Abstract
Human immunodeficiency virus genome dimerization is initiated through an RNA–RNA kissing interaction formed via the dimerization initiation site (DIS) loop sequence, which has been proposed to be converted to a more thermodynamically stable linkage by the viral p7 form of the nucleocapsid protein (NC). Here, we systematically probed the role of specific amino acids of NCp7 in its chaperone activity in the DIS conversion using 2-aminopurine (2-AP) fluorescence and nuclear magnetic resonance spectroscopy. Through comparative analysis of NCp7 mutants, the presence of positively charged residues in the N-terminus was found to be essential for both helix destabilization and strand transfer functions. It was also observed that the presence and type of the Zn finger is important for NCp7 chaperone activity, but not the order of the Zn fingers. Swapping single aromatic residues between Zn fingers had a significant effect on NCp7 activity; however, these mutants did not exhibit the same activity as mutants in which the order of the Zn fingers was changed, indicating a functional role for other flanking residues. RNA chaperone activity is further correlated with NCp7 structure and interaction with RNA through comparative analysis of nuclear magnetic resonance spectra of NCp7 variants, and complexes of these proteins with the DIS dimer.
INTRODUCTION
Human immunodeficiency virus-1 (HIV-1), in its mature form, contains two copies of genomic RNA (gRNA) non-covalently bound as a homodimer through the dimerization initiation site (DIS) (1). Viral RNA dimerization has been shown to be involved in various steps in the viral life cycle such as recombination, reverse transcription and gag translation (2). The DIS is located within the viral packaging sequence in the 5′-untranslated region (5′-UTR) known as the Ψ site (3–8). The Ψ site contains four conserved stem-loop structures (SL1–SL4) that are linked by 4–13 nucleotide single-stranded regions, where the SL1 contains the DIS, SL2 is the major splice donor site, SL3 forms the principal portion of the packaging signal and SL4 contains the AUG translation initiation codon site. Dimerization of the gRNA is initiated through formation of an RNA–RNA kissing complex between the highly conserved palindromic loop sequences of the SL1 stem-loop. In vitro studies of short RNA constructs that mimic the SL1 stem-loop have shown spontaneous formation of RNA–RNA kissing complexes that are stabilized by magnesium (9–13). Further, the RNA–RNA kissing dimer can be converted in vitro to a more thermodynamically stable extended duplex dimer by incubation at a temperature of 55°C or by addition of nucleocapsid protein p7 (NCp7) (14–17). From these studies, it has been shown that this two-step process of first formation of the DIS kissing dimer followed by stem strand exchange to form the extended duplex dimer is dependent on the SL1 stem-loop concentration, the RNA sequence/structure and ionic conditions. The SL1 RNA constructs in these studies demonstrated the essential role of RNA sequence in governing the DIS kissing dimer maturation through overlapping modulation of kissing dimer stability, local structural dynamics and interaction with NCp7. It has been proposed that the conversion of the SL1 kissing dimer linkage to an extended duplex structure also occurs in the context of the genome during budding of the virus particle, and that this process is chaperoned by NCp7 (18,19). In addition to playing a role in the maturation of the gRNA dimer, NCp7 is also essential in other important processes in HIV replication, such as reverse transcription (20–27).
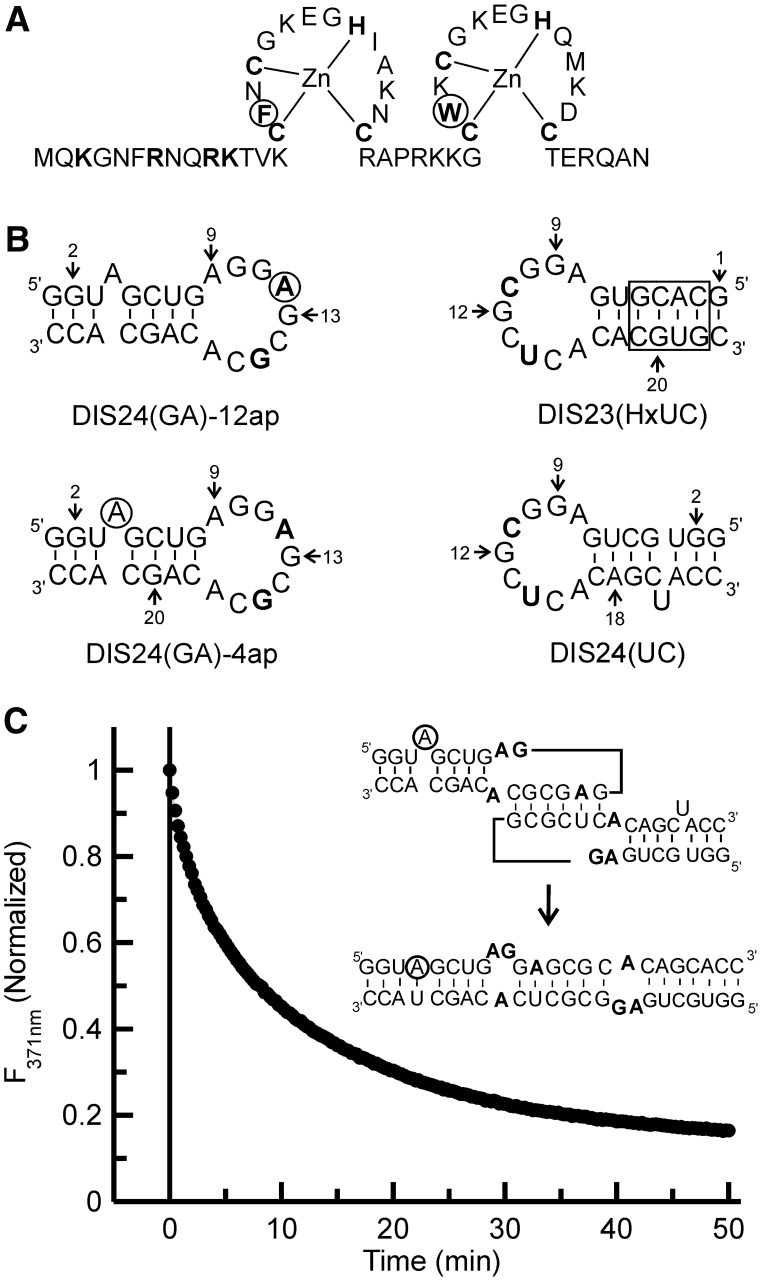
NCp7, the fully processed protein found in mature HIV-1, is a 55-amino acid basic protein that is initially synthesized as a domain of the Gag polyprotein. NCp7 contains two Zinc (Zn) finger motifs of the type Cys-X2-Cys-X4-His-X4-Cys (CCHC) that are linked by a basic seven amino acid sequence, flanked by a highly basic N-terminus and a short six amino acid C-terminus (28). In each Zn finger, a Zn ion is bound in a tetrahedral coordination complex by cysteine thiol side chains and the imidazole ring of histidine. The highly conserved Zn fingers each contain one aromatic residue, phenylalanine in the N-terminal Zn finger (often referred to as the proximal Zn finger), tryptophan in the C-terminal Zn finger (the distal Zn finger), and they differ in only five amino acid positions in the NL4-3 strain, which is used in the current study, as shown in Figure 1A. Three-dimensional structures of the full-length NCp7 (29,30) and an N-terminally truncated version of the protein (NCp7 13–55) (31) have been solved using high-resolution nuclear magnetic resonance (NMR) methods. The full-length NCp7 structure has a disordered N-terminus that has been shown to form a 310 helix on binding Ψ site RNA stem-loops (32,33). In the N-terminally truncated NCp7 structure, the aromatic residues in the Zn fingers were found to be juxtaposed to each other and form a hydrophobic patch (31). This structural positioning of the tryptophan within a hydrophobic patch provides a platform for stacking interactions with guanine bases in single-stranded and loop regions of RNA (34,35).
Figure 1.
(A) NCp7 wild-type protein sequence. Zn coordinating and basic residues in the N-terminus are shown in bold. The aromatic residues Phe16 and Trp37 are in bold and circled. (B) SL1 hairpin constructs. The residues changed from the wild-type sequence in the DIS loop region are bolded, and the adenines that are replaced by 2-AP in the two fluorescent constructs [DIS24(GA)-4ap and DIS24(GA)-12ap] are circled. The DIS24(UC) contains a complementary bulged uracil that forms a base-pair on maturation and the exchanged stem in the DIS23(HxUC), which disfavors formation of the duplex via strand exchange, are boxed. The DIS23(GA) hairpin used in the NMR experiments is the same sequence as the DIS24(GA) constructs with the exception that the bulged adenosine at position 4 is deleted. (C) Representative plot of the DIS kissing complex to extended duplex conversion measured by 2-AP fluorescence emission quenching as a function of time after the addition of wild-type NCp7 to the DIS24(GA)-4ap•DIS24(UC) kissing complex at physiological pH (225 nM NCp7 protein was added to 100 nM of RNA kissing complex). (Inset) Schematic of the DIS kissing complex to extended duplex conversion is shown with the 2-AP position circled.
Owing to NCp7’s multiple roles in HIV replication and its validation as a target for anti-HIV therapies, several groups have studied different aspects of NCp7 function, including, but not limited to, its role in tRNA priming, gRNA packaging and reverse transcription, both in vivo and in vitro (23,28,36–45). These studies have been carried out using the entire gRNA, or model RNA stem-loop structures and have resulted in identification of various residues of NCp7 that are essential for NCp7’s RNA binding and its different chaperone activities. For example, using an NCp7 SSHS/SSHS mutant, where the Zn coordinating cysteines were mutated to serines, and terbium cleavage experiments, it has been shown that the positive charge of the NCp7 alone is sufficient for the annealing of tRNALys,3 to the primer binding site (PBS), but the Zn fingers are needed for efficient destabilization of the tRNA core structure (40). In vivo experiments carried out using Zn finger coordination mutants (H23C and H44C, which change the Zn coordination motif from CCHC to CCCC) have shown that the wild-type Zn fingers are strictly required for efficient reverse transcription and viral DNA protection as well as initial integration events (41,46). Using chemical and enzymatic probing assays, Kanevsky et al. (47) have also shown that NCp7 Zn fingers interact with the guanine residues in the apical loop of Trans activating response element (TAR), whereas the N-terminal basic residues may facilitate binding to the upper stem region. Further, a comprehensive in vivo study of the effect of 40 different mutations of NCp7 on gRNA dimerization has found that each segment of NCp7 (the N-terminus, the two Zn fingers and the linker) plays a role in the efficient gRNA dimerization and packaging (36). Using a fragmentation approach and an in vitro gel assay to dissect the requirements for the Zn fingers and basic regions of NCp7 in its chaperoning activity, it was found that the basic regions were necessary and sufficient for promoting the conversion of SL1 stem-loop DIS kissing dimers (48,49). These studies, however, found that 10 times the concentration of basic peptide relative to NCp7 was needed for comparable activity, showing that the destabilizing function of the Zn fingers, as well as the protein structural context, are necessary for full native chaperone activity.
Studies using NCp7 mutants have also provided details about the role of various highly conserved residues in NCp7’s RNA binding activity. Filter binding assays have shown that the elimination or reduction of positive charge in the N-terminus (particularly residues at positions 3, 7, and 10) resulted in reduced levels of binding to RNA (50). Additionally, investigations of RNA-binding properties of various NCp7 mutants using homopolymeric fluorescently labeled RNA have shown that swapping the Zn finger positions has little or no effect on NCp7’s ability to bind RNA, whereas removal of Zn from the protein, by inducing intra-molecular disulfide bond formation of the Zn coordinating cysteines, results in a loss of non-electrostatic interactions between the protein and the RNA (51). The mutations of the Zn finger motifs also revealed that the N-terminal Zn finger is more essential than the C-terminal finger in determining binding affinity to the RNA. Replacing the aromatic residues in the Zn fingers also resulted in reduction of NCp7’s nucleic acid binding affinity (51,52). The binding interactions of NCp7 to the DIS kissing dimer and their relationship to the mechanism of NCp7 chaperoned DIS maturation have also been investigated by mass spectroscopy (53,54). Through mapping the protein–RNA complexes formed with the isolated SL1 stem-loop and SL1 dimers, these studies showed that binding of NCp7 to the SL1 stem-loop hampered the DIS kissing dimer formation, whereas binding to SL1 stem-bulge in the context of the DIS kissing dimer resulted in a destabilization of the local structure and promotion of the kissing to extended duplex conversion. In contrast, binding of NCp7 to the flanking purines in the DIS loop resulted in a partial inhibition of stem strand exchange in the DIS dimer maturation process, which was attributed to a stabilizing sequestration of these purine nucleotides through the NCp7 interaction that acts at odds with the destabilizing effect of destacking these bases by the protein.
We have previously reported the use of 2-aminopurine (2-AP) as a fluorescent probe to study structural changes of RNA associated with dimerization and NCp7 chaperoned conversion to the mature duplex (16,55,56). 2-AP is an isomorphous base analog of adenine and can form Watson–Crick hydrogen bonding with Uracil. The sensitivity of the quantum yield of 2-AP fluorescence to its microenvironment makes it a useful probe to study structural changes in RNA that result during chaperone mediated refolding.
In the current study, we have used the 2-AP fluorescence assays to systematically investigate several highly functional, as well as loss-of-function, mutants of NCp7 to identify the key amino acid residues involved in NCp7 chaperoned DIS maturation. In addition, comparative NMR spectral analysis was used to probe for structural differences between representatives from the different classes of NCp7 mutants and the wild-type protein. NMR experiments were also used to map differences in the binding interface in complexes formed between wild-type NCp7 and the DIS kissing RNA complex, and those formed with selected NCp7 mutants.
MATERIALS AND METHODS
RNA sample preparation
2-AP incorporated RNA hairpins were synthesized as described previously (16). Briefly, the 2-AP incorporated DIS hairpins (Figure 1B) were synthesized either using standard TOM-protected solid-phase synthesis (57) or commercially obtained from Dharmacon Inc (Lafayette, CO). The standard and 2-AP modified nucleoside phosphoramidites were purchased from Glen Research (Sterling, VA). Unlabeled DIS hairpins were synthesized by in vitro run off transcription using T7 polymerase and standard protocols (58,59). The purification of the RNA was done using standard deprotection followed by gel electrophoresis and electro-elution methods. The purified RNA was extensively dialyzed against standard buffer (1 mM of cacodylate (pH 6.5) and 25 mM of NaCl).
NC mutants
Unlabeled and uniformly 15N labeled wild-type NCp7 were expressed and purified as described previously (32). The mutant forms of NCp7 (derived from pNL4-3) were expressed and purified as described previously (60). Purified NCp7 protein aliquots were stored at −80°C in 25 mM of sodium acetate (pH 6.5), 25 mM of NaCl and 0.1 mM of ZnCl2.
Fluorescence measurements
All the fluorescence experiments were carried out on a SPEX Fluoromax-3 spectrofluorometer (Instruments SA, Edison, NJ) using a 3-mm square quartz cuvette. The buffer conditions and data collection procedure are as described previously (56). Briefly, the kissing complex was first formed by adding 5 mM of MgCl2 to the solution containing DIS24(GA)-4ap and DIS24(UC) in standard buffer. The chaperoning activity of NCp7 wild-type and mutants was determined at 25°C by following the decrease in the fluorescence of 2-AP probe at 371 nm during a period of time, immediately after adding either the wild-type or the mutant form of NCp7 to the pre-formed kissing complex. The time course of the NCp7 chaperoned conversion of the DIS kissing complex to the extended duplex was fit using a single-exponential equation,
| (1) |
where kconv is the observed rate of conversion of kissing complex to extended duplex. The helix destabilization assays using DIS24(GA)-12ap and DIS24(UC) were performed in the same way. The percentage increase in 12ap fluorescence is calculated by taking the difference between the absolute fluorescence of the kissing complex and the fluorescence immediately after addition of the respective protein as shown in equation 2.
| (2) |
where Fp is the percentage increase in the 2-AP fluorescence, Fm is the fluorescence immediately after the addition of protein to the preformed kissing complex, and Fk is the fluorescence of the 2-AP probe in the kissing complex. In both the assays, the concentration of the RNA kissing complex is 100 nM, and NCp7 is added to a final concentration of 225 nM.
NMR spectroscopy
All the NMR experiments were collected on either Bruker 600 MHz DMX spectrometer equipped with 1H/13C/15N or 1H/13C/31P triple resonance probe or Bruker 600 MHz AVANCE spectrometer with a 1H/13C/15N triple resonance, Z-axis gradient Cryoprobe. The data were processed using NMRPipe (61) and analyzed using Sparky (62). Water suppression was achieved using either water flip-back pulse scheme or pulsed field gradients. All data were collected at 25°C, and the proton resonances were referenced to water resonance at 4.7 ppm. Resonance assignments for the imino protons of the DIS hairpins and kissing complex were done using 1D nuclear overhauser effect (NOE) experiments as described previously (55). Backbone assignments of the proteins were done using a series of 2D and 3D NMR experiments, which included 2D 15N-1H HSQC, 3D HSQC-NOESY and 3D HSQC-TOCSY. 3D triple resonances experiments, including a HNCA and a HN(CO)CA, were applied to a 13C, 15N labeled N-terminal mutant of NCp7 to complete the peptide backbone resonance assignments.
RESULTS
Detecting RNA kissing dimer strand transfer activity of NCp7
Conversion of the DIS kissing dimer into an extended duplex involves destabilization of the kissing complex and eventual stem strand exchange between stem-loops. NCp7-chaperoned strand exchange between stem-loops can be observed using DIS hairpin loops, with 2-AP probes inserted into the stem sequence, as has been described previously (Figure 1B) (16). Briefly, incorporation of 2-AP into a bulge position of the DIS {this construct is referred to as [DIS24(GA)-4ap]} allows the formation of the extended duplex in the presence of wild-type and mutants of NCp7 to be monitored. Using this construct, stacking of 2-AP on forming a base pair with the complementary U in the DIS24(UC) hairpin in the extended duplex, leads to a 3–6-fold decrease in the fluorescence yield of the probe. The time-dependent quenching of 2-AP fluorescence in the presence of NCp7 is monitored to obtain the rate of NCp7 chaperoned conversion of kissing duplex to an extended duplex (stem strand transfer ability of NCp7) and the data can be fit with simple exponentials to extract kinetic rates (see Materials and Methods). The NCp7:DIS ratio and concentrations used herein are based on previous work using the same SL1 hairpin model system for the kissing dimer (16), where different NCp7 to DIS dimer ratios were tested to determine the ‘threshold requirement’ for near optimal chaperone activity. The previous work showed that NCp7 bound to this model DIS dimer with a stoichiometry of ∼2:1 (KD ∼ 60 nM), and that a molar ratio greater than ∼2.5 NCp7:DIS dimer (an NCp7 to nt molar ratio of 1–20) did not increase the maturation rate appreciably. Figure 1C shows a representative trace for the 2-AP-detected NCp7 chaperoned conversion of the DIS kissing dimer to mature duplex. A 6-fold decrease in the fluorescence of DIS24(GA)-4ap•DIS24(UC), relative to the initial state, is observed on mixing the kissing complex with two equivalents of wild-type NCp7, which is consistent with previously observed quenching of 2-AP during wild-type NCp7 chaperoned RNA dimer conversion at physiological pH (16). In the chaperone assays, the NCp7 protein concentration is well above the KD reported for wild-type, as well as mutant NCp7 variants, binding to high affinity targets (62) under similar buffer conditions, giving confidence that any such binding sites on the DIS kissing dimer are saturated in the experiments. Nonetheless, for mutants with known weaker binding affinities for RNA and for which chaperone activity was observed to be significantly diminished, the 2-AP assays were repeated at higher concentrations of RNA and protein (200 nM of kissing dimer and 450 nM of NCp7). In these additional assays, the extent of the conversion of kissing complex to extended dimer was found to not change significantly (Supplementary Figure S1).
Comparison of NCp7 wild-type and mutant chaperone activity in DIS maturation
The NCp7 mutants studied here are classified into four categories: Zn finger swap mutants, Zn finger coordination mutants, Zn finger aromatic residue mutants and an N-terminal basic amino acid mutant (Table 1). These mutants were designed to probe the role of the structure and sequence of NCp7 in its chaperone function.
Table 1.
Sequences of NCp7 mutants studied in the present work (the wild-type sequence corresponds to the HIV-1 from pNL4-3)
| NC wild-type | M Q K G N F R N Q R K T V K CF NCG K E GHI A K NC R A P R K R G CW KCG K E GHQ M K DC T E R Q A N |
| Finger swap mutants (36): | |
| NC 1/1 | M Q K G N F R N Q R K T V K CF NCG K E GHI A K NC R A P R K R G CF NCG K E GHI AKNC T E R Q A N |
| NC 2/2 | M Q K G N F R N Q R K T V K CW KCG K E GHQ MKDC R A P R K R G CW KCG K E GHQ M K DC T E R Q A N |
| NC 2/1 | M Q K G N F R N Q R K T V K CW KCG K E GHQ MKDC R A P R K R G CF NCG K E GHI AKNC T E R Q A N |
| Zn coordination mutants (24,47): | |
| H23C | M Q K G N F R N Q R K T V K CF NCG K E GCI A K NC R A P R K R G CW KCG K E GHQ M K DC T E R Q A N |
| H44C | M Q K G N F R N Q R K T V K CF NCG K E GHI A K NC R A P R K R G CW KCG K E GCQ M K DC T E R Q A N |
| H23C/H44C | M Q K G N F R N Q R K T V K CF NCG K E GCI A K NC R A P R K R G CW KCG K E GCQ M K DC T E R Q A N |
| SSHS/SSHS | M Q K G N F R N Q R K T V K SF NSG K E GHI A K NS R A P R K R G SW KSG K E GHQ M K DS T E R Q A N |
| Aromatic and basic residue mutants (34,49): | |
| F16A | M Q K G N F R N Q R K T V K CANCG K E GHI A K NC R A P R K R G CW KCG K E GHQ M K DC T E R Q A N |
| W37A | M Q K G N F R N Q R K T V K CF NCG K E GHI A K NC R A P R K R G CAKCG K E GHQ M K DC T E R Q A N |
| F16A/W37A | M Q K G N F R N Q R K T V K CANCG K E GHI A K NC R A P R K R G CAKCG K E GHQ M K DC T E R Q A N |
| F16W | M Q K G N F R N Q R K T V K CWNCG K E GHI A K NC R A P R K R G CW KCG K E GHQ M K DC T E R Q A N |
| W37F | M Q K G N F R N Q R K T V K CF NCG K E GHI A K NC R A P R K R G CFKCG K E GHQ M K DC T E R Q A N |
| F16W/W37F | M Q K G N F R N Q R K T V K CWNCG K E GHI A K NC R A P R K R G CFKCG K E GHQ M K DC T E R Q A N |
| Nterm (62): | M Q A G N F A N Q A A I I ACF NCG K E GHI A K NC R A P R K R G CW KCG K E GHQ M K DC T E R Q A N |
The residues of the Zn finger motifs are underlined with the Zn binding Cys and His residues in bold. The mutations are shown in italics.
Zn finger swap mutants
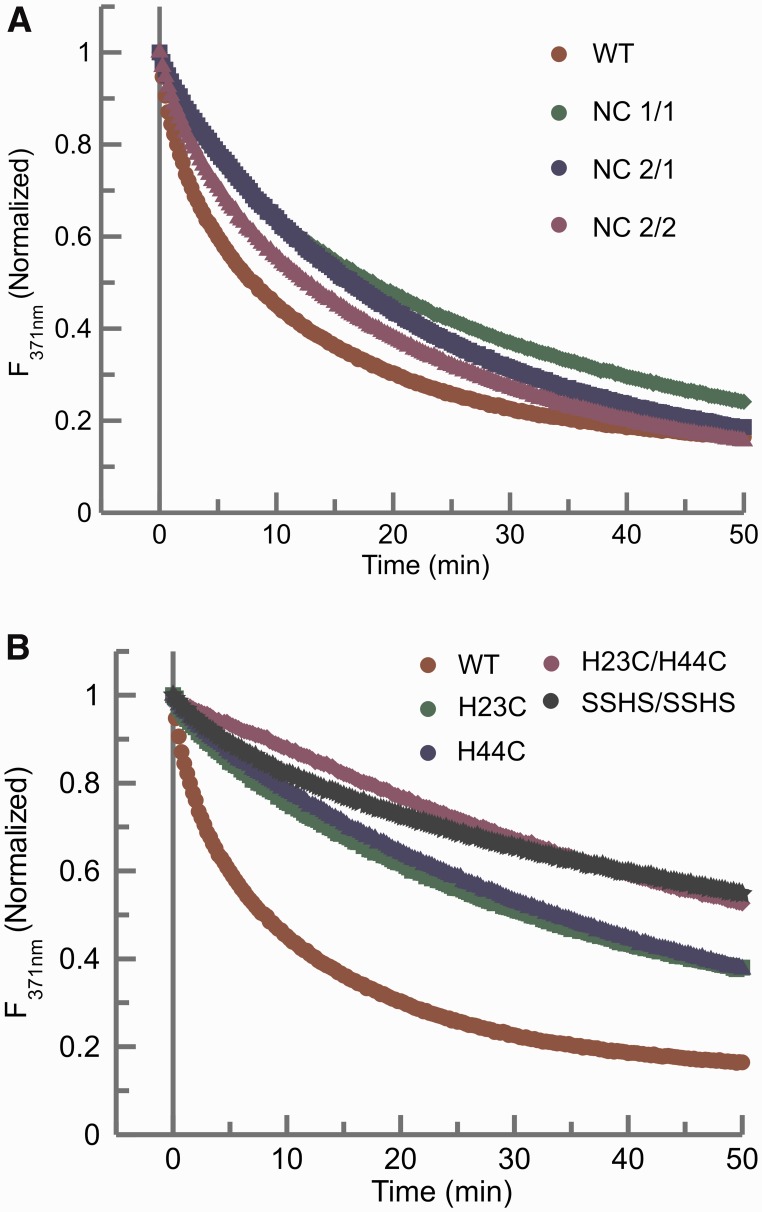
To understand the contribution of each Zn finger in the chaperoning activity of NCp7, three positional variants of Zn fingers were examined. In the NC 1/1 mutant, the N-terminal Zn finger replaces the C-terminal Zn finger; in the NC 2/2 mutant, the N-terminal Zn finger is replaced by the C-terminal Zn finger; finally, in the NC 2/1 mutant, the N-terminal and C-terminal Zn fingers are swapped (38). In the 2-AP assay for chaperone activity, all three Zn finger swap mutants of NCp7 show similar chaperoning activity, with slightly slower kinetic rates of kissing complex to extended duplex conversion when compared with the wild-type protein (Figure 2A and Table 2).
Figure 2.
(A) Comparison of the time dependent 2-AP fluorescence emission quenching plots of the wild-type NCp7 and the Zn finger positional variants of NCp7 (Table 1). NC 1/1 (green); NC 2/2 (purple); NC 2/1 (blue). (B) Comparison of the time dependent 2-AP fluorescence emission quenching plots of the wild-type NCp7 and the Zn finger coordination variants of NCp7: H23C (green), H44C (blue) and H23C/H44C (purple), and SSHS/SSHS (black). In both panels, the NCp7 wild-type curve (red) is plotted as a reference for comparison. In each of these assays, 225 nM of the mutant NCp7 protein was added to 100 nM of RNA kissing complex. The 2-AP quenching curves were fit to an exponential equation to obtain the rates of conversion of kissing complex to extended duplex (Table 2).
Table 2.
Rates of chaperoning activities and loop-loop helix ‘destacking’ of various mutants of NCp7 studied in the present work
| Protein | Strand exchange (min−1)a | Helix destacking (% increase)b |
|---|---|---|
| NC Wt | 0.091 | 116.44 |
| NC 1/1 | 0.046 | 97.92 |
| NC 2/1 | 0.048 | 54.64 |
| NC 2/2 | 0.057 | 50.81 |
| H23C | 0.033 | 18.33 |
| H44C | 0.026 | 40.30 |
| H23C/H44C | n.d.c | 22.66 |
| SSHS/SSHS | n.d. | 17.82 |
| F16A | n.d. | 194.45 |
| W37A | n.d. | 35.36 |
| FAWA | n.d. | 16.37 |
| F16W | 0.061 | 54.21 |
| W37F | 0.037 | 76.26 |
| FWWF | 0.045 | 30.53 |
| N-term | n.d. | 25.08 |
n.d., Not determined.
aTime course of 2-AP (Equation 1), located in the bulge position of DIS24(GA) hairpin, fluorescence quenching fit to a single exponential provided the rates of conversion of kissing to duplex dimer.
bPercentage increase in 2-AP (Equation 2), positioned in the apical loop of the DIS24(GA) hairpin, fluorescence on addition of the respective NCp7 mutant to a preformed kissing complex.
cRates were not fit to an exponential owing to the incompletion of strand conversion within the experimental time of 60 min.
Zn finger coordination mutants
In both the N-terminal and C-terminal Zn fingers of NCp7, Zn is coordinated to the sulfur groups of three cysteines and the imidazole ring of a histidine in a CCHC type of Zn finger. To probe the role of the type of Zn finger architecture (CCHC versus CCCC) and the presence of Zn in NCp7 chaperoning activity, we assayed several mutants of NCp7 in which the Zn finger coordination was changed. The N-terminal, C-terminal, or both Zn fingers were changed to the type Cys-X2-Cys-X4-Cys-X4-Cys (CCCC) by mutating the histidine to cysteine at positions 23, 44 or both (respectively H23C, H44C and H23C/H44C mutants; Table 1) (51). Both single Zn finger coordination mutants, H23C and H44C, show almost a 66% loss of chaperone activity compared with the wild-type protein (Figure 2B), whereas the double mutant (H23C/H44C) in which both the histidines are mutated to cysteines showed an even greater loss of activity compared with the wild-type. To probe the requirement of the Zn finger structures in NCp7’s chaperone activity, we examined an SSHS/SSHS mutant wherein all the Zn coordinating cysteines were mutated to serines (26). Such a mutant is unable to coordinate Zn and therefore does not adopt stable Zn finger structures. For this mutant protein, which is expected to be essentially unstructured, we again observed a dramatic loss of chaperone function compared with the wild-type protein (Figure 2B). In the case of the double mutant (H23C/H44C) and the SSHS/SSHS mutant, only a qualitative comparison could be made with wild-type protein activity owing to the limiting amount of strand conversion (∼40%) within the experimental timeframe.
Zn finger aromatic residue mutants
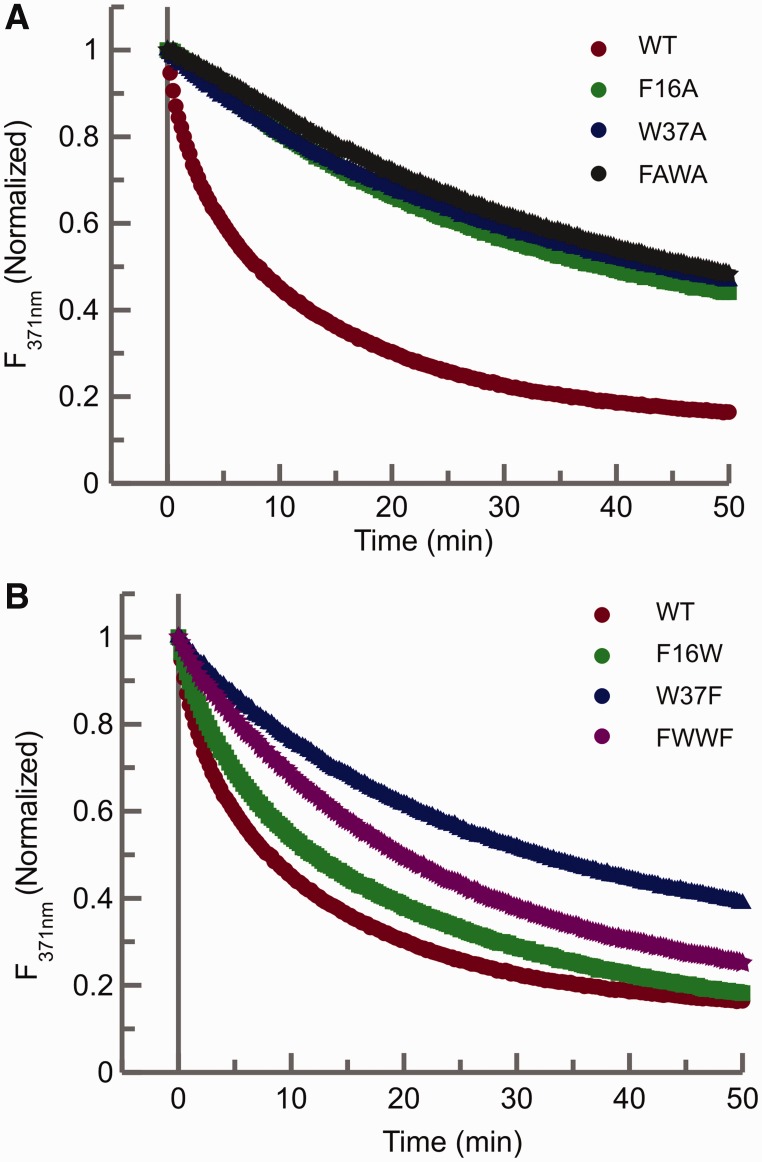
Each Zn finger in the NCp7 contains one aromatic residue, a phenylalanine at position 16 in the N-terminal and tryptophan at position 37 in the C-terminal Zn finger. Mutational studies of these residues have indicated their essential role in the chaperoning activity of NCp7 in the context of other replication functions (36,52). Using the 2-AP fluorescence assays, we again measured chaperone activity for NCp7 proteins with both single and double mutations of these aromatic residues. Mutations were made by replacing the aromatic residue with alanine one at a time (F16A and W37A) to create single mutants or simultaneously (F16A/W37A or FAWA) to create a double mutant. The significance of the identity of the aromatic residue in their respective Zn fingers was also probed by making either single or double swaps of these residues (F16W, W37F and F16W/W37F or FWWF mutants). The chaperoning activity of the F16A, W37A and F16A/W37A mutants are all similarly and greatly reduced as monitored by the 2-AP assay (Figure 3A). In contrast, the aromatic acid residue swap mutants (F16W, W37F and F16W/W37F) show varying degrees of chaperoning activity (Figure 3B). The presence of tryptophan in the N-terminal Zn finger (F16W) in the place of phenylalanine did not significantly affect the chaperone activity of NCp7 and showed a rate similar to the NC 2/2 mutant (Table 2). In contrast, the substitution of the tryptophan residue in the C-terminal finger (W37F mutant) shows a ∼60% loss of function compared with the wild-type and a reduced rate relative to the NC 1/1 mutant (Table 2). The differences in the chaperoning ability of W37F and NC 1/1 indicate a possible structural interplay between residue W37 and other amino acids in the C-terminal Zn finger in the chaperone function of NCp7.
Figure 3.
(A) Comparison of the time dependent 2-AP fluorescence emission quenching plots of the wild-type NCp7 and the aromatic amino acid point mutants of NCp7. Time-dependent fluorescence quenching plots of the wild-type NCp7 (red) and single point alanine mutants of the aromatic residues, F16A (green), W37A (blue) and the double mutant F16A/W37A, named here as FAWA (dark green). (B) Fluorescence decay curves of the aromatic amino acid residue swap mutants: F16W (green), W37F (blue) and FWWF (purple). In both panels, the NCp7 wild-type curve (red circles) is plotted as a reference for comparison. In each of these assays, 225 nM of the mutant NCp7 protein was added to 100 nM of RNA kissing complex. The 2-AP quenching curves were fit to an exponential equation to obtain the rates of conversion of kissing complex to extended duplex (Table 2).
N-terminal mutant of NCp7
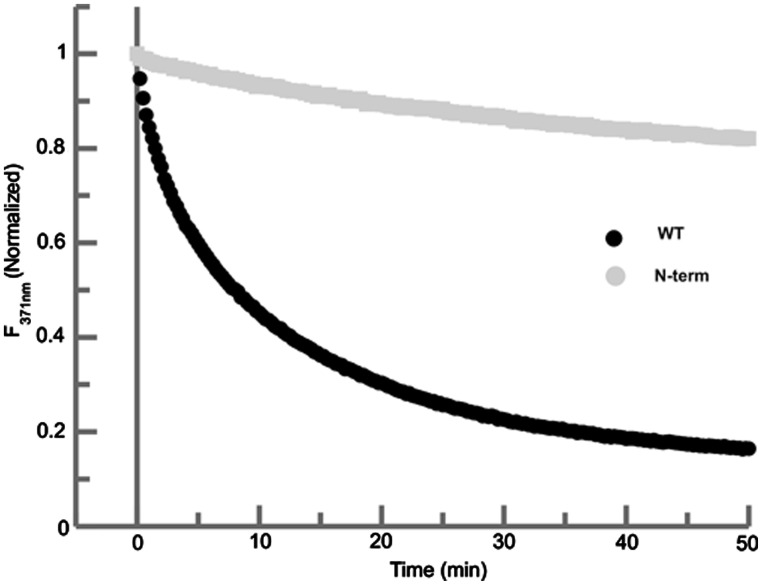
The highly basic N-terminus of NCp7 has been shown to be involved in binding to the RNA (50,63,64). To delineate the role of these basic residues in the chaperoning activity of NCp7 with respect to the conversion of the DIS dimer, we have studied a mutant (N-term) of NCp7 in which all the basic residues, Lys3, Arg7, Arg10, Lys11 and Lys14, in the N-terminus were simultaneously mutated to alanine (Table 1) (63). Using the 2-AP assay, a lack of a significant time-dependent quench was observed for the N-term mutant during the time course of the reaction indicating that the mutation of the basic residues of N-terminus to alanine almost completely abolished the RNA chaperoning activity of NCp7 in the time scale of our experiments (Figure 4).
Figure 4.
Comparison of the time dependent fluorescence decay plots of the wild-type NC (black) and the N-terminal mutant of NCp7 (gray). In this assay, 225 nM of N-terminal mutant of NCp7 protein was added to 100 nM of RNA kissing complex to again be consistent with the wild-type NCp7 2-AP fluorescence assay.
Detecting RNA kissing dimer loop-loop helix ‘destabilization’ by NCp7
Structural changes in the RNA kissing dimer loop-loop helix on binding of NCp7 were probed using complexes formed with a model DIS stem-loop with 2-AP incorporated into the loop [DIS24(GA)-12ap hairpin] and the complementary DIS stem-loop [DIS24(UC)]. In the kissing dimer, the fluorescence of DIS24(GA)-12ap is highly quenched, as it forms a well-stacked base pair with the complementary U in the loop of DIS24(UC). On addition of wild-type NCp7 to the DIS24(GA)-12ap•DIS24(UC) kissing complex (225 nM protein to 100 nM RNA kissing complex), pre-formed in the presence of magnesium, a ∼115% increase in the fluorescence of the 2-AP probe is initially observed. A similar increase in 2-AP fluorescence was previously observed when 2-AP-containing single-stranded hexa and dodecanucleotides were bound by wild-type NCp7 (16,65,66). In the current study, as the 2-AP is positioned in the loop-loop region of DIS dimer, the fluorescence increase likely reflects NCp7-induced destacking of 2-AP, greater exposure to the aqueous solvent and/or a possible loss or gain of non-polar interactions owing to protein binding rather than restriction of the local dynamics of the fluorescence probe in the presence of NCp7.
To confirm that the increase in the 2-AP fluorescence is not a result of the dissociation of the dimer into individual hairpins, we acquired 1D 1H NMR spectra of the kissing complex before and after addition of two equivalents of NCp7. The imino proton region of this spectrum (Supplementary Figure S2A) shows that although chemical shift perturbations are observed, indicative of binding of NCp7 to the kissing complex, the expected signals from the loop-loop helix base pairs are observed even after the addition of NCp7. This observation is consistent with a previous NMR study carried out on SL1 RNA constructs containing the internal loop region, which indicated that the loop-loop helix remains intact in the NCp7 bound kissing dimer, whereas base pairs in the SL1 upper stem helix were observed to be destabilized (17). Additionally, we compared the relative increase of 2-AP fluorescence when NCp7 is added to the kissing complex formed with DIS24(GA)-12ap in the presence of Mg2+ versus the DIS stem-loop [DIS24(GA)-12ap] alone under similar experimental conditions, and found a significantly larger increase in 2-AP fluorescence emission in the latter case. This further supports the interpretation of the fluorescence response of the kissing dimer, DIS24(GA)-12ap•DIS24(UC), on addition of NCp7 as reporting on a destabilization or destacking of the loop-loop helix rather a dissociation of the kissing complex and formation of NCp7 bound stem-loops (Supplementary Figure S2B). Similar steady-state and time-resolved fluorescence studies carried out on TAR and complementary TAR (cTAR) stem-loop structures also found that NCp7 destabilizes (‘melts’) less stable secondary structures initially before proceeding to melt more stable secondary structures (67).
To further probe the changes in the kissing helix in the process of the NCp7 chaperoned conversion of the kissing complex to the duplex dimer, a time-dependent 2-AP fluorescence assay was performed using the DIS24(GA)-12ap•DIS24(UC) kissing complex. In this experiment, an initial increase in the fluorescence was observed (∼115%) on binding of NCp7 to the DIS24(GA)-12ap•DIS24(UC) kissing complex. The 2-AP fluorescence was then observed to decrease in a time-dependent manner, as the extended duplex is formed, and the 2-AP probe is stacked in a 2-AP-U base pair within the loop-loop helix of the duplex dimer, with no further detectable effect of NCp7 on base pairing or stacking (Supplementary Figure S3). The detected rate of formation of the duplex dimer using this 2-AP probe was similar to that measured in the 2-AP stem constructs, suggesting that both probes detected the same process of DIS conversion.
Comparison of NCp7 wild-type and mutant effects on the DIS loop-loop helix
As described earlier in the text for the measurement of strand exchange activity, the same series of NCp7 mutants were systematically examined, using the loop positioned 2-AP probe, for changes imparted to the DIS loop-loop helix on protein binding.
Zn finger swap mutants
Kissing loop-loop helix ‘destacking’ by the Zn finger swap mutants was found to differ significantly. The NC 1/1 mutant caused a similar increase in the 2-AP fluorescence on binding the DIS24(GA)-12ap•DIS24(UC) as compared with the wild-type protein, whereas the other two mutants, NC 2/1 and NC 2/2, showed about half the increase in the 2-AP fluorescence (Table 2). These results suggest that the mode of NCp7 interaction with the loop-loop helix of the kissing dimer depends on the identity of the N- and C-terminal Zn fingers. In particular, when compared with the strand exchange activities of the mutants, the difference in fluorescence increase in these mutants suggests a role for the phenylalanine and tryptophan residues in fingers 1 and 2, respectively, in aligning the fingers in the bound state and/or reorienting the bases in the DIS loop on binding to facilitate NCp7 chaperone activity.
Zinc coordination mutants of NCp7
All three mutants (H23C, H44C and H23C/H44C) result in little increase in the fluorescence of 2-AP on binding to DIS24(GA)-12ap•DIS24(UC), suggesting loss of wild-type NCp7 interaction with the kissing complex and limited ‘destacking’ of the loop-loop helix (Table 2). These results indicate an essential requirement for the presence of CCHC type of Zn fingers for wild-type interaction and destabilization of the DIS loop-loop helix. Similarly, the SSHS/SSHS mutant, wherein the Zn coordinating cysteines have been mutated to serines, again displayed almost no increase in 2-AP fluorescence (Table 2).
Zn finger aromatic residue mutants
Interestingly, the F16A mutant showed twice the initial increase in the fluorescence of DIS24(GA)-12ap•DIS24(UC) (Table 2) when compared with wild-type protein, whereas the other two alanine mutant NCp7 proteins (W37A and F16A/W37A) show only a small increase in 2-AP fluorescence. The increase in the fluorescence of DIS24(GA)-12ap•DIS24(UC) complex in the presence of the F16A mutant indicates a possible lack of π–π interactions between the phenyl ring of F16 and 2-AP that may contribute to the quenching of the 2-AP fluorescence and the reduction in the chaperoning activity of this mutant. The aromatic amino acid swap mutants, F16W and W37F, have similar helix destabilizing activities, as again measured by an increase in the fluorescence of the 2-AP probe, that are slightly less than wild-type. In contrast, the double mutant F16W/W37F showed only a small increase when compared with the wild-type NCp7 (Table 2). These results further show an apparent specificity for the type of aromatic residue at positions 16 and 37, and not simply a non-specific aromatic base stacking interaction, in the cognate NCp7 interaction with the DIS loop.
N-terminal mutant of NCp7
No significant increase in 2-AP fluorescence in DIS24(GA)-12ap• DIS24(UC) was observed for the N-terminal mutant, indicating that binding of this mutant does not result in any significant changes in stacking of the loop-loop helix bases and/or aromatic amino acids of NCp7 with the loop bases (Table 2).
Characterization of NCp7 binding interface with DIS
To survey the interaction of the DIS kissing complex with NCp7, solution NMR experiments were carried out on wild-type NCp7 and its complex with preformed DIS23(GA)•DIS23(HxUC) kissing complex in the absence and presence of low concentrations of MgCl2. Similarly, NMR was used to assess complexes between the DIS kissing complex and mutants of NCp7.
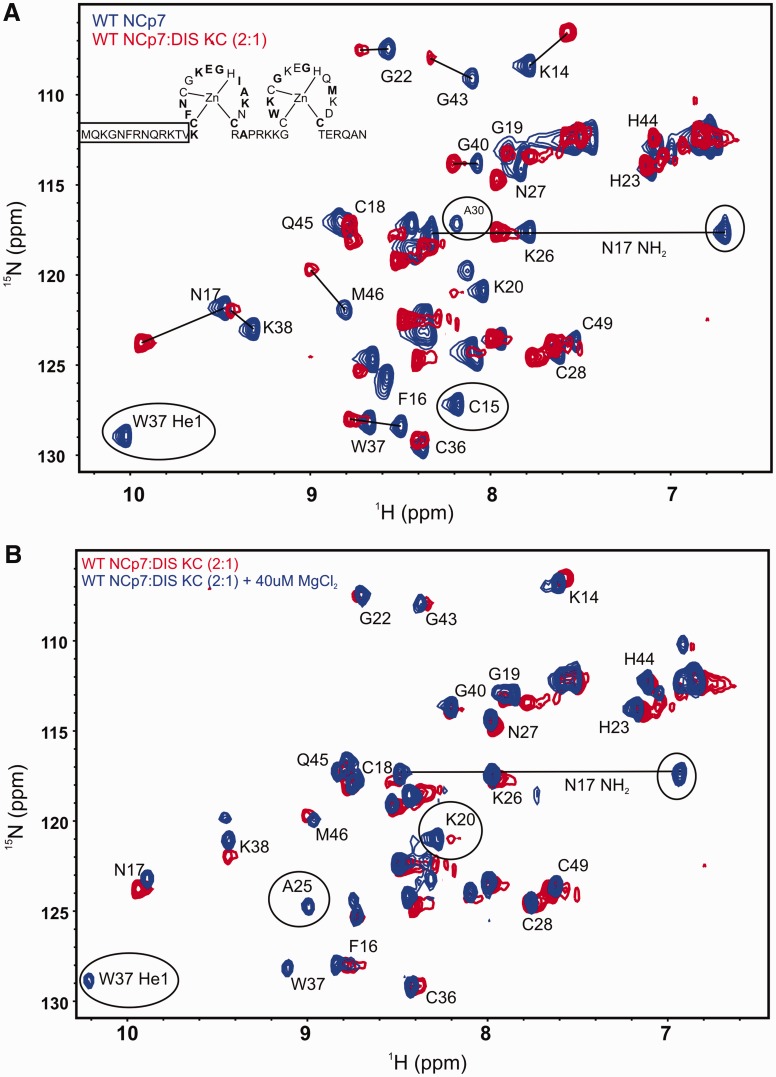
To map the binding interface and structural changes in the wild-type NCp7 on binding to the DIS kissing complex, two equivalents of 15N-labeled NCp7 were mixed with unlabeled DIS RNA kissing complex DIS23(GA)•DIS23(HxUC). The 15N-1H HSQC spectrum of wild-type NCp7 alone is indicative of a well-folded protein, albeit only the amide resonances belonging to the Zn finger motifs are observed in the spectrum, consistent with previous NMR studies on the full-length NCp7 (29). Significant chemical shift perturbation of the protein resonances is observed in the HSQC spectrum on formation of the NCp7-DIS23(GA)•DIS23(HxUC) complex. Resonances belonging to Cys15, Ala30, Trp37, Lys47 and Glu42 backbone amide correlations are broadened beyond detection. The side chain indole NH proton cross peak of Trp37 and amino cross peaks of Asn17 are also broadened beyond detection. Moreover, chemical shift perturbations, in both the 15N and 1H dimensions, are observed for amide resonances belonging to residues Lys14, Phe16, Asn17, Gly22, Lys26, Asn27, Lys38, Gly40, Gly43 and Met46 (Figure 5A). Addition of a small amount of MgCl2 (to a final concentration of 40 µM) significantly improved the quality of the HSQC spectrum of the complex, as evidenced by the sharpening of the cross peaks as well as reappearance of both the backbone and side chain resonances of Trp37 and Asn17, indicating stabilization of the NCp7-DIS kissing complex interaction by Mg2+ (Figure 5B).
Figure 5.
Structural characterization of wild-type NCp7 binding to the DIS kissing complex. (A) An overlay of the 15N-1H HSQC spectrum of the wild-type NCp7 (blue) and its complex with the DIS kissing complex (red). The backbone amide resonances are labeled as well as the side chain indole NH group (He1) of W37 and the side chain amino resonances (NH2) of N17 are shown with a solid line connecting them. Resonances that are broadened beyond detection on binding the RNA are circled, and shifted resonances are indicated with solid lines. The secondary structure of NCp7 is drawn with residues showing significant chemical shift perturbation in bold. The N-terminal residues for which amide resonances are not observed in the HSQC spectrum are boxed. (B) An overlay of the 15N-1H HSQC spectrum of the wild-type NCp7 bound to the DIS kissing complex (red) and the same complex after addition of 40 µM of MgCl2 (blue). The residues whose amide proton resonances either reappear or sharpen on addition of 40 µM of MgCl2 are labeled.
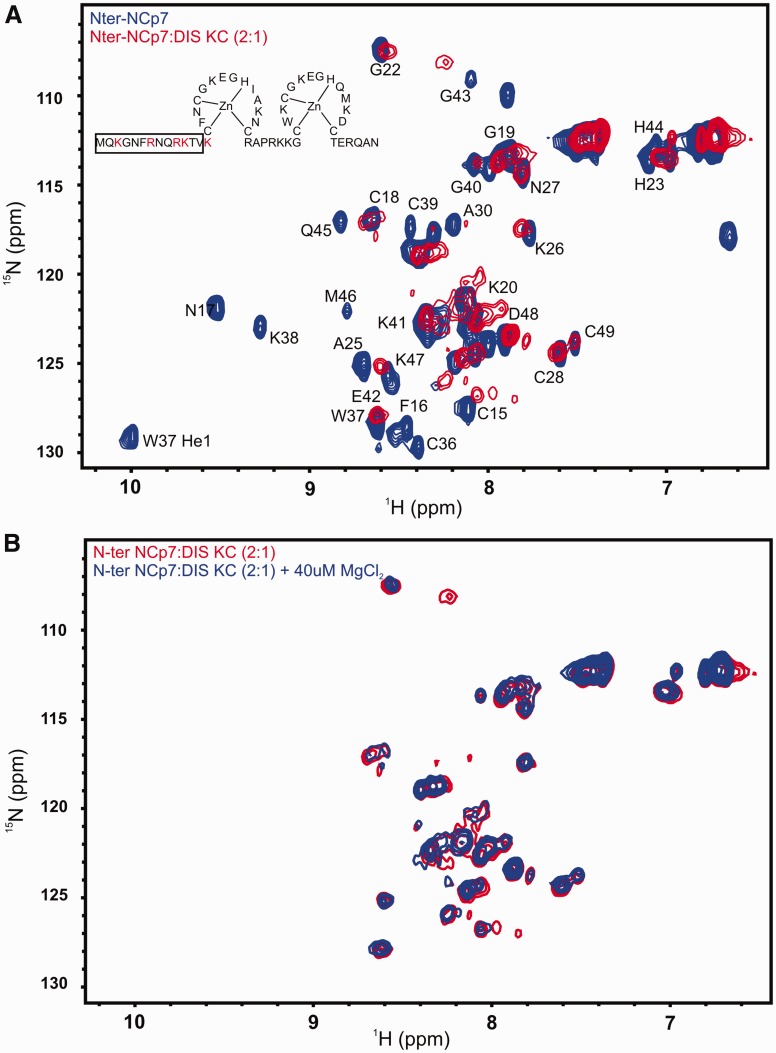
To examine the effect of the lack of N-terminal basic residues on the interaction of NCp7 with the DIS kissing RNA, we similarly measured HSQC spectra of 15N-labeled N-terminal mutant of NCp7 alone and in complex with the DIS23(GA)•DIS23(HxUC) kissing complex. We found that the 15N-1H HSQC spectral pattern of N-terminal mutant is similar to the wild-type NCp7, indicating similar folding of the Zn fingers is maintained even in the absence of positive charge in the N-terminus and that the N-terminus does not interact significantly with the Zn fingers (Supplementary Figure S4). However, the complex of the N-terminal NCp7 mutant with the DIS23(GA)•DIS23(HxUC) kissing complex shows significant differences to the spectra of the complex formed by wild-type NCp7 as might be expected from the fluorescence data. Amide proton resonances of a number of residues are broadened beyond detection in the N-terminal mutant NCp7-RNA complex compared with the wild-type NCp7-RNA complex implying an increase in dynamics and/or a loss of native structural fold of the Zn fingers in the mutant versus the wild-type protein on binding the RNA kissing complex (Figure 6A). As with the wild-type protein, we investigated the effect of Mg2+ on the protein–RNA complex by adding small quantities of Mg2+ to a final concentration of 40 µM. In this case, the addition of Mg2+ did not improve the quality of the NMR spectra, in terms of sharpening the line widths of the amide correlations (Figure 6B).
Figure 6.
(A) An overlay of 15N-1H HSQC spectrum of N-terminal NCp7 (blue) and its complex with the DIS kissing complex (red). The residues that are mutated to alanine are highlighted in red in the structure of NCp7. The backbone amide resonances are labeled besides the side chain indole NH group (He1) of W37, and the side chain amino resonances of N17 (NH2) are shown with a solid line connecting them. The N-terminal residues for which amide resonances are not observed in the HSQC spectrum are boxed. (B) Overlay of 15N-1H HSQC spectrum of N-terminal NCp7 in complex with the DIS kissing complex (red) and the same complex in the presence of 40 µM of MgCl2 (blue).
To further confirm the requirement for folding of the Zn fingers in the DIS loop binding and chaperone activity of NCp7, a 15N-1H HSQC spectrum of the SSHS/SSHS mutant was acquired. This spectrum showed a narrow range of chemical shifts for the 1H resonances and sharp cross peaks indicative of an unfolded protein (Supplementary Figure S5). 15N-1H HSQC spectra were also acquired for the aromatic amino acid residue mutants F16A and W37A, and both mutants show a similar cross peak pattern as the wild-type NCp7, indicating a structural similarity of these mutants with the wild-type, atleast with respect to the Zn finger domains (Supplementary Figure S6).
DISCUSSION
In the present study, we have used 2-AP fluorescence assays and NMR spectroscopy to probe the importance of different NCp7 residues in its function of chaperoning maturation of the SL1 DIS dimer of HIV-1. In addition to the wild-type NCp7, we have examined four different classes of NCp7 mutants: (i) Zn finger swap mutants; (ii) Zn finger coordination mutants; (iii) Zn finger aromatic residue mutants; and (iv) an N-terminal mutant. Using 2-AP fluorescence assays, RNA loop-loop helix ‘destacking’ and strand transfer ability of these mutants could be examined in the context of the model RNA stem-loops (Figure 1) containing the DIS. The foldedness and stability of mutant NCp7 proteins to wild-type were further assessed using NMR spectroscopy, as was the interaction of these proteins with the DIS kissing complex and their subsequent chaperone activity.
Position and identity of individual Zn fingers
Previous analysis of NCp7 mutants have shown that the N-terminal Zn finger of NCp7 plays a critical role in NCp7 function. There are five amino acid differences between the two Zn fingers of NCp7 (in the NL 4-3 strain): phenylalanine to tryptophan (F16 to W37), asparagine to lysine (N17 to K38), isoleucine to glutamine (I24 to Q45), alanine to methionine (A25 to M46) and finally asparagine to aspartate (N27 to D48). These differences result in a slightly higher hydrophobicity of the N-terminal Zn finger than the C-terminal one. In this work, the three Zn finger swap mutants (NC1/1, NC2/1 and NC2/2) show chaperoning activity similar to the wild-type but with slightly slower rates of strand exchange (Figure 2A). However, as assessed by the loop-loop helix 12ap assay, their helix destabilizing activity varied and was slightly different with NC1/1 being the most similar to the wild-type. In this assay, NC2/1 and NC2/2 mutants show a 1.5-fold increase in the fluorescence, for 2-AP at position 12 in the DIS loop, compared with the ∼2-fold increase observed for the wild-type and NC1/1 mutants (Table 2). Interestingly, replacing the phenylalanine in the N-terminal finger with tryptophan results in a chaperone activity similar to that observed for the NC2/2 mutant, but replacing tryptophan in the C-terminal finger with phenylalanine did not yield similar activity as the NC1/1 mutant. These observations suggest that residues other than the aromatic residues in the N-terminal Zn finger may play a role in binding specificity and chaperoning activity of NCp7, perhaps owing to differences in hydrophobicity between the two fingers. The presence of this hydrophobic plateau at the top of the folded Zn fingers was also shown to play an essential role in switching the Primer Binding Sequence (PBS) +/− strand transfer to the kissing loop pathway by stabilizing the stretched conformation of the PBS apical loop, thereby restricting the mobility of the PBS loop dynamics (68). It is also worth noting that the orientation and stacking of Phe16 and Trp37 are considerably different in the wild-type and the swap mutants due to the respective positioning of the aromatic amino acids in the context of the entire protein. These differences may also contribute to the slight differences we have observed in the rates of conversion in the finger swap mutants.
The Zn in NCp7 is present in a CCHC type of coordination motif and is essential for the native chaperone function of the NCp7. The coordination mutants studied here (H23C, H44C and H23C/H44C) all have lower activities in the 2-AP maturation assay when compared with the wild-type (Figure 2B). As shown previously, these mutants retain the ability to bind Zn (69,70). As both the H23C and H44C mutants display similar rates of chaperone activity, we can speculate that wild-type Zn finger coordination may not contribute to the individual fingers’ roles in NCp7 function but serves a common organizational purpose in the structures of the two fingers. As might be expected if the mutations were additive, changing both fingers to the type CCCC leads to a greater loss of NCp7 function, and the behavior of this mutant was similar to the SSHS/SSHS mutant, which does not bind Zn. NMR solution structures of both the H23C and H44A mutants of truncated NCp7 proteins have shown significant structural changes in their respective fingers while showing little or no effect on the other finger (69,70). Fluorescence studies on the H23C mutant, moreover, have indicated an increase in distance between the aromatic centers of Phe16 and Trp37 from 4.8 to 7.1 Å in the native protein to 9–18 Å in this mutant (31,71). The presence of CCCC type of coordination in both fingers may thus lead to a larger increase in the distance between these two aromatic centers and may result in a loss of any hydrophobic interaction between these two residues that is essential for proper functioning of NCp7. Finally, the SSHS/SSHS mutant, which cannot coordinate Zn, shows a highly diminished chaperoning activity. Kawai’s group similarly showed a reduction in the RNA annealing activity of SSHS/SSHS mutant using kissing dimers formed with 39 nt SL1 RNA hairpin constructs that is consistent with the diminished chaperone activity observed in this work (48,49). The 15N-1H HSQC spectrum of this mutant (SSHS/SSHS) is also indicative of an unstructured protein, confirming the role of zinc and its tetrahedral coordination in providing a proper structural scaffold to the NCp7.
Importance of Phe16 and Trp37
The aromatic amino acid residues phenylalanine and tryptophan at positions 16 and 37 are highly conserved in HIV-1 NCp7. Using the residue swap mutants (F16W, W37F, and F16W/W37F) and alanine mutants (F16A, W37A, and F16A/W37A), we have assessed the contribution of each of these residues to the chaperone function of the NCp7 in DIS maturation. Replacing phenylalanine in the N-terminal finger with tryptophan (F16W) resulted in activity similar to the NC2/2 mutant, whereas replacing tryptophan in the C-terminal finger with phenylalanine (W37F) results in a more moderate loss of function compared to the NC1/1 mutant. These results clearly indicate a contribution of neighboring residues in the N-terminal Zn finger (Asn17, Ile24, Ala25 and Asn27) to the functioning of NCp7. The reduced but similar activity of NC2/2 and F16W mutants suggests that the presence of a bulky group at position 16 may hinder proper stacking interactions with the nucleobases, thereby reducing the activity of NCp7. The helix destabilizing properties of both NC2/2 and F16W are similar as is the case with NC1/1 and W37F mutants. The double swap mutant (F16W/W37F) has a slightly lower destabilizing activity than the NC2/1 mutant but a similar chaperoning activity (Figure 3B and Table 2). Again, the presence of a bulky group at position 16 may affect the overall conformation of the N-terminal Zn finger leading to the observed reduced destabilizing activity for these two mutants. Furthermore, reduced chaperone function observed for the alanine mutants (F16A, W37A, F16A/W37A) confirms the importance of having an aromatic residue at both positions in the NCp7 (Figure 3A). A requirement for base stacking of the NCp7 aromatic residues in binding and destabilizing of the DIS loop is consistent with known NCp7-RNA complexes (32–34,72) and the observed increase in the 12ap fluorescence with F16A can be explained by the loss of π–π interactions between the aromatic ring of phenylalanine and the 2-AP base.
Basic residues in the N-terminus
To study the role of the basic residues in the N-terminus, we have used a mutant form of NCp7 in which all the basic residues in the N-terminus were changed to alanine. This mutant neither showed significant helix destabilizing nor strand transfer function. The essential role of the basic region in NCp7’s chaperone activity is consistent with previous work, which showed that peptide fragments representing the basic regions of NCp7 were necessary and sufficient for chaperone activity. These in vitro studies showed a greatly reduced activity for these basic peptides, indicating that the destabilizing function of the zing fingers, as well as the protein structural context, were necessary for full native chaperone activity of NCp7. However, as the previous studies were carried out in the absence of magnesium, which is known to have a significant effect on DIS dimer structure and stability, only a qualitative comparison of these results can be made with the findings in the current study.
Although it did not show measurable chaperone function, we found that the structure of the two Zn fingers in the mutant is similar to the wild-type protein (Supplementary Figure S4). However, the NMR spectra of the complex of this mutant with the kissing RNA dimer suggested differences in the interaction between the mutant protein and RNA when compared with the complex formed by the wild-type protein with the DIS kissing complex. Unlike what was observed for the wild-type NCp7, addition of Mg2+ did not improve the quality of the HSQC spectrum, implying a role for the N-terminus in NCp7 binding to the DIS kissing complex. The highly disordered basic N-terminus of NCp7 forms a 310 helix when bound to RNA stem loops SL2 and SL3 (32,33). NMR structures of NC bound to SL2 and SL3 have indicated possible interactions between the side chains of Phe6 and Lys3 with the nucleobases in the loop region of the RNA, and the authors of these studies concluded that the inherent flexibility of the N-terminus allows for sampling of multiple intra-protein interactions in this region that may facilitate binding to different RNAs. As the N-terminal region of NCp7 was not observed to order in the NMR spectra of the complexes between NCp7 and DIS kissing dimer, we can only speculate that the lack of basic residues in the N-terminal mutant may result in a loss of these interactions and thereby not direct a suitable orientation of NCp7 on the kissing complex that is required for chaperoning activity.
RNA binding affinity and chaperone activity
The 2-AP chaperone assay used in this study measures a combination of NCp7’s RNA binding and chaperone activities, as NCp7 is expected to bind to high affinity regions first (loops and bulges), melt the stems, then continue to bind to the newly liberated single stranded regions, with subsequent annealing of complementary regions. To differentiate between RNA-binding affinities and chaperone activity, binding experiments must be performed in systems where there are no other chaperone-like processes occurring (i.e. strand displacement or annealing). From previous studies with short oligonucleotides (63), the difference in binding to high affinity sequences like TGTG was relatively small (e.g. the N-terminal mutant bound these sequences in the presence of 150 mM of NaCl with an affinity of 60 nM, whereas wild-type protein bound with an affinity of 5 nM). Moreover, difference in high-affinity binding may have little relevance to electrostatic (general nucleic acid) binding that is involved in chaperone activity. In the chaperone assay, the NCp7 protein concentration was well above the KD reported for wild-type, as well as mutant NCp7 variants, giving confidence that any high-affinity interaction(s) was saturated in these experiments. The variability observed in the chaperone activity of the NCp7 mutants is therefore predominantly attributed to modulation of chaperoning interactions and not differences in high-affinity NCp7 binding to the DIS kissing dimer.
Assessing changes in NCp7 when bound to DIS kissing complex
Although no high-resolution structure is available for the complex of NCp7 with either the kissing complex or the extended duplex, a number of NMR structures (73–75) and crystal structures (76) of the DIS kissing dimer have been reported. In these structures, conformational polymorphism is observed, in particularly, for the unpaired purine bases in the DIS loop, which likely represent different conformations that are accessible to the DIS kissing dimer under different experimental conditions. Interestingly, the dynamic polymorphism around these loop purines in the DIS kissing dimer could be correlated with the rate of NCp7 catalyzed maturation of the DIS kissing dimer (55), suggesting a functional role for the observed RNA structural plasticity. Differences have also been observed in the NMR (77), and X-ray (78) determined conformations of the purine junctions in the mature DIS dimer, indicating that even in this dimeric form, the junctions remain plastic. Although structures of NCp7 in complex with either kissing complex or the extended duplex are not available, some structural insights into how NCp7 interacts with nucleic acids are available from the solution structures of full-length and truncated NCp7 bound to RNA stem-loops and DNA oligonucleotides (32–34,72). A common structural theme in these structures is the stacking of unpaired purine residues of the RNA or DNA with the aromatic amino acids (e.g. F16 and W37) in the protein to form so called ‘base grips’.
In the current study, we applied NMR to probe the structural details of the interface between the wild-type NCp7 and the DIS23(GA)•DIS23(HxUC) kissing complex. Significant line broadening and chemical shift perturbation were observed in the complex of wild-type NCp7 with the DIS23(GA)•DIS23(HxUC) kissing complex (Figure 7). The complete loss or significant shift in the cross peaks corresponding to Cys15, Phe16, Asn17 and the loss of chaperoning function of the F16A and F16A/W37A mutant provide corroborating evidence for the involvement of the phenyl ring of Phe16 in NCp7 chaperone function, likely through a stacking interaction with the unpaired purines in the DIS loop. We have also observed significant chemical shift perturbation of residues Lys14, Gly22, Lys26 and Asn27 in the N-terminal Zn finger, indicating a contribution of N-terminal Zn finger in the intermolecular interactions. The absence of evidence for the presence of a 310 helix in the N-terminus of NCp7 on binding the DIS kissing complex suggests that the intramolecular interactions in the N-terminus of NCp7 bound to DIS kissing complex is different from what has previously been observed for NCp7 bound to RNA stem-loops, SL2 and SL3. Interestingly, addition of Mg2+ stabilized the binding interface between NCp7 and the DIS kissing complex as evidenced by a sharpening of the line widths in the spectra. This observation suggests that the previous observation that Mg2+-stabilized kissing complexes were more efficiently chaperoned by NCp7 is related to differences in the RNA–protein binding interaction. The addition of Mg2+, however, did not result in a detectable stabilization of the N-terminus, as no additional cross peaks were observed in these spectra.
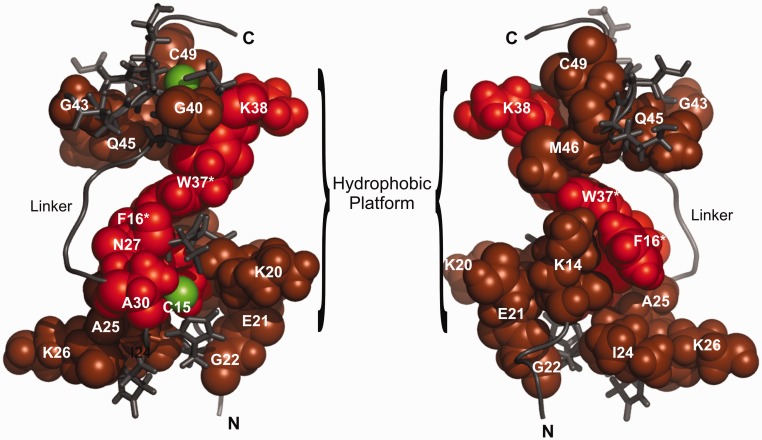
Figure 7.
An NMR chemical shift perturbation (CSP) model of NCp7 bound to the DIS kissing complex. The observed CSP of amide resonances of NCp7 are mapped onto the previously determined NMR structure of NCp7 bound to HIV SL2 (PDB id. 1F6U). Residues whose amide resonances showed a significant CSP in both HN and N dimensions are colored red, and the residues with moderate or less CSP are colored in brown. Zinc atoms are shown in green. The N-terminal, C-terminal and Linker regions are denoted by N, C and L, respectively. The terminal residues on both the N and C terminus are not shown, as they were not observed in the HSQC spectrum. More residues in the proximal Zn finger show significant CSP and both the aromatic residues in the fingers F16 and W37 (highlighted with an asterisks) show a significant CSP on binding the RNA kissing complex. Mutating these residues to alanine results in a severe loss of chaperoning activity of the protein. Interestingly, alanine at position 30, presumed to be at a distant site from the RNA-binding interface, also shows significant CSP on binding the kissing complex, suggesting a conformational change in the linker structure that might reorient the two Zn fingers to accommodate proper stacking interactions between the kissing helix residues of the RNA and the aromatic acid residues of the protein. The hydrophobic patch that facilitates binding of RNA is also indicated.
In conclusion, using 2-AP detected fluorescence assays and NMR spectroscopy, we analyzed the amino acid requirements for the efficient chaperone activity of NCp7 in the maturation of the HIV-1 DIS RNA. Our results demonstrate that the presence, but not the order, of the Zn fingers is essential for proper functioning of NCp7 in chaperoning the DIS maturation process, as is the CCHC type of coordination for both the Zn fingers. The presence of the aromatic residues Phe16 and Trp37 was also found to be critical, and amino acid residues surrounding these sites in the Zn fingers play a coordinated role with these residues. Lastly, the positive charge of the N-terminus appears to properly orient the Zn fingers on binding the RNA, even though this region of NCp7 remains largely disordered in the complex. Though a similar analysis of residues in the linker sequence was not carried out, Kafaie et al.(36) have shown that the Zn linker plays more of a steric rather than electrostatic role and that the identity of five central residues of the linker is not important for gRNA dimerization. Structurally, Mg2+ was found to stabilize the binding interface between NCp7 and the DIS kissing complex, which correlates with previous data indicating that the Mg2+ stablized DIS kissing dimer is chaperoned most efficiently by NCp7 in the maturation process. Overall, the mechanism of NCp7 chaperoned DIS dimer maturation, while sharing many commonalities with other NCp7 functions in viral replication, has unique features that can be attributed to specific RNA-protein structural interactions involved in the DIS dimer refolding process.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6.
FUNDING
National Institutes of Health [GM 59107 to J.P.M. in part]; National Cancer Institute, National Institutes of Health, under contract [HHSN261200800001E] with SAIC-Frederick, Inc. (to R.J.G.); National Institutes of Health graduate training fellowship to UMd (to K.T.B.); NMR instrumentation at IBBR with support from the W.M. Keck Foundation, the National Institutes of Health and the National Institute of Standards and Technology. Funding for open access charge: NIST intramural funds.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Andrea Szakal for insightful discussions of the experiments and critical assessment of the manuscript and Dr. Nese Sari for assistance with the NMR. Certain commercial equipment, instruments and materials are identified in this article to specify the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the material or equipment identified is necessarily the best available for the purpose. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
REFERENCES
- 1.Paillart JC, Marquet R, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of retroviral genomic RNAs: structural and functional implications. Biochimie. 1996;78:639–653. doi: 10.1016/s0300-9084(96)80010-1. [DOI] [PubMed] [Google Scholar]
- 2.Miele G, Mouland A, Harrison GP, Cohen E, Lever AM. The human immunodeficiency virus type 1 5' packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J. Virol. 1996;70:944–951. doi: 10.1128/jvi.70.2.944-951.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clever J, Sassetti C, Parslow TG. RNA secondary structure and binding sites for gag gene products in the 5' packaging signal of human immunodeficiency virus type 1. J. Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clever JL, Miranda D, Jr, Parslow TG. RNA structure and packaging signals in the 5' leader region of the human immunodeficiency virus type 1 genome. J. Virol. 2002;76:12381–12387. doi: 10.1128/JVI.76.23.12381-12387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkowitz RD, Goff SP. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 6.Harrison GP, Miele G, Hunter E, Lever AM. Functional analysis of the core human immunodeficiency virus type 1 packaging signal in a permissive cell line. J. Virol. 1998;72:5886–5896. doi: 10.1128/jvi.72.7.5886-5896.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McBride MS, Schwartz MD, Panganiban AT. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakaguchi K, Zambrano N, Baldwin ET, Shapiro BA, Erickson JW, Omichinski JG, Clore GM, Gronenborn AM, Appella E. Identification of a binding site for the human immunodeficiency virus type 1 nucleocapsid protein. Proc. Natl Acad. Sci. USA. 1993;90:5219–5223. doi: 10.1073/pnas.90.11.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl Acad. Sci. USA. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paillart JC, Westhof E, Ehresmann C, Ehresmann B, Marquet R. Non-canonical interactions in a kissing loop complex: the dimerization initiation site of HIV-1 genomic RNA. J. Mol. Biol. 1997;270:36–49. doi: 10.1006/jmbi.1997.1096. [DOI] [PubMed] [Google Scholar]
- 11.Laughrea M, Jette L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248-271 are dispensable for dimer formation. Biochemistry. 1996;35:1589–1598. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 12.Jossinet F, Paillart JC, Westhof E, Hermann T, Skripkin E, Lodmell JS, Ehresmann C, Ehresmann B, Marquet R. Dimerization of HIV-1 genomic RNA of subtypes A and B: RNA loop structure and magnesium binding. RNA. 1999;5:1222–1234. doi: 10.1017/s1355838299990982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weixlbaumer A, Werner A, Flamm C, Westhof E, Schroeder R. Determination of thermodynamic parameters for HIV DIS type loop-loop kissing complexes. Nucleic Acids Res. 2004;32:5126–5133. doi: 10.1093/nar/gkh841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi KI, Baba S, Chattopadhyay P, Koyanagi Y, Yamamoto N, Takaku H, Kawai G. Structural requirement for the two-step dimerization of human immunodeficiency virus type 1 genome. RNA. 2000;6:96–102. doi: 10.1017/s1355838200991635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muriaux D, De Rocquigny H, Roques BP, Paoletti J. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J. Biol. Chem. 1996;271:33686–33692. doi: 10.1074/jbc.271.52.33686. [DOI] [PubMed] [Google Scholar]
- 16.Rist MJ, Marino JP. Mechanism of nucleocapsid protein catalyzed structural isomerization of the dimerization initiation site of HIV-1. Biochemistry. 2002;41:14762–14770. doi: 10.1021/bi0267240. [DOI] [PubMed] [Google Scholar]
- 17.Mujeeb A, Ulyanov NB, Georgantis S, Smirnov I, Chung J, Parslow TG, James TL. Nucleocapsid protein-mediated maturation of dimer initiation complex of full-length SL1 stemloop of HIV-1: sequence effects and mechanism of RNA refolding. Nucleic Acids Res. 2007;35:2026–2034. doi: 10.1093/nar/gkm097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 19.Feng YX, Copeland TD, Henderson LE, Gorelick RJ, Bosche WJ, Levin JG, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl Acad. Sci. USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lapadat-Tapolsky M, De Rocquigny H, Van Gent D, Roques B, Plasterk R, Darlix JL. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993;21:831–839. doi: 10.1093/nar/21.4.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Druillennec S, Caneparo A, de Rocquigny H, Roques BP. Evidence of interactions between the nucleocapsid protein NCp7 and the reverse transcriptase of HIV-1. J. Biol. Chem. 1999;274:11283–11288. doi: 10.1074/jbc.274.16.11283. [DOI] [PubMed] [Google Scholar]
- 22.Peliska JA, Balasubramanian S, Giedroc DP, Benkovic SJ. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNase H activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 23.Wu W, Henderson LE, Copeland TD, Gorelick RJ, Bosche WJ, Rein A, Levin JG. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J. Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson PE, Turner RB, Wu ZR, Hairston L, Guo J, Levin JG, Summers MF. A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription. Biochemistry. 2000;39:9084–9091. doi: 10.1021/bi000841i. [DOI] [PubMed] [Google Scholar]
- 25.Guo J, Wu T, Kane BF, Johnson DG, Henderson LE, Gorelick RJ, Levin JG. Subtle alterations of the native zinc finger structures have dramatic effects on the nucleic acid chaperone activity of human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 2002;76:4370–4378. doi: 10.1128/JVI.76.9.4370-4378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo J, Wu T, Anderson J, Kane BF, Johnson DG, Gorelick RJ, Henderson LE, Levin JG. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 2000;74:8980–8988. doi: 10.1128/jvi.74.19.8980-8988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darlix JL, Godet J, Ivanyi-Nagy R, Fosse P, Mauffret O, Mely Y. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J. Mol. Biol. 2011;410:565–581. doi: 10.1016/j.jmb.2011.03.037. [DOI] [PubMed] [Google Scholar]
- 28.Berkhout B, Gorelick R, Summers MF, Mely Y, Darlix JL. 6th international symposium on retroviral nucleocapsid. Retrovirology. 2008;5:21. doi: 10.1186/1742-4690-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Summers MF, Henderson LE, Chance MR, Bess JW, Jr, South TL, Blake PR, Sagi I, Perez-Alvarado G, Sowder RC, 3rd, Hare DR, et al. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee BM, De Guzman RN, Turner BG, Tjandra N, Summers MF. Dynamical behavior of the HIV-1 nucleocapsid protein. J. Mol. Biol. 1998;279:633–649. doi: 10.1006/jmbi.1998.1766. [DOI] [PubMed] [Google Scholar]
- 31.Morellet N, Jullian N, De Rocquigny H, Maigret B, Darlix JL, Roques BP. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. EMBO J. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 33.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 34.Morellet N, Demene H, Teilleux V, Huynh-Dinh T, de Rocquigny H, Fournie-Zaluski MC, Roques BP. Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC) J. Mol. Biol. 1998;283:419–434. doi: 10.1006/jmbi.1998.2098. [DOI] [PubMed] [Google Scholar]
- 35.Mori M, Dietrich U, Manetti F, Botta M. Molecular dynamics and DFT study on HIV-1 nucleocapsid protein-7 in complex with viral genome. J. Chem. Inf. Model. 2010;50:638–650. doi: 10.1021/ci100070m. [DOI] [PubMed] [Google Scholar]
- 36.Kafaie J, Song R, Abrahamyan L, Mouland AJ, Laughrea M. Mapping of nucleocapsid residues important for HIV-1 genomic RNA dimerization and packaging. Virology. 2008;375:592–610. doi: 10.1016/j.virol.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 37.Gorelick RJ, Nigida SM, Jr, Bess JW, Jr, Arthur LO, Henderson LE, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorelick RJ, Chabot DJ, Rein A, Henderson LE, Arthur LO. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J. Virol. 1993;67:4027–4036. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drummond JE, Mounts P, Gorelick RJ, Casas-Finet JR, Bosche WJ, Henderson LE, Waters DJ, Arthur LO. Wild-type and mutant HIV type 1 nucleocapsid proteins increase the proportion of long cDNA transcripts by viral reverse transcriptase. AIDS Res. Hum. Retroviruses. 1997;13:533–543. doi: 10.1089/aid.1997.13.533. [DOI] [PubMed] [Google Scholar]
- 40.Hargittai MR, Mangla AT, Gorelick RJ, Musier-Forsyth K. HIV-1 nucleocapsid protein zinc finger structures induce tRNA(Lys,3) structural changes but are not critical for primer/template annealing. J. Mol. Biol. 2001;312:985–997. doi: 10.1006/jmbi.2001.5021. [DOI] [PubMed] [Google Scholar]
- 41.Buckman JS, Bosche WJ, Gorelick RJ. Human immunodeficiency virus type 1 nucleocapsid zn(2+) fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 2003;77:1469–1480. doi: 10.1128/JVI.77.2.1469-1480.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu T, Datta SA, Mitra M, Gorelick RJ, Rein A, Levin JG. Fundamental differences between the nucleic acid chaperone activities of HIV-1 nucleocapsid protein and Gag or Gag-derived proteins: biological implications. Virology. 2010;405:556–567. doi: 10.1016/j.virol.2010.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirambeau G, Lyonnais S, Gorelick RJ. Features, processing states, and heterologous protein interactions in the modulation of the retroviral nucleocapsid protein function. RNA Biol. 2010;7:85–95. doi: 10.4161/rna.7.6.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng YX, Campbell S, Harvin D, Ehresmann B, Ehresmann C, Rein A. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol. 1999;73:4251–4256. doi: 10.1128/jvi.73.5.4251-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski MC, Roques B, Darlix JL. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc. Natl Acad. Sci. USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas JA, Gagliardi TD, Alvord WG, Lubomirski M, Bosche WJ, Gorelick RJ. Human immunodeficiency virus type 1 nucleocapsid zinc-finger mutations cause defects in reverse transcription and integration. Virology. 2006;353:41–51. doi: 10.1016/j.virol.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 47.Kanevsky I, Chaminade F, Ficheux D, Moumen A, Gorelick R, Negroni M, Darlix JL, Fossé P. Specific interactions between HIV-1 nucleocapsid protein and the TAR element. J. Mol. Biol. 2005;348:1059–1077. doi: 10.1016/j.jmb.2005.03.046. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi K, Baba S, Koyanagi Y, Yamamoto N, Takaku H, Kawai G. Two basic regions of NCp7 are sufficient for conformational conversion of HIV-1 dimerization initiation site from kissing-loop dimer to extended-duplex dimer. J. Biol. Chem. 2001;276:31274–31278. doi: 10.1074/jbc.M104577200. [DOI] [PubMed] [Google Scholar]
- 49.Baba S, Takahashi K, Koyanagi Y, Yamamoto N, Takaku H, Gorelick RJ, Kawai G. Role of the zinc fingers of HIV-1 nucleocapsid protein in maturation of genomic RNA. J. Biochem. 2003;134:637–639. doi: 10.1093/jb/mvg200. [DOI] [PubMed] [Google Scholar]
- 50.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Urbaneja MA, Kane BP, Johnson DG, Gorelick RJ, Henderson LE, Casas-Finet JR. Binding properties of the human immunodeficiency virus type 1 nucleocapsid protein p7 to a model RNA: elucidation of the structural determinants for function. J. Mol. Biol. 1999;287:59–75. doi: 10.1006/jmbi.1998.2521. [DOI] [PubMed] [Google Scholar]
- 52.Dorfman T, Luban J, Goff SP, Haseltine WA, Gottlinger HG. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turner KB, Hagan NA, Fabris D. Understanding the isomerization of the HIV-1 dimerization initiation domain by the nucleocapsid protein. J.Mol.Biol. 2007;369:812–828. doi: 10.1016/j.jmb.2007.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hagan NA, Fabris D. Dissecting the protein-RNA and RNA-RNA interactions in the nucleocapsid-mediated dimerization and isomerization of HIV-1 stemloop 1. J. Mol. Biol. 2007;365:396–410. doi: 10.1016/j.jmb.2006.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mihailescu MR, Marino JP. A proton-coupled dynamic conformational switch in the HIV-1 dimerization initiation site kissing complex. Proc. Natl Acad. Sci. USA. 2004;101:1189–1194. doi: 10.1073/pnas.0307966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee HW, Briggs KT, Marino JP. Dissecting structural transitions in the HIV-1 dimerization initiation site RNA using 2-aminopurine fluorescence. Methods. 2009;49:118–127. doi: 10.1016/j.ymeth.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beaucage SL, Caruthers MH. Deoxynucleoside phosphoramidites–A new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981;22:1859–1862. [Google Scholar]
- 58.Stampfl S, Lempradl A, Koehler G, Schroeder R. Monovalent ion dependence of neomycin B binding to an RNA aptamer characterized by spectroscopic methods. Chembiochem. 2007;8:1137–1145. doi: 10.1002/cbic.200700030. [DOI] [PubMed] [Google Scholar]
- 59.Menne S, Asif G, Narayanasamy J, Butler SD, George AL, Hurwitz SJ, Schinazi RF, Chu CK, Cote PJ, Gerin JL, et al. Antiviral effect of orally administered (-)-beta-D-2-aminopurine dioxolane in woodchucks with chronic woodchuck hepatitis virus infection. Antimicrob. Agents Chemother. 2007;51:3177–3184. doi: 10.1128/AAC.00325-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carteau S, Gorelick RJ, Bushman FD. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 62.Goddard TD, Kneller DG. SPARKY 3. San Francisco: University of California; [Google Scholar]
- 63.Fisher RJ, Fivash MJ, Stephen AG, Hagan NA, Shenoy SR, Medaglia MV, Smith LR, Worthy KM, Simpson JT, Shoemaker R, et al. Complex interactions of HIV-1 nucleocapsid protein with oligonucleotides. Nucleic Acids Res. 2006;34:472–484. doi: 10.1093/nar/gkj442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmalzbauer E, Strack B, Dannull J, Guehmann S, Moelling K. Mutations of basic amino acids of NCp7 of human immunodeficiency virus type 1 affect RNA binding in vitro. J. Virol. 1996;70:771–777. doi: 10.1128/jvi.70.2.771-777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Avilov SV, Godet J, Piemont E, Mely Y. Site-specific characterization of HIV-1 nucleocapsid protein binding to oligonucleotides with two binding sites. Biochemistry. 2009;48:2422–2430. doi: 10.1021/bi8022366. [DOI] [PubMed] [Google Scholar]
- 66.Avilov SV, Piemont E, Shvadchak V, de Rocquigny H, Mely Y. Probing dynamics of HIV-1 nucleocapsid protein/target hexanucleotide complexes by 2-aminopurine. Nucleic Acids Res. 2008;36:885–896. doi: 10.1093/nar/gkm1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bernacchi S, Stoylov S, Piemont E, Ficheux D, Roques BP, Darlix JL, Mely Y. HIV-1 nucleocapsid protein activates transient melting of least stable parts of the secondary structure of TAR and its complementary sequence. J. Mol. Biol. 2002;317:385–399. doi: 10.1006/jmbi.2002.5429. [DOI] [PubMed] [Google Scholar]
- 68.Godet J, Ramalanjaona N, Sharma KK, Richert L, de Rocquigny H, Darlix JL, Duportail G, Mely Y. Specific implications of the HIV-1 nucleocapsid zinc fingers in the annealing of the primer binding site complementary sequences during the obligatory plus strand transfer. Nucleic Acids Res. 2011;39:6633–6645. doi: 10.1093/nar/gkr274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Demene H, Dong CZ, Ottmann M, Rouyez MC, Jullian N, Morellet N, Mely Y, Darlix JL, Fournie-Zaluski MC, Saragosti S, et al. 1H NMR structure and biological studies of the His23—>Cys mutant nucleocapsid protein of HIV-1 indicate that the conformation of the first zinc finger is critical for virus infectivity. Biochemistry. 1994;33:11707–11716. doi: 10.1021/bi00205a006. [DOI] [PubMed] [Google Scholar]
- 70.Stote RH, Kellenberger E, Muller H, Bombarda E, Roques BP, Kieffer B, Mely Y. Structure of the His44 —> Ala single point mutant of the distal finger motif of HIV-1 nucleocapsid protein: a combined NMR, molecular dynamics simulation, and fluorescence study. Biochemistry. 2004;43:7687–7697. doi: 10.1021/bi036137u. [DOI] [PubMed] [Google Scholar]
- 71.Morellet N, de Rocquigny H, Mely Y, Jullian N, Demene H, Ottmann M, Gerard D, Darlix JL, Fournie-Zaluski MC, Roques BP. Conformational behaviour of the active and inactive forms of the nucleocapsid NCp7 of HIV-1 studied by 1H NMR. J. Mol. Biol. 1994;235:287–301. doi: 10.1016/s0022-2836(05)80033-6. [DOI] [PubMed] [Google Scholar]
- 72.Bourbigot S, Ramalanjaona N, Boudier C, Salgado GF, Roques BP, Mely Y, Bouaziz S, Morellet N. How the HIV-1 nucleocapsid protein binds and destabilises the (-) primer binding site during reverse transcription. J. Mol. Biol. 2008;383:1112–1128. doi: 10.1016/j.jmb.2008.08.046. [DOI] [PubMed] [Google Scholar]
- 73.Mujeeb A, Clever JL, Billeci TM, James TL, Parslow TG. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat. Struct. Bio. 1998;5:432–436. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 74.Baba STK, Noguchi S, Takaku H, Koyanagi Y, Yamamoto N, Kawai G. Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers. J. Biochem. 2005;138:583–592. doi: 10.1093/jb/mvi158. [DOI] [PubMed] [Google Scholar]
- 75.Kieken F, Paquet F, Brulé F, Paoletti J, Lancelot G. A new NMR solution structure of the SL1 HIV-1Lai loop-loop dimer. Nucleic Acids Res. 2006;34:343–352. doi: 10.1093/nar/gkj427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 2001;8:1064–1068. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 77.Mujeeb A, Parslow TG, Zarrinpar A, Das C, James TL, Clever JL, Billeci TM. NMR structure of the mature dimer initiation complex of HIV-1 genomic RNA. FEBS Lett. 1999;458:387–392. doi: 10.1016/s0014-5793(99)01183-7. [DOI] [PubMed] [Google Scholar]
- 78.Ennifar E, Yusupov M, Walter P, Marquet R, Ehresmann B, Ehresmann C, Dumas P. The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveals an extended duplex with two adenine bulges. J. Mol. Biol. 1999;7:1439–1449. doi: 10.1016/s0969-2126(00)80033-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.