Summary
Cell chemotaxis, such as migration of fibroblasts towards growth factors during development and wound healing, requires precise spatial coordination of signalling events. Phosphoinositides and signalling enzymes involved in their generation and hydrolysis have been implicated in regulation of chemotaxis; however, the role and importance of specific components remain poorly understood. Here, we demonstrate that phospholipase C epsilon (PLCε) contributes to fibroblast chemotaxis towards platelet-derived growth factor (PDGF-BB). Using PLCe1 null fibroblasts we show that cells deficient in PLCε have greatly reduced directionality towards PDGF-BB without detrimental effect on their basal ability to migrate. Furthermore, we show that in intact fibroblasts, signalling events, such as activation of Rac, are spatially compromised by the absence of PLCε that affects the ability of cells to enlarge their protrusions in the direction of the chemoattractant. By further application of live cell imaging and the use of FRET-based biosensors, we show that generation of Ins(1,4,5)P3 and recruitment of PLCε are most pronounced in protrusions responding to the PDGF-BB gradient. Furthermore, the phospholipase C activity of PLCε is critical for its role in chemotaxis, consistent with the importance of Ins(1,4,5)P3 generation and sustained calcium responses in this process. As PLCε has extensive signalling connectivity, using transgenic fibroblasts we ruled out its activation by direct binding to Ras or Rap GTPases, and suggest instead new unexpected links for PLCε in the context of chemotaxis.
Key words: Caenorhabditis elegans, Rab GTPase, Cortical granule exocytosis, Separase
Introduction
Phosphoinositide signalling plays an important role in various aspects of cell motility including cytoskeleton remodelling, regulation of cell adhesion and directional sensing (Kölsch et al., 2008). Directional movement is a property of most cell types during development and is subsequently critical for tissue remodelling and regeneration. Chemotaxis, or migration biased toward a gradient of soluble chemoattractant, has been studied extensively in two model cell types: neutrophils and the slime mould Dictyostelium discoideum. Another example of chemotactic sensing is that of fibroblasts in wound healing. Platelet-derived growth factor (PDGF), produced by platelets and macrophages, forms a gradient in the tissue and serves as a potent chemoattractant and mitogen, thus accelerating the rate of fibroblast invasion into the fibrin clot (Heldin and Westermark, 1999).
In the context of chemotaxis, several components of phosphoinositide signalling have been implicated in regulatory mechanisms. In particular, in D. discoideum and neutrophils the spatial segregation of PtdIns(3,4,5)P3 by the actions of PI3K and PTEN have been well documented and the role of PI3K in chemokinesis and chemotaxis assessed using different in vitro and in vivo conditions (Afonso and Parent, 2011). Links with other important signalling components involved in cell movement and chemotaxis, small GTPases from Ras and Rho family, have also been explored and established in these model systems (Cain and Ridley, 2009). In fibroblasts, chemotaxis to PDGF is also characterised by polarised localisation of PtdIns(3,4,5)P3. However, modelling predicts that localised PtdIns(3,4,5)P3 accumulation primarily reflects receptor occupancy and is not regulated by feedback amplification or inhibition proposed for ameboid cells (Schneider and Haugh, 2005). Indeed, migration of fibroblasts is slower than that of amoeboid cells and although driven by membrane protrusion, it also involves substantial contribution of differential adhesion (Lauffenburger and Horwitz, 1996), suggesting distinct requirements for gradient sensing. Furthermore, it has been shown that robust PDGF sensing requires steeper gradients and a much narrower range of absolute chemoattractant concentration (Schneider and Haugh, 2005).
Another enzyme activity reported to be involved in chemotaxis in D. discoideum and several mammalian cell types is phosphoinositide-specific phospholipase C (PLC). In D. discoideum a single PLC activity, dDPLC, has been implicated in maintaining polarised distribution of PtdIns(4,5)P2 and is suggested to affect localised regulation of PtdIns(3,4,5)P3 production (Kortholt et al., 2007). In mammalian cells, however, there are six families of PLC enzymes (PLCβ, γ, δ, ε, ζ and η), consisting of 13 isozymes in humans, characterised by different regulatory mechanisms and expression pattern (Bunney and Katan, 2011). PLCβ2 and PLCβ3 are abundant isoforms of PLC in hemopoietic cells and respond to agonists to G-protein coupled receptor, including potent chemoattractants fMPL and SDF-α. Studies of neutrophil chemotaxis demonstrated that cells lacking PLCβ2/PLCβ3 enzymes have slightly deficient chemotaxis towards fMLP gradient that is further affected by depletion of PI3K, thus functioning as components of complex and context-dependent regulatory networks (Tang et al., 2011). T cells deficient only in PLCβ2/β3, however, have substantially reduced chemotaxis when analysed in gradients of SDF-1α (Bach et al., 2007). Studies of several breast cancer cell lines demonstrated the involvement of PLC activity in chemotaxis towards EGF (Roussos et al., 2011; Wang et al., 2007) and the chemotaxis of fibroblasts towards PDGF has also been shown to require PLC (Wei et al., 2010). Neither of these studies demonstrated directly which of PLC isoforms are involved in the responses towards growth factor chemoattractant gradient although PLCγ1 enzyme was implicated in chemotactic responses in several earlier studies (Kundra et al., 1994; Rönnstrand et al., 1999). Further complexity of PLC signalling is due to the involvement of several downstream events linked either to changes in PtdIns(4,5)P2 concentrations or generation of second messengers, diacylglycerol and Ins(1,4,5)P3. Studies in cancer cells emphasised reduction of local PtdIns(4,5)P2 and linked this to the activity state of coffilin (Leyman et al., 2009; van Rheenen et al., 2007). In contrast, studies in fibroblasts focused on the role of Ins(1,4,5)P3 production and subsequent, more complex and dynamic changes in calcium levels in protrusions of cells responding to PDGF gradient (Wei et al., 2009). Thus, the role of specific PLC enzymes in chemotaxis, and particularly that towards growth factors, remains to be defined further and new insights are largely dependent on manipulation of expression of specific PLC enzymes and generation of cells from transgenic animals.
The studies reported here use fibroblasts derived from transgenic animals with null or mutated alleles of PLCe1 encoding PLCe enzyme characterised by extensive signalling connectivity, including several small GTPases involved in regulation of cells motility (Bunney and Katan, 2006; Smrcka et al., 2012). We demonstrate that PLCε is required for chemotaxis of fibroblasts to PDGF-BB and contributes to localisation and persistence of signalling in protrusions responding to chemotactic gradient.
Results
PLCε contributes to responses of fibroblasts to PDGF-BB
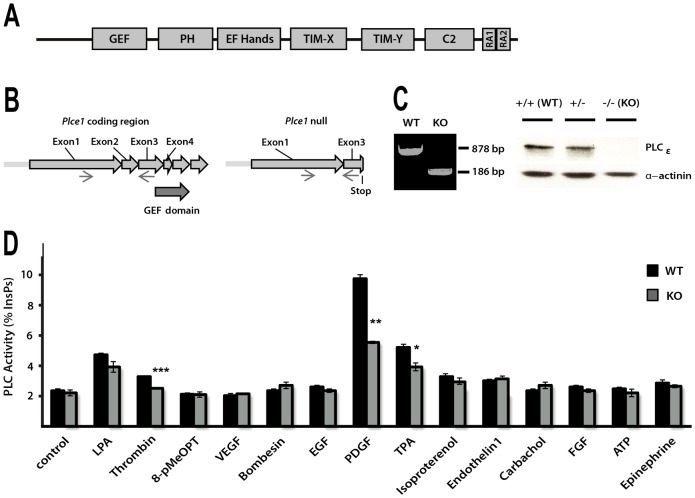
It has been previously established that PLCε1, the only isoform in PLCε family, is not uniformly expressed in different tissues and cell types and, compared to other PLCs, is generally at lower levels (Smrcka et al., 2012). Among cell types, appreciable levels of expression have been detected in fibroblast cell lines and murine fibroblasts (Ikuta et al., 2008; Kelley et al., 2006). Following generation of transgenic mouse strains with altered Plce1 alleles, we isolated mouse embryonic fibroblasts (MEFs) and established that fibroblasts from the Plce1−/− animals (KO) lack a PLCe1 coding region and PLCε protein expression (Fig. 1A–C).
Fig. 1.
Analysis of PLC responses in fibroblasts deficient in PLCε. (A) Schematic diagram shows domain organisation of PLCε consisting of the following domains: Guanine nucleotide Exchange Factor (GEF), Pleckstrin homology (PH), four EF-Hands folds (EF-hands), Phospholipase C catalytic domain (TIM-X and TIM-Y), C2 (C2) and two Ras-association domains (RA1 and RA2; only RA2 is functional in binding of Ras family GTPases). (B) Strategy for generation of PLCε null allele (Plce1−) was based on deletion of exon 2, resulting in frameshift termination in exon 3. (C) Genomic DNA from the WT and PLCε KO MEFs was analysed by RT-PCR using primers in exon 1 and 3 (left panel). Protein extracts from MEFs of indicated genotypes were analysed for PLCε expression by western blotting (right panel). (D) PLC activity in the WT and PLCε KO MEFs (immortalised populations of cells) following stimulation. MEFs were analysed for hydrolysis of 3H-labelled PtdIns(4,5)P2 present in cellular membranes. Asterisks represent significant differences between WT and PLCε KO for stimulation with PDGF-BB, thrombin and TPA (*P<0.05, **P<0.01 and ***P<0.001, t-test). Differences seen for PDGF-BB for these cell populations are consistent with the data for four (n = 4) independently prepared samples of WT and PLCε KO MEFs (P<0.001, ANOVA; see supplementary material Fig. S1A).
Using a number of immortalised cell populations of WT and KO MEFs we established similar, significant differences in response to a potent fibroblast stimulus, PDGF-BB. Notably, PLC responses measured by inositol phosphate production were reduced to about 60% in KO MEFs (n = 4, P<0.001, ANOVA) (supplementary material Fig. S1A). We further extended our analysis using a panel of different extracellular agonists, including growth factors for tyrosine kinase receptors (EGF, FGF and VEGF) and agonists for G-protein coupled receptors (LPA, thrombin, ephinephrine, carbachol, bombesin and ATP) some of which have been previously linked to activation of PLCε (Bunney and Katan, 2006; Smrcka et al., 2012). We also analysed several compounds that could act on potential intracellular mediators of PLCε activation such as 8-pMeOPT (Dzhura et al., 2010). As shown in Fig. 1D, a PDGF-BB appears to be the most potent activator of PLC in MEFs also showing differences in PLC responses between the WT and PLCε KO fibroblasts. Responses to some other agonists, for example thrombin, were also significantly reduced in KO MEFs as reported previously (Kelley et al., 2006).
The finding that PLC activity in PLCε KO fibroblasts was only reduced and not abolished is consistent with the observations that PLCε can be activated together with other PLC enzymes in response to the same stimulus. For example in HEK293T cells stimulated with EGF both PLCγ and PLCε enzymes are activated (Stope et al., 2004) while in fibroblasts stimulated by thrombin, activation includes PLCβ3 and PLCε (Kelley et al., 2006). This differs from overexpression systems where it could be clearly demonstrated that PLCε, as the major expressing PLC, can respond to specific external signals, including PDGF (Kelley et al., 2004; Song et al., 2002). In the experiments shown here (Fig. 1D) the other activated PLC in response to PDGF most likely corresponds to PLCγ1, an isoform that has been clearly linked to PDGF stimulation (Hess et al., 1998; Liao et al., 2006; Wahl et al., 1989). Expression levels and activation of PLCγ1 in response to PDGF-BB remain unchanged in PLCε KO fibroblasts (see further, Fig. 3A).
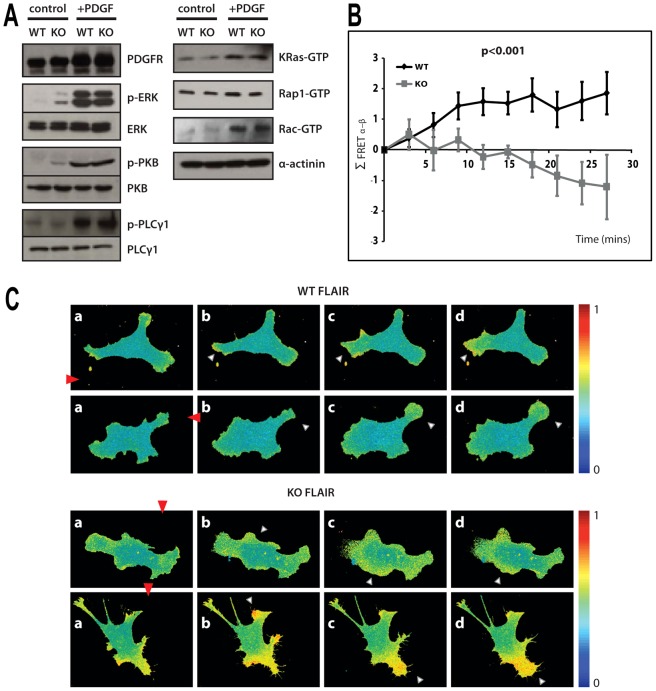
Fig. 3.
Signalling responses to PDGF in the WT and PLCε deficient fibroblasts. (A) The extracts from immortalised pools of WT and KO MEFs without stimulation (control) and following uniform stimulation by PDGF-BB for 5 min (+PDGF), were analysed by western blotting using antibodies to PDGFR, ERK, phospho-ERK, PKB, phospho-PKB, PLCγ1 and phospho-PLCγ1 as indicated in the left panel. The extracts were also subjected to “pull-down” assay using specific, binding domains for GTP-bound forms of Ras, Rap and Rac GTPase and followed by western blotting with antibodies to KRas, Rap1 and Rac. (B) Summary of FRET localisation obtained from MEFs expressing Rac biosensor FLAIR. Plot of the change in difference of FRET index between the membrane regions facing towards (α) and away (β) from the micropipette (Σ FRET α–β) in a number of directionally stimulated MEFs from immortalised cell populations. P<0.001, ANOVA. (C) Illustration of responses to PDGF-BB gradient, generated using micropipette, for the WT (top panels) and KO (bottom panels) MEFs expressing Rac biosensor FLAIR described in B; two individual cells are shown. The cells were monitored before (a, 0 time) and after (b–d, time intervals from 5 to 40 min) the release of PDGF-BB and formation of PDGF-BB gradient. Protrusions with highest FRET are indicated by white triangles and direction of the gradient shown by red triangles. Colour scale indicates the dynamic range of the biosensor response where YPet/mTurquoise emission ratio increases with FRET activation (0, no significant response; 1, maximum value).
Fibroblasts lacking PLCε have impaired chemotaxis in PDGF-BB gradients
As PLC activity has been implicated in chemotaxis towards growth factors (Kundra et al., 1994; Rönnstrand et al., 1999; Roussos et al., 2011; Wang et al., 2007; Wei et al., 2010), we analysed migration of the WT and PLCε KO fibroblasts in gradients of PDGF-BB. Two of the methods that can be applied to generate gradients and monitor cell chemotaxis, Dunn chambers and delivery of PDGF using micropipette, were further adapted and characterised for this analysis (supplementary material Fig. S1C,D).
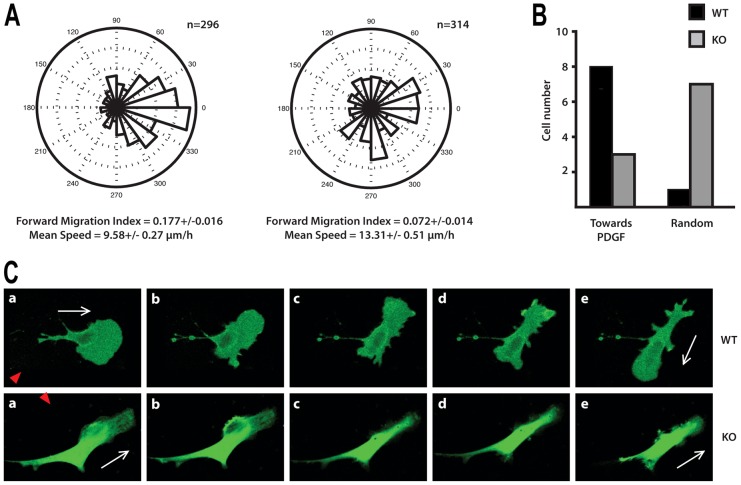
Experiments performed using Dunn chambers demonstrated a clear difference in the number of cells that migrated in the direction of chemotactic gradient (Fig. 2A; supplementary material Fig. S1B). While the majority of the WT cells were biased towards the gradient, KO cells migrated more randomly and had a considerably lower Forward Migration Index (n = 4, P<0.01, ANOVA) (supplementary material Fig. S1B). Interestingly, Mean Speed of migration was slightly enhanced in PLCε KO fibroblasts suggesting that PLCε depletion has no detrimental effect on basal ability of cells to migrate but mainly affects chemotaxis (Fig. 2A).
Fig. 2.
Analysis of fibroblast migration in response to PDGF-BB gradient. (A) Chemotaxis of the WT (left) and PLCε KO (right) MEFs towards PDGF-BB was analysed using Dunn chambers and shown as angular histograms; the number of cells analysed and values calculated for Forward Migration Index and Mean Speed are indicated. Data, using immortalised populations of WT and KO MEFs, represent significant differences between the chemotactic responses of the two data groups; ANOVA (P<0.001). The differences between the WT and PLCε KO MEFs are consistent with the data obtained for four different MEF preparations (n = 4) analysed by ANOVA (P<0.01), see supplementary material Fig. S1B. (B) Summary of properties of the WT (black) and PLCε KO (grey) MEFs using PDGF-BB gradient formed using micropipette. Cell-turning assay was performed by placing a micropipette perpendicular to the direction of the cell movement. Migration of polarised MEFs expressing GFP in the presence of a micropipette perfusing PDGF-BB at a concentration of 100 ng/ml was monitored over a 40 minute period. The number of individual cells analysed from immortalised pools is shown. The analysis of these data by a two-tailed Fisher test to a 2×2 contingency table shows that PLCε KO cells are less responsive to PDGF-BB than WT cells (P = 0.0198). (C) Images illustrating the WT (top panel) and PLCε KO (bottom panel) MEFs monitored before (a, 0 time) and after (b–e, time intervals from 5 to 40 min) the release and formation of PDGF-BB gradient. Red triangles show direction of the gradient and white arrows indicate direction of movement in the absence (in a) and presence (in e) of the PDGF-BB gradient.
We have also analysed the ability of migrating fibroblasts to turn towards PDGF-BB gradients by placing a PDGF-BB containing micropipette perpendicular to cell movement. For this cell-turning study, only fibroblasts with a clear leading protrusion were selected. Analysis and specific examples of the WT and PLCε KO fibroblasts in this assay are shown in Fig. 2B,C. Typically, WT cells formed a new protrusion towards the source, reversed their axes of polarisation and subsequently migrated in direction of higher PDGF-BB concentrations. In contrast, 7 out of 10 KO cells retained their initial direction or formed new protrusions randomly.
Signalling responses to PDGF-BB show differences that are highly localised
A number of intracellular signalling events are activated in fibroblasts following PDGF-BB stimulation. The key network nodes and links include PI3K mediated activation of PKB, activation of Ras and downstream kinases such as ERK and activation of PLCγ1 (Park et al., 2003). Several other small GTPases in addition to Ras, such as Rap, Rho and Rac, also participate in responses to PDGF-BB. Comparison of the WT and PLCε KO fibroblasts following uniform stimulation by PDGF-BB for 5 min did not revealed any major differences in overall activation of the tested components; our analysis covered PDGFR, ERK, PKB, PLCγ1, K-Ras, Rap1 and Rac proteins (Fig. 3A). Analysis of several of these components at later times (not shown) did not reveal any differences between the WT and PLCε KO MEFs.
Localisation or activation of signalling components in many physiological systems is not uniform. Migrating cells and cells responding to chemoattractant are well-documented examples of signalling segregation and compartmentalisation (Berzat and Hall, 2010). One of the components that can be robustly monitored regarding spatial distribution in real time is activation of Rac using FRET biosensors; localised activation of Rac1 has been shown in protrusions of migrating fibroblasts where it has an important role in formation of protrusions and cell movement (Machacek et al., 2009). Using a modified version of one of the previously described biosensors (FLAIR) (Hodgson et al., 2010) we analysed WT and PLCε KO fibroblasts. We selected cells that show Rac activity in several protrusions and monitored the effect of PDGF-BB gradient formed using micropipette in the proximity of these cells. A summary of the change in difference in FRET index between the membrane regions of the cell directed towards and away from the micropipette [based on analysis described in Wei et al. (Wei et al., 2009)] averaged over a number of WT and PLCε KO fibroblasts, is shown in Fig. 3B and the differences illustrated by individual cells in Fig. 3C. In most WT cells, the protrusion size and the Rac activity increase in the protrusion nearest to the high concentrations of PDGF-BB. In contrast, in high proportion of PLCε KO cells a protrusion closest to the PDGF-BB source is enlarged and has high Rac activity only transiently. Subsequently, the main Rac activity and protrusion extension occurs in other parts of the cell regardless of their position in PGDF gradient. Thus, despite the lack of overall differences in responses to uniform PDGF stimulation (Fig. 3A), signalling events in protrusions of fibroblasts lacking PLCε appear to be compromised when tested in the presence of a chemoattractant gradient.
Phospholipase C activity of PLCε is required for normal chemotaxis
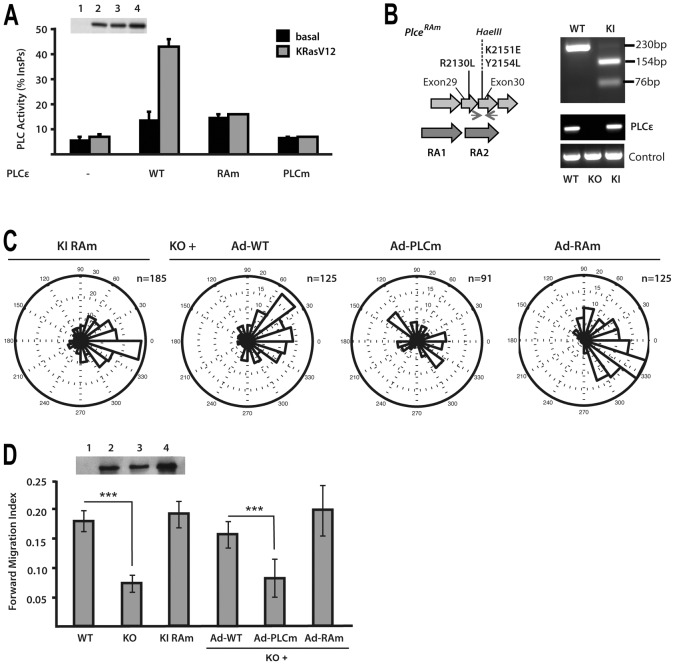
PLCε is a dual function protein, incorporating PLC and GEF enzyme activities. In addition, it has a Ras-association domain (RA2) that mediates interaction with several GTPases from Ras family (H-, K-, N-Ras, Rap1 and Rap2) (Fig. 1A) (Bunney and Katan, 2006; Smrcka et al., 2012). Point mutations that result in specific inactivation of PLC activity and mutations that abolish Ras-binding and activation in transfected cells (Fig. 4A) have been based on previous structural studies (Bunney et al., 2006; Ellis et al., 1998). Mutations in the RA2 domain were also used to generate a transgenic mouse strain expressing RAm PLCε at physiological level (Fig. 4B).
Fig. 4.
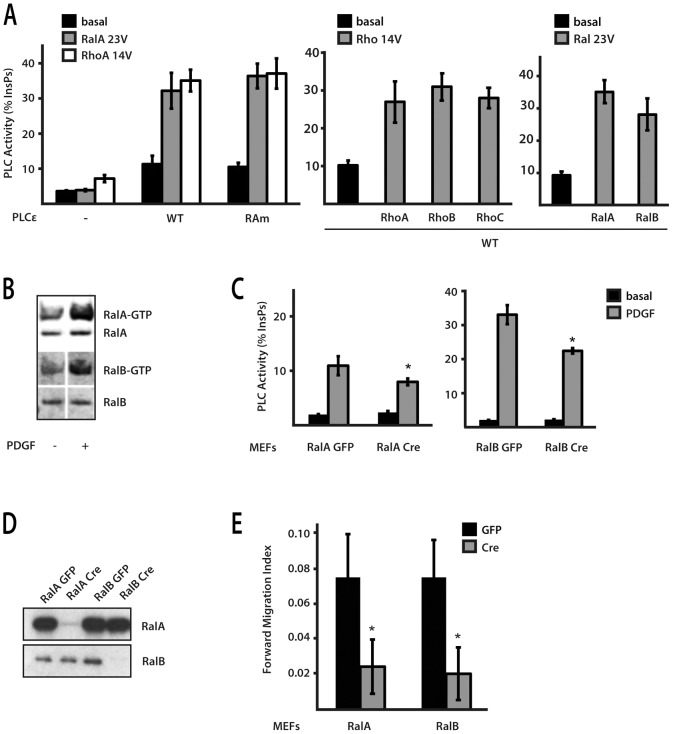
Analysis of PLCε functional domains in the context of chemotaxis. (A) Characterisation of PLCε variants with point mutations in the phospholipase C catalytic domain (PLCm) and point mutations in the RA2 domain (RAm). PLC activity of the WT and RAm and PLCm variants was analysed in COS7 cells in the absence (black) or presence (white) of co-transfected KRasV12. Expression level of PLCε WT and PLCm and RAm variants was analysed by western blotting (inset). (B) Strategy to generate PLCε Ras-binding mutant allele (Plce1RAm) by introducing three point mutations (R2130L, K2151E and Y2154L) in exons 29 and 30; the mutagenesis resulted in insertion of a HaeIII site and generated PCR products were analysed by the restriction site digestion. Expression levels were analysed by PCR, using GAPDH as a control. (C) Analysis of immortalised populations of MEFs expressing RAm PLCε variant and KO MEFs infected with Adenovirus expressing the WT (Ad-WT), PLCm (Ad-PLCm) and RAm (Ad-RAm) variants was performed using Dunn chambers. Expression level of PLCε WT and PLCm and RAm variants in PLCε KO cells was analysed by western blotting (inset). (D) Summary of Forward Migration Index values. Data are from four independent experiments using immortalised pools of MEFs; ***P<0.001, ANOVA.
To assess the importance of the RA2 domain in the context of chemotaxis and potential links with some of the Ras family members that bind this domain, we generated MEFs from Plce1 RAm/RAm mice and analysed their migration using Dunn chambers. These cells displayed clear directional movement towards a PDGF-BB gradient with forward migration index comparable to that of WT fibroblasts (Fig. 4C,D). Next, we used adenoviruses expressing PLCε variants deficient either in PLC activity or in Ras-binding (previously described in Citro et al., 2007; Oestreich et al., 2007) to rescue PDGF-BB-mediated chemotaxis in PLCε null fibroblasts. We observed that the expression of a PLCε variant that cannot bind Ras (Ad-RAm) restored chemotaxix of PLCε KO cells as well as the expression WT PLCε (Fig. 4C,D). This is also consistent with findings that MEFs lacking H-, K- and N-Ras (Drosten et al., 2010) had no significant difference in PLC responses to PDGF-BB stimulation despite a great reduction in the basal rate of cell movement (supplementary material Fig. S2A). Notably, in contrast to PLCε RAm, the variant lacking PLC activity (Ad-PLCm) was not able to rescue compromised chemotaxis of PLCε KO cells (Fig. 4C,D). Based on these observations, PLC activity of PLCε and the consequences of PtdIns(4,5)P2 hydrolysis (at least in part) underlay the role of this protein in chemotaxis. However, regulation of PLCε is not mediated by small GTPases that bind the RA2 domain.
Stimulation of PLCε PLC activity can be mediated by other small GTPases that do not require intact RA2 domain needed for action of Ras and Rap proteins. It has been shown that RhoA and RalA can stimulate PLCε lacking RA2 domain in co-expression or in vitro system (Gandarillas et al., 2009; Kelley et al., 2004; Seifert et al., 2004). The PLCε variant incorporating point-mutations within the RA2 domain used in our study is also fully activated by these small GTPases; we further established that other related family members, RhoB and RhoC and RalB, also stimulate PLCε (Fig. 5A). To elucidate which of these small GTPases could be involved in PLCε activation in the context of chemotaxis, we investigated whether these GTPases are activated in MEFs by PDGF-BB, tested MEFs deficient in specific GTPases and MEFs treated with C. botulinum C3 exoenzyme that impairs function of Rho GTPases by ADP-ribosylation and Ral by direct binding (Just et al., 2010) (Fig. 5B,C; supplementary material Fig. S3). Based on our data and previous reports, Rho GTPases are not readily activated in this system (Monypenny et al., 2009) while the use of MEFs deficient in RhoA was inconclusive due to concomitant upregulation of other GTPases (supplementary material Fig. S2B). In contrast, activation of RalA and RalB was observed in MEFs treated with PDGF-BB (Fig. 5B) and RalA or RalB deficient fibroblasts had both reduced PDGF-BB-induced PLC activation and compromised chemotaxis (Fig. 5C–E), suggesting that Ral proteins could mediate activation of PLCε in this context. This is further supported by the findings that depletion of Ral GTPases by siRNA affects preferentially PLC activation (supplementary material Fig. S3A,B) and directional migration (reduction of Forward Migration Index in WT MEFs by 38%, P<0.01, ANOVA) in WT MEF compared to PLCε KO. Similarly, the C. botulinum C3 exoenzyme, which reduces stimulation of PLC activity by PDGF-BB more potently in WT MEFs, also inhibits Ral activation by PDGF-BB (supplementary material Fig. S3C,D).
Fig. 5.
Analysis of PLCε regulation by different small GTPases. (A) PLC activity of the WT and PLCε RAm variant was analysed in COS7 cells in the absence (black) or presence of either RalAV23 (grey) or RhoAV14 (white) (left panel). PLC activity of the WT PLCε was also tested in the presence of activated forms of RhoA, RhoB, RhoC, RalA and RalB (right panels). (B) Cell extracts from WT MEFs, untreated (−) and treated with PDGF-BB for 5 min (+), were analysed by western blotting using antibodies to RalA and RalB GTPases; total protein and protein present in the “pull-down” with the GST-RalBP1-RBD (GTP-bound forms) were analysed. (C) PLC activity was measured in immortalised cell populations of Rala flox/flox and Ralb flox/flox MEFs following infection with either Ad-GFP or Ad-Cre; untreated MEFs (basal) and MEFs treated with PDGF-BB (PDGF) were analysed. *P<0.05, t-test. (D) Expression of RalA and RalB proteins was analysed in Rala flox/flox and Ralb flox/flox MEFs following infection with either Ad-GFP or Ad-Cre. (E) Forward Migration Index was determined using Dunn chambers; immortalised populations of Rala flox/flox and Ralb flox/flox MEFs, following infection with either Ad-GFP (control) or Ad-Cre, were analysed. The data are from four independent experiments, *P<0.05, ANOVA. Random migration was not affected following infection with Ad-Cre.
Generation of Ins(1,4,5)P3 in cell protrusions can have a role in chemotaxis
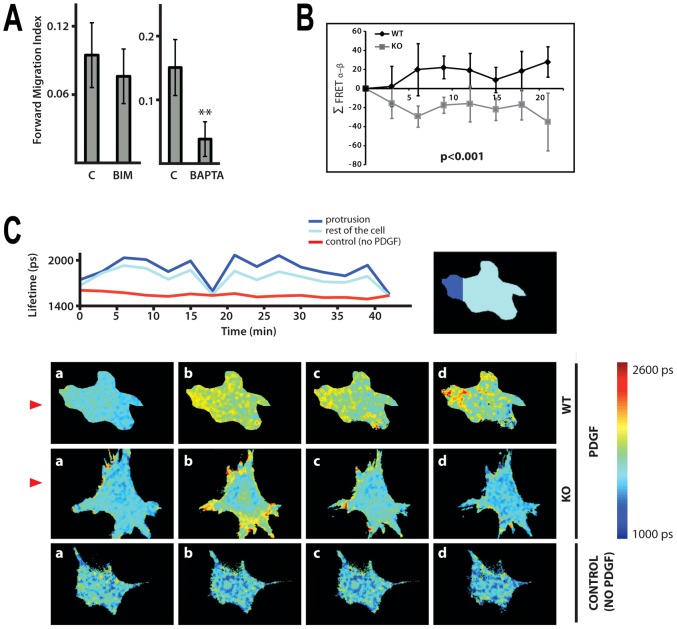
Following our observation that phospholipase C activity of PLCε is required for normal chemotaxis we measured the Forward Migration Index of fibroblasts treated with PKC inhibitor or calcium chelator BAPTA at concentrations that had little or no effect on cell speed; as shown in Fig. 6A, under these conditions addition of BAPTA clearly affected directional motility.
Fig. 6.
Importance of calcium responses in chemotaxis and generation of Ins(1,4,5)P3 in MEFs treated with PDGF. (A) Forward Migration Index of WT MEFs (immortalised cell population) treated with either PKC inhibitor (BIM) or calcium chelator (BAPTA) was determined using Dunn chambers. The data are from four independent experiments, **P<0.01, ANOVA. (B) Summary of FRET localisation obtained from MEFs expressing Ins(1,4,5)P3 biosensor LIBRA. Fluorescence lifetime imaging (FLIM) was used to monitor conformational changes in LIBRA following Ins(1,4,5)P3 binding that results in an increase in donor lifetime. Plot of the change in difference of FRET index between the membrane regions facing towards (α) and away (β) from the micropipette (Σ FRET α–β) in a number of directionally stimulated MEFs from immortalised cell populations. P<0.001, ANOVA. (C) Illustrations for MEFs expressing LIBRA biosensor (described in B) exposed to PDGF-BB gradient generated by perfusion from a micropipette. Change in the lifetime for a single WT MEF (either in the area of protrusion or in the rest of the cell) over 40 min is shown in the top panel; the area of a single cell or its parts was isolated from a background by applying a mask. The bottom panel shows images of the WT and PLCε KO cells taken before (a, 0 time) and after (b–d, time intervals within a 15 min period) the release of PDGF-BB; non-stimulated WT MEFs were used as a control. Direction of the gradient is shown by red triangles.
We further analysed production of PLC-generated second messenger involved in calcium mobilisation, Ins(1,4,5)P3, in fibroblasts exposed to PDGF-BB gradient; we used a modified version of previously described FRET biosensor, LIBRA (Tanimura et al., 2004); the data analysis is shown in Fig. 6B. Despite some impact of expression of LIBRA protein on cell viability (in particular when cells were stimulated with PDGF-BB) we were able to monitor oscillation in Ins(1,4,5)P3 concentrations in cells exposed to PDGF-BB gradient, in some cells over a 40 minute period (Fig. 6C, top). In the WT fibroblasts the greatest change in FLIM-FRET was observed in protrusions closest to the source of PDGF-BB (Fig. 6C, bottom). Similarly as observed when using Rac biosensor FLAIR (Fig. 3B), PLCε KO fibroblasts appear to lack localised changes in Ins(1,4,5)P3 production that are related to the direction of chemotactic gradient (Fig. 6C, bottom). In agreement with the proposed role of PLCε in protrusions of fibroblasts exposed to chemotactic gradient, we also observed that GFP-PLCε was localised to areas of exposed protrusions (data not shown).
Discussion
Despite intensive study and its biological importance the mechanism of chemotaxis remains unclear both more generally and in molecular detail. In particular, the role of cell signalling network either as a critical control mechanism (“compass model”) or a modulator of cell-generated cycle (“pseudopode-based model”), remains to be further characterised and the importance of specific signal transducer components defined within the context of specific cellular responses (Insall, 2010). Within the regulatory network some of the phosphoinositide signalling components and small GTPases have been linked to the interface between transmission of extracellular signals and pseudopode machinery. Here we identified one of the phosphoinositide-specific PLC enzymes, PLCε, as an important component required for regulation of chemotaxis of fibroblasts towards PDGF.
Physiological roles of PLCε have been addressed by generating transgenic animal models and cell lines derived from these animals (Bunney and Katan, 2006; Smrcka et al., 2012). Our strategy was to generate Plce1 null allele and a variant specifically deficient in some of signalling interactions (Figs 1, 4). We subsequently focused on fibroblasts that have not been previously studied with respect to cell migration or growth factor stimulation although being recognised as one of the cell types that express this isoform (Kelley et al., 2006; Ikuta et al., 2008). We found that PLCε KO fibroblasts have reduced PtdIns(4,5)P2 hydrolysis in response to PDGF and compromised chemotaxis towards this chemoattractant despite normal random motility; these cells lack the ability to form stable protrusions towards the PDGF-BB gradient (Figs 1–3). Previous work using PLCε fibroblasts from Plce1 ΔX/ΔX mice, expressing protein deficient in PLC activity, also revealed the importance of PLCε in fibroblast function, notably in the interplay between fibroblasts and cells from the immune system (Ikuta et al., 2008). It has been shown that PLCε is required for secretion of chemokines in response to phorbol ester and consequently for the ability of fibroblasts to mobilise cells of the immune system that constitute the tumour environment. From studies of PLCε KO mice reported so far (Smrcka et al., 2012) it is, however, not clear what other cell types could have impaired directional movement. Nevertheless, supporting evidence for the importance of PLCε in directional cell movement in vivo came from use of transgenic approaches in C. elegans, the first organism where PLCε was identified. Studies in C. elegans have revealed the defect in morphogenesis that involves a programmed series of migrations and fusions of epithelial sheets (Vázquez-Manrique et al., 2008). This process recapitulates many of the traits of wound healing after tissue damage in mammals. Specifically, the analysis of C. elegans using RNAi and mutant strains have shown that depletion of PLC-1/PLCε resulted in slower migration of the ventral epidermal cells. As a consequence, plc-1 loss of function resulted in ruptured embryos with a Gex (gut on exterior) phenotype and lumpy larvae (Vázquez-Manrique et al., 2008).
Among PLC enzymes PLCε is characterised by more complex regulatory interactions and greater degree of signalling connectivity. Notably, several small GTPases important for regulation of cell motility are involved in activation of PLCε (Bunney and Katan, 2006; Smrcka et al., 2012). This high degree of integration into signalling networks provides a scope for various regulatory mechanisms. In the context of chemotaxis to PDGF-BB, regulation of PLCε phospholipase activity is not mediated by direct binding of Ras or Rap proteins to RA2 domain (Fig. 4) and could be mediated by Ral GTPases (Fig. 5). A number of observations that support involvement of Ral proteins in regulation of cell motility, including Ral activation localised to membrane protrusions in response to EGF stimulation (Yoshizaki et al., 2007) and the control of the assembly and localisation of exocyst subunits and associated proteins at the leading edge of motile cells (Parrini et al., 2011), are consistent with the possibility that PLCε could also be regulated by Ral proteins.
Another distinct property of PLCε compared to other PLC enzymes is the capacity to participate in more sustained signal generation (Smrcka et al., 2012). Notably, it was shown that in fibroblasts stimulated by thrombin the immediate increase in Ins(1,4,5)P3 production is due to activation of PLCβ3 while the depletion of PLCε results in reduced Ins(1,4,5)P3 production between 3 and 60 min following stimulation (Kelley et al., 2006). Similar considerations were applied when stimulation of PLCγ and PLCε was observed by the same agonist (EGF) (Stope et al., 2004). The sustained signal generation of PLCε could be important in the context of chemotaxis where persistence of a signal could ensure continuous enlargement of a protrusion in the direction of chemoattractant gradient, the property that is lacking in PLCε KO fibroblasts (Fig. 3). In this respect, the role of PLCε in chemotaxis could differ from that suggested for PLCγ1, in most instances linked to transient activation.
In addition to the difference in responses observed in PLCε KO cells and inhibition of chemotaxis by calcium chelation (Fig. 1D; Fig. 6A), several other findings further support the possibility that PLCε contributes to signalling localised to growing protrusions by regulating Ins(1,4,5)P3 and calcium levels. These include observations that the function of PLCε in chemotaxis requires intact PLC activity (Fig. 4) and that protrusions are characterised by high levels of Ins(1,4,5)P3 generation (Fig. 6C). The recently proposed role for Ins(1,4,5)P3 in chemotaxis could provide one possible mechanism for the Ins(1,4,5)P3-mediated involvement of PLCε (Wei et al., 2009). The proposed model suggests that Ins(1,4,5)P3 receptor in the endoplasmic reticulum functions together with a stretch-activated cation channel (TRPM7) to generate local, dynamic sites of high calcium concentrations described as calcium flickers; the supporting data include the finding that administration of a membrane-permeable Ins(1,4,5)P3 analogue enhances both the calcium flicker activity and the turning process towards PDGF source. Importance of calcium flickers as a mechanism of gradient sensing was further suggested by observations that intracellular loading of EGTA to chelate intracellular calcium slows the turning. Interestingly, after complete abolition of the calcium flicker activity, cells can still migrate at even faster velocity; however, they are no longer able to make turns when exposed to a PDGF gradient (Wei et al., 2009; Wei et al., 2010).
In summary, we have found that PLCε contributes to regulation of fibroblast chemotaxis as an important component of signalling processes localised to cell protrusions that respond to PGDF-BB gradient. As a multifunctional protein with impact on several downstream events, PLCε has extensive signalling connectivity. In the context of chemotaxis it is likely that signalling links include Ral GTPases, as upstream regulators of PLC activity, and Ins(1,4,5)P3-mediated localised changes in calcium concentrations as downstream events affecting pseudopode machinery.
Materials and Methods
Generation of PLCε transgenic animals and isolation of mouse embryonic fibroblasts
Generation of PLCε transgenic mice was based on a standard homologous recombination strategy using stem cell manipulation. Plce1−/− targeting vector was constructed using DNA fragments flanking exon2 region which were cloned into PGKNeo-F2L2DTA plasmid (Addgene plasmid 13445). Plce1RAm targeting vector was constructed using DNA fragments of exon29/30 and using a modified version of PGKNeo-F2L2DTA plasmid where one of the loxP sites was replaced by a polylinker and the other loxP site was mutated by deletion of three nucleotides. The targeting vectors were electroporated into Bruce 4 ES cells and homologous recombination was checked by Southern blot. Positive clones were injected into albino C57BL/6J blastocysts and chimeric males were crossed with Cre or Flp recombinase transgenic mice, as appropriate, in order to remove the PGKNeo cassette. PLCε null allele (Plce1−) was created by exon 2 deletion and frameshift termination in exon 3. RT-PCR, using primers in exon 1 and 3, was used to confirm genotype of Plce1−/− and Plce1+/+ MEFs isolated from corresponding KO and WT embryos. PLCε Ras-binding mutant allele (Plce1RAm) was generated by introducing three point mutations (R2130L, K2151E and Y2154L). This mutagenesis resulted in insertion of a HaeIII and PCR analysis followed by the restriction site digestion was used to confirm Plce1RAm/RAm genotype of isolated MEFs. A similar strategy to the one used for Plce1, using the same plasmids, ES cells, blastocytes and Flp recombinase transgenic mice, was used to generate Rala and Ralb conditional alleles (Peschard et al., 2012).
WT, Plce1−/−, Plce1RAm/RAm, Rala−/− and Ralb−/− MEFs were isolated from C57/Bl6 mouse embryos (after at least four backcrosses to C57/Bl6) at 13.5 days of gestation according to the method described in Michalska (Michalska, 2007).
Cell culture, viruses and transfection
Primary MEFs were cultured at 37°C in 3% O2 and 10% CO2. Primary MEFs were immortalised by infection with retrovirus pBABE-puro SV40 LT (Addgene) and selected in Puromycin at 2.5 µg/ml for 4 days. Immortalised MEFs were cultured in high glucose DMEM medium supplemented with 10% FBS and 2 mM l-glutamine at 37°C and 5% CO2.
Immortalised MEFs with floxed alleles were infected overnight with either Adenovirus expressing only GFP (Ad-GFP) or Cre (Ad-Cre) (Gene Transfer Vector Core) with 5×107 and 2.5×108 PFUs at 50% confluence on a 175 cm dish. 48 hours post-infection cells were trypsinised and, where required, 6×106 cells were first sorted by Facs before further cell culture. Prior to analysis of PLC activity or cell motility in Dunn chambers, the expression of proteins encoded by floxed alleles was analysed by western blotting.
Adenoviruses for transduction of PLCε KO MEFs with recombinant PLCε, described previously (Oestreich et al., 2007), were added in media containing 20 multiplicity of infection of each individual virus; prior to the infection 105 MEFs were let to adhere on a glass coverslip for 3 hours. Following protein expression analysis, cells were analysed in Dunn chambers.
Previously described FRET biosensors FLAIR (Hodgson et al., 2010) and LIBRA (Tanimura et al., 2004) were modified to incorporate the brighter and more photostable mTurquoise fluorophore (Goedhart et al., 2010) in place of CyPet or ECFP, respectively. To create a mTurquoise-Rac1 fusion construct a mTurquoise PCR fragment flanked with the restriction sites NheI and BglII was subcloned into pCyPet-Rac1 digested with same restriction enzymes. The LIBRA probe consists of the N-terminal 20 amino acids from neuromodulin (a membrane targeting signal) followed by the rat IP3R3 domain sandwiched between ECFP and EYFP. To produce mTurquoise-LIBRA a neuromodulin-mTurquoise fusion fragment was created by overlap-extension of these two fragments and subcloned into the LIBRA vector, cut with NheI and BspEI and lacking the neuromodulin domain and ECFP. MEFs were transfected by electroporation using an Nucleofector II device (Lonza Group, Basel, Switzerland) with Ingenio electroporation solution (Mirus Bio LLC, WI, USA) using electroporation sequence A-023. The cells were transfected with 4 µg of plasmid DNA. When transfecting with the FLAIR Rac1 biosensor a ratio of 3∶2 mTurquoise-Rac1:YPet-PBD DNA was used. After electroporation the cells were seeded at a confluency of 30–40% on glass coverslips in DMEM supplemented with 10% FBS. After 3–4 hours the cells expressing LIBRA (consisting of the Ins(1,4,5)P3-binding domain of the type 3 Ins(1,4,5)P3 receptor fused between the FRET pair mTurquoise and YFP) were imaged directly or, for cells expressing FLAIR, media was replaced with 0.5% FBS and starved overnight.
Transfection of primary mouse embryonic fibroblasts (catalogue number M-Fb-481, Lonza Group, Basel, Switzerland) with previously described plasmids encoding GFP-PLCε or GFP (Sorli et al., 2005) was performed using Lonza Mouse/rat hepatocyte Nucleofector solution and seeded in glass-bottom 12 well glass bottom plates (MaTek Corporation, MA, USA) at a density of 105 cells per well. The cells were starved overnight in DMEM supplemented with 0.5% FBS prior to further analysis of cellular localisation.
Expression plasmids for the WT PLCε and its variants, plasmids encoding activated forms of small GTPases and transfection of COS7 cell using Lipofectamine reagent (Invitrogen) were described previously (Bunney et al., 2006; Sorli et al., 2005).
Analysis of protein expression, phosphorylation and activation status of small GTPases
Protein expression and phosphorylation of different signalling proteins was analysed by western blotting using specific antibodies to PLCε (described in Bunney et al., 2006), PLCγ1 (Sigma), phospho-PLCγ1 (phospho-Tyr-783, Cell Signaling), PDGF Rβ (R&D Systems), ERK (Upstate), phospho-ERK (Cell Signaling), PKB (Millipore), phospho-PKB (Cell Signaling), RhoA (Santa Cruz Biotechnology), RhoB and RhoC (Cell Signaling) and CDC42 (Santa Cruz Biotechnology). Antibody to α-actinin (Sigma) was used as a loading control.
Analysis of GTP-bound state of small GTPases was performed as described previously (Jones and Katan, 2007). Briefly, cell lysates were incubated with GST-fusion binding proteins specific to GTP-bound forms, Raf-RBD for Ras, RalGDS-RBD for Rap, nPAK-RBD for Rac and Ral BP1-RBD GST for Ral. The protein present in the “pull-down” was subsequently analysed by western blotting using antibodies to K-Ras (Santa Cruz Biotechnology), Rap1 (BD Biosciences), Rac (Upstate), RalA (BD Transduction Laboratories) or RalB (Santa Cruz Biotechnology).
Analysis of PLC activity
Analysis of inositol phosphate formation in MEFs and COS7 cells was essentially as described previously (Sorli et al., 2005). Briefly, 24 hours after transfection of Cos7 cells and 24 hours after seeding MEFS, cells were labelled with 1.5 µCi/ml myo-[2-3H]inositol (Perkin Elmer). For the treatment with C. botulinum, C3 transferase, MEFs were transfected (by electroporation using an Nucleofector II) with 2 µg/ml of the C3 exoenzyme (Cytoskeleton) 24 h before labelling. After a further 24 hours, the cells were incubated in 1.2 ml inositol-free DMEM, without serum, containing 20 mM LiCl with or without stimulation with indicated compounds for 1 hour. Concentrations were: 25 ng/ml PDGF-BB (Sigma); 2 µM LPA (Sigma); 1.3 U/ml Thrombin (Sigma); 50 nM Bombesin (Sigma); 100 ng/ml EGF (Sigma); 10 µM ATP (Sigma); 500 µM Carbachol (Sigma); 12 µM Epinephrine (Sigma); 100 µM Endothelin1 (Sigma); 50 ng/ml FGF (R&D Systems); 100 ng/ml VEGF (R&D Systems); 100 µM 8-pMeOPT (BIOLOG Life Science Institute); 25 µM Isoproterenol (Sigma); 10 µg/ml TPA (Sigma). The cells were lysed and supernatants and pellets were separated for measurements of inositol phosphates and inositol lipids, respectively. PLC activity is expressed as the total inositol phosphates formed relative to the amount of [3H]inositol in the phospholipid pool. Mass measurements of Ins(1,4,5)P3 were performed using extracts from 2×106 cells, according to a protocol described in detail previously (Rodriguez et al., 2003). Data generation and analysis for transiently transfected COS7 cells was exactly as described in Sorli et al. (Sorli et al., 2005). Similarly, PLC activity measurements in MEFs, unless stated otherwise in figure legends, were from three independent experiments performed in duplicate; bars are the means of three data points, each defined by the average of two technical replicates. The error bars represent s.d.
Dunn chamber analysis
For the Dunn chamber assay 105 cells were seeded on a No. 2 glass coverslip and starved in DMEM supplemented with 0.5% FBS overnight. When applied 20 µM of BAPTA (Merck) or 390 nM of BIM (MERK) was added to the cells 15 min before and during the time of the chamber experiments. The coverslips were placed on the Dunn chamber (Zicha et al., 1997) with a PDGF-BB concentration of 250 ng/ml in the outer annulus. Brightfield images of the cells were recorded in three positions around the bridge region under 4× magnification at 10 minute intervals for 12 hours.
In each image the cells were tracked using the ImageJ plugin Manual Tracking (Fabrice Cordelieres, Institut Curie, France.) Software was written in Matlab (Mathworks, MA, USA) to analyse the cell migration. Under low magnification the curvature of the bridge means that the direction of the gradient can vary substantially across the image. To address this issue the radial gradient was determined by three points on the annulus provided by the user. At each time step the speed and direction of each cell was calculated relative to the local gradient using the cell track information. The Forward Migration (FMI) Index was calculated to quantify the directionality of the cells. The FMI is defined as the ratio of the sum of the displacement of a cell along the axes defined by the chemoattractant gradient to the total path length of the cell. The mean angle of deviation from the chemotactic gradient was calculated to construct angular histograms of the direction of migration of the cell population relative to the chemotactic gradient. A hierarchical unbalanced analysis of variance (ANOVA) test was used to determine the significance of the differences between the FMI of different groups of Dunn chamber experiments (Zicha et al., 1997). This test accounts for the potential variation between individual chamber experiments.
Analysis using chemotactic gradients generated by perfusion
Chemotaxis experiments were performed by continuously perfusing PDGF at a concentration of 0.5 mg/ml from a glass micropipette. Micopipettes were prepared from 0.7 µm borosilicate capillaries using a vertical needle puller (Narishige, Japan). This creates a steep PDGF concentration gradient in the region of the needle. The position of the micropipette was controlled using a three axis hydraulic micromanipulator (Narishige, Japan) and a nominal pressure of 420 hPa applied using a KDS200 syringe pump (KD Scientific Inc., MA, USA).
For FLAIR analysis, transfected cells were imaged on a Zeiss LSM 710 confocal scanning system mounted on an Axio Observer inverted microscope using a 40× oil immersion objective, NA 1.3. The microscope was equipped with an incubator maintained at 37°C. Fluorescence was excited at 405 nm while mTurquoise and YPet emission was simultaneously recorded between 465 and 500 nm and 525–620 nm, respectively. A FRET index was calculated as the ratio between the YPet and mTurquoise emission, as previously reported (Kraynov et al., 2000). YPet is inefficiently excited at 405 nm; cells transfected with YPet-PBD only showed no significant fluorescence above background levels in the acceptor channel when imaged under the same conditions. Control experiments were conducted to ensure there was no significant photobleaching over the time course at the power level used. Transfected cells on No. 1.5 glass coverslips were transferred to a custom-made open top slide holder. The cells were washed with PBS and the media replaced with Phenol-Red free Optimem (Gibco, NY, USA). Cells transfected with both mTurquoise-Rac1 and YPet-PBD were identified in the eyepiece; care was taken to select cells whose morphology was similar to that of untransfected cells. The micropipette was positioned ∼1 µm above the coverslip next to the polarised cell, such that the gradient was not along the axes of polarisation. PDGF perfusion was started and cells were imaged at 2-min intervals over a period of 40 min.
MEFs expressing LIBRA biosensor were imaged on an Olympus IX81 inverted microscope with a Yokogawa CSUX Nipkov spinning disk system providing widefield confocal sectioning. Fluorescence was excited at 420–440 nm using a fibre-laser pumped supercontinuum source (Fianium UK Ltd, SC400-6) and emission recorded at 500–540 nm. Time-gated imaging was provided by a gated optical intensifier (Kentech Instruments, model HRI) with 1 ns gate widths. This was read out using a cooled CCD camera (Hamamatsu Photonics, model ORCA-ER). The HRI was triggered at various delay times after the excitation pulses to temporally sample the fluorescence decay profiles for each pixel in the field of view. Eight gate delays were captured with an integration time of 1 second. A reference instrument response function was recorded using the short lifetime dye DASPI recorded at the same wavelengths. Cells were imaged every 3 min for 40 min. The fluorescence decays were fitted to a single exponential model accounting for the instrument response function using reference reconvolution by in-house software. Temperature was maintained at 37°C using an incubator.
For all imaging experiments the investigator was blind for the genotype and the cells obtained from at least two different populations of immortalised MEFs; about ten cells were analysed for each condition. The analysis of FRET index between the membrane regions facing towards (α) and away (β) from the PDGF-BB source was according to Wei et al. (Wei et al., 2009).
Supplementary Material
Acknowledgments
We are grateful to Alan Smrcka (University of Rochester) for adenoviruses and for the WT and variants of PLCε, Mariano Barbacid (Centro Nacional de Investigaciones Oncológicas) for Rasless MEFs, Cord Brakebusch (University of Copenhagen) for RhoAflox/flox MEFs, Yosuke Tojyo (University of Hokkaido) for FRET biosensor LIBRA, and Theodorus Gadella for pmTurquoise-N1.
Footnotes
Funding
This work is funded by grants from Cancer Research UK [grant numbers A15044 and A10433]; the Biotechnology and Biological Sciences Research Council [grant number BB/H006095/2]; and the Wellcome Trust Value in People Award (to M.K.). Deposited in PMC for release after 6 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.110007/-/DC1
References
- Afonso P. V., Parent C. A. (2011). PI3K and chemotaxis: a priming issue? Sci. Signal. 4, pe22 10.1126/scisignal.2002019 [DOI] [PubMed] [Google Scholar]
- Bach T. L., Chen Q. M., Kerr W. T., Wang Y., Lian L., Choi J. K., Wu D., Kazanietz M. G., Koretzky G. A., Zigmond S.et al. (2007). Phospholipase cbeta is critical for T cell chemotaxis. J. Immunol. 179, 2223–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berzat A., Hall A. (2010). Cellular responses to extracellular guidance cues. EMBO J. 29, 2734–2745 10.1038/emboj.2010.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunney T. D., Katan M. (2006). Phospholipase C epsilon: linking second messengers and small GTPases. Trends Cell Biol. 16, 640–648 10.1016/j.tcb.2006.10.007 [DOI] [PubMed] [Google Scholar]
- Bunney T. D., Katan M. (2011). PLC regulation: emerging pictures for molecular mechanisms. Trends Biochem. Sci. 36, 88–96 10.1016/j.tibs.2010.08.003 [DOI] [PubMed] [Google Scholar]
- Bunney T. D., Harris R., Gandarillas N. L., Josephs M. B., Roe S. M., Sorli S. C., Paterson H. F., Rodrigues–Lima F., Esposito D., Ponting C. P.et al. (2006). Structural and mechanistic insights into ras association domains of phospholipase C epsilon. Mol. Cell 21, 495–507 10.1016/j.molcel.2006.01.008 [DOI] [PubMed] [Google Scholar]
- Cain R. J., Ridley A. J. (2009). Phosphoinositide 3-kinases in cell migration. Biol. Cell 101, 13–29 10.1042/BC20080079 [DOI] [PubMed] [Google Scholar]
- Citro S., Malik S., Oestreich E. A., Radeff–Huang J., Kelley G. G., Smrcka A. V., Brown J. H. (2007). Phospholipase Cepsilon is a nexus for Rho and Rap-mediated G protein-coupled receptor-induced astrocyte proliferation. Proc. Natl. Acad. Sci. USA 104, 15543–15548 10.1073/pnas.0702943104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten M., Dhawahir A., Sum E. Y., Urosevic J., Lechuga C. G., Esteban L. M., Castellano E., Guerra C., Santos E., Barbacid M. (2010). Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J. 29, 1091–1104 10.1038/emboj.2010.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhura I., Chepurny O. G., Kelley G. G., Leech C. A., Roe M. W., Dzhura E., Afshari P., Malik S., Rindler M. J., Xu X.et al. (2010). Epac2-dependent mobilization of intracellular Ca2+ by glucagon-like peptide-1 receptor agonist exendin-4 is disrupted in beta-cells of phospholipase C-epsilon knockout mice. J. Physiol. 588, 4871–4889 10.1113/jphysiol.2010.198424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M. V., James S. R., Perisic O., Downes C. P., Williams R. L., Katan M. (1998). Catalytic domain of phosphoinositide-specific phospholipase C (PLC). Mutational analysis of residues within the active site and hydrophobic ridge of plcdelta1. J. Biol. Chem. 273, 11650–11659 10.1074/jbc.273.19.11650 [DOI] [PubMed] [Google Scholar]
- Gandarillas N. L., Bunney T. D., Josephs M. B., Gierschik P., Katan M. (2009). In vitro reconstitution of activation of PLCepsilon by Ras and Rho GTPases. Methods Mol. Biol. 462, 379–389 10.1007/978-1-60327-115-8_24 [DOI] [PubMed] [Google Scholar]
- Goedhart J., van Weeren L., Hink M. A., Vischer N. O., Jalink K., Gadella T W. (2010). Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nat. Methods 7, 137–139 10.1038/nmeth.1415 [DOI] [PubMed] [Google Scholar]
- Heldin C. H., Westermark B. (1999). Mechanism of action and in vivo role of platelet-derived growth factor. Physiol. Rev. 79, 1283–1316 [DOI] [PubMed] [Google Scholar]
- Hess J. A., Ji Q. S., Carpenter G., Exton J. H. (1998). Analysis of platelet-derived growth factor-induced phospholipase D activation in mouse embryo fibroblasts lacking phospholipase C-gamma1. J. Biol. Chem. 273, 20517–20524 10.1074/jbc.273.32.20517 [DOI] [PubMed] [Google Scholar]
- Hodgson L., Shen F., Hahn K. (2010). Biosensors for characterizing the dynamics of rho family GTPases in living cells. Curr. Protoc. Cell Biol. 46, 14.11.1–14.11.26 10.1002/0471143030.cb1411s46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta S., Edamatsu H., Li M., Hu L., Kataoka T. (2008). Crucial role of phospholipase C epsilon in skin inflammation induced by tumor-promoting phorbol ester. Cancer Res. 68, 64–72 10.1158/0008-5472.CAN-07-3245 [DOI] [PubMed] [Google Scholar]
- Insall R. H. (2010). Understanding eukaryotic chemotaxis: a pseudopod-centred view. Nat. Rev. Mol. Cell Biol. 11, 453–458 10.1038/nrm2905 [DOI] [PubMed] [Google Scholar]
- Jones N. P., Katan M. (2007). Role of phospholipase Cgamma1 in cell spreading requires association with a beta-Pix/GIT1-containing complex, leading to activation of Cdc42 and Rac1. Mol. Cell. Biol. 27, 5790–5805 10.1128/MCB.00778-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just I., Huelsenbeck S. C., Genth H. (2010). Clostridium Botulinum C3 exoenzyme: Rho-inactivating tool in cell biology and a neurotrophic agent. The Open Toxinology Journal 3, 19–23 10.2174/1875414701003010019 [DOI] [Google Scholar]
- Kelley G. G., Reks S. E., Smrcka A. V. (2004). Hormonal regulation of phospholipase Cepsilon through distinct and overlapping pathways involving G12 and Ras family G-proteins. Biochem. J. 378, 129–139 10.1042/BJ20031370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley G. G., Kaproth–Joslin K. A., Reks S. E., Smrcka A. V., Wojcikiewicz R. J. (2006). G-protein-coupled receptor agonists activate endogenous phospholipase Cepsilon and phospholipase Cbeta3 in a temporally distinct manner. J. Biol. Chem. 281, 2639–2648 10.1074/jbc.M507681200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölsch V., Charest P. G., Firtel R. A. (2008). The regulation of cell motility and chemotaxis by phospholipid signaling. J. Cell Sci. 121, 551–559 10.1242/jcs.023333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortholt A., King J. S., Keizer–Gunnink I., Harwood A. J., Van Haastert P. J. (2007). Phospholipase C regulation of phosphatidylinositol 3,4,5-trisphosphate-mediated chemotaxis. Mol. Biol. Cell 18, 4772–4779 10.1091/mbc.E07-05-0407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraynov V. S., Chamberline C., Bokoch C. H., Schwartz M. A., Slabaugh S., Hahn K. M. (2000). Localized Rac activation dynamics visualized in living cells. Science 290, 333–337 10.1126/science.290.5490.333 [DOI] [PubMed] [Google Scholar]
- Kundra V., Escobedo J. A., Kazlauskas A., Kim H. K., Rhee S. G., Williams L. T., Zetter B. R. (1994). Regulation of chemotaxis by the platelet-derived growth factor receptor-beta. Nature 367, 474–476 10.1038/367474a0 [DOI] [PubMed] [Google Scholar]
- Lauffenburger D. A., Horwitz A. F. (1996). Cell migration: a physically integrated molecular process. Cell 84, 359–369 10.1016/S0092-8674(00)81280-5 [DOI] [PubMed] [Google Scholar]
- Leyman S., Sidani M., Ritsma L., Waterschoot D., Eddy R., Dewitte D., Debeir O., Decaestecker C., Vandekerckhove J., van Rheenen J.et al. (2009). Unbalancing the phosphatidylinositol-4,5-bisphosphate-cofilin interaction impairs cell steering. Mol. Biol. Cell 20, 4509–4523 10.1091/mbc.E09-02-0121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H. J., de Los Santos J., Carpenter G. (2006). Contrasting role of phospholipase C-gamma1 in the expression of immediate early genes induced by epidermal or platelet-derived growth factors. Exp. Cell Res. 312, 807–816 10.1016/j.yexcr.2005.11.034 [DOI] [PubMed] [Google Scholar]
- Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G. L., Hahn K. M., Danuser G. (2009). Coordination of Rho GTPase activities during cell protrusion. Nature 461, 99–103 10.1038/nature08242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalska A. E. (2007). Isolation and propagation of mouse embryonic fibroblasts and preparation of mouse embryonic feeder layer cells. Curr. Protoc. Stem Cell Biol. 3, 1C.3.1–1C.3.17 10.1002/9780470151808.sc01c03s3 [DOI] [PubMed] [Google Scholar]
- Monypenny J., Zicha D., Higashida C., Oceguera–Yanez F., Narumiya S., Watanabe N. (2009). Cdc42 and Rac family GTPases regulate mode and speed but not direction of primary fibroblast migration during platelet-derived growth factor-dependent chemotaxis. Mol. Cell. Biol. 29, 2730–2747 10.1128/MCB.01285-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich E. A., Wang H., Malik S., Kaproth–Joslin K. A., Blaxall B. C., Kelley G. G., Dirksen R. T., Smrcka A. V. (2007). Epac-mediated activation of phospholipase C(epsilon) plays a critical role in beta-adrenergic receptor-dependent enhancement of Ca2+ mobilization in cardiac myocytes. J. Biol. Chem. 282, 5488–5495 10.1074/jbc.M608495200 [DOI] [PubMed] [Google Scholar]
- Park C. S., Schneider I. C., Haugh J. M. (2003). Kinetic analysis of platelet-derived growth factor receptor/phosphoinositide 3-kinase/Akt signaling in fibroblasts. J. Biol. Chem. 278, 37064–37072 10.1074/jbc.M304968200 [DOI] [PubMed] [Google Scholar]
- Parrini M. C., Sadou–Dubourgnoux A., Aoki K., Kunida K., Biondini M., Hatzoglou A., Poullet P., Formstecher E., Yeaman C., Matsuda M.et al. (2011). SH3BP1, an exocyst-associated RhoGAP, inactivates Rac1 at the front to drive cell motility. Mol. Cell 42, 650–661 10.1016/j.molcel.2011.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschard P., McCarthy A., Leblanc–Dominguez, V., Yeo M., Guichard S., Stamp G., Marshall C. J. (2012). Genetic deletion of RALA and RALB small GTPases reveals redundant functions in development and tumorigenesis. Curr Biol. 22, 2063–2068 10.1016/j.cub.2012.09.013 [DOI] [PubMed] [Google Scholar]
- Rodriguez R., Matsuda M., Storey A., Katan M. (2003). Requirements for distinct steps of phospholipase Cgamma2 regulation, membrane-raft-dependent targeting and subsequent enzyme activation in B-cell signalling. Biochem. J. 374, 269–280 10.1042/BJ20021778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnstrand L., Siegbahn A., Rorsman C., Johnell M., Hansen K., Heldin C. H. (1999). Overactivation of phospholipase C-gamma1 renders platelet-derived growth factor beta-receptor-expressing cells independent of the phosphatidylinositol 3-kinase pathway for chemotaxis. J. Biol. Chem. 274, 22089–22094 10.1074/jbc.274.31.22089 [DOI] [PubMed] [Google Scholar]
- Roussos E. T., Condeelis J. S., Patsialou A. (2011). Chemotaxis in cancer. Nat. Rev. Cancer 11, 573–587 10.1038/nrc3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I. C., Haugh J. M. (2005). Quantitative elucidation of a distinct spatial gradient-sensing mechanism in fibroblasts. J. Cell Biol. 171, 883–892 10.1083/jcb.200509028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert J. P., Wing M. R., Snyder J. T., Gershburg S., Sondek J., Harden T. K. (2004). RhoA activates purified phospholipase C-epsilon by a guanine nucleotide-dependent mechanism. J. Biol. Chem. 279, 47992–47997 10.1074/jbc.M407111200 [DOI] [PubMed] [Google Scholar]
- Smrcka A. V., Brown J. H., Holz G. G. (2012). Role of phospholipase Cε in physiological phosphoinositide signaling networks. Cell. Signal. 24, 1333–1343 10.1016/j.cellsig.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Satoh T., Edamatsu H., Wu D., Tadano M., Gao X., Kataoka T. (2002). Differential roles of Ras and Rap1 in growth factor-dependent activation of phospholipase C epsilon. Oncogene 21, 8105–8113 10.1038/sj.onc.1206003 [DOI] [PubMed] [Google Scholar]
- Sorli S. C., Bunney T. D., Sugden P. H., Paterson H. F., Katan M. (2005). Signaling properties and expression in normal and tumor tissues of two phospholipase C epsilon splice variants. Oncogene 24, 90–100 10.1038/sj.onc.1208168 [DOI] [PubMed] [Google Scholar]
- Stope M. B., Vom Dorp F., Szatkowski D., Böhm A., Keiper M., Nolte J., Oude Weernink P. A., Rosskopf D., Evellin S., Jakobs K. H.et al. (2004). Rap2B-dependent stimulation of phospholipase C-epsilon by epidermal growth factor receptor mediated by c-Src phosphorylation of RasGRP3. Mol. Cell. Biol. 24, 4664–4676 10.1128/MCB.24.11.4664-4676.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Zhang Y., Xu W., Harden T. K., Sondek J., Sun L., Li L., Wu D. (2011). A PLCβ/PI3Kγ-GSK3 signaling pathway regulates cofilin phosphatase slingshot2 and neutrophil polarization and chemotaxis. Dev. Cell 21, 1038–1050 10.1016/j.devcel.2011.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura A., Nezu A., Morita T., Turner R. J., Tojyo Y. (2004). Fluorescent biosensor for quantitative real-time measurements of inositol 1,4,5-trisphosphate in single living cells. J. Biol. Chem. 279, 38095–38098 10.1074/jbc.C400312200 [DOI] [PubMed] [Google Scholar]
- van Rheenen J., Song X., van Roosmalen W., Cammer M., Chen X., Desmarais V., Yip S. C., Backer J. M., Eddy R. J., Condeelis J. S. (2007). EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J. Cell Biol. 179, 1247–1259 10.1083/jcb.200706206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez–Manrique R. P., Nagy A. I., Legg J. C., Bales O. A., Ly S., Baylis H. A. (2008). Phospholipase C-epsilon regulates epidermal morphogenesis in Caenorhabditis elegans. PLoS Genet. 4, e1000043 10.1371/journal.pgen.1000043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M. I., Olashaw N. E., Nishibe S., Rhee S. G., Pledger W. J., Carpenter G. (1989). Platelet-derived growth factor induces rapid and sustained tyrosine phosphorylation of phospholipase C-gamma in quiescent BALB/c 3T3 cells. Mol. Cell. Biol. 9, 2934–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Eddy R., Condeelis J. (2007). The cofilin pathway in breast cancer invasion and metastasis. Nat. Rev. Cancer 7, 429–440 10.1038/nrc2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Wang X., Chen M., Ouyang K., Song L. S., Cheng H. (2009). Calcium flickers steer cell migration. Nature 457, 901–905 10.1038/nature07577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Wang X., Chen M., Ouyang K., Zheng M., Cheng H. (2010). Flickering calcium microdomains signal turning of migrating cells. Can. J. Physiol. Pharmacol. 88, 105–110 10.1139/Y09-118 [DOI] [PubMed] [Google Scholar]
- Yoshizaki H., Mochizuki N., Gotoh Y., Matsuda M. (2007). Akt-PDK1 complex mediates epidermal growth factor-induced membrane protrusion through Ral activation. Mol. Biol. Cell 18, 119–128 10.1091/mbc.E06-05-0467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zicha D., Dunn G., Jones G. (1997). Analyzing chemotaxis using the Dunn direct-viewing chamber. Methods Mol. Biol. 75, 449–457 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.