SUMMARY
Primary ciliary dyskinesia (PCD) is a genetic disorder, usually autosomal recessive, causing early respiratory disease and later subfertility. Whole exome sequencing may enable efficient analysis for locus heterogeneous disorders such as PCD. We whole exome sequenced one consanguineous Saudi Arabian with clinically diagnosed PCD and normal laterality, to attempt ab initio molecular diagnosis.
We reviewed thirteen known PCD genes and potentially autozygous regions (extended homozygosity) for homozygous exon deletions, non-dbSNP codon, splice-site base variants or small indels. Homozygous non-dbSNP changes were also reviewed exome-wide.
One single molecular read representing RSPH9 p.Lys268del was observed, with no wildtype reads, and a notable deficiency of mapped reads at this location. Among all observations, RSPH9 was the strongest candidate for causality. Searching unmapped reads revealed seven more mutant reads. Direct assay for p.Lys268del (MboII digest) confirmed homozygosity in the affected individual, then confirmed homozygosity in three siblings with bronchiectasis. Our finding in southwest Saudi Arabia indicates that p.Lys268del, previously observed in two Bedouin families (Israel, UAE) is geographically widespread in the Arabian Peninsula. Analogous with cystic fibrosis CFTR p.Phe508del, screening for RSPH9 p.Lys268del (which lacks sentinel dextrocardia) in those at risk would help in early diagnosis, tailored clinical management, genetic counselling and primary prevention.
Keywords: high-throughput nucleotide sequencing, primary ciliary dyskinesia, screening
INTRODUCTION
Primary ciliary dyskinesia (PCD) is a genetic disorder (MIM 242650), usually autosomal recessive, characterised by dysmotile cilia and sperm. It is characterised clinically by respiratory distress and infections from early life, with sinusitis and bronchiectasis. Subfertility is usual, and laterality defects are common. There is considerable locus heterogeneity for PCD, with the principal loci all encoding protein products involved in the assembly and effective beating of motile cilia (respiratory and oviduct) and sperm flagella. DNAH5 (MIM 603335) and DNAH11 (MIM 603339) encode ciliary heavy chain dyneins; DNAI1 (MIM 604336) and DNAI2 (MIM 605483) encode dynein intermediate chains; DNAL1 encodes a dynein light chain (Mazor et al., 2011); and TXNDC3 (MIM 607421) encodes thioredoxin domain-containing protein 3 which is orthologous to the sea urchin IC1 gene which encodes a component of sperm outer dynein arms. RSPH4A (MIM 612647) and RSPH9 (MIM 612648) encode radial spoke head proteins. KTU (MIM 612518) encodes a protein necessary for assembly of the dynein arm. CCDC39 and CCDC40 (Becker-Heck et al., 2011, Merveille et al., 2011) are somehow essential for proper assembly of motile cilia, as is LRRC50 (leucine-rich repeat-containing protein 50) (Loges et al., 2009) which plays a role in cytoplasmic pre-assembly of dynein arms.
However, mutation surveys have indicated generally complex spectra of mutations and the most studied four genes (those encoding dynein chains) collectively account for only a minority of PCD (references in(Reish et al., 2010)). Additionally, linkage studies have pointed to various other chromosomal regions (Geremek & Witt, 2004) although the causal loci at these locations have not been identified. Lastly, PCD may coexist with retinitis pigmentosa (RPGR, which is X-linked) (Badano et al., 2006). There is also genotype-phenotype variation, both in the ultrastructure of the cilium, in the motility patterns of cilia and in the precise clinical phenotype. Motile cilia in the embryonic node determine laterality, and with the dynein chain mutations, laterality becomes randomised (Escudier et al., 2009). This creates the classic picture of situs inversus and PCD first recognised as familial by Kartagener in 1933. However, the laterality defect has not been observed in human mutations or animal models of RSPH9 (Castleman et al., 2009). This leaves a diagnostic uncertainty which is particularly important in the early stages of respiratory disease, when definitive diagnosis and the introduction of careful management can, as in cystic fibrosis, minimise many of the secondary consequences which build up during childhood, including chonic wet cough, recurrent otitis media, nasal congestion, hearing deficit, speech delay, pneumonias and lower lobe bronchiectasis.
A practical problem for PCD molecular diagnostics has been the locus heterogeneity, allelic heterogeneity and large size of the causal genes. Therefore, to date, restricted mutation scanning in some principal genes in highly selected families, has generally been the limit of practice, and even that undertaken only in very few centres. In contrast with conventional Sanger sequencing, ‘next generation’ exome sequencing can acquire near complete human exome information in one pass (Majewski et al., 2011), opening the possibility to efficiently explore the genetic complexity of disorders such as PCD (Berg et al., 2011, Roach et al., 2010). We set out to investigate the application of whole exome sequencing for this research and diagnostic purpose. We describe our initial findings from this study, which suggest scope for RSPH9 p.Lys268del testing both for diagnostic use in “non-CF non-Kartagener’s” chronic respiratory disease in childhood’ in the Arabian Peninsula and research screening of wider groups with respiratory disease with possible patients representing missed “non-CF non-Kartagener’s PCD.’”
METHODS
Ethical approval and consents
Ethical approval for genetic studies in paediatric respiratory disorders was from King Saud University/King Khalid Hospital, Riyadh ethical committee, approval number E-11-448. Family and individual consent was verbal, with the recognition that positive findings would be diagnostically reconfirmed in conjunction with clinical counselling and feedback.
Proband and whole exome sequencing
We exome sequenced one consanguineous (F>=1/16) Saudi Arabian individual with clinically diagnosed PCD and normal laterality, to attempt ab initio molecular diagnosis (in comparison with a normal individual). The proband at age 11 years displayed bronchial asthma and productive cough, with computed tomography features including bronchiectasis of right middle lobe with associated fibroatelectasis and signs consistent with right middle and left lower lobe basal segment infectious bronchioloitis, also opacification of all sinuses with mild mucosal thickening observed in frontal sinus, and moderate enlargement of adenoid gland and both tonsils. Repeated nasal brush sampling for electron microscopy revealed largely metaplastic tissue, presumably reflecting the age combined with repeated infection and inflammation. This itself is characteristic of PCD. High magnification sections of cilia showed a generally normal 9+2 pattern (e.g. Supplementary Figure 1) which in a diagnostic setting were not clearly diagnostic of PCD. There was no younger affected sibling whom we could sample. However, exhaled nitric oxide assay showed a very low level in the proband, a characteristic though unexplained clinical feature of PCD (Karadag et al., 1999). Fractional exhaled nitric oxide measurements were performed according to the present recommendations of the American Thoracic Society using a NOX EVA 4000 chemiluminescence analyzer (SERES-FRANCE, Aix en Provence, France) with a sensitivity of 1 part per billion (ppb). Proband level was 4.0 ppb whereas the reference range is 14 to 17ppb.
DNA was extracted by a salting out procedure from 5ml K-EDTA peripheral venous blood from each sibling, and quantitated by fluorimetry. For targeted exome enrichment, barcoded fragment libraries were prepared from 3ug of blood-isolated and purified DNA, as previously described (Nijman et al., 2010). For each library, 1ug was hybridised for 72h at 47°C with the NimbleGen SeqCap EZ v2 exome probes, and subsequently processed according to manufacturer’s instructions (Roche NimbleGen, Inc., Madison, USA). Clonal amplifications of the fragment libraries on the surface of the sequencing beads were performed on the SOLiD EZ Bead system, following manufacturer’s instructions (Life Technologies Ltd, Paisley, UK). Amplified and templated bead enriched libraries were sequenced on an AB SOLiD sequencing platform with v4 chemistry, generating in excess of 80M quality passed single 50bp reads per sample (Applied Biosystems,Foster City, USA Colour space reads were mapped onto Human Genome Reference Sequence HG19 using the Burrows-Wheeler Aligner (BWA) (Li & Durbin, 2009) mapping software, filtering out reads with mapping scores of zero and generating a BAM file. The BAM file was subsequently parsed with a custom perl script to retain only the subset of reads originating from unique start sites. The Genome Analysis Toolkit (GATK) (McKenna et al., 2010) multiple sequence alignment tool was then deployed to remove alignment artefacts by using local realignment to ensure that the number of mismatching bases is minimized across all the reads. Single Nucleotide variants (SNVs) and small insertions and deletions were called using the GATK toolkit with default parameters and exported in VCF format (Danecek et al., 2011). SNVs were also screened against data publicly available in the dbSNP database (Sayers et al., 2011). Annovar (Wang et al., 2010) was used for VCF annotation, with modifications of the “convert2annovar.pl” code to retain information on allelic read counts and qualities, Excel was used for sorting of the annotations for review based on various criteria (e.g. homozygosity, non-dbSNP/dbSNP, type of variant, gene name, location and quality scores) and Integrative Genomics Viewer (IGV) (Robinson et al., 2011) was used for viewing mapped reads. To identify potential homozygous deletions a custom Python script was developed to plot homozygosity/heterozygosity to visually identify and review homozygous and potential autozygous regions. A second custom Python script was developed to identify bases where the proband displayed 0 reads and the control displayed more than a chosen non-zero number, e.g. 20. We reviewed thirteen genes in which mutations are known to cause PCD, for homozygous exon deletions, non-dbSNP codon, splice site base variants or small indels. We identified autozygous regions and did the same for these. Homozygous non-dbSNP changes were also reviewed exome wide. We also closely re-reviewed all genes with the name starting ‘DNA..’ because these include a number of potentially relevant dynein genes.
Evaluation of RSPH9 p.Lys268del in kindred and local population
RSPH9 p.Lys268del was directly assayed by PCR-RFLP using MboII and primers as described (Castleman et al, 2009). The proband is from a large sibship (detail suppressed for genetic privacy), originating from the southwest part of Saudi Arabia and including several older affected siblings with a clinical diagnosis of PCD, of whom two adults have shown evidence of infertility. None of these exhibits dextrocardia. It was possible to sample three affected siblings, one unaffected sibling and both parents, who are patrilineal first cousins. A local population reference DNA sample (n=260) was set up comprising male and female student volunteers of Saudi Arabian ancestry at King Saud University, Riyadh. Inclusion was voluntary, and informed written consent for anonymised genetic studies was taken in keeping with KSU College of Applied Medical Sciences guidelines.
RESULTS
Exome sequencing performance
Total captured region was 44055647 base pairs. Whole exome sequencing coverage in the proband was to a mean depth of 26x, median 18x. There were 93,905,183 reads of which 49,726,803 (52.95%) were mapped. 19,725 baits (5%) were without read, and 95% of baits gave reads. Similar results were obtained for a control sample.
It should be noted that with SOLiD sequencing compared with, for example, Illumina sequencing, there is no filtering of data prior to mapping; the mapping is the filter. Therefore any read (even if it is from an empty or polyclonal bead) will be in the dataset that is mapped. On a good run, one may get 70% mapping and on a poor run it would be 40%. Therefore this mapping efficiency does not reflect the quality of the reads but more likely it reflects the accuracy of the DNA titration when the machine was loaded. All known PCD genes were generally well covered.
Parallel review of autozygosity, of exome, and of known genes
Long autozygous segments (e.g. greater than 5Mb) were readily identified from the exome sequence - a plot of one chromosome is shown in Figure 1 (for reasons of genetic privacy we do not include the full genome plot). Where shorter segments (down to 2Mb) could be identified from exome data, these were also considered. None of these autozygous segments spanned a gene linked to PCD, nor a reported linkage region (CILD1-CILD13) (Zariwala et al., 2009). However, within these regions, there were numerous potentially plausible candidates, including some with non-dbSNP missense mutations. An example was ARHGAP21 p.H899Y (ARHGAP21 is of interest to ciliary biologists (Hehnly et al., 2009)). Known PCD genes DNAI1 (9p21), DNAH5 (5p), TXNDC3 (7p14), DNAH11 (7p21), DNAI2 (17q25), KTU (14q21.3), RSPH4A (6q22), RSPH9 (6p21), LRRC50 (16q24.1), RPGR, DNAL1, CCDC39 and CCDC40 were systematically examined by several different steps used in parallel. Firstly, they were reviewed for evidence of presence in a shorter (1-5Mb) autozygous region but none was found. Secondly, read depth per exon was viewed in IGV in comparison with an identically processed control DNA but no abnormality and certainly no homozygous absence of reads spanning any exon was detected (this process was also done systematically genome-wide with a custom script). Thirdly, all db-SNP and non-dbSNP single base change homozygosities were reviewed in each gene using an annotated output from the VCF files - there were no stop codons and no homozygous single nucleotide variants with any persuasive case for functional effect. Fourthly, all mapped reads were viewed in each gene in IGV for small insertion/deletion events. From these analyses, only one feature stood out, a single three base pair deletion read in RSPH9 (Fig. 2) not captured in the VCF generation and annotation. This corresponded with a known RSPH9 mutation, p.Lys268del but as it was in a microrepeat (5′-GAAGAA) relatively near the end of a read and unique, it was considered a possible sequencing artefact. However, we also noted a deficiency of mapped reads spanning this region, compared with the control, and therefore remapped using BWA, both using different parameters (which allowed more reads to map but also allowed more artefactual error reads to map) and using a reference sequence modified to correspond with the deletion. In the proband, this identified seven more apparently independent deletion reads and no wild type reads. In the control, the mutated genome reference ablated the mapping of the wildtype reads. Re-analysis of the RSPH9 region for evidence of autozygosity in the proband, indicated a short span of homozygosity involving five loci and an estimated interval of 0.5Mb (Fig. 3).
Figure 1. Plot of part of the exome data for the proband, with known dbSNP loci variant calls (heterozygous in clear circles, homozygous variant in filled circles) plotted to identify autozygous regions.
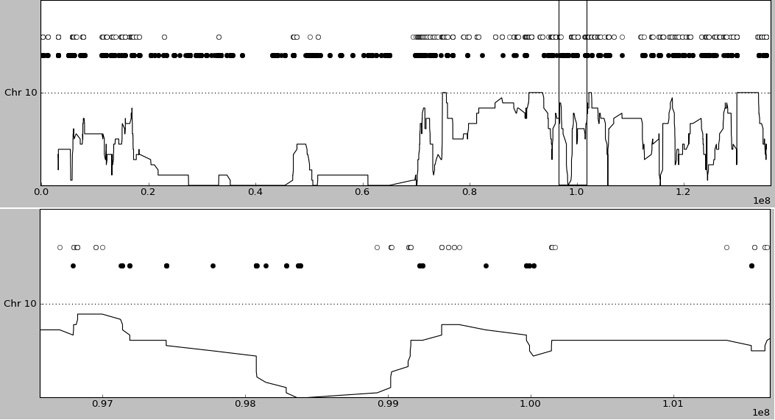
The lower line gives a rolling window average of homozygosity. Upper panel - all of chromosome 10 (~135Mb), note the very large (50Mb) region which is almost entirely homozygous. This reflects autozygosity and the two single isolated heterozygous calls are likely erroneous calls. Lower panel is a zoom of the ~5Mb region marked by vertical bars in the upper panel - note the 2Mb region of homozygosity, likely autozygosity, in the left half of the panel.
Figure 2A. Initial evidence in whole exome sequencing of a 3 basepair deletion in RSPH9.

A single molecular read (upper panel) showing a deletion was observed (horizontal thin line adjacent to thedotted lines flanking the base centred in the viewing panel). Compared with a control (lower panel), there was also a deficiency of mapped reads at this location. The wildtype sequence shows a microrepeat 5′-GAAGAA.
Figure 2B. Remapping using a reference genome in which the 3 basepair deletion had been created.
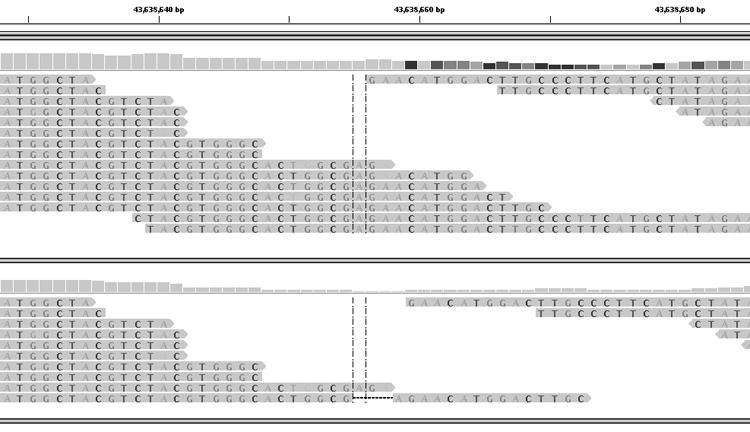
Upper panel shows the remapping of the PCD proband, the lower panel shows the original mapping against normal reference genome.
Figure 3. A plot of variant heterozygosity (upper line) and variant homozygosity (lower line) at known dbSNP loci, in the proband exome sequence on chromosome 6.
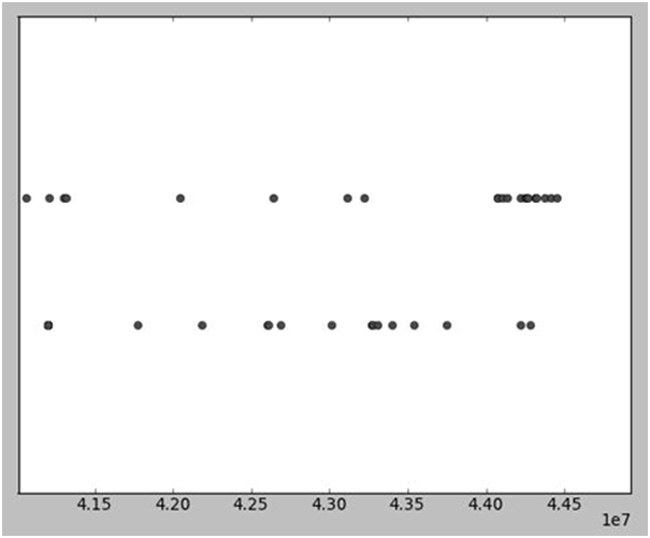
Base locations are on human genome build hg19. RSPH9 is located in the region around 4.36×107. There is apparent homozygosity spanning ~0.5Mb, or possibly up to 0.75Mb but no more.
Direct assay follow-ups in proband, family and local population
Direct assay for RSPH9 p.Lys268del confirmed the exome sequencing data. All three affected sibs were also homozygous for the deletion. The parents, who have no respiratory disease nor infertility, were heterozygous (Fig. 4). In our population sample of student volunteers (n=260) there were no carriers for RSPH9 p.Lys268del.
Figure 4. Direct assay of RSPH9 p.Lys268del in proband and relatives by PCR-RFLP using MboII digest.
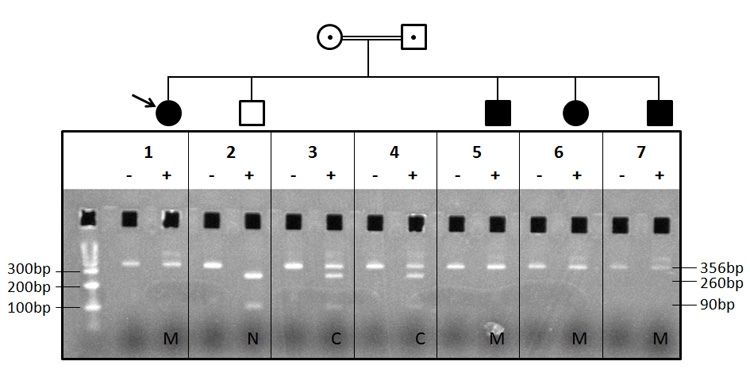
Panel 1 - molecular weight markers, proband amplicon undigested, proband amplicon with MboII. The proband had been whole exome sequenced. All other panels, left track is undigested, right track is digested. Panel 2, an unaffected sibling. Panels 3 and 4, the (healthy) parents. Panels 5, 6 and 7, all represent affected siblings. N, wild type allele homozygote (complete MboII digestion); C, heterozygous carrier (partial MboII digestion); M, homozygous for mutation (no MboII digestion). RSPH9 p.Lys268del resists digestion, wild type allele cuts. A large number of other siblings believed to be healthy, were unavailable for study. The proportion of affected siblings was consistent with single gene autosomal recessive inheritance.
DISCUSSION
We applied whole exome sequencing to a single individual with PCD and identified their causal mutation. The process illustrated strengths and limitations in the use of exome sequencing for ab initio search in a genetically heterogeneous disorder for which there remain numerous uncharacterised genes. Taken together with previous literature, our finding indicates that RSPH9 p.Lys268del is geographically widespread in the Arabian Peninsula: this suggests that a molecular diagnostic strategy may offer practical utility for autosomal recessive bronchiectasis. Along the lines of CFTR p.Phe508del diagnosis in Western Europe, early molecular identification in the absence of definitive clinical features, can lead to a much improved clinical outlook.
Exome sequencing generates a large data set. Dependent on the thresholds of acceptance of variant reads, we observed approximately 15,000 dbSNP variants and 5000 non-dbSNP variants, of which approximately 85% were exonic, the rest mainly intronic in the vicinity of captured exons. Thus, de novo identification of a disease causing mutation in a previously unknown gene, in the absence of search space reduction using linkage information or functional information, would be very difficult. Appropriate pipelines and filtering are essential to define what has been tested and what has been observed. Poorly represented regions, uncertain variant base calls and the recognised difficulties in mapping and identifying reads representing small indels, are present limitations of the technique. Each of these factors limits the possibility of turnkey automation of analysis and demands re-evaluation of analyses using different parameters. Where there is deficiency rather than absence of reads (for whatever reason), higher depth of coverage (e.g. 50x) may help in increasing confidence about variant calls (e.g. where only one or two variant calls were observed). In this instance, the additional information hoped for from the hypothesis of autozygosity (see below), was not obtained. However, the hypothesis of recessive (homozygous) change was an important filter in data analysis. It would have been difficult to make the mutation identification, compared against competing hypotheses, if the mutation had not been in a strong candidate gene. The reason for only observing one deletion read (and no wildtype reads) at the deletion site, reflected the (default) thresholds used for small indel detection. Seven reads in which the 3 nucleotide deletion relative to wildtype was present did not map to the wildtype reference sequence despite otherwise being perfectly matched to the wildtype sequence. Therefore the absence of wildtype reads relative to a control individual was an important second feature which led us to suspect that a real mutation was present rather than there being an artefactual individual read at the 5′-GAAGAA microrepeat site. Present software allows for altering mapping thresholds (achieving more mapped reads at the risk of more mismapped reads) but future methods would benefit from simultaneous evaluation of coverage relative to identical sequencing of a control.
Whole exome sequencing provides the possibility to further investigate digenic or oligogenic effects, such as have been observed in retinitis pigmentosa (Kajiwara et al., 1994); however, we did not observe likely pathogenic variants in any other known PCD nor dynein gene in the proband. Four clinically affected individuals in the large sibship (of whom the others are not believed to be clinically affected by PCD), in contrast with parents, unaffected sibling and population sample, all tested homozygous for RSPH9 p.Lys268del, indicative that this is necessary, and likely sufficient, to cause PCD. Thus it appears that RSPH9 p.Lys268del homozygosity will certainly cause respiratory disease. Since RSPH9 p.Lys268del has not been observed (our search of data at ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20100804/) in 1258 chromosomes in the 1000Genomes Project (and these are of populations other than Arabian, but including European, African and East Asian groups), but has been observed repeatedly in Western Asia (Israel, UAE and now Saudi Arabia), it is likely to represent a founder mutation. If it were a strongly recurrent mutation, then it should appear in European and East Asian populations also. Recessive disease mutations due to consanguinity reside in autozygous segments of the genome, which for offspring of first cousins are mostly larger than 1-2Mb, with a predictable distribution ranging up to many megabases (Woods et al., 2006). However, from the exome sequence of our proband, we were able to deduce, using reliably called SNPs already documented in dbSNP, that the region of homozygosity around RSPH9 p.Lys268del was approximately 0.5Mb (Fig. 3). This suggests, but does not prove, homozygosity through some more remote earlier cousinship (endogamy) rather than through the autozygosity of first cousin parentage. However this could not be resolved without a detailed mapping of recombination breakpoints in the parents and (unavailable) grandparents.
An important parameter for the overall population is the carrier frequency. We undertook prevalence studies in a population sample representing 520 alleles from Saudi Arabia. We did not observe an instance of RSPH9 p.Lys268del heterozygosity. This gives 80% confidence that the allele prevalence q is not greater than 3/1000 in this population. However, under consanguinity, risk is inflated from q2. The additional consanguinity risk is Fq, where F is Wright’s inbreeding coefficient, 0.0625 for offspring of first cousins, and this estimated 30% sector of the population (ignoring second cousin marriages and wider tribal endogamy) therefore face at least a 20-fold increased risk for RSPH9 p.Lys268del (0.0625 divided by less than or equal to 0.003). Our sample represented Saudi Arabian students at King Saud University Riyadh - their ancestries will have been drawn from various parts of Saudi Arabia over the past few generations. Over a timescale of greater than 100 years, many ancestries can be considered to be ‘Bedouin.’ We cannot rule out higher frequency (local founder effects) in less cosmopolitan areas. A previous survey of 160 unrelated males in UAE identified three carriers, which would give q~0.01 (Castleman et al, 2009). Based on these data, absolute risk in an outbred population would be q2=0.0001. If all individuals married their first cousin, absolute risk would be Fq=0.000625 (~1/1700). If 30% marry their first cousin and the rest were fully outbred, absolute risk would be approximately 1/4000. More detail is needed about regional carrier frequency, and the possibility that PCD families without laterality defects may be substantially underdiagnosed needs also to be explored. The present strategies of focus should therefore be firstly on families with a proband developing early life respiratory disease, especially consanguineous families, in whom cascade screening should be encouraged. Further research, if the allele frequency is “high”(e.g. 1%) and the underdiagnosis rate high, may suggest scope to widen screening to at risk (consanguineous) couples or to the broader population. Direct assay of RSPH9 p.Lys268del is straightforward; it is clear that the mutation has high penetrance, and that early classification and appropriate treatment improves life (e.g. asthma can be ruled out, and early infections can be aggressively managed to prevent long term damage from persistence and recurrence). The genetic screen might be used in conjunction with a test of exhaled nitric oxide, which is reduced in PCD (Karadag et al, 1999).
RSPH9 p.Lys268del does not appear to cause situs inversus in families (this study and (Castleman et al, 2009)), whereas this feature much simplifies a definitive diagnosis of PCD. Furthermore, electron microscopy (Supplementary figure 1) in a diagnostic setting showed a generally normal ciliary 9+2 pattern, although subtle difference of dynein arms or radial spokes may be present. It has been noted previously that RSPH9 p.Lys268del (Castleman et al, 2009) produces subtle, if any, phenotype detectable by EM. For these reasons, it is likely that PCD due to RSPH9 p.Lys268del is underdiagnosed. It is possible that it is not fully penetrant, but in the families observed so far, there has been clearcut disease in all those who are genotype positive. In our proband, repeated nasal brush sampling yielded only metaplastic tissue, reflecting their combination of age and recurrent infection and inflammation. Diagnosis of PCD (Zariwala, Knowles, & Leigh, 2009) requires the characteristic clinical phenotype plus either electron microscopic features or mutation identification in one of the known genes. In our described instance the latter prevailed from whole exome sequencing, again illustrating the diagnostic utility possible from whole exome sequencing. However, for new gene identification, ciliary data (more readily obtained early in life prior to secondary epithelial changes) remains an important biological research adjunct. Cystic fibrosis (largely due to CFTR p.Phe508del) occurs in 1/2500 Western Europeans and is screened in couples with close family history to enable reproductive choice and neonatally for the benefits of early recognition and management (McCormick et al., 2002, Sims et al., 2005). PCD is quite analogous to CF, although life expectancy is greater. In PCD, recurrent chest, sinus and middle ear infections from an early age are a risk to survival, growth and development. Bronchiectasis and other complications such as sinus obliteration ensue. However an established diagnosis is likely to lead to a clear plan of management. With good care (clinical guidance, specific motivation, physiotherapy which can be done at home, antibiotics etc), many of the sequelae can be minimised. While the frequency of RSPH9 p.Lys268del in Western Asia is not as high as the frequency of CFTR p.Phe508del in Western Europe, this apparent founder mutation constitutes a high but manageable risk for the large consanguineous subpopulations of Western Asia. At some level, certainly in known risk kindreds, the Wilson and Jungner/ World Health Organisation criteria for a screening programme (http://whqlibdoc.who.int/php/WHO_PHP_34.pdf) are already fulfilled. Cascade screening in extended kindreds may be limited though, by unwillingness to disclose genetic status to second and third degree relatives who may be marriage candidates. A system of access, for example via the hemoglobinopathy screening programme for prospective couples, combined with effective provision of information and of cost efficient assay would be necessary to extend screening to consanguineous, or all, couples. A less parsimonious option, but better than nothing, would be the availability of the assay as a private service, possibly as part of a broader personal genomics service for testing carrier status for a range of high risk genes. This is of potential high utility in societies where first cousin marriage maintains familial ties and a socioeconomic stability. Genetic disease stigmata reduce the chance of a family to participate in their culture of familial and tribal marriages (with good scientific basis to the common reasoning which leads their cousins not to wish to marry them), whereas a gene test for a known disorder can identify at risk couples clearly. While there was initial concern in Saudi Arabia that hemoglobinopathy premarital screening would stigmatise daughters, the programme was well received and surveys indicate a general will to build on this success (AlSulaiman et al., 2008). Lastly, it remains possible that a specific functional intervention in homozygotes for RSPH9 p.Lys268del may in the future be identified.
In summary, our findings illustrate how whole exome sequencing can contribute to research and diagnostics of a monogenic disorder. The specific finding has turned out to be relevant not only to the individual and family studied, but is also catalytic to potential future cascade screening for PCD families in the Arabian Peninsula; and it invites wider frequency surveys in other respiratory diseases and in regional populations.
Supplementary Material
Acknowledgements
We thank the PCD family members and student volunteers for participating in this study.
Funding The development of the population sample was supported by grants from King Abdul-Aziz City for Sciences and Technology, Kingdom of Saudi Arabia and from King Saud University College of Applied Medical Sciences to KKA. The work in the Liverpool Centre for Genomic Research was supported by MRC grant G0900753. PAIG is supported by MRC Centre grant G0600705. CRB is a Wellcome Trust Ph.D student.
Footnotes
Competing interests None
Author Contributions I.N.M.D., K.K.A. and M.M.A. conceived the study. M.M.A. characterised the PCD proband and family. K.K.A. established the population-based reference collection. P.A.I.G. extracted proband and family DNAs. L.R. constructed exome libraries and performed sequencing on the AB LifeTech SOLiD platform. L.L and X.L. developed the analysis pipeline and analyzed the sequence data. N.H. supervised the data generation and analysis. T.R.G. wrote specific scripts to filter and interpret exome data in the context of consanguinity. C.R.B. undertook follow up mutation testing. I.N.M.D. co-ordinated the study, undertook exome analyses, supervised the follow-up laboratory work and drafted the manuscript. All authors commented on, read and approved the final manuscript.
References
- AlSulaiman A, Suliman A, AlMishari M, AlSawadi A, Owaidah TM. Knowledge and attitude toward the hemoglobinopathies premarital screening program in Saudi Arabia: population-based survey. Hemoglobin. 2008;32:531–538. doi: 10.1080/03630260802508384. [DOI] [PubMed] [Google Scholar]
- Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Becker-Heck A, Zohn IE, Okabe N, Pollock A, Lenhart KB, Sullivan-Brown J, McSheene J, Loges NT, Olbrich H, Haeffner K, Fliegauf M, Horvath J, Reinhardt R, Nielsen KG, Marthin JK, Baktai G, Anderson KV, Geisler R, Niswander L, Omran H, Burdine RD. The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation. Nat Genet. 2011;43:79–84. doi: 10.1038/ng.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Evans JP, Leigh MW, Omran H, Bizon C, Mane K, Knowles MR, Weck KE, Zariwala MA. Next generation massively parallel sequencing of targeted exomes to identify genetic mutations in primary ciliary dyskinesia: implications for application to clinical testing. Genet Med. 2011;13:218–229. doi: 10.1097/GIM.0b013e318203cff2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castleman VH, Romio L, Chodhari R, Hirst RA, de Castro, S CP, Parker KA, Ybot-Gonzalez P, Emes RD, Wilson SW, Wallis C, Johnson CA, Herrera RJ, Rutman A, Dixon M, Shoemark A, Bush A, Hogg C, Gardiner RM, Reish O, Greene NDE, O’Callaghan C, Purton S, Chung EMK, Mitchison HM. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am. J. Hum. Genet. 2009;84:197–209. doi: 10.1016/j.ajhg.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudier E, Duquesnoy P, Papon JF, Amselem S. Ciliary defects and genetics of primary ciliary dyskinesia. Paediatr Respir Rev. 2009;10:51–54. doi: 10.1016/j.prrv.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Geremek M, Witt M. Primary ciliary dyskinesia: genes, candidate genes and chromosomal regions. J. Appl. Genet. 2004;45:347–361. [PubMed] [Google Scholar]
- Hehnly H, Longhini KM, Chen JL, Stamnes M. Retrograde Shiga toxin trafficking is regulated by ARHGAP21 and Cdc42. Mol Biol Cell. 2009;20:4303–4312. doi: 10.1091/mbc.E09-02-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara K, Berson EL, Dryja TP. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science. 1994;264:1604–1608. doi: 10.1126/science.8202715. [DOI] [PubMed] [Google Scholar]
- Karadag B, James AJ, Gultekin E, Wilson NM, Bush A. Nasal and lower airway level of nitric oxide in children with primary ciliary dyskinesia. Eur Respir J. 1999;13:1402–1405. doi: 10.1183/09031936.99.13614069. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loges NT, Olbrich H, Becker-Heck A, Haffner K, Heer A, Reinhard C, Schmidts M, Kispert A, Zariwala MA, Leigh MW, Knowles MR, Zentgraf H, Seithe H, Nurnberg G, Nurnberg P, Reinhardt R, Omran H. Deletions and point mutations of LRRC50 cause primary ciliary dyskinesia due to dynein arm defects. Am J Hum Genet. 2009;85:883–889. doi: 10.1016/j.ajhg.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Schwartzentruber J, Lalonde E, Montpetit A, Jabado N. What can exome sequencing do for you? J Med Genet. 2011;48:580–589. doi: 10.1136/jmedgenet-2011-100223. [DOI] [PubMed] [Google Scholar]
- Mazor M, Alkrinawi S, Chalifa-Caspi V, Manor E, Sheffield VC, Aviram M, Parvari R. Primary ciliary dyskinesia caused by homozygous mutation in DNAL1, encoding dynein light chain 1. Am J Hum Genet. 2011;88:599–607. doi: 10.1016/j.ajhg.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick J, Green MW, Mehta G, Culross F, Mehta A. Demographics of the UK cystic fibrosis population: implications for neonatal screening. Eur J Hum Genet. 2002;10:583–590. doi: 10.1038/sj.ejhg.5200850. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merveille AC, Davis EE, Becker-Heck A, Legendre M, Amirav I, Bataille G, Belmont J, Beydon N, Billen F, Clement A, Clercx C, Coste A, Crosbie R, de BJ, Deleuze S, Duquesnoy P, Escalier D, Escudier E, Fliegauf M, Horvath J, Hill K, Jorissen M, Just J, Kispert A, Lathrop M, Loges NT, Marthin JK, Momozawa Y, Montantin G, Nielsen KG, Olbrich H, Papon JF, Rayet I, Roger G, Schmidts M, Tenreiro H, Towbin JA, Zelenika D, Zentgraf H, Georges M, Lequarre AS, Katsanis N, Omran H, Amselem S. CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat Genet. 2011;43:72–78. doi: 10.1038/ng.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman I. c. J., Mokry M, van Boxtel R, Toonen P, de Bruijn E, Cuppen E. Mutation discovery by targeted genomic enrichment of multiplexed barcoded samples. Nat. Methods. 2010;7:913–915. doi: 10.1038/nmeth.1516. [DOI] [PubMed] [Google Scholar]
- Reish O, Slatkin M, Chapman-Shimshoni D, Elizur A, Chioza B, Castleman V, Mitchison HM. Founder mutation(s) in the RSPH9 gene leading to primary ciliary dyskinesia in two inbred Bedouin families. Ann Hum Genet. 2010;74:117–125. doi: 10.1111/j.1469-1809.2009.00559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, Rowen L, Pant KP, Goodman N, Bamshad M, Shendure J, Drmanac R, Jorde LB, Hood L, Galas DJ. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, Feolo M, Fingerman IM, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Lu Z, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Miller V, Mizrachi I, Ostell J, Panchenko A, Phan L, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Slotta D, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Wang Y, Wilbur WJ, Yaschenko E, Ye J. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims EJ, McCormick J, Mehta G, Mehta A. Neonatal screening for cystic fibrosis is beneficial even in the context of modern treatment. J Pediatr. 2005;147:S42–S46. doi: 10.1016/j.jpeds.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CG, Cox J, Springell K, Hampshire DJ, Mohamed MD, McKibbin M, Stern R, Raymond FL, Sandford R, Malik SS, Karbani G, Ahmed M, Bond J, Clayton D, Inglehearn CF. Quantification of homozygosity in consanguineous individuals with autosomal recessive disease. Am J Hum Genet. 2006;78:889–896. doi: 10.1086/503875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zariwala MA, Knowles MR, Leigh MW. Primary Ciliary Dyskinesia. In: Pagon RA, Bird TD, Dolan CR, Stephens K, editors. SourceGeneReviews [Internet] University of Washington, Seattle; Seattle (WA): [1993-. 2007 Jan 24 [updated 2009 Oct 6]]. 2009. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


