveal a repressory function of Pa-cad on Pa-Dl expression in the mid-GZ that matches the wild type expression patterns, since Pa-Dl segmental stripes only form outside the Pa-cad domain (Fig. 4C,F). The Pa-Dl posterior tip expression, which lays adjacent to the posterior arc of Pa-Wnt1, seems to have an immediate requirement for Pa-Wnt1 signalling, while the anterior stripes of Pa-Dl seem to have a delayed, indirect requirement for Pa-Wnt1 regulated through Pa-cad. This explains why the loss of Pa-cad expression in Pa-Wnt1RNAi does not result in the expansion of Pa-Dl expression, as in Pa-Wnt1RNAi there is also no emanating source of Pa-Dl expression from the posterior tip.
Unexpectedly, a functional assessment of N-signalling indicates that this modulating effect of the posterior Wnt-Cad organiser on Pa-Dl expression is reciprocated. Pa-NRNAi embryos at germ band elongation have a reduced and tapered GZ that does not express either Pa-cad (Fig. 6B) or Pa-Wnt1 (Fig. 6D), implying that Pa-Dl, and/or other components or targets of N-signalling may be required to maintain the posterior Pa-Wnt1 expression, and hence, Pa-cad. This inference is corroborated by embryo culture experiments. When embryos were cultured with the N-inhibitor DAPT there was a noticeable reduction in Pa-cad expression (Fig. 6G) and the arc of Pa-Wnt1 is reduced from a 4-cell wide band to a 2-cell wide stripe (Fig. 6J). These results confirm the role of Pa-Dl in maintaining Pa-Wnt1, and Pa-cad, expression in a feedback mechanism in the posterior GZ. The reduction, but not loss, of both Pa-Wnt1 and Pa-cad expression can be explained by the incomplete block of N-activity in these experiments. For example, the segmentation process is perturbed but not entirely blocked, as shown by new but incomplete segmental stripes of Pa-Wnt1 (Fig. 6J) and Pa-en (Pueyo et al., 2008) appearing during DAPT culture. Importantly, this hypomorphy allows a normal-sized GZ to remain and consequently shows that the gene expression effects in Pa-NRNAi experiments are not purely a secondary consequence of the lack of expressing cells (Pueyo et al., 2008).
Fig. 6. N-signalling affects Pa-cad and Pa-Wnt1 during germ band elongation.
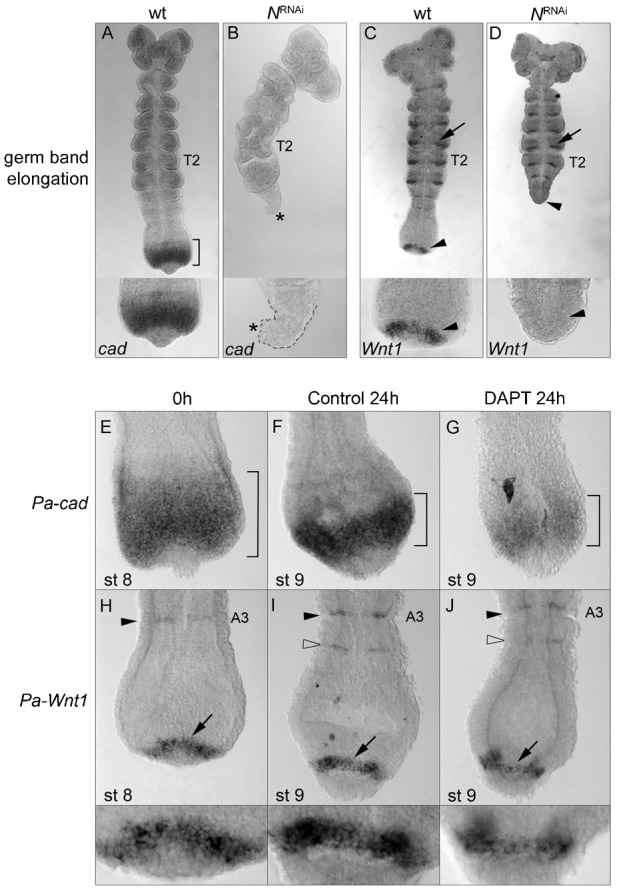
(A) Wild type Pa-cad expression (bracket). (B) NRNAi embryos have a reduced GZ (outline, inset) completely devoid Pa-cad expression (*). (C) Wild type Pa-Wnt1 expression. (D) In NRNAi embryos there is no posterior Pa-Wnt1 expression (arrowhead), while the anterior segmental stripes remain unaffected (arrow; compare with C). (E–J) Effects of the N-inhibitor DAPT on posterior gene expression. (E–G) Pa-cad expression in the 0-hour control embryos (bracket, E) is unaffected in embryos cultured for 24 hours in DMSO control (bracket, F), but is reduced after 24 hours in DAPT culture (bracket, G). (H–J) Pa-Wnt1 is in segmental stripes (arrowhead) and a posterior arc (arrow) in embryos prior to culture (0-hour control, H) and after 24 hours in DMSO control culture, in which a new Pa-Wnt1 stripe forms in the anterior GZ (open arrowhead, I). (J) 24-hour DAPT cultured embryos show a reduction of the Pa-Wnt1 posterior arc from four to two cells in width and the new A4 stripe only partially formed (open arrowhead). T2: second thoracic segment; A3: third abdominal segment.
Discussion
A Wnt-dependent posterior organiser is present in Periplaneta americana
Our results reveal the presence of a posterior organiser regulating posterior development and segmentation in the short germ band insect, Periplaneta americana. Shortly after cellularisation and before the sequential addition of segments takes place, Pa-Wnt1 is observed in two clusters of cells in the posterior-most part of the embryo and Pa-cad is strongly detected in nearby cells covering what will become the GZ (Fig. 7A). Our experiments suggest that Pa-Wnt1 expression at this stage activates or maintains Pa-cad, which in turn is essential for the proper patterning of the embryo. Since this patterning includes the establishment of a GZ from which most of the segments will form during germ band elongation, perturbation of Pa-Wnt1 and Pa-cad at this early stage precludes the proper development of much of the embryo. In this sense, this Pa-Wnt1+Pa-Cad module is operationally similar to the ‘posterior organiser’ observed in other animals. Wnt-signalling knock-down experiments in other arthropods, such as the spider Achaearanea and the insects Gryllus, Oncopeltus, and Tribolium all show similar phenotypes to Periplaneta Wnt1RNAi embryos: posterior truncations and a reduced GZ (Angelini and Kaufman, 2005; Bolognesi et al., 2008; McGregor et al., 2008; Miyawaki et al., 2004). These results suggest a conserved role of Wnt-signalling in establishing and/or maintaining the GZ, including the activation of caudal (McGregor et al., 2008; Shinmyo et al., 2005) as in Periplaneta. In vertebrates, Wnt and cad/Cdx genes are also involved in posterior patterning and their regulatory interactions appear to be conserved as well. For instance, Wnt-signalling activates cad/Cdx expression in the posterior end of zebrafish and mouse embryos (Ikeya and Takada, 2001; Shimizu et al., 2005) and this activation is essential for the proper development of the animal. In fact, it has been proposed that most metazoans display a Wnt-dependent posterior organiser and that this must be a conserved ancestral feature of animal development (Martin and Kimelman, 2009). Our results are consistent with such a model.
Fig. 7. Model depicting the regulatory interactions of the posterior gene network in Periplaneta americana.
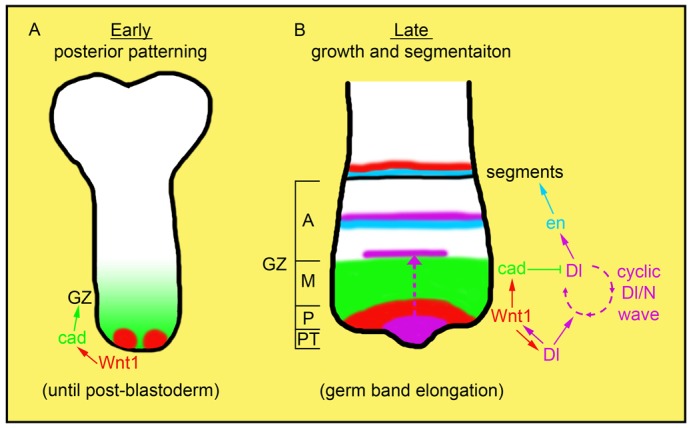
(A) In the posterior region of the post-blastoderm embryo there are two clusters of cells expressing Pa-Wnt1 (red). Pa-Wnt1 activates the expression of zygotic Pa-cad (green) in the neighbouring cells. This activation is required for the posterior organisation and establishment of the growth zone (GZ). Pa-Dl has no functional role at the posterior end at this stage. (B) During germ band elongation the embryo grows from the posterior and new segments are laid down sequentially from the GZ. The arc of Pa-Wnt1 at the posterior GZ (P) has two main functions: 1) maintaining Pa-cad expression in a broad domain (green) in the mid-GZ (M) and 2) promoting Pa-Dl expression (purple) in the posterior tip (PT). Upon activation, Pa-Dl forms a positive feedback loop with Pa-Wnt1, thereby maintaining the expression of each other. As germ band elongation proceeds cyclic waves of Pa-Dl expression emerge from the posterior tip and pass through the Pa-cad domain (dashed arrow). During this process Pa-cad dampens the expression of Pa-Dl, thus inhibiting the formation of segmental stripes but allowing the wave of expression to travel through. Pa-Dl stripes form outside the Pa-cad domain in the anterior GZ (A) and activate segmental stripes of Pa-en (blue). Pa-en then activates a segment polarity gene network involving Pa-Wnt1 eventually leading to segment border formation. Thus, the interaction between the posterior gene network and the N-dependent oscillator regulates growth and segmentation in Periplaneta during germ band elongation.
Despite early Pa-Wnt1 expression, we observe no effect on the formation of the most anterior segments in Pa-Wnt1RNAi embryos, suggesting that formation of these segments is Wnt1 independent or that there could be other, functionally redundant, Wnt ligands implicated at this stage (Bolognesi et al., 2008; Janssen et al., 2010; McGregor et al., 2008). Interestingly, formation of these anterior segments does not seem to involve N-signalling either (Pueyo et al., 2008), but a separate head-segmentation mechanism.
Control of segmentation by the posterior organiser
Altogether our results indicate that there exist interdependent and dual roles of the Wnt and N-signalling pathways during posterior development and segmentation in Periplaneta (Fig. 7B). Pa-Wnt1 signalling modulates the cyclic expression of Pa-Dl during germ band elongation in two opposite ways: it locally promotes Pa-Dl in the nearby posterior tip, while indirectly and via Pa-cad, dampens Pa-Dl expression in the mid-GZ. The combined effect of these two inputs is that the posterior tip source from which the cyclic waves of Pa-Dl originate is maintained, but the waves are only allowed to coalesce into a stripe at the anterior GZ. Reciprocally, Pa-Dl expression at the posterior tip has two distinct roles during posterior segmentation. On the one hand, it gives rise to the cyclical waves that precede and promote the sequential formation of segments in the anterior GZ (Pueyo et al., 2008). On the other hand, constant N-signalling in the posterior tip of the embryo maintains the non-cyclic expression of Pa-Wnt1 in nearby cells during germ band elongation, allowing for maintenance of the GZ until all the segments are laid down.
This model links with the earlier findings of Pueyo et al. (Pueyo et al., 2008), and explains some of the features reported. Pa-NRNAi embryos rarely display disruptions of segmentation in head or T1 segments, despite the generation of neurogenic phenotypes in these places (Jiménez and Campos-Ortega, 1982). Our results also highlight the relevance of the posterior tip expression of Pa-Dl and other members of the N-pathway (Pueyo et al., 2008) during N-mediated segmentation. The N-signalling stripes promote segment formation in the anterior GZ but N-signalling also has a role in the maintenance of the GZ itself. This structure is highly reduced in Pa-NRNAi embryos, involving a reduction of cell division and a mild increase in cell death (Pueyo et al., 2008). Following our results, this requirement for GZ maintenance can now be attributed to Pa-Dl expression at the posterior tip, which is essential to maintain Pa-Wnt1 and Pa-cad.
Pa-cad appears as the most direct agent studied here involved in maintaining a GZ of appropriate size. Pa-Cad could act through a stimulation of cell division, or through a repression of differentiation as suggested by the lack of Pa-Dl, Pa-en and Pa-Wnt1 segmental stripes in the Pa-cad domain. In a unified hypothesis, these cellular and gene regulation effects can be traced to the role of Pa-Cad in dampening Pa-Dl expression throughout the posterior GZ. This partial repression still allows the progression of N-signalling waves through the Pa-cad domain, but only allows the formation of segmental N-signalling stripes outside it. Since these stripes of N-signalling eventually lead to segment formation and differentiation, the expression of Pa-cad might be essential to maintain an undifferentiated, actively dividing cell population at the GZ that can continue to bud off new segments. Loss of Pa-Cad eventually allows high Pa-Dl levels across the posterior region of the embryo, producing widespread N-signalling, and hence, differentiation of all GZ cells and truncation of the embryo. In this model, as in normal development, patterning and growth are inextricably linked. The patterning activity of the Wnt posterior organiser sets up a GZ, which is then needed to provide cells for the formation and patterning of new segments by N. N itself maintains the posterior organiser, and hence the GZ, thus completing the circle.
The posterior segmentation gene network in other arthropods
Comparing Wnt, Cad and N roles in different arthropods allows us to examine the possible conservation of a ‘posterior segmentation gene network’ (Wnt→cad→N). Assessment of N-signalling has revealed dynamic expression of N-signalling members in the GZ in two spider species, Achaearanea tepidariorum and Cupiennius salei (Oda et al., 2007; Schoppmeier and Damen, 2005; Stollewerk et al., 2003). Knockdown of N-signalling members via RNAi leads to loss of posterior segmentation in both species (Oda et al., 2007; Schoppmeier and Damen, 2005; Stollewerk et al., 2003), but some differences were reported in the different studies. In Cupiennius-NRNAi, only the segmentation of posterior segments was affected, while Achaearanea-DlRNAi and NRNAi not only affected posterior segmentation but also had effects earlier in the formation of the caudal lobe (GZ). These differences could be due to the RNAi techniques used. In Cupiennius dsRNA was injected into the eggs at blastula stage whereas in Achaearanea RNAi was injected in the mothers, which produced stronger and earlier phenotypes hinting that zygotic RNAi may be less efficient at interfering with N-signalling. Indeed, the Achaearanea results are similar to our maternal Pa-NRNAi, whereas the Cupiennius results are more similar to our DAPT embryo culture. In Achaearanea, N-signalling is revealed as necessary for caudal expression in the GZ during later stages (Oda et al., 2007), and is similarly regulated by Wnt-signalling (McGregor et al., 2008). Thus, there seems to exist a connection between N and Wnt-signalling in the establishment of the GZ and posterior germ band elongation in spiders similar to the one in Periplaneta, as well as striking similarities in the regulatory interactions between cad, N and Wnt-signalling pathways during these processes.
This view is supported by a recent report in Gryllus where maternal Gb-DlRNAi and Gb-NRNAi embryos display truncated phenotypes with reduced GZ similar to those observed in Achaearanea and Periplaneta (Mito et al., 2011). The authors suggest that posterior segmentation might be controlled by parallel mechanisms, one via stripes of N-signalling and the other through cad and Wnt-signalling; however, we view these pathways to be dynamically linked in Gryllus as in Periplaneta. At early stages, Gb-wg and Gb-cad are not regulated by N-signalling, whereas during germ band elongation Gb-N signalling at the posterior tip maintains Gb-wg and, therefore, Gb-cad, which feeds back onto Gb-Dl (Mito et al., 2011). As in Periplaneta, these regulatory interactions can allow for sustained growth by maintaining a properly sized and functional GZ. Another study in Gryllus (Kainz et al., 2011) used zygotic Gb-DlRNAi and observe defects mostly in the development of the nervous system (a universal function of N-signalling in arthropods that is most sensitive to loss or partial loss, as shown in Drosophila (Mohr, 1924; Van Breugel and Langhout, 1983)), but partial loss of segment markers in only a small minority of embryos. These results could be explained as in spiders where zygotic RNAi produced weaker phenotypes compared to maternal RNAi.
Overall, the different studies of Wnt and N-signalling in different arthropods can be explained by the two functions of Wnt and Cad in maintaining 1) the GZ and 2) Delta expression, and by the two functions of N-signalling 1) maintaining posterior Wnt1 expression, and 2) triggering segment formation in the anterior GZ. Different perturbations in different species (zygotic versus maternal RNAi; embryo culture) reveal different aspects of these functions, but in doing so they offer a temporal window to the regulatory intricacies of the posterior gene segmentation network.
Evolution of the posterior gene network
Our findings in Periplaneta, along with studies in other arthropods, suggest that there exists conservation in both the developmental roles and in the regulatory interactions among the posterior segmentation network. The easiest explanation is that these different arthropods must have inherited such a network from their last common arthropod ancestor. This hypothesis begs the question whether conservation of the posterior gene network can be pushed back further in time. In vertebrates Wnt-signalling regulates cyclic expression of N-signalling members in the presomitic mesoderm (PSM) (Dunty et al., 2008; Gibb et al., 2009). Recently, it has been shown that this Wnt-dependent regulation of N-signalling oscillations seems partly mediated through Cdx, as the expression of the mouse Delta homologue Delta-like1 (Dll1) is disrupted in the PSM in Cdx mutants, and Cdx protein binds to the regulatory regions of Dll1 (Grainger et al., 2012). Furthermore, patterns of expression of this posterior gene network have also been found in the GZ of several different species of annelids, the third segmented phyla, including segmental stripes of Delta and Notch and posterior expression of Wnt and cad (Cho et al., 2010; de Rosa et al., 2005; Janssen et al., 2010; Rivera and Weisblat, 2009; Thamm and Seaver, 2008).
Curiously, vestiges of a Wnt-Cad organiser remain in insects that seem to have lost the requirement for N-mediated segmentation (Tautz, 2004). In the long germ band insect Drosophila a posterior stripe of Dm-wg is expressed in the blastoderm, independent of its segment polarity expression and related to Dm-cad (Vorwald-Denholtz and De Robertis, 2011; Wu and Lengyel, 1998). While a Wnt-Cad posterior organiser may exist in Tribolium (Beermann et al., 2011; Bolognesi et al., 2008; Copf et al., 2004), the sequential addition of posterior segments occurs through a cyclical mechanism that involves pair-rule genes (Choe et al., 2006; Sarrazin et al., 2012), but is not yet completely understood.
In summary, the available data strongly suggest that at least two components of the regulatory gene network, Wnt→cad, were involved in posterior organisation and growth in the last common bilaterian ancestor, the Urbilateria (De Robertis, 1997; Martin and Kimelman, 2009). In addition, following reports from annelids (Rivera and Weisblat, 2009; Thamm and Seaver, 2008), it seems likely that the third component of this gene network, N signalling, would also have been involved in the Urbilateria. However, data are as yet unavailable from partially segmented (metameric) phyla and thus we cannot rule out the possibility that each of the segmented clades convergently and independently recruited a N-mediated segmentation mechanism (Chipman, 2010). However, this would require a) an ancestral in-built tendency of Cad to modulate Delta, b) a predisposition for the Notch pathway to form oscillatory clocks, and c) some unknown constraint of selective advantage leading to the repeated recruitment of these pathways in different phyla, in the face of other genes that could fulfil similar roles. In our view, the most parsimonious explanation remains that this posterior gene regulatory network evolved once and is ancestral. Perhaps the Urbilateria developed by posterior elongation (Jacobs et al., 2005) and contained some kind of serially repeated structures, added sequentially from the posterior end of the animal (Balavoine and Adoutte, 2003; Couso, 2009; De Robertis, 2008), and generated by the interplay between a N-oscillator and the Wnt-Cad posterior organiser.
Supplementary Material
Acknowledgments
Thanks to A. Hurst and R. Phillips for technical assistance and A. Maartens and lab members for manuscript comments. This work was supported by a Wellcome Trust Senior Fellowship to J.P.C. (Ref. 087516) and a University of Sussex GTA studentship to J.E.C.
Footnotes
Competing interests: The authors have no competing interests to declare.
References
- Anderson D. T. (1972a). The development of hemimetabolous insects. Developmental Systems: Insects, Vol. 1 (ed. Counce S J, Waddington C H.), pp. 96–162 London: Academic Press. [Google Scholar]
- Anderson D. T. (1972b). The development of homometabolous insects. Developmental Systems: Insects, Vol. 1 (ed. Counce S J, Waddington C H.), pp. 166–241 London: Academic Press. [Google Scholar]
- Angelini D. R., Kaufman T. C. (2005). Functional analyses in the milkweed bug Oncopeltus fasciatus (Hemiptera) support a role for Wnt signaling in body segmentation but not appendage development. Dev. Biol. 283, 409–423 10.1016/j.ydbio.2005.04.034 [DOI] [PubMed] [Google Scholar]
- Aulehla A., Herrmann B. G. (2004). Segmentation in vertebrates: clock and gradient finally joined. Genes Dev. 18, 2060–2067 10.1101/gad.1217404 [DOI] [PubMed] [Google Scholar]
- Balavoine G., Adoutte A. (2003). The segmented Urbilateria: a testable scenario. Integr. Comp. Biol. 43, 137–147 10.1093/icb/43.1.137 [DOI] [PubMed] [Google Scholar]
- Beermann A., Prühs R., Lutz R., Schröder R. (2011). A context-dependent combination of Wnt receptors controls axis elongation and leg development in a short germ insect. Development 138, 2793–2805 10.1242/dev.063644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognesi R., Farzana L., Fischer T. D., Brown S. J. (2008). Multiple Wnt genes are required for segmentation in the short-germ embryo of Tribolium castaneum. Curr. Biol. 18, 1624–1629 10.1016/j.cub.2008.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawengsaksophak K., de Graaff W., Rossant J., Deschamps J., Beck F. (2004). Cdx2 is essential for axial elongation in mouse development. Proc. Natl. Acad. Sci. USA 101, 7641–7645 10.1073/pnas.0401654101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Dodge M. E., Tang W., Lu J., Ma Z., Fan C. W., Wei S., Hao W., Kilgore J., Williams N. S.et al. (2009). Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 5, 100–107 10.1038/nchembio.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman A. D. (2010). Parallel evolution of segmentation by co-option of ancestral gene regulatory networks. Bioessays 32, 60–70 10.1002/bies.200900130 [DOI] [PubMed] [Google Scholar]
- Cho S. J., Vallès Y., Giani V. C., Jr, Seaver E. C., Weisblat D. A. (2010). Evolutionary dynamics of the wnt gene family: a lophotrochozoan perspective. Mol. Biol. Evol. 27, 1645–1658 10.1093/molbev/msq052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe C. P., Miller S. C., Brown S. J. (2006). A pair-rule gene circuit defines segments sequentially in the short-germ insect Tribolium castaneum. Proc. Natl. Acad. Sci. USA 103, 6560–6564 10.1073/pnas.0510440103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copf T., Schröder R., Averof M. (2004). Ancestral role of caudal genes in axis elongation and segmentation. Proc. Natl. Acad. Sci. USA 101, 17711–17715 10.1073/pnas.0407327102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couso J. P. (2009). Segmentation, metamerism and the Cambrian explosion. Int. J. Dev. Biol. 53, 1305–1316 10.1387/ijdb.072425jc [DOI] [PubMed] [Google Scholar]
- Couso J. P., Bate M., Martínez–Arias A. (1993). A wingless-dependent polar coordinate system in Drosophila imaginal discs. Science 259, 484–489 10.1126/science.8424170 [DOI] [PubMed] [Google Scholar]
- Davis G. K., Patel N. H. (2002). Short, long, and beyond: molecular and embryological approaches to insect segmentation. Annu. Rev. Entomol. 47, 669–699 10.1146/annurev.ento.47.091201.145251 [DOI] [PubMed] [Google Scholar]
- De Robertis E. M. (1997). Evolutionary biology. The ancestry of segmentation. Nature 387, 25–26 10.1038/387025a0 [DOI] [PubMed] [Google Scholar]
- De Robertis E. M. (2008). The molecular ancestry of segmentation mechanisms. Proc. Natl. Acad. Sci. USA 105, 16411–16412 10.1073/pnas.0808774105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rosa R., Prud'homme B., Balavoine G. (2005). caudal and even-skipped in the annelid Platynereis dumerilii and the ancestry of posterior growth. Evol. Dev. 7, 574–587 10.1111/j.1525-142X.2005.05061.x [DOI] [PubMed] [Google Scholar]
- Dearden P. K., Akam M. (2001). Early embryo patterning in the grasshopper, Schistocerca gregaria: wingless, decapentaplegic and caudal expression. Development 128, 3435–3444. [DOI] [PubMed] [Google Scholar]
- Dequéant M. L., Pourquié O. (2008). Segmental patterning of the vertebrate embryonic axis. Nat. Rev. Genet. 9, 370–382 10.1038/nrg2320 [DOI] [PubMed] [Google Scholar]
- Dunty W. C., Jr, Biris K. K., Chalamalasetty R. B., Taketo M. M., Lewandoski M., Yamaguchi T. P. (2008). Wnt3a/β-catenin signaling controls posterior body development by coordinating mesoderm formation and segmentation. Development 135, 85–94 10.1242/dev.009266 [DOI] [PubMed] [Google Scholar]
- Farzana L., Brown S. J. (2008). Hedgehog signaling pathway function conserved in Tribolium segmentation. Dev. Genes Evol. 218, 181–192 10.1007/s00427-008-0207-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb S., Zagorska A., Melton K., Tenin G., Vacca I., Trainor P., Maroto M., Dale J. K. (2009). Interfering with Wnt signalling alters the periodicity of the segmentation clock. Dev. Biol. 330, 21–31 10.1016/j.ydbio.2009.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger S., Lam J., Savory J. G., Mears A. J., Rijli F. M., Lohnes D. (2012). Cdx regulates Dll1 in multiple lineages. Dev. Biol. 361, 1–11 10.1016/j.ydbio.2011.09.034 [DOI] [PubMed] [Google Scholar]
- Grossmann D., Scholten J., Prpic N. M. (2009). Separable functions of wingless in distal and ventral patterning of the Tribolium leg. Dev. Genes Evol. 219, 469–479 10.1007/s00427-009-0310-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland L. Z. (2002). Heads or tails? Amphioxus and the evolution of anterior–posterior patterning in deuterostomes. Dev. Biol. 241, 209–228 10.1006/dbio.2001.0503 [DOI] [PubMed] [Google Scholar]
- Ikeya M., Takada S. (2001). Wnt-3a is required for somite specification along the anteroposterior axis of the mouse embryo and for regulation of cdx-1 expression. Mech. Dev. 103, 27–33 10.1016/S0925-4773(01)00338-0 [DOI] [PubMed] [Google Scholar]
- Jacobs D. K., Hughes N. C., Fitz–Gibbon S. T., Winchell C. J. (2005). Terminal addition, the Cambrian radiation and the Phanerozoic evolution of bilaterian form. Evol. Dev. 7, 498–514 10.1111/j.1525-142X.2005.05055.x [DOI] [PubMed] [Google Scholar]
- Janssen R., Le Gouar M., Pechmann M., Poulin F., Bolognesi R., Schwager E. E., Hopfen C., Colbourne J. K., Budd G. E., Brown S. J.et al. (2010). Conservation, loss, and redeployment of Wnt ligands in protostomes: implications for understanding the evolution of segment formation. BMC Evol. Biol. 10, 374 10.1186/1471-2148-10-374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. J., Aerne B. L., Smithers L., Haddon C., Ish–Horowicz D., Lewis J. (2000). Notch signalling and the synchronization of the somite segmentation clock. Nature 408, 475–479 10.1038/35044091 [DOI] [PubMed] [Google Scholar]
- Jiménez F., Campos–Ortega J. A. (1982). Maternal effects of zygotic mutants affecting early neurogenesis in Drosophila. Dev. Genes Evol. 191, 191–201 10.1007/BF00848335 [DOI] [PubMed] [Google Scholar]
- Kainz F., Ewen–Campen B., Akam M., Extavour C. G. (2011). Notch/Delta signalling is not required for segment generation in the basally branching insect Gryllus bimaculatus. Development 138, 5015–5026 10.1242/dev.073395 [DOI] [PubMed] [Google Scholar]
- Lenoir–Rousseaux J. J., Lender T. (1970). Table de développement embryonnaire de Periplaneta americana (L.) insecte, Dictyoptere. Bulletin de la Societe Zoologique de France. 95, 737–751. [Google Scholar]
- Liu P. Z., Kaufman T. C. (2005). Short and long germ segmentation: unanswered questions in the evolution of a developmental mode. Evol. Dev. 7, 629–646 10.1111/j.1525-142X.2005.05066.x [DOI] [PubMed] [Google Scholar]
- Lohnes D. (2003). The Cdx1 homeodomain protein: an integrator of posterior signaling in the mouse. Bioessays 25, 971–980 10.1002/bies.10340 [DOI] [PubMed] [Google Scholar]
- Marie B., Bacon J. P. (2000). Two engrailed-related genes in the cockroach: cloning, phylogenetic analysis, expression and isolation of splice variants. Dev. Genes Evol. 210, 436–448 10.1007/s004270000082 [DOI] [PubMed] [Google Scholar]
- Martin B. L., Kimelman D. (2009). Wnt signaling and the evolution of embryonic posterior development. Curr. Biol. 19, R215–R219 10.1016/j.cub.2009.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez–Arias A. (1993). Development and patterning of the larval epidermis of Drosophila. The Development Of Drosophila Melanogaster, (ed. Bate M, Martinez–Arias) A, pp. 517–608 Plainview, NY: Cold Spring Harbour Laboratory Press. [Google Scholar]
- Martinez–Arias A., Lawrence P. A. (1985). Parasegments and compartments in the Drosophila embryo. Nature 313, 639–642 10.1038/313639a0 [DOI] [PubMed] [Google Scholar]
- McGregor A. P., Pechmann M., Schwager E. E., Feitosa N. M., Kruck S., Aranda M., Damen W. G. (2008). Wnt8 is required for growth-zone establishment and development of opisthosomal segments in a spider. Curr. Biol. 18, 1619–1623 10.1016/j.cub.2008.08.045 [DOI] [PubMed] [Google Scholar]
- McGregor A. P., Pechmann M., Schwager E. E., Damen W. G. (2009). An ancestral regulatory network for posterior development in arthropods. Commun. Integr. Biol. 2, 174–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito T., Shinmyo Y., Kurita K., Nakamura T., Ohuchi H., Noji S. (2011). Ancestral functions of Delta/Notch signaling in the formation of body and leg segments in the cricket Gryllus bimaculatus. Development 138, 3823–3833 10.1242/dev.060681 [DOI] [PubMed] [Google Scholar]
- Miyawaki K., Mito T., Sarashina I., Zhang H. J., Shinmyo Y., Ohuchi H., Noji S. (2004). Involvement of Wingless/Armadillo signaling in the posterior sequential segmentation in the cricket, Gryllus bimaculatus (Orthoptera), as revealed by RNAi analysis. Mech. Dev. 121, 119–130 10.1016/j.mod.2004.01.002 [DOI] [PubMed] [Google Scholar]
- Mohr O. L. (1924). A genetic and cytological analysis of a section deficiency involving four units of the X-chromosome in Drosophila melanogaster. Mol. Gen. Genet. 32, 108–232. [Google Scholar]
- Mooi R., David B. (2008). Radial symmetry, the anterior/posterior axis, and echinoderm Hox genes. Annu. Rev. Ecol. Evol. Syst. 39, 43–62 10.1146/annurev.ecolsys.39.110707.173521 [DOI] [Google Scholar]
- Niehrs C. (2010). On growth and form: a Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development 137, 845–857 10.1242/dev.039651 [DOI] [PubMed] [Google Scholar]
- Nüsslein–Volhard C., Wieschaus E. (1980). Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801 10.1038/287795a0 [DOI] [PubMed] [Google Scholar]
- Oda H., Nishimura O., Hirao Y., Tarui H., Agata K., Akiyama–Oda Y. (2007). Progressive activation of Delta-Notch signaling from around the blastopore is required to set up a functional caudal lobe in the spider Achaearanea tepidariorum. Development 134, 2195–2205 10.1242/dev.004598 [DOI] [PubMed] [Google Scholar]
- Patel N. H., Martin–Blanco E., Coleman K. G., Poole S. J., Ellis M. C., Kornberg T. B., Goodman C. S. (1989). Expression of engrailed proteins in arthropods, annelids, and chordates. Cell 58, 955–968 10.1016/0092-8674(89)90947-1 [DOI] [PubMed] [Google Scholar]
- Peel A., Akam M. (2003). Evolution of segmentation: rolling back the clock. Curr. Biol. 13, R708–R710 10.1016/j.cub.2003.08.045 [DOI] [PubMed] [Google Scholar]
- Prud'homme B., de Rosa R., Arendt D., Julien J. F., Pajaziti R., Dorresteijn A. W., Adoutte A., Wittbrodt J., Balavoine G. (2003). Arthropod-like expression patterns of engrailed and wingless in the annelid Platynereis dumerilii suggest a role in segment formation. Curr. Biol. 13, 1876–1881 10.1016/j.cub.2003.10.006 [DOI] [PubMed] [Google Scholar]
- Pueyo J. I., Lanfear R., Couso J. P. (2008). Ancestral Notch-mediated segmentation revealed in the cockroach Periplaneta americana. Proc. Natl. Acad. Sci. USA 105, 16614–16619 10.1073/pnas.0804093105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford N., Olson P. D. (2011). Wnt gene loss in flatworms. Dev. Genes Evol. 221, 187–197 10.1007/s00427-011-0370-8 [DOI] [PubMed] [Google Scholar]
- Rivera A. S., Weisblat D. A. (2009). And Lophotrochozoa makes three: Notch/Hes signaling in annelid segmentation. Dev. Genes Evol. 219, 37–43 10.1007/s00427-008-0264-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan J. F., Mazza M. E., Pang K., Matus D. Q., Baxevanis A. D., Martindale M. Q., Finnerty J. R. (2007). Pre-bilaterian origins of the Hox cluster and the Hox code: evidence from the sea anemone, Nematostella vectensis. PLoS ONE 2, e153 10.1371/journal.pone.0000153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin A. F., Peel A. D., Averof M. (2012). A segmentation clock with two-segment periodicity in insects. Science 336, 338–341 10.1126/science.1218256 [DOI] [PubMed] [Google Scholar]
- Savory J. G., Mansfield M., Rijli F. M., Lohnes D. (2011). Cdx mediates neural tube closure through transcriptional regulation of the planar cell polarity gene Ptk7. Development 138, 1361–1370 10.1242/dev.056622 [DOI] [PubMed] [Google Scholar]
- Schoppmeier M., Damen W. G. M. (2005). Suppressor of Hairless and Presenilin phenotypes imply involvement of canonical Notch-signalling in segmentation of the spider Cupiennius salei. Dev. Biol. 280, 211–224 10.1016/j.ydbio.2005.01.024 [DOI] [PubMed] [Google Scholar]
- Shimizu T., Bae Y. K., Muraoka O., Hibi M. (2005). Interaction of Wnt and caudal-related genes in zebrafish posterior body formation. Dev. Biol. 279, 125–141 10.1016/j.ydbio.2004.12.007 [DOI] [PubMed] [Google Scholar]
- Shinmyo Y., Mito T., Matsushita T., Sarashina I., Miyawaki K., Ohuchi H., Noji S. (2005). caudal is required for gnathal and thoracic patterning and for posterior elongation in the intermediate-germband cricket Gryllus bimaculatus. Mech. Dev. 122, 231–239 10.1016/j.mod.2004.10.001 [DOI] [PubMed] [Google Scholar]
- Stollewerk A., Schoppmeier M., Damen W. G. M. (2003). Involvement of Notch and Delta genes in spider segmentation. Nature 423, 863–865 10.1038/nature01682 [DOI] [PubMed] [Google Scholar]
- Takada S., Stark K. L., Shea M. J., Vassileva G., McMahon J. A., McMahon A. P. (1994). Wnt-3a regulates somite and tailbud formation in the mouse embryo. Genes Dev. 8, 174–189 10.1101/gad.8.2.174 [DOI] [PubMed] [Google Scholar]
- Tautz D. (2004). Segmentation. Dev. Cell 7, 301–312 10.1016/j.devcel.2004.08.008 [DOI] [PubMed] [Google Scholar]
- Thamm K., Seaver E. C. (2008). Notch signaling during larval and juvenile development in the polychaete annelid Capitella sp. I. Dev. Biol. 320, 304–318 10.1016/j.ydbio.2008.04.015 [DOI] [PubMed] [Google Scholar]
- Van Breugel F. M. A., Langhout B. V. Z. (1983). The Notch locus of Drosophila hydei: alleles, phenotypes and functional organization. Genetics 103, 197–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Ven C., Bialecka M., Neijts R., Young T., Rowland J. E., Stringer E. J., Van Rooijen C., Meijlink F., Nóvoa A., Freund J. N.et al. (2011). Concerted involvement of Cdx/Hox genes and Wnt signaling in morphogenesis of the caudal neural tube and cloacal derivatives from the posterior growth zone. Development 138, 3451–3462 10.1242/dev.066118 [DOI] [PubMed] [Google Scholar]
- van den Akker E., Forlani S., Chawengsaksophak K., de Graaff W., Beck F., Meyer B. I., Deschamps J. (2002). Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development 129, 2181–2193. [DOI] [PubMed] [Google Scholar]
- Vorwald–Denholtz P. P., De Robertis E. M. (2011). Temporal pattern of the posterior expression of Wingless in Drosophila blastoderm. Gene Expr. Patterns 11, 456–463 10.1016/j.gep.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. H., Lengyel J. A. (1998). Role of caudal in hindgut specification and gastrulation suggests homology between Drosophila amnioproctodeal invagination and vertebrate blastopore. Development 125, 2433–2442. [DOI] [PubMed] [Google Scholar]
- Young T., Rowland J. E., van de Ven C., Bialecka M., Novoa A., Carapuco M., van Nes J., de Graaff W., Duluc I., Freund J. N.et al. (2009). Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev. Cell 17, 516–526 10.1016/j.devcel.2009.08.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


