Summary
Target of rapamycin (TOR), an evolutionarily conserved serine/threonine protein kinase, plays pivotal roles in several important cellular processes in eukaryotes. In the fission yeast Schizosaccharomyces pombe, TOR complex 1 (TORC1), which includes Tor2 as a catalytic subunit, manages the switch between cell proliferation and differentiation by sensing nutrient availability. However, little is known about the direct target of TORC1 that plays key roles in nutrient-dependent TORC1 signaling in fission yeast. Here we report that in fission yeast, three AGC kinase family members, named Psk1, Sck1 and Sck2, which exhibit high homology with human S6K1, are phosphorylated under nutrient-rich conditions and are dephosphorylated by starvation conditions. Among these, Psk1 is necessary for phosphorylation of ribosomal protein S6. Furthermore, Psk1 phosphorylation is regulated by TORC1 in nutrient-dependent and rapamycin-sensitive manners in vivo. Three conserved regulatory motifs (the activation loop, the hydrophobic and the turn motifs) in Psk1 are phosphorylated and these modifications are required for Psk1 activity. In particular, phosphorylation of the hydrophobic motif is catalyzed by TORC1 in vivo and in vitro. Ksg1, a homolog of PDK1, is also important for Psk1 phosphorylation in the activation loop and for its activity. The TORC1 components Pop3, Toc1 and Tco89, are dispensable for Psk1 regulation, but disruption of pop3+ causes an increase in the sensitivity of TORC1 to rapamycin. Taken together, these results provide convincing evidence that TORC1/Psk1/Rps6 constitutes a nutrient-dependent signaling pathway in fission yeast.
Key words: TORC1, Target of rapamycin, S6 kinase, Nutrient response, Rapamycin, Signal transduction
Introduction
Eukaryotic cells operate numerous signal transduction events mediated by protein phosphorylation to respond promptly to environmental changes. Target of rapamycin (TOR), a highly conserved serine/threonine protein kinase of the phosphatidylinositol kinase-related kinase family, plays pivotal roles in controlling various cellular processes such as cell growth, cell cycle and protein synthesis in response to nutrient availability, growth factors, cellular energy status and stress conditions. TOR is included as a catalytic subunit in two distinct multiprotein complexes, TOR complex 1 (TORC1) and TORC2. TORC1 signaling promotes anabolic processes such as protein synthesis, transcription and ribosome biogenesis, and prevents catabolic processes such as autophagy, whereas TORC2 appears to be involved in actin organization and cell survival. An immunosuppressant and anticancer drug, rapamycin, preferentially inhibits TORC1 activity (Wullschleger et al., 2006; Sengupta et al., 2010).
The fission yeast Schizosaccharomyces pombe possesses two TOR genes, tor1+ and tor2+ that encode catalytic subunits of TORC2 and TORC1, respectively. TORC1, which consists of a raptor homolog Mip1, an mLST8 homolog Pop3 (also known as Wat1), Toc1 and Tco89 in addition to Tor2, plays a crucial role in the switch between cell proliferation and differentiation by sensing nitrogen source availability. In contrast, TORC2 is composed of Tor1, a rictor homolog Ste20, Sin1, Pop3 and Bit61 participates in cell proliferation under stress conditions, entry into sexual differentiation under nitrogen starvation, and is involved in leucine uptake (Alvarez and Moreno, 2006; Hayashi et al., 2007; Kawai et al., 2001; Matsuo et al., 2003; Matsuo et al., 2007; Uritani et al., 2006; Weisman and Choder, 2001; Weisman et al., 2005; Weisman et al., 2007). Rhb1, a fission yeast homolog of Rheb, which is a member of the Ras superfamily G-protein, and the complex of Tsc1 and Tsc2, a counterpart of the mammalian TSC1–TSC2 complex that acts as a GTPase-activating protein for Rheb, regulate TORC1 activity as upstream factors (Aspuria and Tamanoi, 2008; Matsumoto et al., 2002; Nakase et al., 2006; Urano et al., 2005; van Slegtenhorst et al., 2004; Yang et al., 2001; Murai et al., 2009). It has recently been reported that the Rab-family G-protein, Ryh1, which is a homolog of human Rab6, is involved in the activity of TORC2 (Tatebe et al., 2010).
One of the important issues with regard to TOR signaling is to define direct targets of TOR complexes to gain an understanding of how upstream signals can be transmitted through TOR complexes. Several serine/threonine protein kinases in the AGC (protein kinase A/protein kinase G/protein kinase C) kinase family, which include S6K, AKT (also known as PKB), SGK and PKC in mammals and their homologs in other eukaryotes, are directly phosphorylated, and their activities are regulated by either TOR complex (Jacinto and Lorberg, 2008); in mammalian systems, S6K is phosphorylated and activated by mammalian TORC1 (mTORC1), whereas AKT, SGK and PKC are modified by mTORC2 (Laplante and Sabatini, 2009). In fission yeast, TORC2 directly regulates the phosphorylation and function of Gad8, which is a member of the AGC kinase family (Matsuo et al., 2003; Ikeda et al., 2008). However, direct target of TORC1 has not been identified.
To gain insights into direct downstream targets of TORC1 in fission yeast, we have combined the study on AGC kinases in fission yeast with our previous study that identified Rps6 as a counterpart of ribosomal protein S6 (Nakashima et al., 2010). The latter study established that phosphorylation of Rps6 is mediated by TORC1 in a process that is dependent on availability of nutrients such as ammonium and glucose, and that the Tsc1–Tsc2 complex and Rhb1 participate in its phosphorylation (Nakashima et al., 2010). Fission yeast contains AGC kinases including Psk1, Sck1 and Sck2. There was a suggestion that Sck1 and Sck2 are counterparts of budding yeast Sch9 that has recently been characterized as an S6 kinase in budding yeast (Jin et al., 1995; Fujita and Yamamoto, 1998; Urban et al., 2007). We show here that Rps6 is a target of Psk1, based on the finding that Rps6 phosphorylation is regulated by Psk1 in a nutrient signal-dependent manner. We also show that Psk1 itself is directly phosphorylated, and its involvement in phosphorylation of Rps6 is regulated by TORC1 in nutrient-dependent and rapamycin-sensitive fashions. In addition, Ksg1, a kinase of the PDK1 homolog, is also implicated in Psk1 phosphorylation and is critical for its activity. These results point to the significance of phosphorylation of Psk1 for nutrient-dependent signal transduction in fission yeast.
Results
Phosphorylation of S6K1 homologs in response to nitrogen source availability
To identify the S6K candidates that would directly phosphorylate ribosomal protein (Rp) S6 and that should be downstream of TORC1 in fission yeast, we first searched using the BLAST program for homologs of p70 S6K1, the predominant kinase of S6 in humans. This analysis showed that fission yeast has at least four potential S6K1 homologs, namely Psk1, Sck1, Sck2 and Gad8, which belong to the AGC kinase family (Fig. 1A; supplementary material Fig. S1) (Mukai et al., 1995; Jacinto and Lorberg, 2008). Phylogenetic analysis reveals that Psk1 exhibits the highest homology with S6K1, whereas Gad8 is similar to human AKT (Fig. 1A), which is a substrate of mTORC2 (Sarbassov et al., 2005). Indeed, some phosphorylation sites in Gad8 are catalyzed by TORC2 (Matsuo et al., 2003; Ikeda et al., 2008), suggesting that Gad8 is a counterpart of AKT in fission yeast.
Fig. 1.
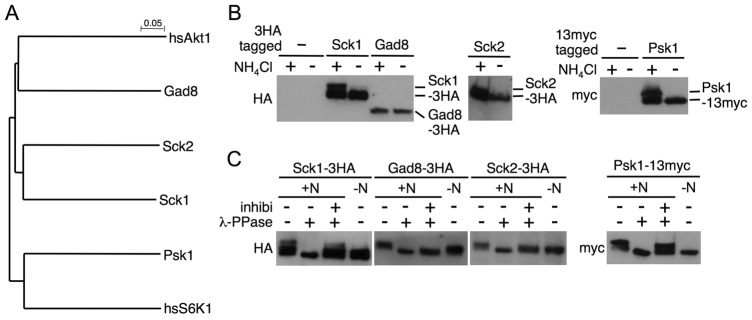
Nitrogen source-dependent phosphorylation of S6K homologs in fission yeast. (A) A phylogenetic tree of fission yeast homologs of human S6K1 was generated using the ClustalW program. (B) Cells of strains JUp1204 (non-tagged), AN0151 (Sck1-3HA), AN0176 (Gad8-3HA), AN0153 (Sck2-3HA) and AN0180 (Psk1-13myc) were cultured in EMM, then washed and starved in EMM-N for 1 hour. After the starvation, ammonium (final concentration, 0.5%; +) or water (−) was added to cells and they were incubated for 30 minutes. (C) Protein extracts from ammonium-stimulated cells as described in B were incubated with λ-phosphatase in the presence or absence of its inhibitors. (B,C) Protein extracts were subjected to immunoblotting with the indicated antibodies.
We next examined whether these kinases are phosphorylated in response to ammonium as the sole nitrogen source in Edinburgh minimal medium (EMM), because we previously showed that TORC1 is activated in ammonium-rich conditions and conversely inactivated in an ammonium-free medium (Nakashima et al., 2010). For this purpose, we constructed strains in which the epitope-tagged S6K homologs (three tandem copies of influenza hemagglutinin (HA) or 13 tandem copies of myc at their carboxyl termini) are chromosomally expressed under the control of their native promoters and detected them by immunoblotting. Mobility shifts of epitope-tagged Sck1, Sck2 and Psk1 were observed after the re-addition of ammonium, whereas the migration of Gad8–3HA was not altered in media with or without ammonium (Fig. 1B). Consistent with our previous results (Nakashima et al., 2010), Rps6 (fission yeast S6) is phosphorylated under ammonium stimulation in these strains (supplementary material Fig. S2). The λ-phosphatase assay revealed that the mobility shift of the three kinases, Sck1, Sck2 and Psk1, was due to phosphorylation (Fig. 1C). These results suggest that Sck1, Sck2 and Psk1 are phosphorylated in nitrogen-rich conditions and, conversely, dephosphorylated at least in part under nitrogen starvation, whereas Gad8 is constitutively phosphorylated. Thus, Sck1, Sck2 and Psk1 appear to be candidates for S6K.
Psk1 is necessary for phosphorylation at Ser235/236 in ribosomal protein S6
To determine a bona fide kinase of Rps6, we assessed Rps6 phosphorylation in deletion mutants of those S6K candidates using an anti-phospho-Akt substrate (PAS) antibody that can recognize phosphorylation at Ser235/236 in Rps6, which is regulated by TORC1 signaling in response to nutrient conditions (Nakashima et al., 2010). The corresponding serine residues of S6 are directly phosphorylated by S6K in other eukaryotes (Ruvinsky and Meyuhas, 2006). As shown in Fig. 2A, Rps6 phosphorylation under nitrogen-rich conditions in sck1Δ and in sck2Δ was comparable to that in the wild type, and its phosphorylation in both the single mutants disappeared under nitrogen starvation as it did in the wild type. Furthermore, the double disruption of sck1+ and sck2+ and the deletion of gad8+ had no significant effect on the nutrient-dependent Rps6 phosphorylation (data not shown). In contrast, Rps6 phosphorylation in psk1Δ was not detected even in nitrogen-rich conditions (Fig. 2A). Fission yeast has two distinct Rps6 gene products, Rps601 and Rps602, and the phosphorylation of both is regulated by TORC1 (Nakashima et al., 2010). In psk1Δ, expression of myc-tagged Rps602 under control of its own promoter was comparable to that in the wild type (Fig. 2B). These results suggest that the deletion of psk1+ results in failure to phosphorylate Rps6. To further investigate whether Psk1 phosphorylates Rps6 protein in vitro, we examined the kinase activity of Psk1 using recombinant Rps602 as a substrate. In this experiment, we utilized a Psk1 mutant (Thr415Glu), a phospho-mimetic mutant of its hydrophobic motif, as the mutant exhibited higher activity than the wild-type protein. As shown in Fig. 2C, the Psk1 protein phosphorylated Rsp6 in vitro. However, the Rsp6 mutant that has two potential serine phosphorylation sites changed to alanine was not phosphorylated by Psk1. In addition, a kinase-dead mutant (Lys120Ala) of Psk1 did not phosphorylate Rps6. Taken together, these results suggest that Psk1 is the primary kinase responsible for Rps6 phosphorylation in response to nutrient conditions, although at this point, the possibility that a Psk1-associated kinase whose activity depends on Psk1 is involved in the phosphorylation is not excluded. In the following study, we focused on Psk1 to investigate its relationship to TORC1.
Fig. 2.
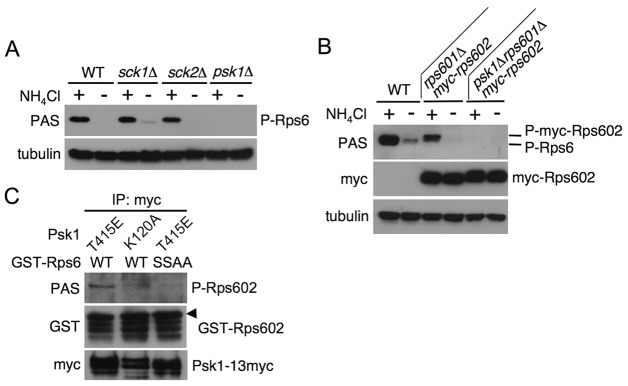
Identification of Psk1 as the major kinase to phosphorylate Rps6. (A–C) Proteins were probed with the indicated antibodies. (A) JUp1204 (WT), AN0170 (sck1Δ), AN0203 (sck2Δ) and AN0133 (psk1Δ) cells were cultured in EMM (+), washed, and then incubated in EMM-N for 30 minutes (−). (B) JUp1204 (WT), AN0129 (rps601Δmyc-rps602) and AN0168 (psk1Δrps601Δmyc-rps602) cells were washed and incubated in EMM with (+) or without (−) ammonium for 20 minutes. (C) AN0216 (T415E) and AN0219 (K120A) cells were grown in EMM and harvested. Myc-tagged Psk1 was immunoprecipitated and an in vitro Psk1 kinase assay was carried out using GST–Rps602 proteins as substrates, as described in Materials and Methods. Phosphorylation of Rps602 was evaluated by immunoblotting. The arrowhead denotes a band corresponding to phosphorylated Rps602 probed with the PAS antibody.
Phosphorylation and the activity of Psk1 are regulated by TORC1 in nutrient-dependent and rapamycin-sensitive manners
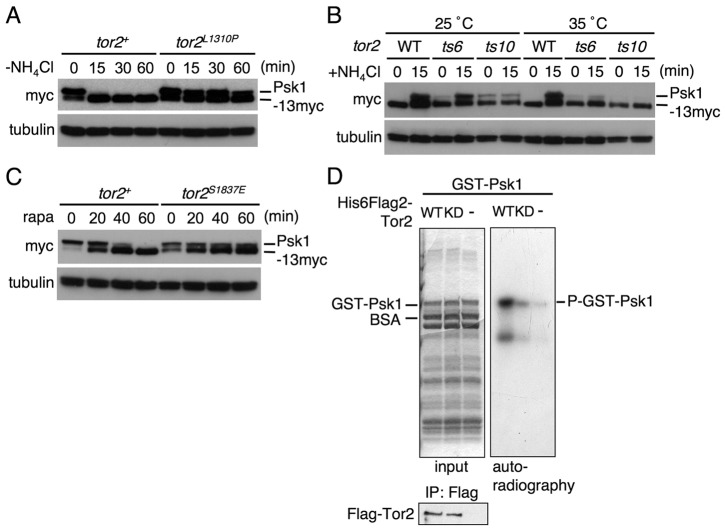
To determine whether TORC1 participates in the ammonium-dependent Psk1 phosphorylation, we constructed a series of strains in which Psk1-13 myc is expressed chromosomally in the tor2 mutant background that includes the activating, the temperature sensitive (ts), or the rapamycin-resistant mutation. Phosphorylation status of Psk1 in those tor2 mutants was examined as in Fig. 1B by assessing its mobility shift. As shown in Fig. 3A, Psk1 phosphorylation, identified as the upper band in tor2+ wild-type cells, was decreased rapidly after the shift to nitrogen starvation, whereas in the tor2L1310P active mutant (Urano et al., 2007), its phosphorylation was sustained for at least 60 minutes after the shift to the starvation. Conversely, downregulation of tor2 function using the two different tor2-ts mutations (ts6 and ts10) (Matsuo et al., 2007) at the non-permissive temperature (35°C) diminished induction of Psk1 phosphorylation by ammonium re-addition following nitrogen starvation (Fig. 3B). Rapamycin prevents Rps6 phosphorylation through inhibition of TORC1 (Nakashima et al., 2010), raising the possibility that this macrolide inhibits Psk1 phosphorylation. As expected, Psk1 phosphorylation in the wild-type cells was decreased by 60 minutes after addition of rapamycin to a level similar to that seen in nitrogen-starved cells, whereas phosphorylation of Psk1 was sustained for at least 60 minutes in the rapamycin-resistant tor2S1837E mutant (Nakashima et al., 2010) (Fig. 3C). These results suggest that TORC1 regulates Psk1 phosphorylation in response to nitrogen source availability and that rapamycin prevents its phosphorylation by inhibiting TORC1.
Fig. 3.
Psk1 is a downstream target of TORC1. (A–C) Proteins were probed with the indicated antibodies. (A) AN0182 (tor2+) and AN0184 (tor2L1310P) cells (0 minute) were washed and incubated in EMM-N for the indicated times. (B) AN0180 (WT), AN0217 (tor2-ts6) and AN0218 (tor2-ts10) cells were grown in EMM at 25°C. For treatment at the non-permissive temperature, cell culture was shifted to 35°C for 1 hour before cells were transferred to pre-warmed EMM-N. After incubation in EMM-N for 1 hour at either 25°C or 35°C, ammonium was added (0 minutes) to cells to 0.5% and incubated for 15 minutes. (C) AN0181 (tor2+) and AN0185 (tor2S1837E) cells were treated with 200 nM rapamycin for the indicated times. (D) In vitro kinase assay of Tor2 was carried out as described in Materials and Methods. The gel was dried and autoradiographed (the right panel). The left panel (input) shows the Coomassie Brilliant Blue staining. Flag-Tor2 represents the immunoblotting of the immunoprecipitated Flag-Tor2 protein.
The Tsc1–Tsc2 complex regulates Rps6 phosphorylation by negatively regulating TORC1. We further examined whether Psk1 acts as a downstream factor of the TSC1/2-TORC1 signaling pathway. Consistent with our previous results (Nakashima et al., 2010), Rps6 phosphorylation was maintained in both the tor2L1310P active and the tsc2 null mutants even after the shift to nitrogen starvation, whereas deletion of psk1+ in those mutants abolished phosphorylation of the ribosomal protein in the presence and absence of ammonium (supplementary material Fig. S3A). In contrast, the deletion of psk1+ did not suppress resistance of tsc2Δ cells to canavanine, a toxic arginine analog (van Slegtenhorst et al., 2004) (supplementary material Fig. S3B). These results suggest that Psk1 is at least one of the downstream factors of TSC1/2-TORC1 signaling.
Psk1 is phosphorylated by TORC1 in vitro
To examine whether Psk1 is directly phosphorylated by TORC1, we performed an in vitro kinase assay using Tor2 (TORC1) as the enzyme and recombinant Psk1 as the substrate. As shown in Fig. 3D, immunopurified wild-type Tor2 (WT) strongly phosphorylated Psk1, suggesting that Psk1 is a direct substrate of TORC1. Meanwhile, only a small amount of radioactive Psk1 was seen in the immunopurified fraction of the Tor2 kinase-dead mutant (KD; Fig. 3D). Because TORC1 is known to form homodimers in mammals and in budding yeast (Wullschleger et al., 2005; Takahara et al., 2006, Urano et al., 2007), exogenously expressed Tor2 kinase-dead mutant in wild-type cells probably forms a heterodimer with endogenous wild-type Tor2, thereby acquiring a weak activity to phosphorylate Psk1.
Deletion of pop3+ but not of toc1+ and tco89+, all of which are components of TORC1, increases the sensitivity to rapamycin of Psk1 regulation by TORC1
As described above, TORC1 consists of several proteins, such as Mip1, Pop3, Toc1 and Tco89, in addition to Tor2 (Alvarez and Moreno, 2006; Matsuo et al., 2007; Hayashi et al., 2007). Of these, Mip1 and Tor2 are essential for cell proliferation (Shinozaki-Yabana et al., 2000), implying that Mip1 plays a crucial role in TORC1 function similar to its counterparts, raptor and Kog1, in mammals and budding yeast, respectively. Pop3 is known to be included also in TORC2 and to be required for TORC2-mediated phosphorylation of Gad8 (Alvarez and Moreno, 2006; Hayashi et al., 2007; Ikeda et al., 2008). However, the role of Pop3 as well as Toc1 and Tco89 in TORC1 function remains unclear. To examine whether these three TORC1 components play a role in phosphorylating TORC1 downstream factors, we constructed deletion mutants of these genes. The pop3 disruptant is known to be viable (Kemp et al., 1997; Ochotorena et al., 2001). The toc1 and tco89 genes were also dispensable for cell proliferation (data not shown). Levels of Psk1-13myc protein in the pop3 disruptant were somewhat lower than those in the other strains, but no significant differences in the regulation of phosphorylation of Psk1 and Rps6 in response to ammonium conditions were observed in the gene disruptants of these TORC1 components compared with those in the wild type (Fig. 4A), suggesting that Pop3, Toc1 and Tco89 are dispensable for nutrient-dependent TORC1 activity at least for the modulation of Psk1 and Rps6 phosphorylation.
Fig. 4.
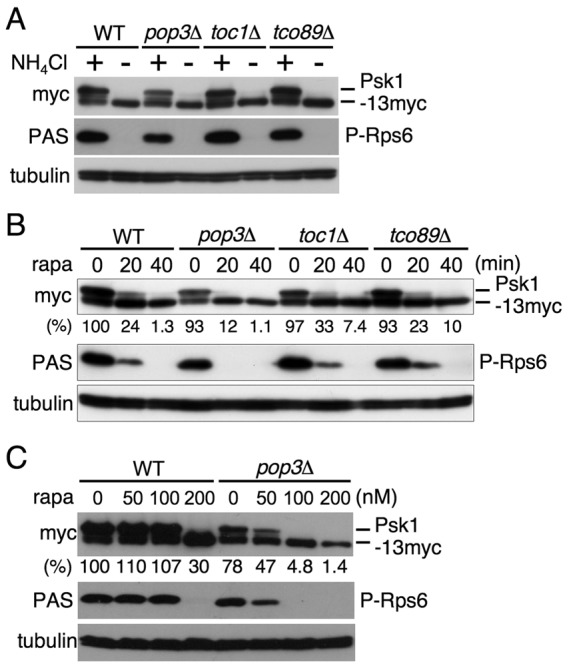
Effect of gene disruptions of the TORC1 components on phosphorylation of the TORC1 downstream factors. (A–C) Proteins were probed with the indicated antibodies. (A) AN0179 (WT), AN0233 (pop3Δ), AN0237 (toc1Δ) and AN0238 (tco89Δ) cells in EMM (+) were washed and cultured in EMM-N for 15 minutes (−). (B) Cells as described in A were treated with 150 nM rapamycin (rapa) for the indicated times. (C) AN0179 (WT) and AN0233 (pop3Δ) cells were cultured with the indicated concentrations of rapamycin for 1 hour. For B and C the relative reduction of Psk1 phosphorylation (%) was estimated by densitometry using ImageJ software and normalized to tubulin expression. The amount of Psk1 phosphorylation in wild type before addition of rapamycin (0 minutes) was set as 100.
In contrast, disruption of pop3+ but not of toc1+ and tco89+ led to increased sensitivity of the phosphorylation of TORC1 downstream factors to rapamycin inhibition, compared with those in the wild type in both time-course and dose experiments (Fig. 4B,C). This may suggest that Pop3 influences the interaction of Tor2 with the rapamycin–FKBP12 complex.
Predicted phosphorylation sites in the conserved regulatory motifs in Psk1 are important for its kinase activity
In the case of mammalian S6K, its activation requires phosphorylation of serine or threonine residues in three important regulatory motifs, namely the activation loop (T-loop) in the kinase domain, the hydrophobic motif (HM) in the C-terminal tail region, and the turn motif (TM) located in the linker region close to the HM. Of these, phosphorylation in the T-loop in S6K is catalyzed by phosphoinositide-dependent protein kinase 1 (PDK1), whereas phosphorylation in the HM and the TM are mediated by mTORC1. In particular, the phosphorylated HM site is important for phosphorylation and activation of S6K by PDK1 (Jacinto and Lorberg, 2008; Pearce et al., 2010). In fission yeast, these regulatory motifs are conserved in Psk1 as well as in the other AGC kinases described here (Fig. 5A; supplementary material Fig. S1).
Fig. 5.
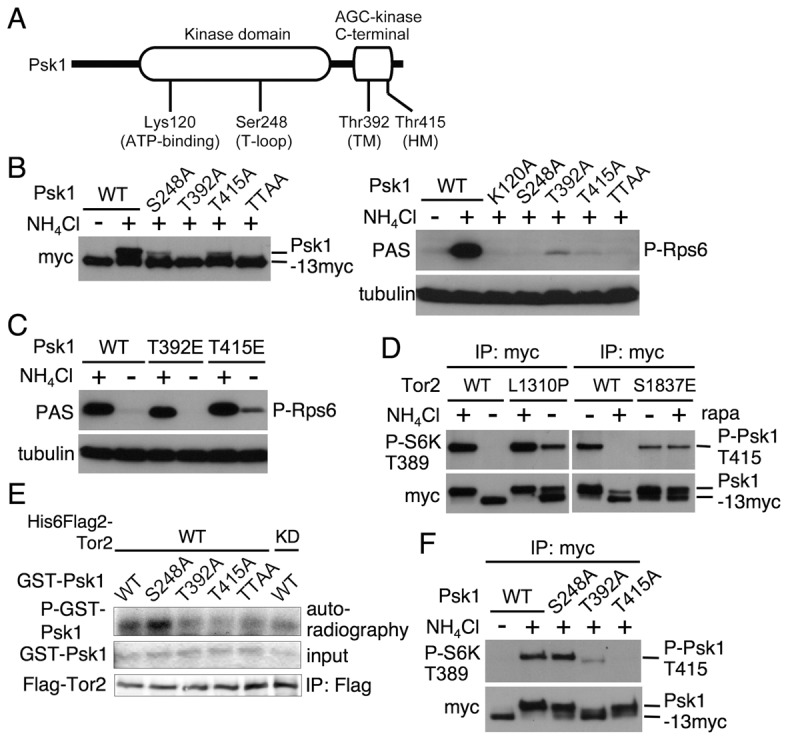
Role of the predicted phosphorylation sites of Psk1 in its kinase activity. (A) Schematic diagram of Psk1 that has a kinase domain in the middle region and an AGC-kinase C-terminal domain. The predicted amino acid sequence indicates Lys120 at the ATP-binding site, Ser248 in the activating loop (T-loop), Thr392 in the turn motif (TM) and Thr415 in the hydrophobic motif (HM), as shown in supplementary material Fig. S1. (B–F) Proteins were probed with the indicated antibodies. (B) AN0179 (WT), AN0219 (K120A), AN210 (S248A), AN211 (T392A) and AN0212 (T415A) cells were cultured in EMM (+). Another portion of the AN0179 culture was washed and cultured in EMM-N for 20 minutes. (C) AN0179 (WT), AN0213 (T392E) and AN0216 (T415E) cells in EMM (+) were washed and cultured in EMM-N for 20 minutes (−). (D) The left panels: AN0182 (WT) and AN0184 (L1310P) cells in EMM (+) were washed and cultured in EMM-N for 30 minutes. The right panels: AN0181 (WT) and AN0185 (S1837E) in EMM were treated with DMSO (−) or 200 nM rapamycin (+) and incubated for 1 hour. Psk1-13myc was immunoprecipitated and subjected to immunoblotting. (E) In vitro phosphorylations of GST–Psk1 proteins as indicated by Tor2 was carried out as described in Fig. 3D. (F) AN0179 (WT), AN210 (S248A), AN211 (T392A) and AN0212 (T415A) cells were cultured in EMM (+). Another portion of the AN0179 culture was washed and cultured in EMM-N for 20 minutes.
We therefore examined phosphorylation of these regulatory motifs and their role in Psk1 activity. To this end, we constructed a series of psk1 mutants in which the serine or threonine residues corresponding to the predicted phosphorylation sites in the three regulatory motifs were substituted with alanines, and then we checked phosphorylation levels and activities of these Psk1 mutants by examining mobility shifts and Rps6 phosphorylation, respectively. As shown in Fig. 5B, even under nitrogen-rich conditions, mutations of Ser248Ala in the T-loop and Thr415Ala in the HM significantly decreased phosphorylation of Psk1. Furthermore, a mutation of Thr392Ala in the TM as well as double mutations of Thr392Ala and Thr415Ala (TTAA) dramatically decreased phosphorylation of the kinase, which was strikingly similar to that observed in nitrogen-starved cells. As for Psk1 activity, Rps6 phosphorylation in both the single T-loop (Ser248Ala) and the double TM (Thr392Ala)/HM (Thr415Ala) mutants was severely impaired to a level similar to that seen in the Psk1 kinase-dead (Lys120Ala) mutant. However, phosphorylation of Rps6 in both the single mutants of the TM (Thr392Ala) and the HM (Thr415Ala) was detectable but was substantially lower than that in the wild type (the right panels in Fig. 5B). Conversely, a phospho-mimetic mutation of the HM (Thr415Glu) somewhat attenuated downregulation of Psk1 activity under nitrogen starvation, because Rps6 phosphorylation was detected under the starvation conditions (Fig. 5C). Taken together, these results suggest that, similar to S6K, Psk1 is phosphorylated in the three regulatory motifs depending on nitrogen source availability and that these phosphorylation events are important for its kinase activity.
We next examined whether TORC1 regulates phosphorylation of the HM (Thr415) in Psk1, depending on nutrient conditions. We first found that a phosphospecific antibody against the phosphorylated HM (Thr389) in mammalian S6K, an anti-phospho-p70 S6K (Thr389) antibody, recognized phosphorylation of wild-type Psk1 immunopurified from protein extract of fission yeast cells harvested under nitrogen-rich conditions, whereas this antibody did not detect non-phosphorylated HM mutant (Thr415Ala) immunopurified from cells grown under the same conditions (supplementary material Fig. S4). These results suggest the utility of this phosphospecific antibody to monitor Thr415 phosphorylation in the HM in Psk1. Additionally, Thr415 phosphorylation in Psk1 disappeared under nitrogen starvation (supplementary material Fig. S4), suggesting that Thr415 phosphorylation is regulated in response to nitrogen source availability. Using this antibody, we also found that downregulation of Thr415 phosphorylation under nitrogen starvation was markedly suppressed in the tor2L1310P active mutant (Fig. 5D) and that rapamycin blocked Thr415 phosphorylation, whereas this inhibition was completely suppressed in the tor2S1837E rapamycin-insensitive mutant (Fig. 5D). Furthermore, an in vitro kinase assay of immunopurified Tor2 revealed that phosphorylation levels of recombinant Psk1 proteins mutated in the TM (T392A), the HM (T415A), as well as in both these residues (TTAA) were significantly less than those of the wild-type and the T-loop (S248A) mutant proteins (Fig. 5E). These findings suggest that phosphorylation of the HM (Thr415) and possibly also of the TM (Thr392) in Psk1 are regulated by TORC1 in nutrient-dependent and rapamycin-sensitive manners.
We further examined the effect of phosphorylation in other regulatory motifs on the modification in the HM in Psk1. As shown in Fig. 5F, Thr415 phosphorylation in Psk1 mutated in the TM (Thr392Ala) was markedly decreased even under nutrient-rich medium, whereas its phosphorylation in the T-loop mutant (Ser248Ala) was comparable to that in the wild type, suggesting that phosphorylation of the TM (Thr392) but not of the T-loop (Ser248), is important for the HM (Thr415) phosphorylation. We also noticed that mutation in the TM (Thr392Ala) resulted in loss of its mobility shift, representing the lowest phosphorylation level as observed in nitrogen starvation (Fig. 5B). Therefore, phosphorylation in the TM may also affect the modification of the T-loop in addition to that of the HM.
Ksg1, a kinase homologous to PDK1, is required for phosphorylation and activity of Psk1
In fission yeast, Ksg1 is known as a homolog of PDK1 (Niederberger and Schweingruber, 1999). We therefore examined the role of Ksg1 in phosphorylation and activity of Psk1 using the temperature-sensitive (ts) mutants of two distinct ksg1 alleles (ts208 and ts358). As shown in Fig. 6A, induction of Psk1 phosphorylation by re-adding ammonium following nitrogen starvation was markedly attenuated in both the ksg1-ts mutants, not only at the non-permissive temperature (35°C) but also at the permissive temperature (25°C). Correspondingly, Psk1 activity to catalyze Rps6 phosphorylation after re-feeding ammonium was abolished in both ksg1 mutants at both temperatures (Fig. 6A). We next examined whether Ksg1 phosphorylates Psk1 in vitro and found that GST–Ksg1 purified from bacteria was able to phosphorylate recombinant wild-type Psk1 protein as well as its mutant that harbors T/A mutations in both the TM and the HM (TTAA), but it could not phosphorylate the T-loop mutant (S248A; Fig. 6B). These results suggest that Ksg1 is important for phosphorylation, particularly in the T-loop, and for activity of Psk1.
Fig. 6.
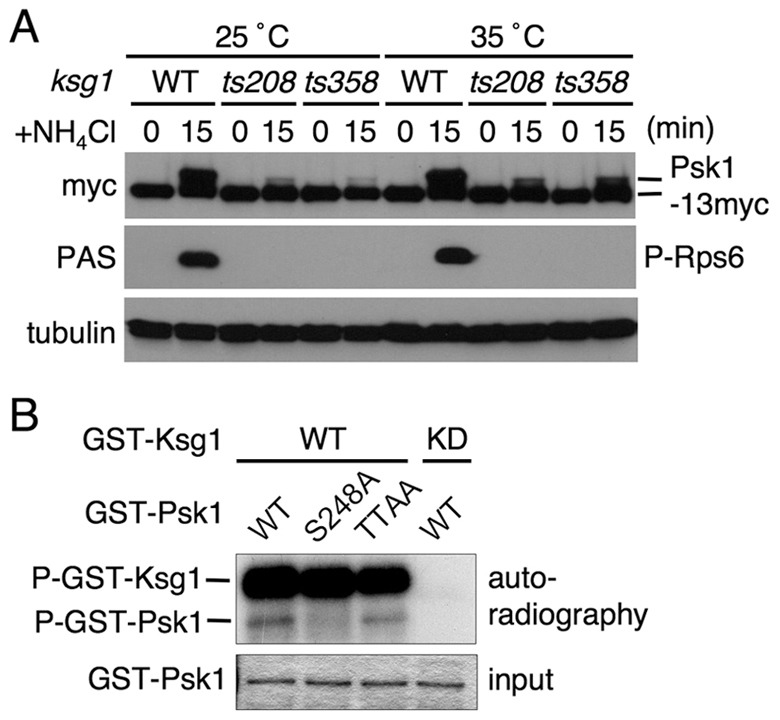
Ksg1, a PDK1 homolog, is required for phosphorylation and activity of Psk1. (A) Thermal treatment of AN0180 (WT), AN0243 (ksg1-ts208) and AN245 (ksg1-ts358) cells was carried out as described in Fig. 3B. After incubation of cells in EMM-N for 1 hour at 25°C or 35°C (0 minute), ammonium was added to 0.5% and the cells were incubated for a further 15 minutes. Proteins were probed with the indicated antibodies. (B) In vitro phosphorylation of GST–Psk1 proteins, as indicated, by GST–Ksg1 was carried out as described in the Materials and Methods. The gel was dried and autoradiographed (upper panel). The lower panel (input) shows the Coomassie Brilliant Blue staining.
The TORC1-dependent phosphorylation of Psk1 is regulated by glucose and glutamine as well as ammonium
We further examined the effect of other nutrients, such as glucose and amino acids, on the regulation of Psk1 phosphorylation and its activity, because we previously found that TORC1-dependent Rps6 phosphorylation is downregulated by glucose starvation (Nakashima et al., 2010). Psk1 phosphorylation was substantially decreased when cells were grown in a medium containing either a low concentration of glucose (glucose starvation) or 2-deoxy-glucose (2-DG), which is a transportable, but non-metabolizable glucose analog that inhibits glucose metabolism (Fig. 7A), suggesting that the TORC1-dependent Psk1 regulation responds to glucose availability. It is of interest that the phosphorylation of Psk1 and Rps6 under glucose starvation, but not under nitrogen starvation, also increased by 60 minutes (Fig. 7B).
Fig. 7.
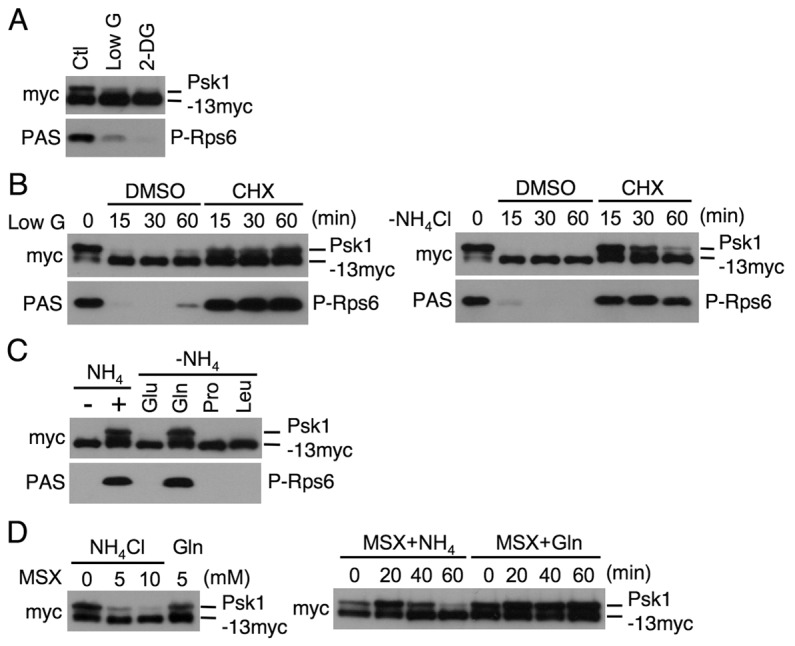
The TORC1 signaling responds to the availability of glucose and glutamine. (A–D) Proteins were probed with the indicated antibodies. (A) AN0179 cells were washed and incubated in EMM, EMM low glucose (Low G), or EMM+2-DG for 15 minutes. (B) AN0180 (0 minute) cells were washed and incubated with either EMM low glucose (LowG) or EMM-N with DMSO or cycloheximide (CHX; 50 µg/ml) for the indicated times. (C) After incubation in EMM-N for 1 hour (−), AN0179 cells were incubated in EMM (+) or EMM containing either 20 mM glutamic acid (Glu), glutamine (Gln), proline (Pro) or leucine (Leu) instead of ammonium for 15 minutes. (D) Left panel: AN0179 cells were incubated in EMM in the absence or presence of 5 or 10 mM L-methionine sulfoximine (MSX) for 30 minutes or in EMM plus 20 mM glutamine instead of ammonium (Gln) with 5 mM MSX. Right panel: AN0179 cells were transferred to EMM (+NH4) or EMM plus 20 mM glutamine (Gln) with 5 mM MSX and incubated for the indicated times.
It has been shown that TORC1-dependent phosphorylation of substrates, such as S6K and Sch9 in mammalian cells and in budding yeast, respectively, is sustained when cells are treated with cycloheximide, which blocks protein synthesis, even under nutrient starvation (Iiboshi et al., 1999; Beugnet et al., 2003; Urban et al., 2007). As expected, cycloheximide significantly suppressed dephosphorylation of Psk1 and Rps6 under glucose or nitrogen starvation (Fig. 7B), suggesting that similar to other eukaryotes, TORC1 activity in fission yeast is also maintained by the addition of cycloheximide under nutrient starvation.
It is well known that amino acids, especially leucine, are important cues in the cellular environment for the regulation of TORC1 activity (Avruch et al., 2009). We therefore examined the effect of short-term incubation with several amino acids on TORC1 activity by measuring phosphorylation of the TORC1 downstream factors. Cells were stimulated with glutamic acid, glutamine, proline or leucine instead of ammonium for 15 minutes following nitrogen starvation (Fig. 7C). Glutamic acid and glutamine are good nitrogen sources, whereas proline is a poor one for cell proliferation. Of these, glutamine was sufficient to induce phosphorylation of both Psk1 and Rps6 to a level comparable to that seen with ammonium stimulation, whereas the other amino acids showed little effect on phosphorylation (Fig. 7C). L-Methionine sulfoximine (MSX) is a glutamine synthetase inhibitor that provokes glutamine depletion in cells resulting in a decrease in TORC1 activity in budding yeast (Crespo et al., 2002). Similar to the previous findings observed in budding yeast, the treatment with L-methionine sulfoximine led to dose- and time-dependent reductions in Psk1 phosphorylation in the medium containing ammonium, whereas addition of glutamine instead of ammonium suppressed these inhibitions (Fig. 7D). These results suggest that extracellular and intracellular glutamine effectively upregulates TORC1 signaling.
TORC1 catalyzes nutrient-dependent phosphorylation of Sck1 and Sck2
As shown in Fig. 1, phosphorylation levels of Sck1 and Sck2 were altered depending on nitrogen conditions. To investigate the possibility that Sck1 and Sck2 are downstream effectors of TORC1, we examined in vitro kinase assays of Tor2 using recombinant Sck1 and Sck2 proteins as substrates. Similar to Psk1 (Fig. 3D), Sck1 and Sck2 were also phosphorylated by TORC1 in vitro (Fig. 8A). The phosphorylation was much less when a kinase-dead mutant of Tor2 was used. Further support for TORC1 being involved in the phosphorylation of Sck1 is shown in Fig. 8B. In this experiment, we examined Sck1 phosphorylation using tor2 mutants. While the intensity of the top band decreased as a result of starvation in the wild-type cells, this band remained in the tor2 active mutant sample (left panels). In addition, phosphorylation of Sck1 was inhibited by rapamycin treatment in the wild type, but not in the tor2 rapamycin-resistant mutant (Fig. 8B, right panels). The level of modification before rapamycin addition in the tor2 mutant was lower than that in the wild type (right panels). Similar low activity of the rapamycin-resistant mutant was observed for Psk1 (Fig. 3C, Fig. 5D). A preliminary study with Sck2 also suggested that Sck2 phosphorylation is decreased in the wild-type cells after shifting to nitrogen starvation and that this decrease is partially suppressed in the tor2 active mutant (unpublished results).
Fig. 8.
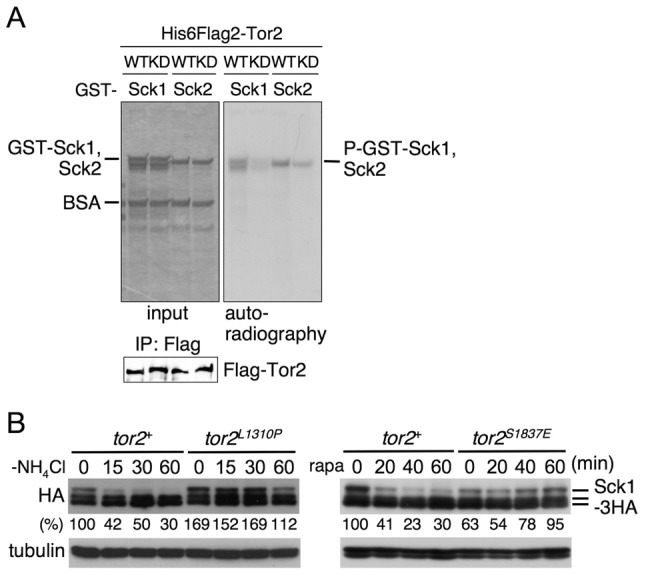
Sck1 and Sck2 might also be partly involved in TORC1 signaling. (A) In vitro phosphorylation of Sck1 and Sck2 by Tor2 carried out as described in Fig. 3D. (B) AN0163 (tor2+), AN0164 (tor2+), AN0166 (tor2L1310P), AN0167 (tor2S1837E) cells were nitrogen-starved and treated with rapamycin as described in Fig. 3A,C. Proteins were probed with the indicated antibodies. The relative reduction of Sck1 phosphorylation (%) was estimated by densitometry using ImageJ software, and normalized to tubulin expression. The amount of Sck1 phosphorylation in wild type before nitrogen starvation or treatment with rapamycin (0 minutes) was set as 100 (B).
It has been shown that sck1+ and sck2+ have a redundant function with pka1+, which encodes a catalytic subunit of cyclic AMP-dependent protein kinase (Jin et al., 1995; Fujita and Yamamoto, 1998). Therefore, we examined whether psk1+ has a redundant function with pka1+. Unlike overexpression of sck1+ or sck2+, exogenously expressed psk1+ from the thiamine-repressible nmt81 promoter (Basi et al., 1993) failed to suppress slow growth (30°C) and cold-temperature sensitivity (25°C) in pka1Δ on minimal medium (supplementary material Fig. S5). On synthetic defined (SD) medium, on which plasmid gene expression was repressed, growth of pka1Δ cells carrying the plasmid encompassing the kinase gene was comparable to that of the cells having an empty vector (supplementary material Fig. S5). Taken together, these results show that psk1+ has no redundant function with pka1+.
Discussion
In this study, we present evidence that an AGC kinase Psk1 functions as a downstream effector of TORC1 in fission yeast. This microbe has several AGC kinases homologous to human S6K1, such as Psk1, Sck1, Sck2 and Gad8. Of these, Psk1 is the closest S6K1 homolog and phosphorylated Rps6 in vivo and in vitro (Fig. 1A, Fig. 2A,C). Furthermore, Psk1 itself was highly phosphorylated under nitrogen-rich conditions, and its phosphorylation was at least partially catalyzed by TORC1 in vivo and in vitro (Figs 3, 5). Rapamycin inhibited Psk1 phosphorylation as well as Rps6 phosphorylation by blocking TORC1 activity (Fig. 3C). Therefore, Psk1 acts like S6K to mediate the nutrient-dependent TORC1 signaling to its substrate(s).
We further showed that the TORC1 components Toc1 and Tco89, as well as Pop3, are dispensable for cell proliferation and have little effect on the regulation of Psk1 by TORC1, at least in response to nutrient conditions (Fig. 4). In mouse studies as well as in studies using mouse embryonic fibroblasts lacking mLST8 (a homolog of Pop3), it has been shown that mLST8 is necessary for mTORC2 activity but not for that of mTORC1 (Guertin et al., 2006). Similar results were obtained in fission yeast. In contrast, there is no homolog of Tco89 in vertebrates. Unlike Tco89 in fission yeast that is dispensable for Psk1 phosphorylation, this homolog in budding yeast is required for TORC1 activity to phosphorylate Sch9, a counterpart of S6K (Binda et al., 2009). There are no Toc1 homologs in mammals and in budding yeast (Hayashi et al., 2007). Therefore, the function of each TORC1 component may have diverged during evolution.
Psk1 contains three regulatory motifs that are conserved among the AGC kinase families: the activation loop (T-loop), the hydrophobic motif (HM) and the turn motif (TM) (supplementary material Fig. S1; Fig. 5A). Site-directed mutagenesis analyses in the regulatory motifs revealed that the conserved serine or threonine residue in these motifs was phosphorylated and that these phosphorylations are required for Psk1 activity to phosphorylate Rps6 (Fig. 5B). We also demonstrate by in vitro kinase assay of Tor2 and immunoblotting using an anti-phospho-S6K (Thr389) antibody that TORC1 catalyzes and regulates Thr415 phosphorylation in the HM of Psk1 in nitrogen-dependent and rapamycin-sensitive manners (Fig. 5D,E). Furthermore, TORC1 appears to also regulate phosphorylation in the TM, because mutation of the TM (T392A) in the recombinant Psk1 protein significantly decreased its phosphorylation by TORC1 in vitro (Fig. 5E). Additionally, phosphorylation status of the TM mutant in cells, which was detected by loss of its mobility shift, was very similar to that of the wild-type Psk1 under nitrogen starvation or when treated with rapamycin, where TORC1 is inactive (Fig. 5B). These modifications of Psk1 controlled by TORC1 are consistent with those in S6K in other eukaryotes (Jacinto and Lorberg, 2008).
We also found that Ksg1, a protein kinase homologous to PDK1, is engaged in phosphorylation in the T-loop (Ser248) of Psk1 and is required for the activity of Psk1 (Fig. 6), similar to the T-loop phosphorylation in S6K by PDK1 in mammalian cells (Jacinto and Lorberg, 2008). We have previously demonstrated that phosphorylation and function of Gad8, which exhibits the highest homology with AKT, are regulated by TORC2 and Ksg1 (Matsuo et al., 2003). Therefore, like other eukaryotes, fission yeast possesses the signaling pathways involving two different AGC kinases that are separately regulated by the two TOR complexes and the PDK1 counterpart.
Phosphorylation of mammalian S6K occur in a stepwise manner; namely, phosphorylation of the TM in S6K is required for phosphorylation in the HM and then the modified HM participates in the phosphorylation of the T-loop by PDK1 (Jacinto and Lorberg, 2008; Pearce et al., 2010). In fission yeast, it seems likely that phosphorylation of the three regulatory motifs in Psk1 are also closely related to one another and arise in a stepwise manner because mutation of Thr392Ala in the TM caused a marked decrease in Thr415 phosphorylation in the HM (Fig. 5F). In addition, phosphorylation in the T-loop in Psk1 may require phosphorylation of HM by TORC1, because phosphorylation status of the T-loop mutant (Ser248Ala) resembled that of the HM mutant (Thr415Ala; Fig. 5B).
Similar to phosphorylation of Rps6 as previously described (Nakashima et al., 2010), Psk1 phosphorylation was sensitive to glucose availability (Fig. 7A). However, cycloheximide attenuated dephosphorylation of Psk1 and Rps6 by nitrogen or glucose starvation (Fig. 7B). Similarly, in mammalian cells, cycloheximide as well as other protein synthesis inhibitors, puromycin and anisomycin, attenuates the decrease in mTORC1 activity to phosphorylate its substrates (Iiboshi et al., 1999; Beugnet et al., 2003). Those translation inhibitors are likely to suppress consumption of amino acids by protein synthesis, thereby maintaining the pools of intracellular amino acids. In budding yeast, cycloheximide causes hyperphosphorylation of Sch9, which is catalyzed by TORC1. In this case, elevated intracellular amino acid pools are observed under amino acid depletion (Urban et al., 2007; Binda et al., 2009). We further showed that TORC1-dependent Psk1 phosphorylation is controlled by extracellular and intracellular glutamine (Fig. 7C,D). Similarly, a subset of TORC1 function is regulated in response to intracellular glutamine levels in budding yeast (Crespo et al., 2002). Therefore, TORC1, at least in part, may react to intracellular storage of amino acids directly or indirectly. It has recently been reported that in mammalian systems that when amino acids are made available by Rag G-proteins that interact with raptor (which is a component of mTORC1) mTORC1 is translocated to the surface of lysosomes where the kinase is activated by Rheb G-protein. This translocation of mTORC1 appears to be necessary for its activation by amino acids (Sancak et al., 2008). In fission yeast, there are two homologous proteins of the Rag G-proteins: Gtr1 and Gtr2. Recently, it has been reported that these Rag proteins appear to participate also in regulation of TORC1 by amino acids (Valbuena et al., 2012).
We have shown here that TORC1 controls phosphorylation and function of Psk1 and possibly of Sck1 and Sck2, and that Psk1 is the major kinase for phosphorylation of the fission yeast S6 proteins but Sck1 and Sck2 have no effect on the phosphorylation. By contrast, it has been shown that the loss of Tor2 (TORC1) function leads to entry into sexual differentiation (Alvarez and Moreno, 2006; Matsuo et al., 2007; Uritani et al., 2006). In a preliminary study, we detected increased mating efficiency by disruption of psk1+, although this was seen in the background of disruption of sck1+ and sck2+ (data not shown). Further work is needed to clarify how these AGC kinases function downstream of TORC1 to affect various physiological aspects of fission yeast cells.
In the current study, the direct substrate of the fission yeast TORC1 was identified. We also showed that the phosphorylations catalyzed by TORC1 could be easily monitored, providing a convenient assay to examine the activation of TORC1 signaling. Employing these tools should be helpful to investigate molecules in nutrient–TORC1 signaling in fission yeast.
Materials and Methods
Yeast strains, growth media and general methods
Fission yeast strains used in this study are listed in supplementary material Table S1. Cells were grown exponentially at 30°C, except in the experiments using temperature-sensitive mutants. Media used were: yeast extract with supplements (YES) medium; Edinburgh minimal medium (EMM), supplemented with 200 mg/l adenine when necessary, which contained 2% glucose as a carbon source and 0.5% ammonium chloride as a nitrogen source (Moreno et al., 1991); minimal medium (MM); or synthetic defined (SD) medium (Watanabe et al., 1988). EMM-N, a nitrogen-free version, and EMM low glucose (Low G: 3% glycerol and 0.1% glucose), a glucose depletion version (Kohda et al., 2007), were employed as starvation media. EMM+2-DG contained 2% 2-deoxyglucose (Sigma) instead of glucose. EMM+glutamic acid, EMM+glutamine, EMM+proline or EMM+leucine, contained 20 mM of each amino acid instead of ammonium, and were used within a week after preparation. General and molecular genetic techniques followed standard protocols (Moreno et al., 1991).
Antibodies and reagents
Both polyclonal antibodies recognizing phospho-Akt substrates (PAS) and phospho-p70 S6K (Thr389) were purchased from Cell Signaling Technology. Anti-GST polyclonal, anti-FLAG (M2) and anti-α-tubulin (B5-1-2) antibodies, and L-methionine sulfoximine (MSX) were purchased from Sigma. Anti-myc (9E10) antibody was purchased from Santa Cruz Biotechnology and Covance. Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG antibody and protein-G–Sepharose were purchased from GE Healthcare Bio-Sciences. HRP-conjugated donkey anti-rabbit IgG antibody and rapamycin were purchased from Pierce and Calbiochem, respectively.
Construction of modified strains and gene expression plasmids
Direct chromosomal integration methods were used to integrate either the 3xHA-hphMX or 13xmyc-hphMX cassette before the terminal codons of genes encoding the AGC kinases, and for gene disruptions by replacing the individual open reading frames (ORF) with either the kanMX or hphMX cassette (Bähler et al., 1998; Sato et al., 2005). To construct the psk1 mutation alleles, a DNA fragment containing psk1+-13xmyc-hphMX and its 5′ and 3′ UTR regions was amplified by PCR employing the genomic DNA of the psk1+-13xmyc-hphMX strain as a template, and then cloned into pCR2.1. The regulatory motif sites in psk1+ were replaced with alanine (K120A, S248A, T392A and T415A) or with glutamic acid (T392E and T415E) by site-directed mutagenesis using pCR2.1-psk1+-13xmyc-hphMX, and the sequence was confirmed. The substituted DNA fragments were amplified by PCR and integrated by homologous recombination.
To construct expressing plasmids of psk1+, sck1+ and sck2+, ORFs of these kinases were amplified by PCR and cloned into pREP81-HA, which possesses the thiamine repressible nmt1 promoter (Basi et al., 1993). To construct plasmids expressing GST–rps602+ and GST–rps602SS235236AA, a DNA fragment containing rps602+ or rps602SS235236AA was amplified by PCR and cloned into pGEX. ORFs containing psk1+, the psk1 mutants, sck1+ and sck2+ were amplified by PCR and cloned into pGEX-KG.
Protein preparation, phosphatase treatment and immunoblotting
Cultures were mixed with trichloroacetic acid (TCA; final concentration 6%) and put on ice for at least 5 minutes. After centrifugation, cell pellets were washed twice with cold ethanol, and dried. Cells were disrupted in buffer A [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 10% glycerol, 0.2% NP-40, 20 mM β-glycerophosphate, 0.1 mM Na3VO4, 10 mM p-nitrophenyl phosphate (p-NPP), 10 mM NaF, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF) and protease inhibitor cocktail (Complete EDTA-free; Roche)] with glass beads. Whole-cell extracts were mixed with 3× SDS sample buffer, and boiled for 5 minutes.
For phosphatase treatment, TCA-quenched cells were broken in urea buffer [50 mM Tris-HCl (pH 7.5), 6 M urea, 5 mM EDTA, 1% SDS, 10 mM NaN3, 10 mM sodium pyrophosphate, 10 mM β-glycerophosphate, 0.1 mM Na3VO4, 10 mM p-NPP, 10 mM NaF, 1 mM DTT, 1 mM PMSF and protease inhibitor cocktail] with glass beads by vortexing three times for 5 minutes each at RT and incubation for 10 minutes at 65°C. After centrifugation, supernatants were diluted 20-fold into a reaction buffer with 2 mM MnCl2 and then incubated with λ-phosphatase (400 IU; New England Biolabs) at 30°C for 40 minutes with or without the phosphatase inhibitors (50 mM EDTA, 10 mM Na3VO4 and 50 mM NaF).
To assess phosphorylation at Thr415 in Psk1 with the anti-phospho-p70 S6 kinase (Thr389) antibody, cells in 40 ml cultures were quenched with TCA and were broken in 140 µl of urea buffer with glass beads as described above. Cell extracts were diluted gradually with 1100 µl of buffer A and centrifuged at room temperature. Supernatants were incubated with anti-myc antibody and protein-G–Sepharose at 4°C for 2.5 hours. Immunoprecipitates were washed three times with buffer A without protease inhibitors.
Cell extracts or immunoprecipitates were separated by SDS-PAGE and then immunoblotted with the primary antibodies. After incubation with HRP-conjugated secondary antibodies, proteins were detected using the ECL plus detection system (GE Healthcare Bio-Sciences).
In vitro kinase assay
GST-fused proteins were expressed in the Escherichia coli BL21 (DE3) strain and purified. Cells expressing psk1T415E-13xmyc or psk1K120A-13xmyc were broken with glass beads in buffer A with 20 µg/ml leupeptin. After centrifugation at 10,000 g for 15 minutes and subsequently at 14,000 g for 20 minutes at 4°C, supernatants were incubated with anti-myc antibody and protein-G–Sepharose at 4°C for 2.5 hours, and immunoprecipitates were washed twice with buffer A without protease inhibitors and then washed twice with washing buffer [50 mM MOPS-KOH (pH 7.2), 10 mM β-glycerophosphate, 0.1 mM Na3VO4, 10 mM MgCl2]. The Psk1 kinase assay was carried out as follows: the immunoprecipitates were incubated with 1.5 µg of GST–Rps602 wild-type or GST–Rps602SS235236AA as a substrate in buffer Kpsk1 [50 mM MOPS-KOH (pH 7.2), 10 mM β-glycerophosphate, 0.1 mM Na3VO4, 10 mM MgCl2, 100 µM ATP] for 1 hour at 30°C.
For the Tor2 kinase assay in vitro, cells were transformed with either pREP41-His6Flag2-tor2+ (Matsuo et al., 2007) or pREP41-His6Flag2-tor2KD (kinase dead; D2140A). Tor2 or Tor2KD was immunoprecipitated as described previously (Matsuo et al., 2007). After washing three times with buffer B [50 mM Tris-HCl (pH 7.6), 150 mM KCl, 5 mM EDTA, 1 mM DTT, 10% glycerol, 0.2% NP-40, 20 mM β-glycerophosphate, 0.1 mM Na3VO4, 15 mM p-NPP and 1 mM PMSF], immunoprecipitates were washed twice with Ktor buffer [20 mM HEPES-KOH (pH 7.5), 1 mM DTT, 20 mM β-glycerophosphate, 0.1 mM Na3VO4, 15 mM p-NPP, 10 mM MnCl2]. Then, an in vitro kinase assay was carried out as follows: the immunoprecipitates were incubated with 2 µg of GST–Psk1, GST–Psk1 mutants, GST–Sck1, or GST–Sck2 as substrates and 2 µg BSA in Ktor buffer. The reaction was initiated by adding 25 µM cold ATP and 5 µCi [γ-32P]ATP and terminated by adding SDS sample buffer after incubation for 35 min at 32°C.
In vitro kinase assay of Ksg1 using GST–Psk1 wild type and its mutants as substrates was carried out essentially as described previously (Matsuo et al., 2003).
Supplementary Material
Acknowledgments
We thank other members in our laboratories and Dr Ushio Kikkawa at Kobe University for helpful discussions and for technical assistance, and Drs Susan Forsburg, M. Ernst Schweingruber and Takashi Toda for providing yeast strains and plasmids.
Footnotes
Funding
This work was supported by the National Institutes of Health [grant number CA41996 to F.T.]; Grants-in-Aid for Scientific Research [grant numbers 23570223 to A.Y., and 21227007 to M.Y.] and Grants-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science [grant number 23770229 to A.N.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.111146/-/DC1
References
- Alvarez B., Moreno S. (2006). Fission yeast Tor2 promotes cell growth and represses cell differentiation. J. Cell Sci. 119, 4475–4485 10.1242/jcs.03241 [DOI] [PubMed] [Google Scholar]
- Aspuria P. J., Tamanoi F. (2008). The Tsc/Rheb signaling pathway controls basic amino acid uptake via the Cat1 permease in fission yeast. Mol. Genet. Genomics 279, 441–450 10.1007/s00438-008-0320-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avruch J., Long X., Ortiz–Vega S., Rapley J., Papageorgiou A., Dai N. (2009). Amino acid regulation of TOR complex 1. Am. J. Physiol. Endocrinol. Metab. 296, E592–E602 10.1152/ajpendo.90645.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- Basi G., Schmid E., Maundrell K. (1993). TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123, 131–136 10.1016/0378-1119(93)90552-E [DOI] [PubMed] [Google Scholar]
- Beugnet A., Tee A. R., Taylor P. M., Proud C. G. (2003). Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem. J. 372, 555–566 10.1042/BJ20021266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binda M., Péli–Gulli M. P., Bonfils G., Panchaud N., Urban J., Sturgill T. W., Loewith R., De Virgilio C. (2009). The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 35, 563–573 10.1016/j.molcel.2009.06.033 [DOI] [PubMed] [Google Scholar]
- Crespo J. L., Powers T., Fowler B., Hall M. N. (2002). The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA 99, 6784–6789 10.1073/pnas.102687599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Yamamoto M. (1998). S. pombe sck2+, a second homologue of S. cerevisiae SCH9 in fission yeast, encodes a putative protein kinase closely related to PKA in function. Curr. Genet. 33, 248–254 10.1007/s002940050333 [DOI] [PubMed] [Google Scholar]
- Guertin D. A., Stevens D. M., Thoreen C. C., Burds A. A., Kalaany N. Y., Moffat J., Brown M., Fitzgerald K. J., Sabatini D. M. (2006). Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell 11, 859–871 10.1016/j.devcel.2006.10.007 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Hatanaka M., Nagao K., Nakaseko Y., Kanoh J., Kokubu A., Ebe M., Yanagida M. (2007). Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes Cells 12, 1357–1370 10.1111/j.1365-2443.2007.01141.x [DOI] [PubMed] [Google Scholar]
- Iiboshi Y., Papst P. J., Kawasome H., Hosoi H., Abraham R. T., Houghton P. J., Terada N. (1999). Amino acid-dependent control of p70(s6k). Involvement of tRNA aminoacylation in the regulation. J. Biol. Chem. 274, 1092–1099 10.1074/jbc.274.2.1092 [DOI] [PubMed] [Google Scholar]
- Ikeda K., Morigasaki S., Tatebe H., Tamanoi F., Shiozaki K. (2008). Fission yeast TOR complex 2 activates the AGC-family Gad8 kinase essential for stress resistance and cell cycle control. Cell Cycle 7, 358–364 10.4161/cc.7.3.5245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E., Lorberg A. (2008). TOR regulation of AGC kinases in yeast and mammals. Biochem. J. 410, 19–37 10.1042/BJ20071518 [DOI] [PubMed] [Google Scholar]
- Jin M., Fujita M., Culley B. M., Apolinario E., Yamamoto M., Maundrell K., Hoffman C. S. (1995). sck1, a high copy number suppressor of defects in the cAMP-dependent protein kinase pathway in fission yeast, encodes a protein homologous to the Saccharomyces cerevisiae SCH9 kinase. Genetics 140, 457–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M., Nakashima A., Ueno M., Ushimaru T., Aiba K., Doi H., Uritani M. (2001). Fission yeast tor1 functions in response to various stresses including nitrogen starvation, high osmolarity, and high temperature. Curr. Genet. 39, 166–174 10.1007/s002940100198 [DOI] [PubMed] [Google Scholar]
- Kemp J. T., Balasubramanian M. K., Gould K. L. (1997). A wat1 mutant of fission yeast is defective in cell morphology. Mol. Gen. Genet. 254, 127–138 10.1007/s004380050400 [DOI] [PubMed] [Google Scholar]
- Kohda T. A., Tanaka K., Konomi M., Sato M., Osumi M., Yamamoto M. (2007). Fission yeast autophagy induced by nitrogen starvation generates a nitrogen source that drives adaptation processes. Genes Cells 12, 155–170 10.1111/j.1365-2443.2007.01041.x [DOI] [PubMed] [Google Scholar]
- Laplante M., Sabatini D. M. (2009). mTOR signaling at a glance. J. Cell Sci. 122, 3589–3594 10.1242/jcs.051011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S., Bandyopadhyay A., Kwiatkowski D. J., Maitra U., Matsumoto T. (2002). Role of the Tsc1-Tsc2 complex in signaling and transport across the cell membrane in the fission yeast Schizosaccharomyces pombe. Genetics 161, 1053–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T., Kubo Y., Watanabe Y., Yamamoto M. (2003). Schizosaccharomyces pombe AGC family kinase Gad8p forms a conserved signaling module with TOR and PDK1-like kinases. EMBO J 22, 3073–3083 10.1093/emboj/cdg298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T., Otsubo Y., Urano J., Tamanoi F., Yamamoto M. (2007). Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol. Cell. Biol. 27, 3154–3164 10.1128/MCB.01039-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Mukai H., Miyahara M., Takanaga H., Kitagawa M., Shibata H., Shimakawa M., Ono Y. (1995). Identification of Schizosaccharomyces pombe gene psk1+, encoding a novel putative serine/threonine protein kinase, whose mutation conferred resistance to phenylarsine oxide. Gene 166, 155–159 10.1016/0378-1119(95)00553-1 [DOI] [PubMed] [Google Scholar]
- Murai T., Nakase Y., Fukuda K., Chikashige Y., Tsutsumi C., Hiraoka Y., Matsumoto T. (2009). Distinctive responses to nitrogen starvation in the dominant active mutants of the fission yeast Rheb GTPase. Genetics 183, 517–527 10.1534/genetics.109.105379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase Y., Fukuda K., Chikashige Y., Tsutsumi C., Morita D., Kawamoto S., Ohnuki M., Hiraoka Y., Matsumoto T. (2006). A defect in protein farnesylation suppresses a loss of Schizosaccharomyces pombe tsc2+, a homolog of the human gene predisposing to tuberous sclerosis complex. Genetics 173, 569–578 10.1534/genetics.106.056895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A., Sato T., Tamanoi F. (2010). Fission yeast TORC1 regulates phosphorylation of ribosomal S6 proteins in response to nutrients and its activity is inhibited by rapamycin. J. Cell Sci. 123, 777–786 10.1242/jcs.060319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederberger C., Schweingruber M. E. (1999). A Schizosaccharomyces pombe gene, ksg1, that shows structural homology to the human phosphoinositide-dependent protein kinase PDK1, is essential for growth, mating and sporulation. Mol. Gen. Genet. 261, 177–183 10.1007/s004380050955 [DOI] [PubMed] [Google Scholar]
- Ochotorena I. L., Hirata D., Kominami K., Potashkin J., Sahin F., Wentz–Hunter K., Gould K. L., Sato K., Yoshida Y., Vardy L.et al. (2001). Conserved Wat1/Pop3 WD-repeat protein of fission yeast secures genome stability through microtubule integrity and may be involved in mRNA maturation. J. Cell Sci. 114, 2911–2920 [DOI] [PubMed] [Google Scholar]
- Pearce L. R., Komander D., Alessi D. R. (2010). The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11, 9–22 10.1038/nrm2822 [DOI] [PubMed] [Google Scholar]
- Ruvinsky I., Meyuhas O. (2006). Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem. Sci. 31, 342–348 10.1016/j.tibs.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar–Peled L., Sabatini D. M. (2008). The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005). Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 10.1126/science.1106148 [DOI] [PubMed] [Google Scholar]
- Sato M., Dhut S., Toda T. (2005). New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast 22, 583–591 10.1002/yea.1233 [DOI] [PubMed] [Google Scholar]
- Sengupta S., Peterson T. R., Sabatini D. M. (2010). Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 40, 310–322 10.1016/j.molcel.2010.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki–Yabana S., Watanabe Y., Yamamoto M. (2000). Novel WD-repeat protein Mip1p facilitates function of the meiotic regulator Mei2p in fission yeast. Mol. Cell. Biol. 20, 1234–1242 10.1128/MCB.20.4.1234-1242.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara T., Hara K., Yonezawa K., Sorimachi H., Maeda T. (2006). Nutrient-dependent multimerization of the mammalian target of rapamycin through the N-terminal HEAT repeat region. J. Biol. Chem. 281, 28605–28614 10.1074/jbc.M606087200 [DOI] [PubMed] [Google Scholar]
- Tatebe H., Morigasaki S., Murayama S., Zeng C. T., Shiozaki K. (2010). Rab-family GTPase regulates TOR complex 2 signaling in fission yeast. Curr. Biol. 20, 1975–1982 10.1016/j.cub.2010.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano J., Comiso M. J., Guo L., Aspuria P. J., Deniskin R., Tabancay A. P., Jr, Kato–Stankiewicz J., Tamanoi F. (2005). Identification of novel single amino acid changes that result in hyperactivation of the unique GTPase, Rheb, in fission yeast. Mol. Microbiol. 58, 1074–1086 10.1111/j.1365-2958.2005.04877.x [DOI] [PubMed] [Google Scholar]
- Urano J., Sato T., Matsuo T., Otsubo Y., Yamamoto M., Tamanoi F. (2007). Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. Proc. Natl. Acad. Sci. USA 104, 3514–3519 10.1073/pnas.0608510104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H.et al. (2007). Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26, 663–674 10.1016/j.molcel.2007.04.020 [DOI] [PubMed] [Google Scholar]
- Uritani M., Hidaka H., Hotta Y., Ueno M., Ushimaru T., Toda T. (2006). Fission yeast Tor2 links nitrogen signals to cell proliferation and acts downstream of the Rheb GTPase. Genes Cells 11, 1367–1379 10.1111/j.1365-2443.2006.01025.x [DOI] [PubMed] [Google Scholar]
- Valbuena N., Guan K. L., Moreno S. (2012). The Vam6 and Gtr1-Gtr2 pathway activates TORC1 in response to amino acids in fission yeast. J. Cell Sci. 125, 1920–1928 10.1242/jcs.094219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Slegtenhorst M., Carr E., Stoyanova R., Kruger W. D., Henske E. P. (2004). Tsc1+ and tsc2+ regulate arginine uptake and metabolism in Schizosaccharomyces pombe. J. Biol. Chem. 279, 12706–12713 10.1074/jbc.M313874200 [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Lino Y., Furuhata K., Shimoda C., Yamamoto M. (1988). The S.pombe mei2 gene encoding a crucial molecule for commitment to meiosis is under the regulation of cAMP. EMBO J. 7, 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman R., Choder M. (2001). The fission yeast TOR homolog, tor1+, is required for the response to starvation and other stresses via a conserved serine. J. Biol. Chem. 276, 7027–7032 10.1074/jbc.M010446200 [DOI] [PubMed] [Google Scholar]
- Weisman R., Roitburg I., Nahari T., Kupiec M. (2005). Regulation of leucine uptake by tor1+ in Schizosaccharomyces pombe is sensitive to rapamycin. Genetics 169, 539–550 10.1534/genetics.104.034983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman R., Roitburg I., Schonbrun M., Harari R., Kupiec M. (2007). Opposite effects of tor1 and tor2 on nitrogen starvation responses in fission yeast. Genetics 175, 1153–1162 10.1534/genetics.106.064170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Oppliger W., Hall M. N. (2005). Molecular organization of target of rapamycin complex 2. J. Biol. Chem. 280, 30697–30704 10.1074/jbc.M505553200 [DOI] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M. N. (2006). TOR signaling in growth and metabolism. Cell 124, 471–484 10.1016/j.cell.2006.01.016 [DOI] [PubMed] [Google Scholar]
- Yang W., Tabancay A. P., Jr, Urano J., Tamanoi F. (2001). Failure to farnesylate Rheb protein contributes to the enrichment of G0/G1 phase cells in the Schizosaccharomyces pombe farnesyltransferase mutant. Mol. Microbiol. 41, 1339–1347 10.1046/j.1365-2958.2001.02599.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.