Summary
The small ubiquitin-like modifier (SUMO) ligase PIAS1 (Protein Inhibitor of Activated Stat-1) has been shown to play a role in cellular stress response by SUMOylating several proteins that are involved in DNA repair, apoptosis and transcription. In this paper, we show that PIAS1 regulates ultraviolet (UV)-induced apoptosis by recruiting Death-associated protein 6 (Daxx) to PIAS1-generated SUMO-foci. Cells that ectopically express PIAS1, but not other PIASes, show increased sensitivity to UV irradiation, suggesting that PIAS1 has a distinct function in UV-dependent apoptosis. Domain analysis of PIAS1 indicates that both PIAS1 SUMO-ligase activity and the specific localization of PIAS1 through its N-terminal and C-terminal domains are essential for UV-induced cell death. Daxx colocalizes with PIAS1-generated SUMOylated foci, and the reduction of Daxx using RNAi alleviates UV-induced apoptosis in PIAS1-expressing cells. PIAS1-mediated recruitment of Daxx and apoptosis following UV irradiation are dependent upon the Daxx C-terminal SUMO-interacting motif (SIM). Overall, our data suggest that the pro-apoptotic protein Daxx specifically interacts with one or more substrates SUMOylated by PIAS1 and this interaction leads to apoptosis following UV irradiation.
Key words: PIAS1, SUMO, Apoptosis, UV irradiation, Daxx
Introduction
The small ubiquitin-like modifier (SUMO) proteins regulate the activity, localization and/or stability of various cellular proteins through a reversible covalent modification or noncovalent interactions with their substrates (Geiss-Friedlander and Melchior, 2007). Three well-characterized SUMO isoforms, numbered 1 to 3, exist in vertebrates, and the gene for a fourth potential SUMO isoform (SUMO-4) has been identified; however, it is unclear whether SUMO-4 is expressed in all cells and whether it can be conjugated to other proteins. SUMO-1, SUMO-2 and SUMO-3 covalently attach to a substrate by forming an isopeptide linkage with one or more specific lysine residues on the protein. In addition, SUMO-2 and SUMO-3, which share 95% sequence identity, are known to form poly-SUMO chains (Johnson, 2004). SUMO modification involves the sequential transfer of a processed active SUMO protein from an E1 SUMO-activating enzyme to an E2 SUMO-conjugating enzyme and the subsequent covalent conjugation to the substrate in a reaction that involves a SUMO-E3 ligase. In their substrate-conjugated form, SUMO proteins can interact with other proteins containing a SUMO-interacting motif (SIM), which is characterized by ΨΨXΨ or ΨXΨΨ residues (where Ψ is a hydrophobic amino acid) and is often flanked by negatively charged residues (Song et al., 2004; Wilkinson and Henley, 2010). Therefore, protein modification by SUMO can lead to new protein-protein interaction modules.
Since the first SUMO-conjugated protein was identified, the repertoire of SUMO-modified substrates has steadily increased. The diversity of SUMO substrates is presumably governed by a small number of SUMO E3 ligases that exist in cells. The PIAS (Protein Inhibitor of Activated Stat) family of SUMO E3 ligases contains homologs in both vertebrates and invertebrates and is characterized by the presence of a zinc-binding SP-RING (Siz/PIAS-RING) domain (Palvimo, 2007; Rytinki et al., 2009). In addition, PIAS family members all contain an N-terminal SAP (scaffold attachment factor, acinus and PIAS) domain that is involved in DNA binding and protein-protein interactions, a PINIT domain that has been proposed to be required for subcellular localization and substrate interactions (Yunus and Lima, 2009), a SUMO-interacting motif (SIM), and a variable C-terminal serine-threonine-rich region. Mammals contain five major PIAS isoforms, PIAS1, PIAS2α/Xα and 2β/Xβ, PIAS3 and PIAS4/y. PIAS proteins act as adaptor proteins that bridge the SUMO-conjugated E2 enzyme and the substrate in the SUMOylation reaction. The SP-RING finger domain is essential for the SUMO-ligase activity of PIAS proteins. It has also been suggested that PIAS proteins promote SUMO paralog selection (Azuma et al., 2005; Rytinki et al., 2009). Various cellular stress stimuli have been demonstrated to induce global protein modification by SUMO-2/3, which suggests that cells may use SUMO-modification as a protective response (Saitoh and Hinchey, 2000). PIAS1 and PIAS4 have been implicated in DNA double-strand break repair that depends on the SUMO-2/3 conjugation of BRCA1 (breast cancer susceptibility protein 1) and the SUMO-1 conjugation of 53BP1 (Tumor suppressor p53 binding protein 1), respectively (Galanty et al., 2009). PIAS proteins are also known to promote apoptosis. PIAS1 has been shown to initiate apoptosis in response to the presence of reactive oxygen species by JNK (c-Jun N-terminal kinase) activation (Leitao et al., 2011).
Daxx (Death-associated protein 6) is another PML-(promyelocytic leukemia protein) associated protein that has been shown to participate in UV-induced cell death via the activation of the JNK pathway (Khelifi et al., 2005). Daxx is a well-characterized SUMO-interacting protein and interacts with SUMO-modified proteins via its N- and C-terminal SIMs (Escobar-Cabrera et al., 2011). Daxx acts as a transcriptional corepressor for a number of transcriptional factors, including ETS-1 and NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells). Daxx may also regulate chromatin dynamics by binding to histone deacetylases and chromatin remodeling proteins (Shih et al., 2007). Daxx interacts with the SUMOylated form of the PML protein (Ishov et al., 1999), and the sequestration of Daxx in PML bodies suppresses its transcription inhibition function (Li et al., 2000). Various lines of evidence suggest that Daxx can act as both a pro- and an anti-apoptotic protein. Daxx is known to be involved in the axin-mediated activation of p53 transcription, which leads to apoptosis (Li et al., 2007), and has been shown to suppress anti-apoptotic gene transcription by repressing NFκB transcriptional activity (Croxton et al., 2006). Conversely, RNAi-induced Daxx silencing has been shown to increase apoptosis (Michaelson and Leder, 2003) and sensitize cells to cellular stressors, such as UV and TNF-α (Tumor Necrosis Factor α) (Chen and Chen, 2003).
In this study, we present our finding that the exogenous expression of PIAS1, but not other members of the PIAS family of SUMO E3 ligases, increases UV-induced apoptosis in HeLa cells. We demonstrate that PIAS1's SUMO-ligase activity is essential for increased UV sensitivity and that increased apoptosis is mediated by the recruitment of Daxx to SUMOylated PIAS1 foci. Additionally, we show that the N-terminus of PIAS1 governs its localization in the cell and substrate selectivity, which in turn regulates the recruitment of Daxx and apoptosis. We also show that the recruitment of Daxx to SUMOylated foci depends on its C-terminal SIM. Daxx mutants lacking the C-terminal SIM fail to form foci in cells and do not lead to UV-induced apoptosis in PIAS1-expressing cells. Based on these results, we propose a novel, specific role for PIAS1-mediated SUMOylation in sensitizing cells to UV damage through Daxx.
Results
PIAS1-transfected cells display increased sensitivity to UV irradiation
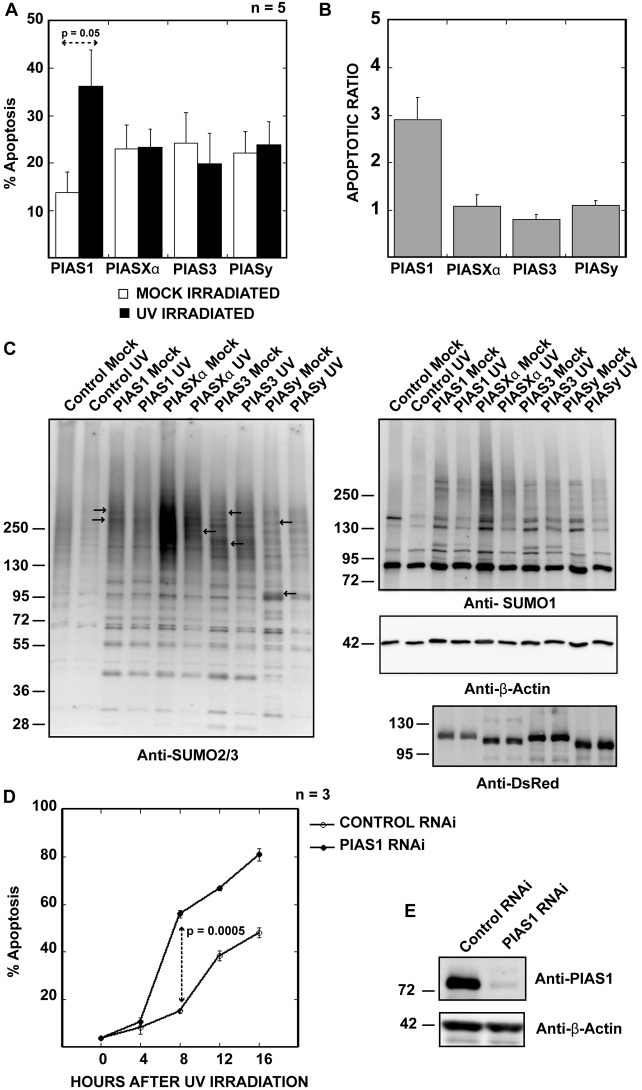
To understand the specific roles of the PIAS family of SUMO ligases in the DNA damage response, we expressed C-terminal mCherry-tagged PIAS1, PIAS2α/Xα, PIAS3 or PIAS4/y in HeLa cells and subjected the cells to 30 J/m2 of UV irradiation 24 hours after transfection. Four hours post-UV irradiation, apoptotic cells were counted based on their aberrant nuclear morphology and fragmentation. Fixed cells were also stained with an anti-Parp1 antibody (supplementary material Fig. S1A). Parp1 is a known substrate of caspase-3 during apoptosis (Tewari et al., 1995). In non-apoptotic cells, Parp1 is present only in the nucleus, whereas in apoptosis-initiating cells, cleaved Parp1 can be detected in the cytoplasm. Based on nuclear morphology and Parp1 staining, we determined that less than 2% of the untransfected cells underwent apoptosis within 4 hours after irradiation. In cells expressing PIAS family proteins, 10–20% of each group of PIAS-expressing cells were apoptotic in the absence of UV irradiation. Four hours after irradiation, the PIAS1-expressing cells contained significantly more apoptotic cells compared with the other PIAS-expressing cells; hereafter, we refer to this phenotype as ‘UV-hypersensitive apoptosis’ (Fig. 1A). To generalize this phenotype, we further confirmed PIAS1-mediated UV-hypersensitive cell death by counting apoptotic cells based on annexin V staining in both HeLa and Hct116 cells. In both types of cells, PIAS1 expression showed UV-hypersensitive apoptosis by an annexin V-based assay at 4 hours after UV irradiation (supplementary material Fig. S1B,C). We also compared UV irradiation-induced apoptosis in untransfected HeLa and Hct116 cells (supplementary material Fig. S1D). Both cell lines showed a significant increase in cell death between 8 and 12 hours after UV irradiation. Thus, PIAS1 expression appears to change the kinetics of apoptosis, and cells exogenously expressing PIAS1 initiate apoptosis early. This increase of apoptotic cells in PIAS1-expressing cells could be suppressed by treating cells with caspase-3 inhibitors before and during recovery from the UV-treatment (supplementary material Fig. S1E). In the mock-irradiated condition, the PIAS1-expressing cells displayed better survival when compared with the cells expressing the other PIASes, which indicates that PIAS1 expression is less toxic to HeLa cells than the expression of the other PIAS proteins (Fig. 1A). We calculated the apoptotic ratio by normalizing UV irradiation-induced apoptosis to the apoptosis in mock-irradiated cells. As shown in Fig. 1B, PIAS1-expressing cells displayed more than twice the amount of apoptosis compared with other PIAS-expressing cells 4 hours after irradiation. These results suggest a specific role for PIAS1 in UV-induced apoptosis.
Fig. 1.
PIAS1 overexpression sensitizes HeLa cells to UV irradiation. (A) PIAS1-expressing cells undergo increased UV irradiation-induced apoptosis. HeLa cells were UV irradiated (30 J/m2) or mock irradiated 24 hours after transfection with the PIASes. After a 4-hour recovery period, the numbers of dead and surviving cells were counted based on their nuclear morphologies and Parp1 staining patterns. The plotted values are the average of five independent experiments. The error bars represent 1 SEM. (B) The differences between UV-irradiation induced apoptosis in various PIAS isoform-expressing cells are emphasized by plotting the ratio of the percentage of apoptotic cells after UV irradiation over the percentage of apoptotic cells in mock-irradiated samples, as calculated for A. (C) PIAS SUMO ligases have distinct substrates in cells. HeLa cells expressing mCherry-fused PIASes were sorted from untransfected cells, and SUMOylation was analyzed by immunoblotting using either the anti-SUMO-1 or the anti-SUMO-2/3 antibody. Some of the distinct SUMO-2/3 bands have been indicated by arrows. The anti-DsRed antibody marks the mCherry-tagged PIASes, and anti-β-actin antibody was used as a loading control. (D) PIAS1 RNAi-treated HeLa cells show increased apoptosis after UV irradiation. HeLa cells were treated with PIAS1 siRNA or control siRNA for 48 hours before treatment with 30 J/m2 UV irradiation. Apoptotic cells were counted by Annexin V staining every 4 hours. PIAS1 siRNA-treated HeLa cells show an almost 40% increase in apoptotic cells 8 hours after UV irradiation compared with control siRNA treated cells. The plotted values are an average of three independent experiments, and the error bars represent 1 SEM. (E) PIAS1 RNAi reduces the amount of PIAS1 in HeLa cells to less than 10% of the control RNAi treatment.
Exogenously expressed PIAS proteins show distinct isoform-specific localization in cells (supplementary material Fig. S2A). We examined whether all PIAS proteins have similar SUMO ligase activity in transfected cells. To compare the SUMOylation profiles of the PIAS proteins, we enriched PIAS-expressing cells based on the mCherry signal using a cell sorter (MoFlo XDP, Beckman-Coulter) from both mock-irradiated and UV-irradiated samples. The SUMOylation profile of the sorted cells was analyzed by immunoblotting with either anti-SUMO-1 or anti-SUMO-2/3 antibodies (Fig. 1C), and anti-β-actin was used as a loading control. All of the PIAS isoforms increased the levels of SUMO-1 modification of several proteins that ranged in molecular weight from 95 kD to 300 kD. However, no significant difference was observed in the pattern of SUMO-1-modified proteins between the cells that expressed PIAS1 and those that expressed other PIASes, suggesting that SUMO-1 modification may not be relevant to UV-hypersensitive apoptosis. Conversely, the SUMO-2/3 immunoblot showed clear differences in the SUMOylation profile upon the expression of various PIAS proteins. Each of the PIAS proteins produced a number of unique SUMO-2/3 modified bands (indicated with an arrow in Fig. 1C), indicating that each PIAS protein has a unique subset of substrates for SUMO-2/3 modification. The molecular weights of most of the unique bands are greater than 130 kD. PIAS2α/Xα appears to give the most intense SUMOylation signals, although it does not show UV-hypersensitive apoptosis. This suggests that increasing SUMOylation in cells alone does not contribute to UV hypersensitivity. We did not observe any significant differences in the SUMOylation pattern between the mock and UV irradiated cells in any of the samples. In general, HeLa cells appeared to slightly repress SUMOylation in response to UV irradiation. These results indicate that all PIAS proteins promote the SUMOylation of their SUMO-2/3-specific substrates in cells, and PIAS1-specific substrates may contribute to the sensitization of cells to UV irradiation and apoptosis induction.
PIAS proteins have been shown to self-SUMOylate in vitro and when both the PIAS proteins and SUMO are overexpressed in cells (Kotaja et al., 2002; Nakagawa and Yokosawa, 2002; Schmidt and Müller, 2002), but PIAS SUMOylation has not been detected in cells overexpressing PIAS alone. We sought to examine whether self-SUMOylation of PIAS1 contributes to the UV-hypersensitive apoptosis phenotype by analyzing exogenously expressed PIASes for self-SUMOylation. Although we observed faint higher-molecular-weight bands that are presumably a SUMOylated form of PIASes in PIASXα- and 3-expressing cells, we could not detect a similar level of higher molecular weight bands of PIAS1 in either mock or UV-irradiated cells (supplementary material Fig. S2B).
To further understand the role of PIAS1 in UV-induced apoptosis, we treated HeLa cells with PIAS1 RNAi for 48 hours, and then the cells were UV-irradiated (Fig. 1D,E). PIAS1 RNAi significantly reduced the levels of PIAS1 protein in HeLa cells (Fig. 1E). We counted the number of apoptotic cells at 4-hour intervals. Interestingly, we observed increased apoptosis in PIAS1-knockdowned cells 8 hours after UV irradiation. Thus, either overexpression or the reduction in the amount of PIAS1 sensitizes cells to UV irradiation. However, the kinetics of apoptosis induction appears to be different; cells exogenously expressing PIAS1 show significant cell death 4 hours after UV irradiation, whereas PIAS1 knockdown cells show increased apoptosis at 8 hours after UV irradiation. In this study, we focused on elucidating the molecular mechanism of UV hypersensitivity elicited by PIAS1 overexpression.
PIAS1's SUMO ligase activity is required for UV-sensitivity
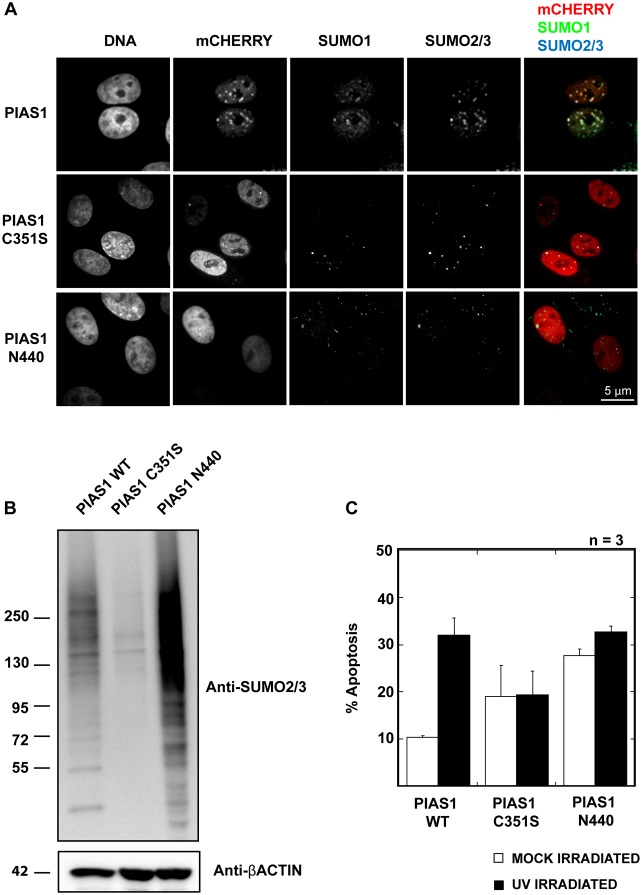
PIAS family SUMO E3 ligases have been shown to regulate a number of cellular pathways independent of their SUMO ligase activity (Rytinki et al., 2009). PIASes are known to regulate the activity of other proteins by altering their localization via direct interactions that do not depend on the presence of a functional SP-RING domain. For example, PIAS1 has been shown to regulate apoptosis-related proteins, such as p53 and Msx1, independent of its SUMO ligase activity (Megidish et al., 2002; Lee et al., 2006; Song and Lee, 2011). To determine whether PIAS1's SUMO ligase activity is required for UV-hypersensitive apoptosis, we expressed the PIAS1 C351S mutant and a PIAS1 N440 deletion mutant in HeLa cells and compared the rate of UV-hypersensitive apoptosis to that in cells expressing wild-type PIAS1 (PIAS1wt). The C351S mutant contains a mutation in the SP-RING domain that disrupts RING finger formation; therefore, it lacks SUMO E3 ligase activity (Lee et al., 2003). The PIAS deletion mutant lacks the C-terminal SIM domain that has been shown to increase the affinity of PIAS for SUMO, although it is not required for SUMOylation, in vitro (Yunus and Lima, 2009). PIAS1wt and the two mutants did not show similar localization in the nucleus. PIAS1 forms numerous (>30) small foci in the nucleus at low expression levels and comparatively fewer (<10) but larger foci at high expression levels. Additionally, PIAS1 shows a more distinct colocalization with SUMO-2/3 than with SUMO-1. In contrast, the PIAS1 C351S mutant has a more homogenous nuclear distribution and forms very few foci (Fig. 2A). PIAS1 N440 also shows a homogenous nuclear distribution and does not form clear foci.
Fig. 2.
PIAS1's SUMOylation activity is required for UV sensitivity. (A) The ligase-dead mutant (PIAS1 C351S) and the SIM domain truncation mutant (PIAS N440) do not exhibit nuclear punctate localization that is shown by wild-type PIAS1. C-terminal mCherry-tagged PIAS1wt, PIAS1 C351S or PIAS1 N440 were expressed in HeLa cells for 24 hours. Fixed cells were stained with anti-SUMO-1 and anti-SUMO-2/3 antibodies. The images were acquired using a confocal microscope using a 100× objective. Scale bar: 5 µm. (B) Altered SUMOylation profile in PIAS1-mutant-expressing cells. The cells expressing mCherry-tagged PIAS1 C351S and PIAS1 N440 were sorted, and their SUMO-2/3 profile was compared with those of PIAS1-expressing cells by immunoblotting using the anti-SUMO-2/3 antibody. (C) Wild-type PIAS1, but neither PIAS1 C351S nor PIAS1 N440, promotes increased apoptosis upon UV irradiation. The plotted values are an average of three independent experiments. The error bars represent 1 SEM. The percentage of apoptotic cells was determined, as explained in Fig. 1A.
As we predicted, PIAS1 C351S did not increase the SUMO-2/3 modification of cellular proteins. Rather, it suppressed most SUMO-2/3 modifications and acted in a dominant-negative manner (Fig. 2B). PIAS1 N440, however, showed higher SUMOylation activity than PIAS1wt. Our results are in agreement with earlier studies showing that the SIM domain of yeast PIAS homolog Siz1 is dispensable for specific substrate recognition and site selective SUMOylation of PCNA (Yunus and Lima, 2009). HeLa cells expressing PIAS1 C351S and PIAS1 N440 showed more cell death compared with PIAS1wt-expressing cells. Four hours after UV irradiation, cells expressing either of the PIAS1 mutants did not show elevated apoptosis, unlike the PIAS1wt-expressing cells (Fig. 2C). This result indicates that the SUMO ligase activity of PIAS1 is required for UV-hypersensitive apoptosis and that neither the inhibition of SUMOylation by the PIAS1 C351S nor the aberrant SUMOylation by PIAS1 N440 contribute to UV-hypersensitive apoptosis.
PIAS1 colocalizes with Daxx in cells
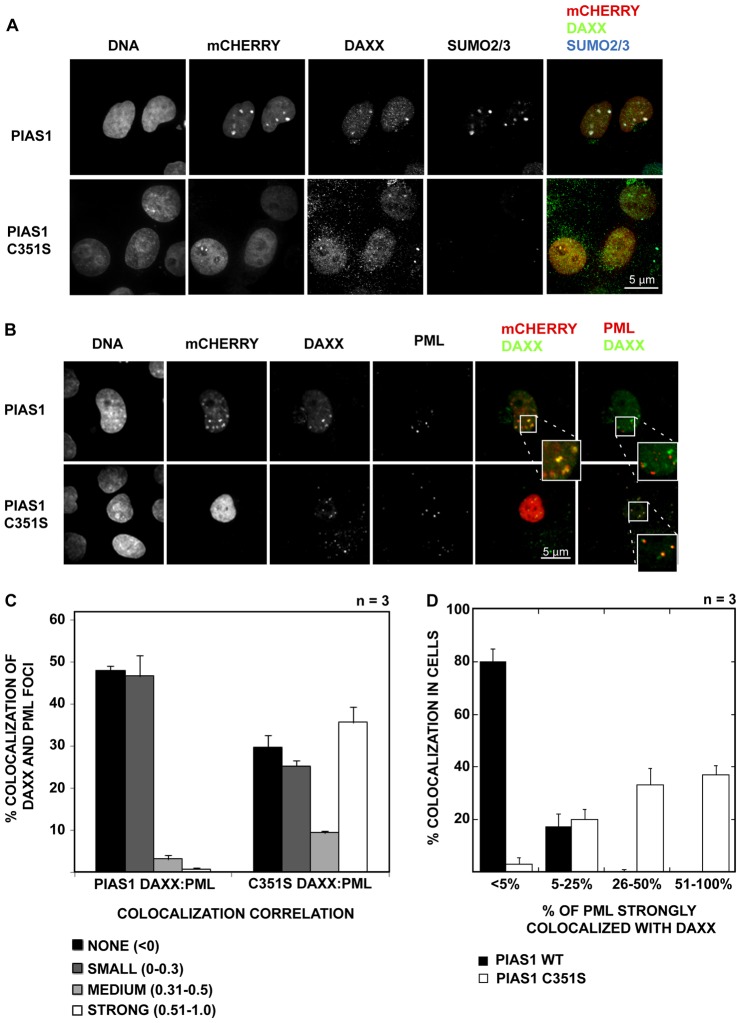
Because PIAS1 promotes the SUMOylation of specific cellular proteins, we aimed to identify the protein(s) that interact with SUMOylated substrates in PIAS1-expressing cells. Daxx was one of the logical candidates for this approach because it is a known pro-apoptotic protein that contains two SUMO-interaction motifs (one at each terminus) (Escobar-Cabrera et al., 2011). We immunostained PIAS1-transfected, mock- or UV-treated cells with an anti-Daxx antibody. In untransfected cells, Daxx showed small punctate localization in the nucleus, which largely coincided with promyelocytic leukemia nuclear bodies (PML-NBs). Daxx localization was altered in the PIAS1-expressing cells. In ∼20 to 30% of the PIAS1-expressing cells, Daxx colocalized with some of the PIAS1 foci (Fig. 3A). This colocalization was more easily observed in the high-expression PIAS1 foci in both the mock-irradiated and UV-irradiated cells. We also compared Daxx localization in cells expressing other PIASes. Although the expression of other PIASes also modified Daxx localization in cells, none of the PIASes showed significant colocalization with Daxx (supplementary material Fig. S3A). In addition, PIAS1 colocalized with Daxx in both mock-treated and UV-treated cells (supplementary material Fig. S3B).
Fig. 3.
PIAS1 colocalizes with Daxx in cells and regulates its localization to PML bodies. (A) Daxx colocalizes with the SUMOylated foci in PIAS1-expressing cells, but not in PIAS1 C351S-expressing cells. PIAS1-mCherry and PIAS1 C351S-mCherry fusions were expressed in HeLa cells. The cells were stained with an anti-Daxx antibody 24 hours after transfection. The stained samples were imaged using confocal microscopy with a 100× objective. Scale bar: 5 µm. (B) PIAS1 expression prevents Daxx from colocalizing with PML bodies. PIAS1- or PIAS1 C351S-transfected cells were stained with anti-Daxx and anti-PML antibodies. Daxx colocalizes with PML bodies in PIAS1 C351S-expressing HeLa cells but not in PIAS1-expressing cells. Scale bar: 5 µm. PIAS1-Daxx and PIAS1-PML colocalization is enlarged (inset). (C) Quantitative measurement of colocalization correlation between Daxx and PML in PIAS1 versus PIAS1 C351S expressing cells. PIAS1- or PIAS1 C351S-expressing cells were stained with anti-Daxx and anti-PML antibodies. Over 90 images were acquired from three independent experiments using a confocal microscope. Pearson's correlation between Daxx and PML was calculated in the transfected cells from the images using CellProfiler v2.0 software. (D) A cell-by-cell count of PML and Daxx colocalization. The percentage of PML spots in each transfected nucleus showing a strong (>0.5) Pearson's correlation with Daxx was calculated using the data in (C) and plotted against the percentage of cells showing strong colocalization between PML and Daxx.
Daxx is known to localize to PML-NBs in cells, and this localization requires PML SUMOylation. The colocalization of PML and Daxx has been proposed to regulate Daxx's corepressor function (Li et al., 2000). We therefore examined whether the PML/Daxx interaction was altered by PIAS1 expression. By immunofluorescence staining, we observed that Daxx and PML show very little colocalization in PIAS1-expressing cells. However, in PIAS1 C351S-expressing cells, PML and Daxx largely remain colocalized (Fig. 3B). We calculated the colocalization between PML and Daxx in PIAS1wt- and PIAS1 C351S-expressing cells using the cell image analysis software CellProfiler v2.0 (Carpenter et al., 2006). The correlation between each focus of PML and Daxx was calculated using Pearson's correlation coefficient. As shown in Fig. 3C, in PIAS1 C351S-expressing cells, over 35% of Daxx foci strongly correlate with the PML foci. In contrast, PIAS1wt-expressing cells display a strong correlation in less than 5% of Daxx and PML foci. To further evaluate PML/Daxx interaction in each cell, we counted the number of cells showing a strong correlation (greater than 0.5) coefficient between PML and Daxx. As shown in Fig. 3D, in nearly 80% of the PIAS1-expressing cells, less than 5% PML foci showed a strong correlation with Daxx. In contrast, nearly 40% of the C351S-expressing cells showed greater than 50% of the PML foci strongly colocalizing with Daxx, and almost an equal number of cells showed between 26 and 50% of the PML foci colocalizing with Daxx. This suggests that the inhibition of SUMOylation by PIAS1 C351S expression does not disrupt the PML/Daxx interaction, but increased SUMOylation by PIAS1wt promotes the dissociation of Daxx from PML. We further examined whether the expression of PIAS1 C351S disrupts PML SUMOylation. We did not see any changes in PML profile between PAIS1wt and PIAS1 C351S expressing cells (supplementary material Fig. S4), suggesting PML SUMOylation does not play a critical role in the observed Daxx relocation. These observations strongly suggest that Daxx preferentially localizes to PIAS1-induced SUMO foci, which may then lead to UV-hypersensitive apoptosis.
The PIAS1 N-terminus is required for UV-induced hypersensitivity
The SUMO substrate selectivity of PIASes can stem from their distinct localizations within the cell (Azuma et al., 2005; Ryu and Azuma, 2010). We have shown previously that PIASy localization is determined by its N-terminal SAP domain. We hypothesized that the PIAS1 N-terminus may similarly be important for the correct localization and substrate selectivity of PIAS1 and thus for the UV-hypersensitivity displayed by PIAS1-expressing cells. To confirm our hypothesis, we designed two N-terminus truncation mutants and an N-terminus domain-swap mutant of PIAS1 (Fig. 4A). The PIAS1 61C mutant has the first 61 amino acids deleted and thus lacks the SAP domain; the PIAS1 132C mutant has the first 132 amino acids deleted and starts just N-terminus of the PINIT domain. The domain-swap mutant (NXα PIAS1 132C) has the N-terminal 155 amino acids of PIASXα fused before the PIAS1 132C truncation mutant. All mutants also carry an mCherry tag at the C-terminus. We expressed these PIAS1 mutants in HeLa cells and sorted the cells as described earlier to compare the SUMOylation profile for the mutants and PIAS1wt by immunoblotting for SUMO-2/3. SUMO-2/3 modifications appeared less efficient in the mutants, with the deletion mutants lacking many of the higher-molecular-weight bands. Moreover, the PIAS1 N-terminus deletion mutants did not induce apoptosis in UV-treated cells (Fig. 4C). The basal cell death in cells expressing the PIAS1 mutants was higher in the mock-irradiated samples, but there was no significant increase in apoptosis 4 hours after UV irradiation, unlike PIAS1wt. More significantly, fusing the N-terminus of PIASXα to the PIAS1 132C truncation mutant did not restore the UV hypersensitivity of cells. Once again, we calculated the apoptotic ratio by normalizing UV irradiation-induced apoptosis to apoptosis in the mock-irradiated cells, and we observed threefold less apoptosis in PIAS1 N-terminus deletion mutants-expressing cells when compared with cells expressing PIAS1wt (Fig. 4D). These results indicate the importance of the PIAS1 N-terminus domain in substrate selection and in UV hypersensitivity. In contrast to PIAS1wt, the two PIAS1 truncation mutants showed strong colocalization with PML-NBs and disrupted Daxx distribution in cells (Fig. 5). PIAS1 61C showed weak colocalization with Daxx in the PML bodies, but PIAS1 132C failed to colocalize with Daxx. The NXα PIAS1 132C mutant showed much reduced localization with the PML bodies, but also failed to localize significantly with Daxx (Fig. 5). These results indicate that the N-terminus of PIAS1 is required for correct localization in the nucleus and for recruitment of Daxx to SUMOylated foci.
Fig. 4.
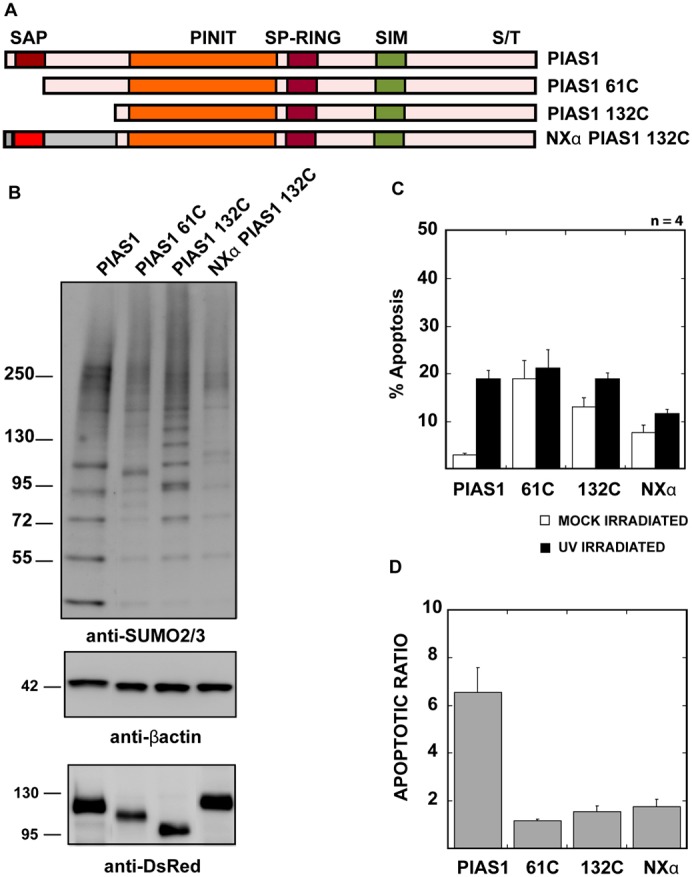
PIAS1 N-terminus SAP domain is essential for correct localization and UV hypersensitivity. (A) Diagrammatic representation of PIAS1 N-terminus truncation and domain swap mutant design. The truncation and the PIASXα domain swap mutants were designed with a C-terminal mCherry fusion, as described in the Materials and Methods section. (B) The PIAS1 N-terminus is required for correct substrate modification. PIAS1wt or truncation/swap mutant-expressing cells were sorted based on mCherry signal and SUMOylation profile was analyzed by immunoblotting with anti-SUMO-2/3 antibody. Anti-DsRed antibody marks the mCherry-tagged PIAS1 or PIAS1 truncation mutants and anti-β-actin antibody was used as a loading control. (C) PIAS1 mutants lacking PIAS1 N-terminus do not increase apoptosis on UV irradiation. The plotted values are an average of four independent experiments. The error bars represent 1 SEM. The percentage of apoptotic cells was determined, as explained in Fig. 1A. (D) The apoptotic ratio was determined as explained in Fig. 1B using the values calculated for C.
Fig. 5.
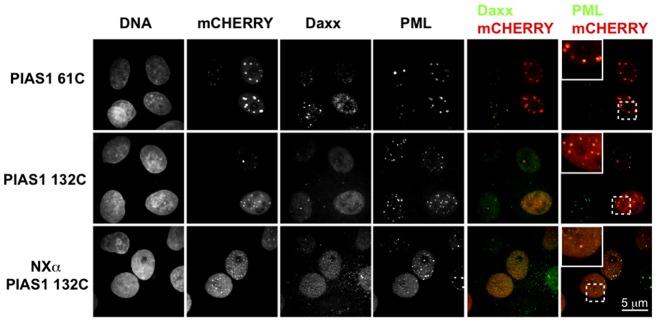
PIAS1 N-terminus mutants localize preferentially to PML-bodies but do not colocalize with Daxx. C-terminal mCherry-tagged PIAS1 61C, 132C and NXα 132C mutants were expressed in HeLa cells for 24 hours. Fixed cells were stained with anti-Daxx and anti-PML antibodies. Images were acquired using a confocal microscope using a 100× objective. Scale bar: 5 µm.
The depletion of Daxx alleviates PIAS1-induced apoptosis
Because we observed that PIAS1-mediated SUMOylation alters the localization of Daxx, we examined whether Daxx acts as a pro-apoptotic protein downstream of PIAS1. We utilized Daxx RNAi to elucidate the role of Daxx in PIAS1-mediated UV-hypersensitive apoptosis. We used two different siRNAs against Daxx and depleted ∼70% of Daxx within 48 hours of treatment using either siRNA (Fig. 6A). Under these conditions, we expressed PIAS1-mCherry fusion protein to measure UV-hypersensitive apoptosis. In the Daxx depleted cells, PIAS1-mediated UV-hypersensitive apoptosis was significantly reduced (Fig. 6B). Compared with the control siRNA-treated cells, the cells that were treated with Daxx siRNAs showed 50% less apoptosis following UV irradiation. There was a slight increase in non-UV-induced apoptosis in the PIAS1-expressing cells that were treated with Daxx siRNA. This result indicates that Daxx plays a critical role in PIAS1-mediated UV-hypersensitive apoptosis and acts downstream of PIAS1-induced SUMOylation.
Fig. 6.
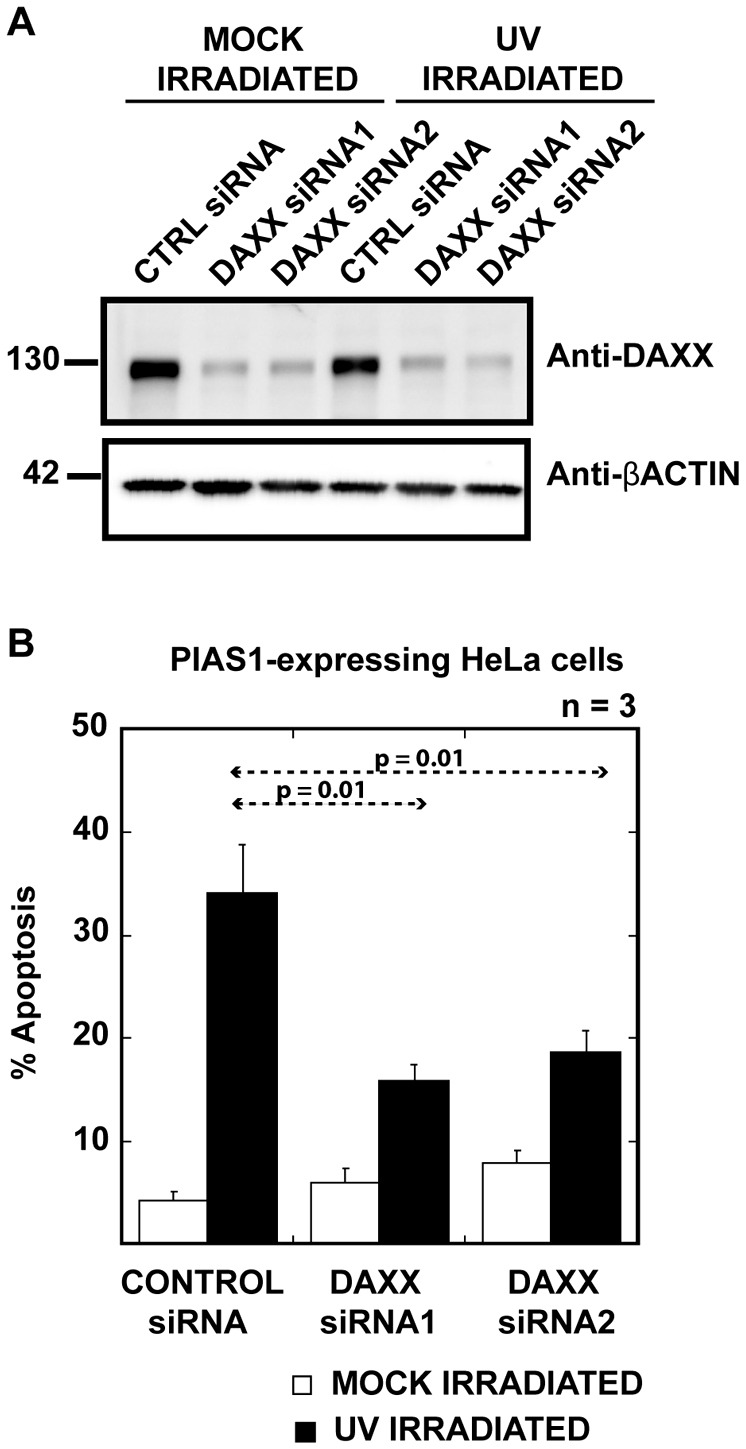
Daxx depletion alleviates UV sensitivity in PIAS1-expressing cells. (A) Treatment with either siRNA reduced cellular Daxx by nearly 70%. Daxx was depleted from HeLa cells using two different siRNAs against Daxx. mCherry-tagged PIAS1 was transfected into HeLa cells 24 hours after the Daxx siRNA treatment. siRNA-treated HeLa cell extracts were immunoblotted with anti-Daxx antibody. Anti-β-actin antibody was used as a loading control. The intensity of the Daxx signals was measured by Image station 400R (Kodak). (B) The expression of Daxx siRNA, but not the control siRNA, significantly reduced apoptosis in cells expressing PIAS1 after UV treatment. Daxx siRNA-treated, PIAS1-expressing cells were UV irradiated, and the apoptotic cells were counted, as described above, 48 hours after Daxx depletion. The plotted values are an average of three independent experiments. The error bars represent 1 SEM.
Taken together, these data indicate that PIAS1 has a distinct role in regulating Daxx activity among the PIAS family of SUMO ligases. PIAS1-specific SUMOylation alters Daxx's function by recruiting Daxx to SUMOylated proteins, which allows Daxx to mediate UV-hypersensitive apoptosis.
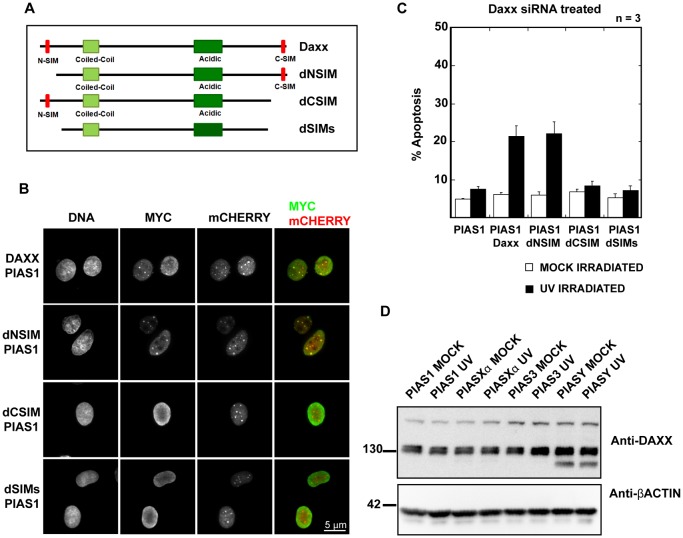
The Daxx C-terminus SIM domain is important for PIAS1-mediated UV hypersensitivity
Daxx has two SUMO interacting motifs (SIM), one at each terminus. The SIMs enable Daxx to interact with SUMOylated proteins including PML and a number of transcription factors. The two SIM domains have been shown to display different affinities for SUMO (Escobar-Cabrera et al., 2011). To understand the requirement of either Daxx SIM domain for PIAS1-mediated UV hypersensitivity, we expressed RNAi-resistant myc-tagged Daxx (supplementary material Fig. S5A,B) and Daxx SIM deletion mutants (Fig. 7A) with PIAS1 in HeLa cells treated with Daxx siRNA. The exogenously expressed Daxx and the Daxx N-terminal SIM deletion mutant (dNSIM) form distinct nuclear foci that colocalize with PIAS1 (Fig. 7B). The Daxx C-terminal SIM deletion mutant (dCSIM) and the Daxx deletion mutant lacking both the SIMs (dSIMs) fail to form distinct foci and thus do not show colocalization with the PIAS1 foci (Fig. 7B). As previously shown in Fig. 6, PIAS1 expression alone in Daxx siRNA-treated cells did not increase apoptosis following UV irradiation (Fig. 7C). We observed a more than twofold increase in the percentage of apoptotic cells when RNAi-resistant Daxx was expressed in addition to PIAS1 in Daxx siRNA-treated cells. The expression of Daxx lacking the dNSIM mutant also increased apoptosis to a similar extent. However, the expression of Daxx lacking the C-terminus SIM (dCSIM) or both the SIMs (dSIMs) failed to increase apoptosis in PIAS1-expressing cells. These results indicate that the Daxx C-terminus SIM is important for PIAS1-mediated hypersensitivity to UV irradiation. Daxx is known to be SUMOylated in cells. Over 15 different lysines in Daxx can be modified, primarily by SUMO-1 (Lin et al., 2006). Using immunoblotting, we investigated whether any of the PIASes can act as a SUMO E3 ligase for Daxx. We could not detect higher-molecular-weight bands in any of the PIAS-transfected cells, which indicates that Daxx is not a major SUMOylated protein in PIAS-expressing cells (Fig. 7D). Together, these results indicate that Daxx is recruited to PIAS1 foci by an interaction with PIAS1-SUMOylated substrates via its C-terminal SIM domain and that this recruitment of Daxx, and probably not Daxx SUMOylation itself, is important for PIAS1-mediated UV-induced apoptosis in cells.
Fig. 7.
The Daxx C-terminal SIM domain is required for PIAS1-mediated UV hypersensitivity. (A) Diagrammatic representation of Daxx SIM domain truncation mutants. The truncation mutants were designed as described in the Materials and Methods section. The Daxx dNSIM mutant lacks the first 17 amino acids, dCSIM lacks the final 20 amino acids, and the dSIMs mutant lacks both the first 17 amino acids and the last 20 amino acids, thus eliminating both SIM domains from Daxx. All Daxx constructs have an N-terminal Myc-tag. (B) The Daxx C-terminal SIM domain is essential for proper Daxx localization. HeLa cells transfected with the bicistronic vector carrying PIAS1-mCherry and Myc-Daxx or Myc-Daxx truncations. Fixed cells were stained with anti-Myc-tag antibody. The images were acquired using a confocal microscope with a 100× objective. Scale bar: 5 µm. (C) The replacement of endogenous Daxx with the C-terminal SIM-deletion mutants rescues UV-hypersensitivity phenotype in PIAS1-expressing cells. Apoptotic cells were counted as described in Fig. 1A. The plotted values are an average of three independent experiments. The error bars represent 1 SEM. (D) Daxx does not appear to be a major SUMOylation substrate. HeLa cells were transfected with each of the PIAS isoforms. Mock- and UV-treated cells were sorted based on mCherry signal and the lysates were immunoblotted with anti-Daxx antibody. Anti-β-actin was used as a loading control.
Discussion
Many different stress stimuli lead to the SUMOylation of several cellular proteins that act in the damage repair pathway or lead to apoptosis (Saitoh and Hinchey, 2000; Rytinki et al., 2009). Members of the PIAS family of SUMO E3 ligases have been shown to have distinct substrates and contribute to SUMO paralog selectivity in response to various stress pathways (Rytinki et al., 2009). Various pieces of evidence indicate that PIAS1 is an important regulator of the cellular stress response. Galanty et al. (Galanty et al., 2009) have shown that PIAS1 stimulates double strand break repair by modifying BRCA1 with SUMO-2/3. PIAS1 has also been shown to participate in cellular responses to oxidative stress by regulating the JNK pathway, and PIAS1 knockdown prevents reactive oxygen species-induced apoptosis (Liu and Shuai, 2001; Leitao et al., 2011). PIAS1 also SUMOylates p53 and leads to an increase in p53 transcriptional activity (Kahyo et al., 2001; Megidish et al., 2002). In this study, we describe a specific role of PIAS1 in regulating apoptosis in response to UV irradiation. The overexpression of PIAS1 in HeLa cells leads to increased sensitivity to UV irradiation and results in an increase in cell death four hours after UV irradiation. The expression of other members of the PIAS family does not lead to increased apoptosis induced by UV irradiation. PIAS1's SUMO ligase activity is required for inducing hypersensitivity to UV irradiation; a ligase inactive mutant of PIAS1 fails to elicit UV hypersensitivity in cells. Each of the PIASes displays a distinct SUMO-substrate profile and localization in the nucleus, but only PIAS1 expression leads to increased apoptosis following UV irradiation. This indicates that substrates unique to PIAS1 participate in the UV-induced cell death response. We observe an increase in basal apoptosis (in the absence of UV) on ectopic expression of other PIASes as well as PIAS1 mutants that do not appear to increase apoptosis after UV irradiation. Further experiments will be required to understand the mechanism of this apoptosis; however, we expect it to be clearly distinct from Daxx-mediated apoptosis that we see in PIAS1-expressing cells on UV irradiation because the other PIASes do not recruit Daxx to SUMO foci (supplementary material Fig. S3; Fig. S5). The identification of the specific substrates of each PIAS isoforms will be essential to further understand the role of PIAS-mediated SUMOylation in stress-related apoptosis and in spontaneous apoptosis induced by ectopic expression of PIASes.
Reindle et al. (Reindle et al., 2006) have shown that in the case of the yeast SUMO E3 ligases Siz1 and Siz2, the localization of the ligases dictates the substrate selectivity. Each of the PIASes shows a unique nuclear distribution and appears to have a unique subset of substrates. This localization-dependent regulation of SUMO ligase activity is consistent with our published results showing that PIAS4/y mediates mitotic chromosomal SUMOylation in Xenopus egg extract assays (Azuma et al., 2005; Ryu and Azuma, 2010). Because the UV-hypersensitivity phenotype is specific to PIAS1, we surmised that PIAS1's distinctive localization and the resulting substrate selectivity are responsible for the PIAS1-specific cellular response to UV irradiation. This hypothesis is supported by the results that the ligase-dead PIAS1 C351S mutant, the PIAS1 SIM deletion mutant N440, and the PIAS1 N-terminus truncation mutants do not display the same localization pattern as wild-type PIAS1 and do not activate apoptosis in response to UV irradiation. Therefore, the PIAS1-mediated UV-hypersensitive apoptosis is dependent on PIAS1-specific SUMO substrates. We have previously shown that PIASy's centromeric localization at the mitotic chromosome requires its N-terminus SAP domain (Ryu and Azuma, 2010). We show a similar requirement of the PIAS1 N-terminus for its correct localization and SUMOylation activity. Mutants lacking the PIAS1 SAP domain localize preferentially to PML bodies and do not display the same SUMOylation profile as PIAS1wt. The PIAS1 N-terminus domain is specifically required for UV hypersensitivity because a substitution of the domain with the equivalent domain from PIASXα fails to elicit similar hypersensitivity to UV irradiation.
Only PIAS1-SUMOylated foci recruit the apoptotic protein Daxx. Daxx is a well-known apoptosis regulator that acts primarily as a transcriptional regulator, although its status as a pro- or anti-apoptotic factor is debated (Michaelson, 2000; Salomoni and Khelifi, 2006; Shih et al., 2007). Daxx contains two well-characterized SUMO-interacting motifs (SIM) at its termini that allow it to interact with SUMOylated proteins in cells (Santiago et al., 2009; Chang et al., 2011; Escobar-Cabrera et al., 2011). In response to UV irradiation, Daxx appears to act as a pro-apoptotic protein downstream of PIAS1. We confirmed the involvement of Daxx using a knockdown approach. PIAS1-induced UV sensitivity is alleviated when Daxx levels are reduced using siRNA against Daxx. We also show that of the two Daxx SIMs, the C-terminus SIM is specifically required for PIAS1-induced UV sensitivity. Overall, our data show that the SUMOylation-dependent regulation of Daxx is critical for sensitizing cells to UV irradiation, which is consistent with the previously proposed model (Mukhopadhyay and Matunis, 2011).
Daxx is known to localize to PML bodies in cells (Ishov et al., 1999). The PML-NB recruitment of Daxx has been shown to attenuate its repressive activity toward several promoters (Li et al., 2000). In PIAS1-expressing cells, Daxx does not localize to PML bodies; instead, it relocates to PIAS1 SUMOylation foci. Because a large number of known SUMOylated proteins are transcription factors, and many SUMOylated transcription factors are known targets of Daxx-mediated transcriptional repression, PIAS1 may mediate the SUMO-2/3 modification of transcription factors, and Daxx may act as a corepressor in a SUMOylation-dependent manner. Therefore, Daxx may suppress the production of anti-apoptotic factors and sensitize cells to cellular stressors, such as UV irradiation. However, we did not observe any significant differences in the expression of the Bcl2 family of apoptotic proteins (supplementary material Fig. S6) in the PIAS1-expressing cells. Daxx has been shown to repress the transcription of anti-apoptotic proteins downstream of NFκB (Croxton et al., 2006), and it would be interesting to examine whether this pathway is also targeted downstream of PIAS1 during UV-hypersensitive apoptosis. Another intriguing target is the JNK pathway because both Daxx and PIAS1 have been shown to regulate the JNK pathway (Khelifi et al., 2005; Leitao et al., 2011).
One aspect of the PIAS1-mediated UV-hypersensitive response that we did not pursue further in this study is that PIAS1 knockdown also leads to increased UV sensitivity, albeit the kinetics of apoptosis induction appears to be different from that of PIAS1 overexpression. This difference in the kinetics of apoptosis induction may reflect different UV response pathways in cells that are regulated by PIAS1-mediated SUMOylation. We speculate that different substrates targeted by PIAS1 for SUMOylation may be involved in apoptosis in the two conditions, and apoptosis induced due to PIAS1 knockdown may not involve Daxx. In either case, PIAS1 appears to be an important regulator of cell survival following UV stress.
In conclusion, we show that PIAS1 sensitizes cells to UV stress-induced apoptosis through the recruitment of Daxx to PAIS1-specific SUMOylated substrates (Fig. 8). Daxx itself does not appear to be a substrate of PIAS1, but interacts with SUMOylated substrates via its C-terminal SIM domain. The identification of the specific PIAS1 substrates, their mechanism of interaction with Daxx and Daxx's downstream targets will be important to promote our understanding of PIAS1-mediated cell death in response to UV irradiation.
Fig. 8.
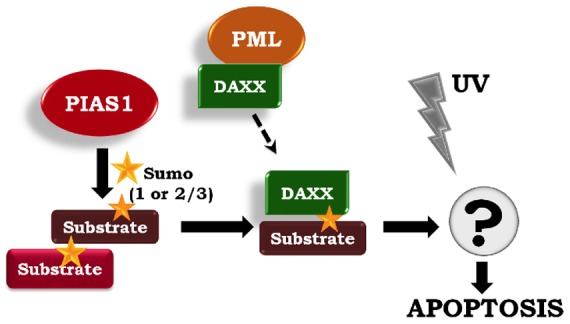
PIAS1-directed SUMO-modified substrates recruit Daxx to mediate UV-hypersensitive apoptosis. Based on our data, we propose a model wherein the ectopic expression of PIAS1 promotes SUMOylation of specific substrates. One or more of these SUMOylated proteins recruit Daxx. The Daxx C-terminal SIM domain is necessary for this interaction. The Daxx-mediated corepression of certain transcription factors may then contribute to apoptosis after UV irradiation.
Materials and Methods
DNA constructs and cell transfection
mCherry DNA was amplified and cloned into the pCDNA His maxC vector (Invitrogen, Carlsbad, CA) using NotI and XhoI restriction enzymes to create a pCDNA4 His maxC mCherry vector. DNA sequences encoding PIAS1, PIAS2/Xα, PIAS3 and PIAS4/y were amplified and subcloned into the pCDNA4 His maxC mCherry vector using EcoRI and NotI restriction enzymes; this produced a C-terminal mCherry fusion for the PIAS proteins. The PIAS1 C351S mutant was generated using the QuikChange II XL Site-Directed Mutagenesis kit from Agilent Technologies (Santa Clara, CA). PIAS1 N- and C-terminus truncation mutants were generated by amplifying PIAS1 from bp184 onwards (61C), bp397 onwards (132C) or up to bp1320 (N440) and cloning into the pCDNA4 His maxC mCherry vector, as described for PIAS1. PIASXα DNA coding for the first 155 amino acids was amplified and inserted into the pCDNA4 His maxC PIAS1 132C mCherry construct using an EcoRI restriction site to create the NXα PIAS1 132C fusion protein construct.
Daxx cDNA was amplified from HeLa cells and cloned between NheI and EcoRI sites in the pEGFPC1 vector (Clontech Laboratories Inc., Mountain View, CA). The Daxx RNAi-resistant mutant was generated by introducing 5 silent mutations in the Daxx DNA [(620-aaaaagaattggatctttcagagttagatga-650, the mutations are underlined] that disrupt siRNA binding using the Quikchange II XL Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA). The Daxx SIM deletion mutants were created by amplifying Daxx DNA coding for amino acids 18–740 (dNSIM), 1–720 (dCSIM) and 18–720 (dSIMs). A Myc-tagged RNAi-resistant Daxx and Daxx deletion mutants were cloned into MCS-A of the bicistronic pIRES vector (Clontech Laboratories Inc., Mountain View, CA). HeLa and Hct116 cells were grown to 50% confluence in McCoy's medium (Mediatech Inc., Manassas, VA) that was supplemented with 10% FBS and transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA), per manufacturer's instructions.
UV irradiation and apoptotic assays
The transfected HeLa cells were either mock irradiated or irradiated with 30 J/m2 of UV using an Entela UV crosslinker. In brief, the cells were washed twice with phosphate buffer saline (PBS) and then irradiated in PBS. After irradiation, the PBS was removed and replaced with growth medium. The apoptotic cells were counted based on aberrant nuclear morphology and Parp-1 staining at the indicated time after UV irradiation. Cells grown on coverslips were fixed with 4% paraformaldehyde and permeabilized using 0.2% Triton X-100. The cells were then stained with an anti-Parp1 antibody and Hoechst 33342 dye (which stains DNA) for immunofluorescence analysis. The apoptotic cells were counted based on DNA morphology and the redistribution of Parp1 signal into the cytoplasm. The apoptotic cells were also counted based on Annexin V staining using the Alexa Fluor 488-conjugated Annexin V (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Cell sorting and western blot analysis
HeLa cells that were transfected with mCherry-tagged PIAS family members were sorted from the untransfected cells using a Beckman Coulter MoFlo XDP sorter (Fort Collins, CO) with a 488-nm excitation filter and a 620/29 nm emission filter. The sorting was performed 4 hours after the mock or UV irradiation. The sorted cells were subjected to an immunoblotting analysis, and the protein levels were normalized by the intensity of β-actin signals detected by anti-β-actin antibody (Cell Signaling Technology, Danvers, MA).
Immunofluorescence analysis
Transfected and treated cells were fixed with 4% paraformaldehyde and permeabilized using 0.2% Triton X-100. The cells were then blocked for 15 min with 5% BSA and 2.5% fish gelatin prepared in PBS-T (PBS containing 0.1% Tween20). After washing, the cells were stained for 1 hour with the appropriate antibody following the manufacturer's dilution instructions, which was followed by staining with an Alexa dye-tagged secondary antibody (Invitrogen, Carlsbad, CA) and Hoechst 33342 dye. The cells were mounted using Vectashield mounting medium (Vector Laboratories, Burlingame, CA), and imaged using a DSU-10 spinning disk (Yokugawa-type) confocal microscope that was equipped with an Olympus PlanApo100X objective (with a numerical aperture of 1.45), Hamamatsu EMCCD and the Slidebook imaging package (Intelligent Imaging Innovations, Denver, CO). An anti-human Daxx antibody (D7810) was purchased from Sigma, and a monoclonal anti-PML antibody (N19) and monoclonal anti Myc-tag antibody (NE10) were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). The other antibodies used in this study were generated in the lab (Ryu et al., 2010).
Daxx and PIAS1 silencing using siRNAs
siRNAs against human Daxx were obtained from Invitrogen (Carlsbad, CA) (Daxx siRNA1: DAXX-HSS102654, Daxx siRNA2: DAXX-HSS175936). HeLa cells were transfected with 200 pmol of either siRNA using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) using the reverse transfection method, as described by the manufacturer. The cells were plated to obtain a next-day confluency of 50%. Twenty-four hours after the siRNA transfection, the cells were transfected with pCDNA4 His maxC PIAS1 mCherry or the bicistronic pIRES construct expressing PIAS1 and Myc-tagged RNAi resistant Daxx or Daxx truncation mutants. The cells were UV irradiated 48 hours after the siRNA treatment as described above, and the apoptotic cells were counted 4 hours later. siRNA against PIAS1 was also obtained from Invitrogen (PIAS1 siRNA: PIAS1VHS41400). Cells were transfected with 200 pmol PIAS1 siRNA and UV irradiated 48 hours after transfection. The depletion efficiencies of the target proteins were estimated by immunoblotting followed by measuring the intensity of the bands with Image Station 4000R (Kodak).
Colocalization correlation calculations
The colocalization correlation between PML and Daxx was calculated using 60 confocal images acquired from three independent experiments as described above. CellProfiler v2.0 (Carpenter et al., 2006) cell image analysis software was used to calculate colocalization using Pearson's correlation. The Otsu Adaptive threshold was used to identify and select all nuclei expressing mCherry. The correlation was calculated between Daxx and PML spots within the selected nuclei.
Supplementary Material
Acknowledgments
We appreciate the information on the RNAi procedure from A. Arnaoutov (NICHD/NIH) and the information on the anti-PML antibody from David Davido (University of Kansas). We thank H. Shinogle (Microscopy and Analytical Imaging Laboratory at University of Kansas) for her assistance with cell sorting and confocal microscopic analysis. We thank H. Ryu and M. Azuma for the critical reading of the manuscript. We also thank NPG Language Editing for editorial assistance.
Footnotes
Funding
This project was supported in part by a general research fund from the University of Kansas; and by the National Institutes of Health (National Institute of General Medical Sciences) [grant number GM80278]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.110825/-/DC1
References
- Azuma Y., Arnaoutov A., Anan T., Dasso M. (2005). PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 24, 2172–2182 10.1038/sj.emboj.7600700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I. H., Friman O., Guertin D. A., Chang J. H., Lindquist R. A., Moffat J.et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. C., Naik M. T., Huang Y. S., Jeng J. C., Liao P. H., Kuo H. Y., Ho C. C., Hsieh Y. L., Lin C. H., Huang N. J.et al. (2011). Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol. Cell 42, 62–74 10.1016/j.molcel.2011.02.022 [DOI] [PubMed] [Google Scholar]
- Chen L. Y., Chen J. D. (2003). Daxx silencing sensitizes cells to multiple apoptotic pathways. Mol. Cell. Biol. 23, 7108–7121 10.1128/MCB.23.20.7108-7121.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxton R., Puto L. A., de Belle I., Thomas M., Torii S., Hanaii F., Cuddy M., Reed J. C. (2006). Daxx represses expression of a subset of antiapoptotic genes regulated by nuclear factor-kappaB. Cancer Res. 66, 9026–9035 10.1158/0008-5472.CAN-06-1047 [DOI] [PubMed] [Google Scholar]
- Escobar–Cabrera E., Okon M., Lau D. K., Dart C. F., Bonvin A. M., McIntosh L. P. (2011). Characterizing the N- and C-terminal Small ubiquitin-like modifier (SUMO)-interacting motifs of the scaffold protein DAXX. J. Biol. Chem. 286, 19816–19829 10.1074/jbc.M111.231647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y., Belotserkovskaya R., Coates J., Polo S., Miller K. M., Jackson S. P. (2009). Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462, 935–939 10.1038/nature08657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss–Friedlander R., Melchior F. (2007). Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 10.1038/nrm2293 [DOI] [PubMed] [Google Scholar]
- Ishov A. M., Sotnikov A. G., Negorev D., Vladimirova O. V., Neff N., Kamitani T., Yeh E. T., Strauss J. F., 3rd, Maul G. G. (1999). PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147, 221–234 10.1083/jcb.147.2.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. S. (2004). Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 10.1146/annurev.biochem.73.011303.074118 [DOI] [PubMed] [Google Scholar]
- Kahyo T., Nishida T., Yasuda H. (2001). Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8, 713–718 10.1016/S1097-2765(01)00349-5 [DOI] [PubMed] [Google Scholar]
- Khelifi A. F., D'Alcontres M. S., Salomoni P. (2005). Daxx is required for stress-induced cell death and JNK activation. Cell Death Differ. 12, 724–733 10.1038/sj.cdd.4401559 [DOI] [PubMed] [Google Scholar]
- Kotaja N., Karvonen U., Jänne O. A., Palvimo J. J. (2002). PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 22, 5222–5234 10.1128/MCB.22.14.5222-5234.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. M., Kang H. J., Lee H. R., Choi C. Y., Jang W. J., Ahn J. H. (2003). PIAS1 enhances SUMO-1 modification and the transactivation activity of the major immediate-early IE2 protein of human cytomegalovirus. FEBS Lett. 555, 322–328 10.1016/S0014-5793(03)01268-7 [DOI] [PubMed] [Google Scholar]
- Lee H., Quinn J. C., Prasanth K. V., Swiss V. A., Economides K. D., Camacho M. M., Spector D. L., Abate–Shen C. (2006). PIAS1 confers DNA-binding specificity on the Msx1 homeoprotein. Genes Dev. 20, 784–794 10.1101/gad.1392006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitao B. B., Jones M. C., Brosens J. J. (2011). The SUMO E3-ligase PIAS1 couples reactive oxygen species-dependent JNK activation to oxidative cell death. FASEB J. 25, 3416–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Leo C., Zhu J., Wu X., O'Neil J., Park E. J., Chen J. D. (2000). Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell. Biol. 20, 1784–1796 10.1128/MCB.20.5.1784-1796.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wang X., Wu X., Rui Y., Liu W., Wang J., Wang X., Liou Y. C., Ye Z., Lin S. C. (2007). Daxx cooperates with the Axin/HIPK2/p53 complex to induce cell death. Cancer Res. 67, 66–74 10.1158/0008-5472.CAN-06-1671 [DOI] [PubMed] [Google Scholar]
- Lin D. Y., Huang Y. S., Jeng J. C., Kuo H. Y., Chang C. C., Chao T. T., Ho C. C., Chen Y. C., Lin T. P., Fang H. I.et al. (2006). Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24, 341–354 10.1016/j.molcel.2006.10.019 [DOI] [PubMed] [Google Scholar]
- Liu B., Shuai K. (2001). Induction of apoptosis by protein inhibitor of activated Stat1 through c-Jun NH2-terminal kinase activation. J. Biol. Chem. 276, 36624–36631 10.1074/jbc.M101085200 [DOI] [PubMed] [Google Scholar]
- Megidish T., Xu J. H., Xu C. W. (2002). Activation of p53 by protein inhibitor of activated Stat1 (PIAS1). J. Biol. Chem. 277, 8255–8259 10.1074/jbc.C200001200 [DOI] [PubMed] [Google Scholar]
- Michaelson J. S. (2000). The Daxx enigma. Apoptosis 5, 217–220 [DOI] [PubMed] [Google Scholar]
- Michaelson J. S., Leder P. (2003). RNAi reveals anti-apoptotic and transcriptionally repressive activities of DAXX. J. Cell Sci. 116, 345–352 10.1242/jcs.00234 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D., Matunis M. J. (2011). SUMmOning Daxx-mediated repression. Mol. Cell 42, 4–5 10.1016/j.molcel.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa K., Yokosawa H. (2002). PIAS3 induces SUMO-1 modification and transcriptional repression of IRF-1. FEBS Lett. 530, 204–208 10.1016/S0014-5793(02)03486-5 [DOI] [PubMed] [Google Scholar]
- Palvimo J. J. (2007). PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem. Soc. Trans. 35, 1405–1408 10.1042/BST0351405 [DOI] [PubMed] [Google Scholar]
- Reindle A., Belichenko I., Bylebyl G. R., Chen X. L., Gandhi N., Johnson E. S. (2006). Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J. Cell Sci. 119, 4749–4757 10.1242/jcs.03243 [DOI] [PubMed] [Google Scholar]
- Rytinki M. M., Kaikkonen S., Pehkonen P., Jääskeläinen T., Palvimo J. J. (2009). PIAS proteins: pleiotropic interactors associated with SUMO. Cell. Mol. Life Sci. 66, 3029–3041 10.1007/s00018-009-0061-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Azuma Y. (2010). Rod/Zw10 complex is required for PIASy-dependent centromeric SUMOylation. J. Biol. Chem. 285, 32576–32585 10.1074/jbc.M110.153817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Al–Ani G., Deckert K., Kirkpatrick D., Gygi S. P., Dasso M., Azuma Y. (2010). PIASy mediates SUMO-2/3 conjugation of poly(ADP-ribose) polymerase 1 (PARP1) on mitotic chromosomes. J. Biol. Chem. 285, 14415–14423 10.1074/jbc.M109.074583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H., Hinchey J. (2000). Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275, 6252–6258 10.1074/jbc.275.9.6252 [DOI] [PubMed] [Google Scholar]
- Salomoni P., Khelifi A. F. (2006). Daxx: death or survival protein? Trends Cell Biol. 16, 97–104 10.1016/j.tcb.2005.12.002 [DOI] [PubMed] [Google Scholar]
- Santiago A., Godsey A. C., Hossain J., Zhao L. Y., Liao D. (2009). Identification of two independent SUMO-interacting motifs in Daxx: evolutionary conservation from Drosophila to humans and their biochemical functions. Cell Cycle 8, 76–87 10.4161/cc.8.1.7493 [DOI] [PubMed] [Google Scholar]
- Schmidt D., Müller S. (2002). Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. USA 99, 2872–2877 10.1073/pnas.052559499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih H. M., Chang C. C., Kuo H. Y., Lin D. Y. (2007). Daxx mediates SUMO-dependent transcriptional control and subnuclear compartmentalization. Biochem. Soc. Trans. 35, 1397–1400 10.1042/BST0351397 [DOI] [PubMed] [Google Scholar]
- Song Y. J., Lee H. (2011). PIAS1 negatively regulates ubiquitination of Msx1 homeoprotein independent of its SUMO ligase activity. Mol. Cells 32, 221–226 10.1007/s10059-011-1020-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y. (2004). Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. USA 101, 14373–14378 10.1073/pnas.0403498101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari M., Quan L. T., O'Rourke K., Desnoyers S., Zeng Z., Beidler D. R., Poirier G. G., Salvesen G. S., Dixit V. M. (1995). Yama/CPP32 beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell 81, 801–809 10.1016/0092-8674(95)90541-3 [DOI] [PubMed] [Google Scholar]
- Wilkinson K. A., Henley J. M. (2010). Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 428, 133–145 10.1042/BJ20100158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunus A. A., Lima C. D. (2009). Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell 35, 669–682 10.1016/j.molcel.2009.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.