Abstract
Interlock is a structural element in DNA G-quadruplexes that can be compared with the commonly used complementary binding of ‘sticky ends’ in DNA duplexes. G-quadruplex interlocking can be a basis for the assembly of higher-order structures. In this study, we formulated a rule to engineer (3 + 1) interlocked dimeric G-quadruplexes and established the folding topology of the designed DNA sequences by nuclear magnetic resonance spectroscopy. These interlocked G-quadruplexes are very stable and can serve as compact robust scaffolds for various applications. Different structural elements can be engineered in these robust scaffolds. We demonstrated the anti-HIV inhibition activity of the newly designed DNA sequences.
INTRODUCTION
Guanine-rich (G-rich) nucleic acid sequences can adopt special structures called G-quadruplexes based on stacking of G•G•G•G tetrads (1–6). In nature, G-rich DNA and RNA sequences are found in many important genomic regions, such as the telomeres, oncogenic promoters and RNA 5′-untranslated regions (1–6). G-quadruplexes formed by these natural sequences have been established as attractive anticancer targets (7,8). On the other hand, some engineered G-rich oligonucleotides, obtained by in vitro selection (9,10) or rational design (11–13) and capable of forming G-quadruplexes (14–16), possess interesting biological activities, such as anticoagulant (9), anticancer (11–13) and anti-HIV activities (10). G-quadruplexes can also have potential applications in chemistry, material sciences and nanotechnology (1). For example, these scaffolds can be used as a drug delivery system or a basis for the design of new materials and nanodevices; they can also be used to guide charge transfer or to support catalysts (17–23). Studies have shown that G-quadruplex structures are highly polymorphic (1–6). Therefore, it is important to control their different structural elements and folding topologies for various applications.
Interlock (16,24,25) is a structural element in G-quadruplexes that can be compared with the commonly used complementary binding of ‘sticky ends’ in DNA duplexes. G-quadruplex interlocking can be a basis for higher-order structures, such as G-wires (26). In the structure of interlocked G-quadruplexes (16,24,25), G-tetrads at the interface between the two subunits are completed by (i) two guanines from each subunit (in ‘2 + 2’ type) (24,25) or (ii) three guanines from one subunit and one guanine from another subunit (in ‘3 + 1’ type) (16).
The 93del sequence d(GGGGTGGGAGGAGGGT), a HIV-1 integrase inhibitor (10), forms in K+ solution an extremely stable ‘3+1’-type interlocked dimeric G-quadruplex with totally six G-tetrad layers (16) (Figure 1a). Dimer formation is achieved through mutual pairing of G1 from one subunit with G2, G6 and G13 of the other subunit. In each subunit, there are (i) four parallel-stranded G-tracts (or legs) and (ii) three single-residue double-chain-reversal loops bridging two or three G-tetrad layers. The first leg is long with four Gs; the second and fourth legs are medium with three Gs; the third one is short with only two Gs. The 5′-end guanine from the long leg (first) of one subunit compensates the short leg (third) of the other subunit in completing the G-tetrads at the interlocking interface (Figure 1a). Here, we aim to determine a rule to control this G-quadruplex interlocking motif.
Figure 1.
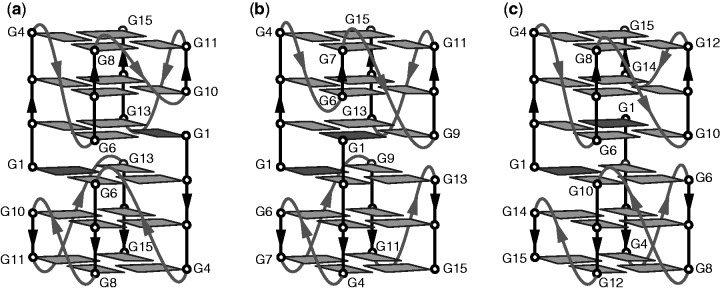
Schematic structures of dimeric (3 + 1) interlocked G-quadruplexes: (a) 93del, (b) s2 and (c) s4. Anti and syn guanines are shown in light and dark grey, respectively.
METHODS
Sample preparation
Unlabeled and site-specific labeled DNA oligonucleotides were chemically prepared using products from Glen Research and Cambridge Isotope Laboratories. Samples were purified following the protocol from Glen Research and then dialyzed successively against KCl and water solution. DNA oligonucleotides were dissolved in a solution containing 70 mM potassium chloride and 20 mM potassium phosphate, pH 7.0. DNA concentration was expressed in strand molarity using a nearest-neighbor approximation for the extinction coefficients of the unfolded species.
Nuclear magnetic resonance spectroscopy
Nuclear magnetic resonance (NMR) experiments were performed on Bruker Avance spectrometers operating at 600 or 700 MHz for 1H. Spectra were recorded at 25°C, unless otherwise specified. Experiments in H2O used the jump-and-return (JR)-type water suppression for detection. Guanine resonances were assigned unambiguously by using site-specific low-enrichment 15N labeling, site-specific 2H labeling, through-bond correlations at natural abundance ([13C,1H] JRHMBC, [13C,1H] HSQC and [1H,1H] TOCSY) and NOESY. Thymine resonances were identified by the through-bond H6-CH3 correlations from TOCSY spectra. The folding topologies and other structural information of G-quadruplexes were obtained from NOESY spectra.
Protein preparation
The pET28b plasmid containing the mutated core domain of HIV-1 integrase, IN50-212(F185H), was expressed in Escherichia coli strain BL21 (DE3-Gold) as described previously (16). The cells were grown at 37°C in BL medium containing 50 µg/ml of kanamycin until the 600-nm optical density reached 0.5–0.8, then induced by adding 0.5 mM IPTG and let grow in shaking culture for 3 h. The cells were harvested by centrifugation at 8000 rpm for 10 min at 4°C. The cell pellet from 1 L culture was suspended in 20 ml of lysis buffer (25 mM HEPES, pH 7.5, 0.5 M NaCl, 2 mM 2-mercaptoethanol, 5 mM imidazole, 0.5% Triton X-100, 1 mM PMSF). The cells were sonicated and the debris was discarded by centrifugation at 14 000 rpm for 20 min at 4°C. The collected supernatant was filtered through a 0.2-µm filter and applied to a HisTrap™ HP column. The column was then washed successively by elution buffer (25 mM HEPES, pH 7.5, 0.5 M NaCl, 2 mM 2-mercaptoethanol, 10% glycerol), which contained 20, 150, 400 and 800 mM imidazole, respectively, with five column-volumes for each imidazole concentration. The protein was collected at 400 mM imidazole fraction and dialyzed overnight at 4°C in a buffer containing 25 mM HEPES, pH 7.5, 0.5 M NaCl, 1 mM DTT, 1 mM EDTA, 10% (wt/vol) glycerol. The sample was frozen in liquid nitrogen and stored at −80°C.
HIV-1 integrase activity test
The reaction mixture contained 20 mM HEPES (pH 7.5), 10 mM MnCl2, 30 mM NaCl, 10 mM DTT, 0.05% Nonidet-P40, 600 nM HIV − 1 integrase and 200 nM DB-Y1. The DB-Y1 substrate (5′-TGCTAGTTCTAGCAGGCCCTTGGGCCGGCGCTTGCGCC-3′) used in the reaction (purchased from 1st BASE Pte. Ltd., Singapore) was labeled with 6-FAM at the 5′-end. After incubating at 37°C for 90 min, the reaction was stopped by adding an equal volume of 98% deionized formamide, 10 mM EDTA, pH 8.0 and heated at 90°C for 3 min. For inhibition tests, the inhibitor was added into the mixture and incubated for 30 min before adding DB-Y1. The product was analyzed by electrophoresis on a 15% polyacrylamide gel supplemented with 7 M urea and viewed under 495-nm light.
RESULTS AND DISCUSSION
Rational design of G-rich sequences that form (3 + 1) interlocked G-quadruplexes
We propose that in general a (3 + 1) interlocked G-quadruplex might be formed by sequences containing one long G-tract, one short G-tract and two medium G-tracts. In the next section, we demonstrate by NMR that (3 + 1) interlocked dimeric G-quadruplexes (Figure 1) can be formed by sequences s2 or s4 (Table 1), where the first G-tract is long (with four Gs) and, respectively, the second or fourth G-tract is short (with two Gs), whereas the remaining G-tracts are medium (with three Gs). Single-residue linkers were chosen to connect G-tracts because they could form stable double-chain-reversal loops bridging two or three G-tetrad layers (16).
Table 1.
Sequences used in this study
| Name | Sequence (5′-3′) |
|---|---|
| 93del | GGGGTGGGAGGAGGGT |
| s3 | GGGGTGGGTGGTGGGT |
| s2 | GGGGTGGTGGGTGGGT |
| s2-A | GGGGAGGTGGGTGGGT |
| s4 | GGGGTGGGTGGGTGGT |
| s4-A | GGGGTGGGTGGGAGGT |
| s4-Br | BrGGGGTGGGTGGGTGGT |
| s1R | TGGTGGGTGGGTGGGG |
| s2R | TGGGTGGTGGGTGGGG |
| s3R | TGGGTGGGTGGTGGGG |
| s3F | GGGGTGGGTGGGTGGGT |
We also ask whether (3 + 1) interlocked G-quadruplexes could be formed by sequences containing one long G-tract (four Gs), one short G-tract (two Gs) and two medium G-tracts (three Gs) but with the 3′-end fourth G-tract being long (instead of the 5′-end first G-tract) (sequences s1R, s2R and s3R; Table 1). Another question is whether a short G-tract is required to control the (3 + 1) interlocking motif; in other words, could a sequence with the first G-tract long (four Gs) and three medium G-tracts (three Gs) (sequence s3F, Table 1) form an interlocked G-quadruplex?
Verification of (3 + 1) interlocked G-quadruplex formation by NMR
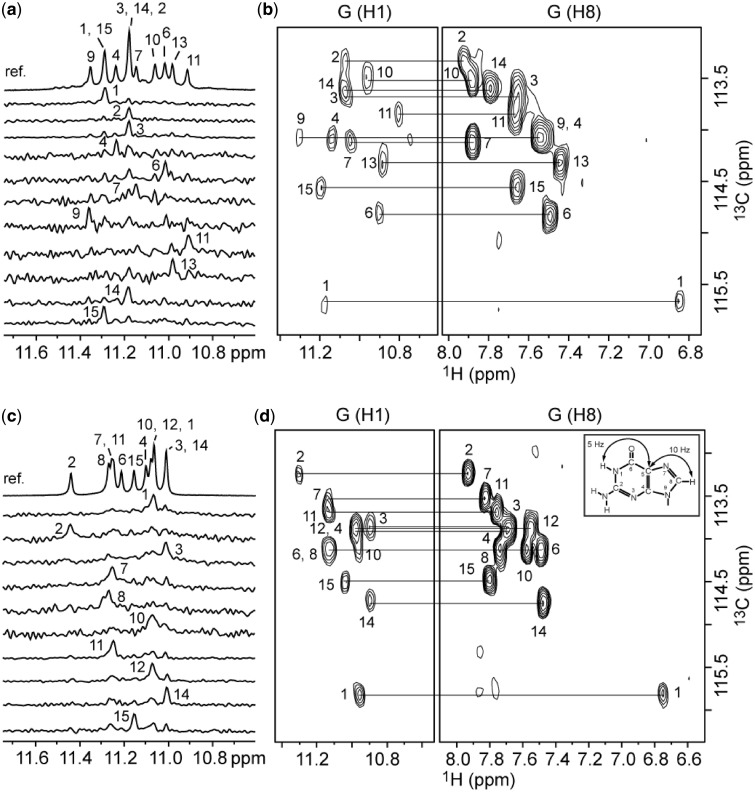
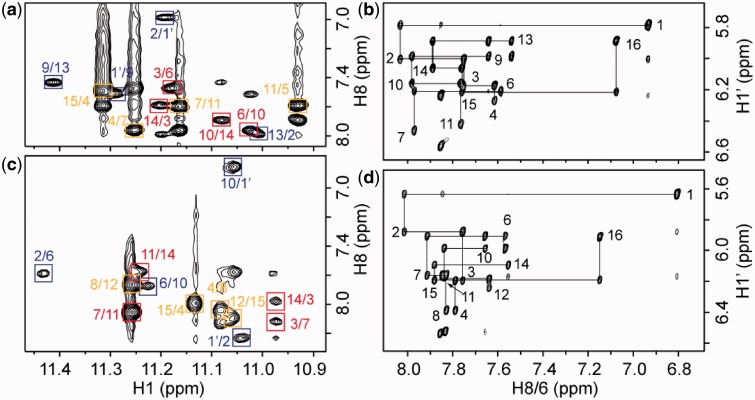
We used NMR to establish the interlocked G-quadruplex topologies of s2 and s4. Guanine imino and H8 protons were unambiguously assigned by using site-specific low-enrichment 15N labeling, site-specific 2H labeling and through-bond correlations at natural abundance (27–30) (Figure 2 and Supplementary Figure S1). With the help of these unambiguous assignments and other through-bond correlation experiments (COSY, TOCSY, [13C-1H]-HMBC and [13C-1H]-HSQC) (data not shown), the classical H8/H6-H1′ NOE sequential connectivity could be traced for four G-stretches (Figure 3). The intensity of intraresidue H8-H1′ Nuclear Overhauser Effect (NOE) cross-peaks indicated a syn glycosidic conformation for G1 and anti conformations for the other guanines (Supplementary Figure S2). Specific NOEs between imino and H8 protons established G-tetrad alignments (Figure 3) and confirmed the designed folding topology of s2 and s4 (Figure 1). Each structure comprises six G-tetrad layers formed by interlocking of two parallel-stranded G-quadruplex subunits. The folding topologies of 93del, s2 and s4 differ from each other by the relative positions of the two interlocking points: in 93del these two points are across the G-tetrad core diagonal, whereas in s2 and s4 these two points are of different adjacent configurations (Figure 1). These proposed folding topologies are consistent with deuterium exchange data, which showed that imino protons of the middle G-tetrads are well protected from the exchange with solvent (Supplementary Figure S3; data not shown). These interlocked G-quadruplexes are all extremely stable, as some imino protons persist in D2O solution even after 5 min at 100°C (Supplementary Figure S3).
Figure 2.
Resonance assignments of s2 and s4 in K+ solution at 25°C. (a and c) Imino protons of s2 and s4, respectively, were assigned by site-specific 2% 15N-labeling. (b and d) H8 protons of s2 and s4, respectively, were assigned by through-bond correlations between imino and H8 protons via 13C5 at natural abundance, using long-range J couplings shown as insert in (d).
Figure 3.
Determination of the G-quadruplex folding topology for the s2 and s4 sequences in K+ solution. (a and c) The imino-H8 proton region of NOESY spectra (mixing time, 200 ms) of s2 and s4, respectively at 25°C. The characteristic guanine imino-H8 cross-peaks for G-tetrads (colored red, yellow and blue, respectively) are framed and labeled with the imino proton assignment in the first position and that of the H8 proton in the second position. (b and d) The H8/6-H1′ proton region of NOESY spectra (mixing time, 300 ms) of s2 and s4, respectively at 25°C. The assignments and H8/6-H1′ NOE sequential connectivities are shown.
In contrast to the situation of s2 and s4, NMR imino proton spectra of sequences with the fourth G-tract being long (instead of the first one) (sequences s1R, s2R and s3R, Table 1) did not indicate the formation of a well-defined G-quadruplex (Supplementary Figure S4). For a sequence with one long (first G-tract) and three medium G-tracts (sequence s3F, Table 1), the imino proton spectrum (Supplementary Figure S4) indicated the formation of more than one G-quadruplex conformations, which could be different possible interlocked or non-interlocked G-quadruplexes. This result shows that the presence of a short G-tract is important to control the formation of a well-defined (3 + 1) interlocked G-quadruplex.
Engineering of structural elements in (3 + 1) interlocked G-quadruplexes
Different structural elements can be engineered in these robust interlocked scaffolds. In s2 and s4, all the non-G linkers are a thymine (T) residue, whereas in 93del two linkers are an adenine (A) residue (Table 1). In the latter structure, the adenine before the short leg (namely, A9) participates in the formation of a A•(G•G•G•G) pentad (Supplementary Figure S5). We attempted to design a A•(G•G•G•G) pentad in the s2 and s4 interlocked G-quadruplexes. The 1D NMR spectra of the corresponding sequences, s2-A and s4-A, where the T residue before the short leg was substituted by an A, indicated the formation of A•(G•G•G•G) pentads in these structures (Supplementary Figure S6). The formation of these pentads was further supported by NOESY experiments, e.g. the formation of the A5•(G3•G6•G10•G14) pentad in s2-A was supported by the detection of a NOE cross-peak between the A5(H8) and G6(H8) protons (Supplementary Figure S7). Reversely, A-to-T substitutions of 93del (i.e. sequence s3, Table 1) abolished the A•(G•G•G•G) pentad but retained the general folding topology (Supplementary Figure S6).
In these (3 + 1) interlocked G-quadruplex topologies, G1 adopts a syn conformation. NMR data (Supplementary Figure S8) showed that this residue in s4 could be substituted by a BrG, favoring a syn conformation in G-quadruplexes (31,32), without altering the general folding topology.
HIV-1 integrase inhibition activity of (3 + 1) interlocked G-quadruplexes
We demonstrated the inhibition activity of the three newly engineered sequences s2, s3 and s4 against HIV-1 integrase in the reverse ‘disintegration’ assays (33,34). Our results (Figure 4) indicated that these molecules have an HIV-1 integrase inhibition activity comparable with that of 93del and higher than that of negative controls (a duplex DNA or a G-quadruplex DNA formed by a human telomeric sequence). Note, however, that this conclusion should only be taken as qualitative, because the sample concentrations used in the assays were in the micromolar range, much higher than the HIV-1 integrase inhibition activity of 93del (10).
Figure 4.
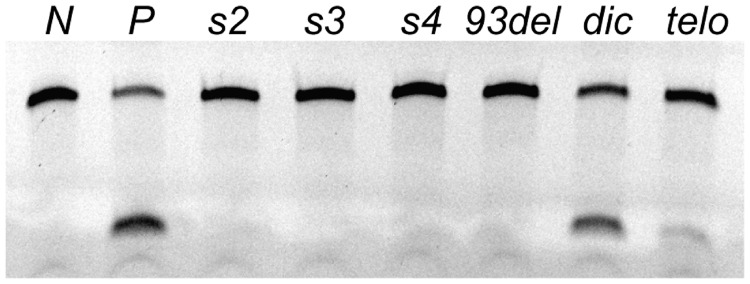
HIV-1 integrase inhibition disintegration assays. Lanes ‘N’ and ‘P’ are controls without and with HIV-1 integrase alone, respectively. Other lanes are inhibition assays with added inhibitors as indicated, where dic is the Dickerson d(CGCGAATTCGCG) dodecamer and telo is the human telomeric sequence d[TT(GGGTTA)3GGGA]. The concentrations of the enzyme, substrate and inhibitors were 0.6, 0.2 and 3.0 µM, respectively. The DNA substrate (dumbbell DB-Y1) and disintegration product (see ‘Methods’ section) are monitored as bands of 38 nt (top band) and 14 nt (bottom band), respectively (33,34).
CONCLUSION
We have engineered three different (3 + 1) interlocked dimeric G-quadruplexes. These structures are very stable and can serve as compact robust scaffolds for various applications. Different structural elements can be added in these robust interlocked scaffolds. The principle for interlocked G-quadruplex formation can be used for the assembly of higher-order structures.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–8.
FUNDING
Singapore Biomedical Research Council, Singapore Ministry of Education and Nanyang Technological University (to A.T.P.). Funding for open access charge: Nanyang Technological University.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 2.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phan AT, Kuryavyi V, Patel DJ. DNA architecture: from G to Z. Curr. Opin. Struct. Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Phan AT. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 7.Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH. Inhibition of human telomerase by a G-quadruplex-interactive compound. J. Med. Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 8.Balasubramanian S, Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded-DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 10.de Soultrait VR, Lozach PY, Altmeyer R, Tarrago-Litvak L, Litvak S, Andréola ML. DNA aptamers derived from HIV-1 RNase H inhibitors are strong anti-integrase agents. J. Mol. Biol. 2002;324:195–203. doi: 10.1016/s0022-2836(02)01064-1. [DOI] [PubMed] [Google Scholar]
- 11.Jing N, Li Y, Xiong W, Sha W, Jing L, Tweardy DJ. G-quartet oligonucleotides: a new class of signal transducer and activator of transcription 3 inhibitors that suppresses growth of prostate and breast tumors through induction of apoptosis. Cancer Res. 2004;64:6603–6609. doi: 10.1158/0008-5472.CAN-03-4041. [DOI] [PubMed] [Google Scholar]
- 12.Qi H, Lin C-P, Fu X, Wood LM, Liu AA, Tsai Y-C, Chen Y, Barbieri CM, Pilch DS, Liu LF. G-quadruplexes induce apoptosis in tumor cells. Cancer Res. 2006;66:11808–11816. doi: 10.1158/0008-5472.CAN-06-1225. [DOI] [PubMed] [Google Scholar]
- 13.Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009;86:151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultze P, Macaya RF, Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 15.Martino L, Virno A, Randazzo A, Virgilio A, Esposito V, Giancola C, Bucci M, Cirino G, Mayol L. A new modified thrombin binding aptamer containing a 5′-5′ inversion of polarity site. Nucleic Acids Res. 2006;34:6653–6662. doi: 10.1093/nar/gkl915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phan AT, Kuryavyi V, Ma J-B, Faure A, Andréola M-L, Patel DJ. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. Proc. Natl Acad. Sci. USA. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang YC, Sen D. A contractile electronic switch made of DNA. J. Am. Chem. Soc. 2010;132:2663–2671. doi: 10.1021/ja908508j. [DOI] [PubMed] [Google Scholar]
- 18.Chinnapen DJF, Sen D. A deoxyribozyme that harnesses light to repair thymine dimers in DNA. Proc. Natl Acad. Sci. USA. 2004;101:65–69. doi: 10.1073/pnas.0305943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wieland M, Hartig JS. Turning inhibitors into activators: A hammerhead ribozyme controlled by a guanine quadruplex. Angew. Chem. Int. Ed. Engl. 2006;45:5875–5878. doi: 10.1002/anie.200600909. [DOI] [PubMed] [Google Scholar]
- 20.Tang Z, Goncalves DPN, Wieland M, Marx A, Hartig JS. Novel DNA catalysts based on G-quadruplex recognition. Chembiochem. 2008;9:1061–1064. doi: 10.1002/cbic.200800024. [DOI] [PubMed] [Google Scholar]
- 21.Cai J, Shapiro EM, Hamilton AD. Self-assembling DNA quadruplex conjugated to MRI contrast agents. Bioconjug. Chem. 2009;20:205–208. doi: 10.1021/bc8004182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li T, Wang EK, Dong SJ. Potassium-lead-switched G-quadruplexes: A new class of DNA logic gates. J. Am. Chem. Soc. 2009;131:15082–15083. doi: 10.1021/ja9051075. [DOI] [PubMed] [Google Scholar]
- 23.Alberti P, Bourdoncle A, Sacca B, Lacroix L, Mergny JL. DNA nanomachines and nanostructures involving quadruplexes. Org. Biomol. Chem. 2006;4:3383–3391. doi: 10.1039/b605739j. [DOI] [PubMed] [Google Scholar]
- 24.Majumdar A, Kettani A, Skripkin E, Patel DJ. Pulse sequences for detection of NH2… N hydrogen bonds in sheared G. A mismatches via remote, non-exchangeable protons. J. Biomol. NMR. 2001;19:103–113. doi: 10.1023/a:1008311624772. [DOI] [PubMed] [Google Scholar]
- 25.Krishnan-Ghosh Y, Liu DS, Balasubramanian S. Formation of an interlocked quadruplex dimer by d(GGGT) J. Am. Chem. Soc. 2004;126:11009–11016. doi: 10.1021/ja049259y. [DOI] [PubMed] [Google Scholar]
- 26.Miyoshi D, Karimata H, Wang Z-M, Koumoto K, Sugimoto N. Artificial G-wire switch with 2,2′-bipyridine units responsive to divalent metal ions. J. Am. Chem. Soc. 2007;129:5919–5925. doi: 10.1021/ja068707u. [DOI] [PubMed] [Google Scholar]
- 27.Phan AT, Patel DJ. A site-specific low-enrichment 15N,13C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J. Am. Chem. Soc. 2002;124:1160–1161. doi: 10.1021/ja011977m. [DOI] [PubMed] [Google Scholar]
- 28.Phan AT. Long-range imino proton-13C J-couplings and the through-bond correlation of imino and non-exchangeable protons in unlabeled DNA. J. Biomol. NMR. 2000;16:175–178. doi: 10.1023/a:1008355231085. [DOI] [PubMed] [Google Scholar]
- 29.Huang XN, Yu PL, LeProust E, Gao XL. An efficient and economic site-specific deuteration strategy for NMR studies of homologous oligonucleotide repeat sequences. Nucleic Acids Res. 1997;25:4758–4763. doi: 10.1093/nar/25.23.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim KW, Alberti P, Guédin A, Lacroix L, Riou J-F, Royle NJ, Mergny J-L, Phan AT. Sequence variant (CTAGGG)n in the human telomere favors a G-quadruplex structure containing a G.C.G.C tetrad. Nucleic Acids Res. 2009;37:6239–6248. doi: 10.1093/nar/gkp630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorgan. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 32.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chow SA, Vincent KA, Ellison V, Brown PO. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 34.Do NQ, Lim KW, Teo MH, Heddi B, Phan AT. Stacking of G-quadruplexes: NMR structure of a G-rich oligonucleotide with potential anti-HIV and anticancer activity. Nucleic Acids Res. 2011;39:9448–9457. doi: 10.1093/nar/gkr539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.