Abstract
Active DNA demethylation is an important part of epigenetic regulation in plants and animals. How active DNA demethylation is regulated and its relationship with histone modification patterns are unclear. Here, we report the discovery of IDM1, a regulator of DNA demethylation in Arabidopsis. IDM1 is required for preventing DNA hypermethylation of highly homologous multicopy genes and other repetitive sequences that are normally targeted for active DNA demethylation by Repressor of Silencing 1 and related 5-methylcytosine DNA glycosylases. IDM1 binds methylated DNA at chromatin sites lacking histone H3K4 di- or trimethylation and acetylates H3 to create a chromatin environment permissible for 5-methylcytosine DNA glycosylases to function. Our study reveals how some genes are indicated by multiple epigenetic marks for active DNA demethylation and protection from silencing.
Epigenetic control of gene expression involves dynamic regulation of DNA methylation and histone modification marks (1, 2). A DNA repair–based mechanism functions in active DNA demethylation in plants and possibly in animals as well (3). In plants, active DNA demethylation is initiated by a family of 5-methylcytosine DNA glycosylases that includes ROS1, DME, DML2, and DML3 (3). Although a large body of literature concerns the regulation of DNA methylation by histone modification patterns (1, 2), little is known about the regulation of active DNA demethylation.
We developed a polymerase chain reaction (PCR)–based marker for reporting DNA methylation status at the 3′ region of At1g26400 (also referred to as DT-77)(4). This marker suggests that the At1g26400 region is hypermethylated in ros1 mutant alleles (Fig. 1A). We screened a population of homozygous transferred DNA (T-DNA) insertion lines of Arabidopsis for DNA hypermethylation mutants. Two of the identified mutants (Fig. 1A), referred to as idm1 (for increased DNA methylation 1), have T-DNA insertions in the gene At3g14980 (fig. S1A). Bisulfite sequencing showed that, compared to the Col wild-type control, the idm1 and ros1-4 mutants have higher levels of DNA methylation in all sequence contexts at DT-77 (Fig. 1B). There is also increased CG and CHG (where H is C, A, or T) methylation at the At4g18650 gene promoter, which was previously shown to be targeted for demethylation by ROS1 (5) (Fig. 1C). ROS1 was originally identified because it is required for preventing the RD29A-LUC and 35S-NPTII trans-genes from transcriptional silencing (6). From an independent genetic screen for mutants impaired in the prevention of silencing of the 35S-NPTII transgene, we found the idm1-3 mutation that changes glutamic acid-451 to a premature stop codon and causes the silencing of 35S-NPTII but not the RD29A-LUC transgene (fig. S2).
Fig. 1.
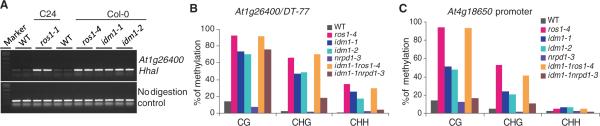
Identification and characterization of the idm1 mutants. (A)Analysis showing the effects of idm1 mutations on At1g26400 (B) and At4g18650 (C) of DNA methylation level at the At1g26400 locus by chop-PCR (4). Un-DNA methylation in different sequence contexts and genetic interactions digested DNA is shown as acontrol. (B and C) Bisulfite sequencing data with ros1 or nrpd1. WT, wild type.
Double-mutant analysis indicated that the DNA hypermethylation phenotypes of ros1-4 and idm1-1 are not additive (Fig. 1, B and C, and fig. S3A). These results suggest that IDM1 and ROS1 may function in the same genetic pathway to prevent DNA hypermethylation. Quantitative reverse transcription PCR analysis showed that ROS1 or ROS3 expression is not reduced in the idm1 mutants (fig. S4A). In addition, like ROS1 (3), IDM1 expression level was reduced in RNA-directed DNA methylation (RdDM) pathway mutants (fig. S4B), which suggests that IDM1 expression is sensitive to non-CG methylation. The hypermethylation in idm1-1 at CHG and CHH sequence contexts could be suppressed by the nrpd1-3 mutation (7), suggesting that the methylation at both DT-77 and At4g18650 promoter is caused by RdDM (Fig. 1, B and C).
Southern blot analysis showed that idm1 does not affect the DNA methylation at 5S or 45S ribosomal DNA or at the 180–base pair centromeric repeat (fig. S5). We compared the genome-wide DNA methylation profiles of idm1-1 and wild-type plants. There was no significant difference between idm1-1, ros1-4, or ros1-3dml2-1dml3-1 (rdd) (8) and wild-type plants in their overall genome methylation patterns (fig. S6). We identified 1098, 4991, and 9290 loci where DNA methylation levels are increased significantly in idm1-1, ros1-4, and rdd, respectively (tables S1 to S3). In contrast, only 75, 106, and 1052 loci were found to have a decreased DNA methylation in idm1-1, ros1-4, and rdd, respectively (tables S4 to S6). Out of the 1098 hypermethylated loci in idm1-1 (referred herein as DT loci for demethylation target loci), seven were selected for validation by individual locus bisulfite sequencing and were all confirmed to be hypermethylated in idm1-1 as well as ros1-4 or rdd (fig. S3). The DT loci are distributed across the five chromosomes (fig. S7A), are enriched in small RNAs (fig. S7B), and the majority of them overlap with repeats (table S1 and fig. S8). Genic loci account for ~80% of the DT loci in idm1-1 (fig. S7C). The hypermethylation in genic regions in idm1-1 mutant plants is not restricted to CGs (Fig. 1, B and C, and fig. S3). More than half of these hypermethylated genic loci in idm1-1 are in multigene families where the member genes are highly homologous.
About 52% and 63% of the 1098 hyper-methylated loci in idm1-1 are also hypermethylated in the ros1-4 and rdd mutants, respectively, based on the methylome analysis (table S1 and fig. S7D). As a comparison, the same analysis showed that ~81% (as opposed to a theoretical 100%) of the hypermethylated loci in ros1-4 overlap with those in rdd (table S2 and fig. S7D). Three loci that were identified as hypermethylated in idm1-1 but not ros1-4 or rdd were analyzed by individual locus bisulfite sequencing (fig. S3B). The analysis shows that all of them are hypermethylated in ros1 or rdd as well as in idm1. In addition, four other loci that were identified as hypermethylated in the rdd mutant but not in idm1-1 in the DNA methylome study were tested by individual locus bisulfite sequencing and found hypermethylated in idm1-1 (Fig. 1C and fig. S3). These results suggest that the actual number of hypermethylated loci in idm1-1 is larger and the percentage of overlap with the hypermethylated loci in the rdd mutant is likely higher. It thus appears that the majority of the loci hypermethylated in idm1 are also hypermethylated in the rdd mutant. This is consistent with the idm1ros1 double-mutant analysis that indicated that IDM1 and ROS1(andDML2andDML3) function in the same DNA demethylation pathway.
The expression levels of 15 genes that have increased DNA methylation in or near the genes in idm1-1 and ros1-4 mutants were examined. We found that seven of the tested genes show a substantial reduction in their transcript levels in the mutants (fig. S8). Our results suggest that, like ROS1, IDM1 is critical for preventing the transcriptional silencing of some transgenes and endogenous genes.
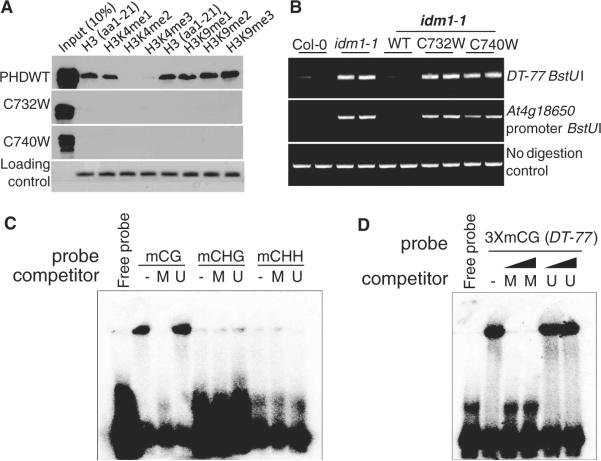
The tissue pattern of IDM1 expression is similar to those of DML2 and ROS1 (fig. S9). IDM1 is predicted to have a MBD domain, a PHD finger domain and an N-acetyltransferase domain (fig. S1D). We found that the PHD finger of IDM1 specifically binds the N-terminal tail of histone H3 and that the binding is inhibited by H3K4 di- or trimethylation (fig. S10A). H3R2 methylation also appears to inhibit the binding (fig. S10A). In vitro affinity pull-down assays confirmed that the PHD finger of IDM1 recognizes H3 N-terminal tail and that the recognition is prevented by H3K4 di- or trimethylation (Fig. 2A). In contrast, H3K9 methylation does not inhibit the interaction (Fig. 2A and fig. S10A). C732W or C740W mutation (fig. S11A) abolishes the binding of IDM1 to histone H3 (Fig. 2A). Expression of the wild-type but not the C732W or C740W mutant version of IDM1 under its native promoter could complement the DNA hypermethylation phenotype of the idm1-1 mutant (Fig. 2B and fig. S10B). The results suggest that histone H3 binding is important for IDM1 function in vivo. IDM1 binds to the DNA containing CG methylation, and the binding is competitively blocked by unlabeled methylated, but not unmethylated, DNA of the same sequence (Fig. 2C and fig. S12). We also tested DNA with CG methylation corresponding to the DT-77 locus and found that IDM1 binds specifically to methylated DT-77 DNA (Fig. 2D).
Fig. 2.
Functional analysis of the PHD finger and MBD domains of IDM1. (A) Western blot analysis of the PHD finger of IDM1 in a peptide pull-down assay. Coomassie blue–stained biotinylated histone peptides are shown as a loading control. (B) DNA hypermethylation phenotypes of idm1-1 plants transformed with WT or mutant forms of IDM1. (C) Electrophoretic mobility shift assay (EMSA) of IDM1-N (amino acids 1 to 400) binding to methylated oligonucleotides (fig. S12). -, no competitor; M, methylated; U, unmethylated. (D) EMSA showing IDM1-N (amino acids 1 to 400) binding to a methylated oligonucleotide probe corresponding to the DT-77 locus.
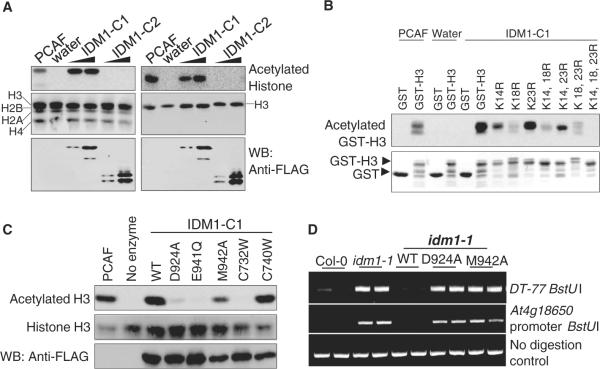
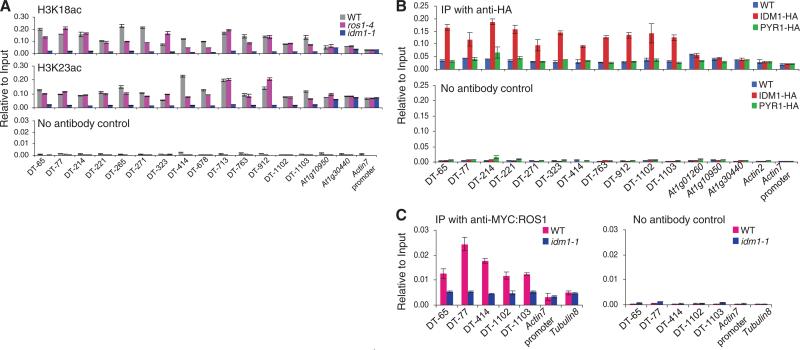
We expressed and purified IDM1 fragments containing the putative HAT and PHD domains (IDM1-C1) or the HAT domain only (IDM1-C2) (fig. S13, A and B) from insect cells. Using core histones, histone H3 (Fig. 3A) and oligonucleo-some (fig. S13C) as substrates, we found that IDM1-C1, but not IDM1-C2, has an acetyltransferase activity on histone H3. K14, K18, and K23 of H3 are acetylated by IDM1-C1 in vitro (Fig. 3B). Mass spectrometry analysis of the in vitro acetylated histone H3 confirmed that K18 and K23, as well as K14, are acetylated and K18 and K23 are the main targets of acetylation by IDM1-C1 (fig. S13, D and E). D924A, E941Q, or M942A mutation in conserved residues in the HAT domain abolished or diminished the in vitro acetyltransferase activity of IDM1-C1 (Fig. 3C). Expression of the D924A or M942A mutated IDM1 under its native promoter failed to complement the DNA hypermethylation phenotypes of the idm1-1 mutant (Fig. 3D and fig. S10B). The results suggest that the histone H3 acetylation activity is necessary for IDM1 function in preventing DNA hypermethylation in plants. The C732W, but not the C740W, mutation in the PHD domain required for H3 interaction also abolished the acetyltransferase activity of IDM1-C1 (Fig. 3C), suggesting that the PHD domain can affect the acetyltransferase activity of IDM1 independently of its role in binding histone H3. Chromatin immunoprecipitation (ChIP) assays showed that acetylated histone H3K18 and H3K23 marks are reduced considerably in idm1-1 mutant plants specifically at the DT loci (Fig. 4A), suggesting that IDM1 is critical for H3K18 and H3K23 acetylation in vivo.
Fig. 3.
Histone acetyltransferase activity of IDM1. (A) HAT assays. (Top) Autoradiograph for acetylated histone. (Middle) Coomassie blue–stained membrane picture. (Bottom) Western blot detection of IDM1 with antibody to FLAG. p300-CBP-associated factor (PCAF) was used as a positive control. (B) HAT assay results using recombinant histone H3 mutated at different lysine positions as substrates. (C) HAT assay results using histone H3 as substrate and using WT and mutated forms of IDM1-C1 proteins as enzymes. (D) DNA hypermethylation phenotypes of idm1-1 plants transformed with WT or mutant forms of IDM1.
Fig. 4.
Histone acetylation marks and IDM1 and ROS1 association with chromatin. (A) H3K18 acetylation (ac) and H3K23ac levels at the DT loci and control regions. ChIP was performed with antibodies to H3K18ac and H3K23ac. (B) Association of IDM1 protein with DT loci. ChIP was performed in WT, PYR1:3HA, and IDM1:3HA:YFP transgenic plants with antibody to hemagglutinin. (C) Effect of idm1 on ROS1 protein association with DT loci. ChIP was performed in MYC:ROS1/ros1-4 (WT) and MYC:ROS1/idm1-1 plants using antibody to MYC. The ChIP signal was quantified as relative to input DNA. The no-antibody precipitates served as a negative control. Standard errors were calculated from three technical repeats.
ChIP assays also indicated that IDM1 is enriched at all of the DT loci tested (Fig. 4B). The DT loci have CG methylation (Fig. 1, B and C, fig. S3, and table S1), and several of them were found to have low levels of H3K4 dimethylation by ChIP assays (fig. S14). In contrast, IDM1 is not enriched in the highly expressed At1g01260, At1g10950, and At1g30440 genes (Fig. 4B) that have abundant CG methylation (9)and ahigh level of H3K4 dimethylation. DT loci in general correspond to sequences with low H3K4 mono-, di-, and trimethylation, relative to a comparison group of CG methylated and expressed genes (10) (fig. S15). The results are consistent with the notion that IDM1 binds to methylated DNA through its MBD domain but, because of its PHD finger domain, the binding is prevented at loci where there is a high level of H3K4 di- or trimethylation. Transposons are characterized by high CG methylation and low H3K4 di- or trimethylation (10). However, IDM1 does not affect the methylation status of most transposons, which are associated with heterochromatin (fig. S7C). This discrimination against heterochromatic regions could potentially be explained by the inhibition of IDM1 interaction with histone H3 by H3R2 methylation (fig. S10A), which is known to be associated with heterochromatin in yeast (11). The DNA hypermethylation phenotypes of idm1 mutants and the HAT activity and DNA-and histone-binding characteristics suggest that IDM1 may be important in selectively binding to the DT loci and creating a permissible chromatin environment for DNA demethylation enzymes. Consistent with this notion, ChIP results show that ROS1 protein is enriched at the DT loci in wild-type plants but that the enrichment is reduced to basal levels of nonspecific binding in idm1 mutant plants (Fig. 4C).
Active DNA demethylation in plants is achieved through a base excision repair pathway that is initiated by the ROS1 subfamily of 5-methylcytosine DNA glycosylases (3). Our results here suggest that IDM1 controls the DNA methylation levels of a subset of loci targeted by the 5-methylcytosine DNA glycosylases. It is critical for preventing DNA hypermethylation and transcriptional silencing of some transgenes and endogenous genes with repetitive sequences or highly homologous genes in multi-gene families. We showed that IDM1 is a his-tone H3 acetyltransferase that is capable of recognizing methylated DNA through its MBD domain and recognizing unmethylated histone H3K4 through its PHD domain. Thus, IDM1 can recognize multiple epigenetic features at some demethylation target loci and create acetylated H3K18 and H3K23 marks, which then may be recognized by DNA demethylation enzymes or their interacting partner proteins (fig. S16) because IDM1 does not appear to colocalize with ROS1 (fig. S17).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants R01GM070795 and R01GM059138 to J.-K.Z. and by the Chinese Academy of Sciences (CAS), China. We thank B. Stevenson for technical assistance and X. Zhang for the gift of glutathione S-transferase H3 (amino acids 1 to 57) construct. Sequence data are available in the Gene Expression Omnibus database (GEO accession GSE33071).
Footnotes
Supplementary Materials www.sciencemag.org/cgi/content/full/336/6087/1445/DC1 Materials and Methods Figs. S1 to S17 Tables S1 to S7 References (12–20)
References and Notes
- 1.Tariq M, Paszkowski J. Trends Genet. 2004;20:244. doi: 10.1016/j.tig.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Law JA, Jacobsen SE. Nat. Rev. Genet. 2010;11:204. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu JK. Annu. Rev. Genet. 2009;43:143. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Materials and methods are available as supplementary materials on Science Online.
- 5.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. Curr. Biol. 2007;17:54. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 6.Gong Z, et al. Cell. 2002;111:803. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 7.Onodera Y, et al. Cell. 2005;120:613. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Penterman J, et al. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6752. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lister R, et al. Cell. 2008;133:523. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirmizis A, et al. Nature. 2007;449:928. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.