Abstract
The human telomeric DNA sequence with four repeats can fold into a parallel-stranded propeller-type topology. NMR structures solved under molecular crowding experiments correlate with the crystal structures found with crystal-packing interactions that are effectively equivalent to molecular crowding. This topology has been used for rationalization of ligand design and occurs experimentally in a number of complexes with a diversity of ligands, at least in the crystalline state. Although G-quartet stems have been well characterized, the interactions of the TTA loop with the G-quartets are much less defined. To better understand the conformational variability and structural dynamics of the propeller-type topology, we performed molecular dynamics simulations in explicit solvent up to 1.5 μs. The analysis provides a detailed atomistic account of the dynamic nature of the TTA loops highlighting their interactions with the G-quartets including formation of an A:A base pair, triad, pentad and hexad. The results present a threshold in quadruplex simulations, with regards to understanding the flexible nature of the sugar-phosphate backbone in formation of unusual architecture within the topology. Furthermore, this study stresses the importance of simulation time in sampling conformational space for this topology.
INTRODUCTION
Telomeric DNA, containing guanine rich tandem repeats, is found at the end of all eukaryotic species, and is associated with a number of specialized proteins, forming the telomere. Its function is to protect chromosomal DNA from degradation, end-to-end fusions and recombination (1). Telomeres shorten progressively due to the end-replication problem, a consequence of the inability of DNA polymerases to fully replicate the 3′-ends of chromosomal DNA (2,3). Human telomeric DNA consists of repeats of the hexanucleotide sequence d(TTAGGG)n. A minimum of four repeats can fold into four-stranded intramolecular structures termed G-quadruplexes (4). These structures have been shown to play a role in telomere regulation and therefore have been proposed as a potential target for therapeutic intervention by small molecule ligands (4,5).
The folding and formation of human G-quadruplex structures have been studied by a variety of biophysical and chemical probe methods (6–9). The central unit of G-quadruplexes is a hydrogen-bonded array of four guanine bases stacked on top of another to form the G-quartet stem. The highly electronegative channel running along the axis of the stem is stabilized by monovalent cations (10). The TTA linker sequence is arranged diagonally, external to the helical-like stacks of the G-quartet stem, forming the loops. At least six intramolecular G-quadruplex structures with different topologies, formed from the human telomeric sequence, have been reported to date, including the basket, chair, 3 + 1 hybrid and propeller form (11,12). The elucidation of NMR solution structures reveals that the sequence can switch between parallel, antiparallel and hybrid conformations depending on the precise sequence, metal ions, solvent conditions, oligonucleotide concentration and possibly other factors (13).
The 22-mer sequence, d[AG3(T2AG3)3] adopts a distinct topology in concentrated K+ solution, as observed in the crystal structure (PDB id:1KF1) (10). The loops are in an open conformation in a propeller-like arrangement, consistent with the folding of a parallel-stranded G-quadruplex. The crystal structure contains three loops: Loop1 (Thy5-Thy6-Ade7), Loop2 (Thy11-Thy12-Ade13) and Loop3 (Thy17-Thy18-Ade19), which link the bottom (3′-end) and the top (5′-end) of the three-stacked G-quartets. The adenine in each TTA sequence is swung back forming a Thy-Ade-Thy stack (10). Recently, experiments carried out under molecular crowding conditions have demonstrated that the 22-mer sequence also adopts a similar propeller-type topology in the (non-crystalline) solution state (12). The environment inside a cell is molecularly crowded owing to macromolecular concentration of up to 40% (w/v) (14). Therefore, it can be speculated that human telomere quadruplexes can adopt a propeller-type topology under physiological conditions. The biological relevance of this topology can be established from the reports that it greatly decreases telomerase activity and processivity and therefore is a potential target for anti-cancer therapy (15). Furthermore, all G-quadruplex structures co-crystallized in combination with small molecule ligands/drugs also show propeller-type topology (16,17). This is unlike the other forms observed by NMR methods for native telomeric sequences, suggesting that the preferred favoured topology for ligand binding is the intramolecular parallel-stranded propeller-type (18). In the light of these findings, we have used this topology in the present molecular dynamics (MD) simulation and aim to provide, for the first time, an extended analysis of quadruplex loop structural dynamics on a microsecond timescale.
The MD simulations approach has provided much information regarding the dynamic behaviour of DNA in solution, and an increasing number of studies have used this approach with quadruplex nucleic acids (12,19–21). Although X-ray crystallography provides detailed atomic-level information of a quasi-static state, generalization cannot be made that crystal structures necessarily represent the major conformations. The X-ray structure of the propeller-type quadruplex reveals that although the dihedral angles for G-quartets are constrained in the range of B-DNA, the dihedral angles for loops vary from A-, B- and Z-DNA forms (22). The sugar-phosphate backbone has considerable potential for conformational variability within the quadruplex. Thus, although the G-quartet stem is considered to be well characterized by experimental and theoretical approaches, conformational plasticity and structural dynamics of the single-stranded loops as well as interactions of loops with the G-quartets are much less understood. As loops are likely dynamical, static structures alone cannot provide details of conformational transitions occurring within a quadruplex. Our earlier 15 ns MD simulations of the same sequence confirmed that the G-quartet stem is a highly stable structure. The simulations revealed some flexibility in the loop region in line with other studies (20,23). However, no interactions of loops with the G-quartet stem were noted, and it is likely that sampling of the conformational space of the loops was not exhaustive (20). Owing to the recent improvements in force fields and wide availability of high-end processing power, microsecond (μs) long simulations of nucleic acids are becoming affordable and often dramatically change the picture obtained by shorter simulations and older force fields (24–28).
In this study, we present a detailed analysis of a continuous 1.5 μs long MD simulation of a quadruplex formed from the human 22-mer telomeric repeat. The study focuses on the conformational sampling and the dynamic nature of the connecting loops. The folding of the loops external to the G-quartet stem are of particular interest, as they present an interface for interactions with proteins, other nucleic acids and ligands. In addition, the topology and the conformation adopted by the loops have profound consequences for small-molecule drug design and development. The present simulation represents a threshold in G-quadruplex simulations, with regards to testing the force field for simulating four-stranded structures, the role of cations, dynamic flexibility and structural variability of parallel stranded propeller-type topology G-quadruplexes.
MATERIALS AND METHODS
MD simulations
The crystal structure of the parallel stranded propeller-type quadruplex formed from the human telomeric sequence d[AG3(T2AG3)3] (PDB id:1KF1) was used as the starting structure for the simulation (10). The X-ray structure contains a vertical alignment of K+ ions along the axis within the electronegative core of the structure. The ions, arranged in a square antiprismatic coordination, are sandwiched between the quartets and were retained in positions as in the crystal structure. The MD simulations were carried out using AMBER11 software using the Parmbsc0 version of the Cornell et al. force field (29–31). The structure was explicitly solvated with TIP3P water molecules in a periodic box (65 Å × 64 Å × 68 Å) whose boundaries extended at least 10 Å from any solute atom. Additional 19 K+ counter ions were added to the system to neutralize the charge on the quadruplex backbone, whose parameters were adapted from Hazel et al. (32).
The protocol for energy minimization and MD in explicit solvent was adopted from Haider et al. (20). The initial round of equilibration with explicit solvent and ions involved 1000 steps of steepest descent, followed by 1000 steps of conjugate gradient energy minimization. A 300 ps MD equilibration was performed in which the quadruplex was constrained, whereas the solvent and ions were allowed to equilibrate. The system was gently heated from 0 K to 300 K with a time step of 0.5 ps. This was followed by subsequent rounds of MD simulation, at constant pressure and 300 K for 200 ps. The constraints were gradually relaxed, until no constraints were applied to the system. The final MD production run was performed at 300 K using the particle mesh ewald summation method with a charge grid spacing of ∼1.0 Å (33). A cut-off of 10 Å for non-bonded Lennard–Jones interactions was used, and the non-bonded pair list updated every 20 steps. The SHAKE algorithm was used to constrain hydrogen atoms with a tolerance of 0.0005 Å and a 2 fs time step (34). The final production run was carried out without any restraints on the system for a total of 1.5 μs. Frames collected every 2 ps made up the trajectory and were analysed using the ptraj module in Ambertools 1.4 (35). The simulation were run on GPU clusters and lasted ∼45 days.
Clustering
Clustering analysis can detect and classify objects, described by structural data, into different groups based on structural similarity (36). To identify structural clusters from a trajectory, a root mean squared deviation (RMSD) parameter was used, where the pairwise distances measured as coordinates between structures are defined by a cut-off value reflecting the range of conformations and their relative populations. The algorithm generates centroids describing each cluster and then gives an RMSD for each structure in the trajectory with respect to each identified cluster. Clustering was carried out after removal of the first 300 ns, which were attributed to the equilibration phase. The frames were extracted at a time interval of 6 ps yielding 50 000 frames. The MMTSB toolkit code (29) was used with a cut-off of 3.0 Å.
Data representation
The backbone angles α, β, γ, ε, δ, ζ and the glycosidic torsion angle χ were monitored using the ptraj module of AMBER (35). Deoxyribose puckering pseudorotation angles (p) and amplitudes (A) have been determined following the definitions of Altona and Sundaralingam (37), using the same reference state for P = 0.0 degrees. The torsion, pseudorotation angles and amplitudes were extracted from the AMBER trajectory and plotted using MATLAB ®(2012a,The Mathworks City, USA). The structural illustrations were prepared using VIDA (open eye scientific software, Santa Fe, New Mexico), VMD (38) and PyMol (http://www.pymol.org) programs. The clustering data were plotted using the Xmgrace program (http://plasma-gate.weizmann.ac.il/Grace).
RESULTS
Structural dynamics on a microsecond timescale differs dramatically from shorter simulations
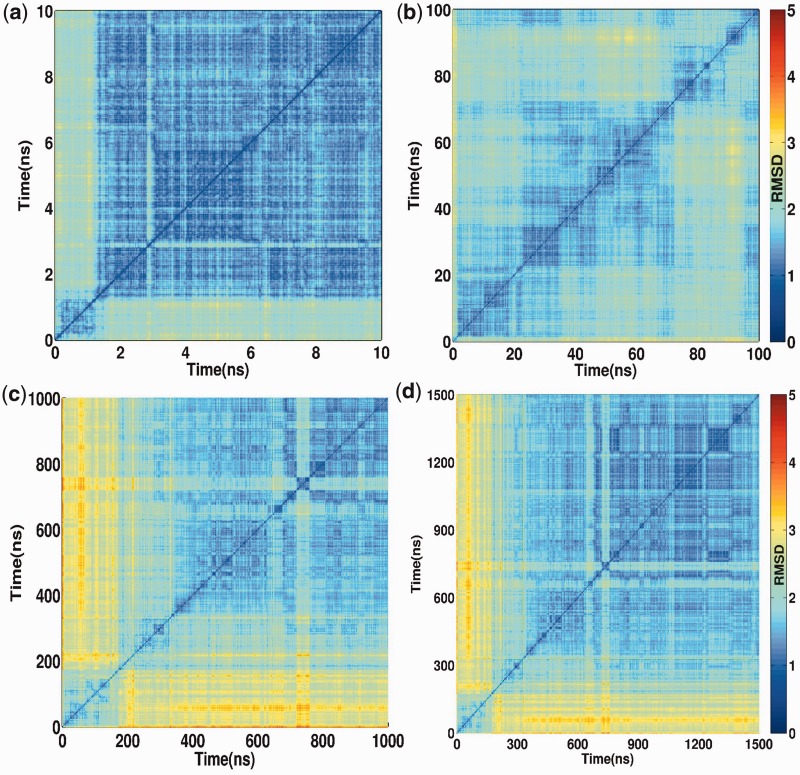
The structural integrity of the quadruplex persisted during the complete course of the lengthy 1.5 µs of MD simulation. To analyse the importance of the timescale on which the propeller-type structures equilibrate and vary, the pairwise RMSD were assessed over both short and long simulation times (Figure 1). Most of the simulations of G-quadruplex systems published to date were performed on a time scale from 1 to 100 ns (12,20,23,39–42). One of the principal results of our work is that such simulations are insufficient to properly characterize structural dynamics of the G-quadruplex loops. As shown in Figure 1, different timescales provide dramatically different qualitative description of the structural dynamics. In fact, we show that the first ∼300 ns of our simulation give a misleading picture of the loop structural dynamics. Only after this period does the system settle down. In addition, although the loops establish few characteristic stable interactions (mainly Ade1-Ade13 and Thy17-Gua15, see later in the text), they sample a rich ensemble of transient geometries. The loop geometry thus cannot be characterized by a single structure. Examination of the trajectory in short bins reveals that the structure can adopt stable conformations that can last several nanoseconds. Although such structures might be easily interpreted as ‘stable’ in short simulations, considerisng them in the context of a long µs or longer simulation, these metastable structures are transient, and some of them can be only marginally populated in sufficiently long simulations. We have therefore treated the first 300 ns as equilibration and carried out all analyses (except when explicitly stated otherwise) presented later in the text on the trajectory portion beyond this point.
Figure 1.
Conformational dynamics of human parallel stranded propeller-type quadruplex DNA on different timescale assessed by pairwise RMSD matrices. The minimum and maximum was assigned to a linear colour bar to indicate the variation in RMSD. Owing to large sample size, the trajectory for (a) 0–10 ns was sampled at δt = 1 ps; (b) 0–100 ns was sampled at δt = 10 ps; (c) 0–1000 ns was sampled at δt = 100 ps; (d) 0–1500 ns was sampled at δt = 150 ps. The matrix of 10 ns trajectory shows relaxation of coordinates after first ∼2 ns. The high RMSD (yellow stripes) in 100 ns matrix showed that the trajectory in this duration was still in equilibration. It is evident from matrix (c and d) that the trajectory equilibrates at ∼300 ns. A large conformational transition observed at ∼720 ns is owing to the formation of a transient triad.
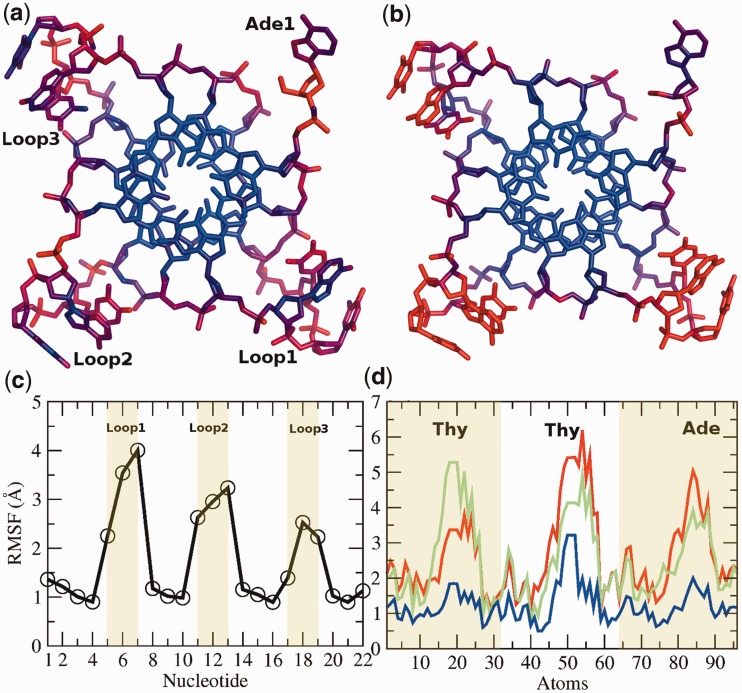
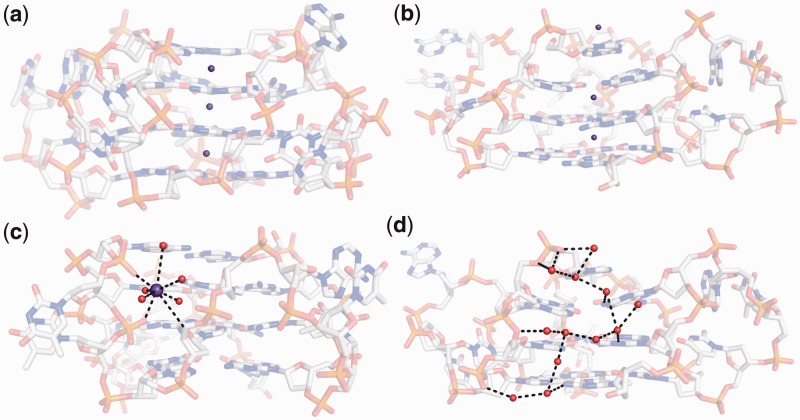
The average RMSD of the all-atom structure (4.5 Å) is significantly greater than that of the backbone (3.4 Å) and can be attributed to the wobbling effect of bases and the flexible nature of the TTA loops (Supplementary Figure S1). The stacked G-quartet stem is the most stable sub-segment of the structure. To highlight regions of stability and similarity between the crystal and the simulated structure via MD, wire figures coloured by temperature factors and RMSF have been illustrated in Figure 2. The stable (blue) central G-quartets are surrounded by more mobile TTA loops (red). Plotting RMSF values, calculated from the simulation and comparing with the B-factors in the crystal structure, highlights similar thermal fluctuations.
Figure 2.
Thermal mobility in the parallel stranded propeller-type quadruplex. A comparison of atomic fluctuations in the (a) crystal structure and (b) MD simulation is represented as wireframe. The mobility distribution was calculated using B-factors from the crystal structure (PDB id 1KF1) and averaged RMSF from the MD simulation. The stable central G-quartets (blue) are surrounded by more mobile loops (red). This is also illustrated in (c). The mobility of the loops is independent of each other. (d) Loop1 (red) and Loop2 (green) exhibit higher mobility than the Loop3 (blue).
The three loops show non-equivalent dynamics
Following the flexibility of the loop structure during the course of the simulation and comparing them with the X-ray structure reveals significant conformational changes. However, the loop conformational rearrangement did not have any great impact on the structure of the central G-quartet stem. The RMSF analysis indicates that the mobility of each of the simulated loops is independent of one another (Figure 2c and d). Although the three loops might appear at first sight to be equivalent, they occur in different structural contexts, which make their dynamics distinct. The most important factor leading to non-equivalence of the loop dynamics is the flanking Ade1 nucleotide. Ade13 from loop2 forms hydrogen bonds with the flanking Ade1 and end caps the quadruplex at one end. This interaction once formed is retained throughout the simulation. The stable conformation of Ade13 on top of the terminal quartet stretches loop2, and as a result of this arrangement, the Thy11-Ade13-Thy12 stacking conformation, as observed in the crystal structure, is permanently lost. Bases from the other two loops sample an ensemble of transient loop geometries, which can be characteristically stable for several nanoseconds. For instance, the first thymines in loops 1 and 3 (Thy5 and Thy17) dynamically interact with the middle quartet to form pentad and hexad architecture (see later in the text). Moreover, Ade7 also rotates around the χ-torsion angle to interact via π-π stacking with Thy6 and via edge-to-face with Thy5. We also observe in the simulation that the loops interact with the quartets to adopt distinct global geometries, and long simulation times are required to explore the conformational space appropriately.
Sugar pucker and the backbone angles
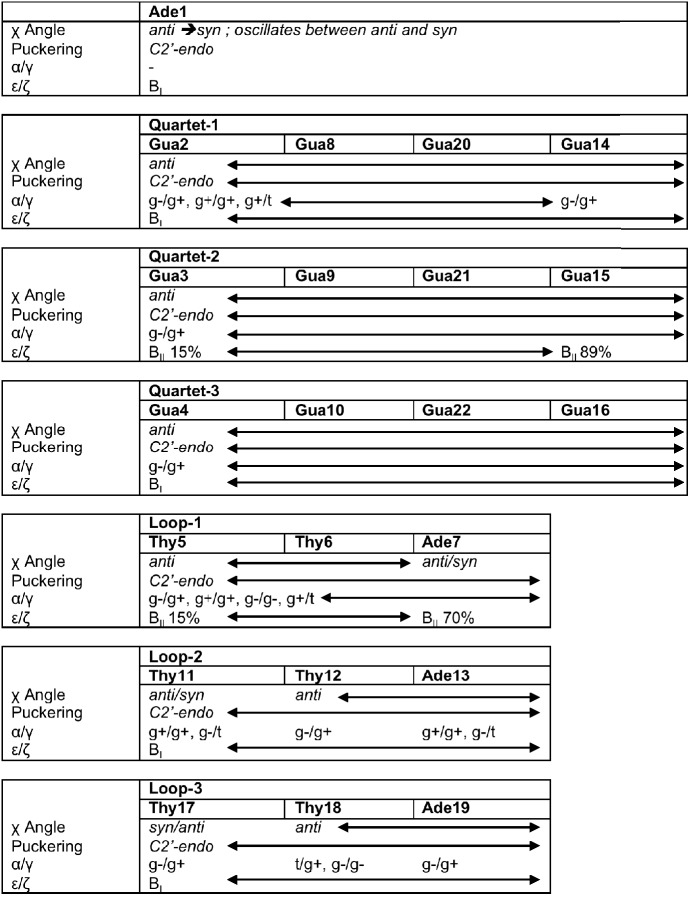
The furanose ring in nucleic acids is conformationally flexible (43). The backbone torsion angles and sugar conformations in DNA arrange to present an interface for proteins and other molecules to interact (44). The backbone conformation of the quadruplex has been analysed based on the following degrees of freedom: χ angle, sugar puckering, α/γ and ε/ζ crankshaft motions. The χ angle describes the relative glycosyl orientation and is in either anti (180°–360°) or syn (0°–180°) conformation. The sugar pucker describes the extent to by which the furanose ring in nucleic acids deviates from a plane. The α and γ angles occupy trans (t) and gauche (g) ± regions, and α/γ sampling is useful to validate the force fields. For example, the α/γ crankshaft motions indefinitely trapped in non-canonical regions of α/γ conformational space lead to force-field-dependent artefacts (45). The nucleic acid backbone is considered to be in the BI form when the ε and ζ difference is negative and in the BII form when it is positive (46). The above-described conformational classes are typically discussed for B-DNA duplexes, but they also have relevance to the quadruplex backbone. A summary of conformational variability of all nucleotides is presented in Figure 3, and descriptive analysis is presented in the Supplementary section.
Figure 3.
Conformational variability in the backbone of the parallel stranded propeller-type quadruplex. The arrows indicate similar values for different bases.
Terminal capping by A:A pair and A:A:A triad
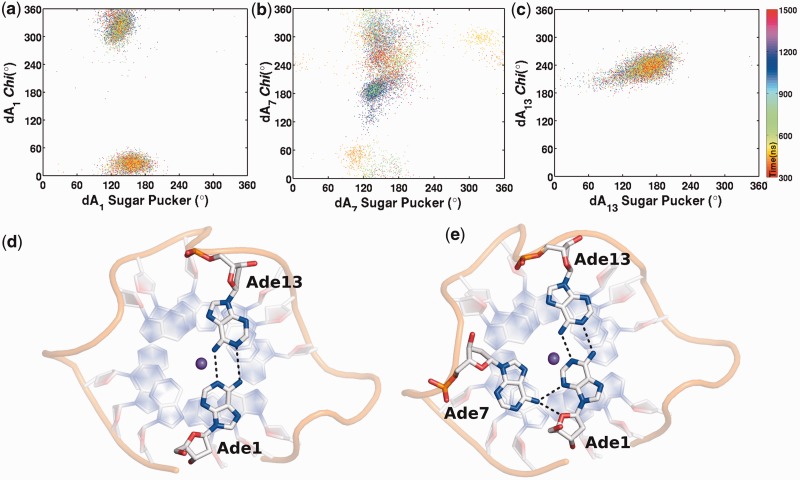
The 5′ adenine (Ade1) in the crystal structure protrudes away from the core into the solvent (Figure 2a). It adopts an anti conformation and does not interact with any other part of the quadruplex. In our previous work, we had removed this nucleotide assuming its conformation to be a crystallization artefact that might affect the stability of the quadruplex (20). An NMR solution structure of a parallel-stranded quadruplex formed from human telomeric DNA sequence has subsequently been determined under molecular crowding conditions (12). It adopts the same fold as the X-ray structure; however, the 5′ adenine is stacked on the adjacent terminal G-quartet. Based on this observation, we decided to retain Ade1 in the starting structure. Within the first few nanoseconds of the production run, the glycosidic χ angle of Ade1 changed from anti to syn conformation. This leads to formation of an intramolecular hydrogen bond between the 5′-OH group and N3. An analogous hydrogen bond has been observed in experimental structures that have a guanine nucleotide at 5′ termini in a syn conformation (47). Such an intramolecular hydrogen bond has also been shown to strongly stabilize the syn conformation of the first guanine in the G-quartet stems in the absence of any upstream nucleotide (48). The χ angle switches back to an anti conformation (∼470 ns), which is accompanied by a change in sugar pucker from C2′-endo to C1′-exo. The χ-angle then oscillates between anti and syn positions during the latter course of the simulation (Figure 4a). The flexibility of the sugar phosphate backbone allows Ade1 to form π-π stacking interactions with guanines on the terminal quartet. Stacking is observed with the adjacent Gua2 or across with the neighbouring Gua20. The hydrogen bond between 5′-OH-N3 is lost when Ade1 is in anti conformation. We observe that owing to stochastic backbone movements, Ade1 can flip back into the solvent. However, this is an unfavourable and only transient event, as Ade1 subsequently flips back to stack on the G-quartet stem. Ade13 also moves to the top of the quartet to form reverse Watson–Crick hydrogen bonds with Ade1 (Figure 4d). The syn conformation of Ade1 and anti conformation of Ade13 facilitates head-to-head base pairing between Ade1 and Ade13 (Supplementary Figure S10a). Ade7 from Loop1 also comes in-plane with the Ade1:Ade13 pair to form an A:A:A triad (Figure 3e). It bonds to Ade1 by side-by-side trans-sheared A:A pairing, which appears to be the only plausible configuration for the A:A:A structure observed in the simulation (49). Quadruplex loops are highly flexible structures, and the π-stacking interactions of Ade7 with the first quartet are accompanied by stereochemically permissible distortions in the backbone angles of the loop residues. To achieve the chain turn-back, the γ torsion angle changes from g+ to trans, allowing the sheared A:A bonding to take place (Supplementary Figure S7) (50).
Figure 4.
Time development of A:A base pair and A:A:A triad on the terminal quadruplex. Scattergram of glycosidic chi angle versus sugar pucker angle of (a) Ade1, (b) Ade7 and (c) Ade13. (d) The syn conformation of Ade1 and anti conformation of Ade13 facilitates reverse Watson and Crick A:A base pairing on terminal quartet. (e) Ade7 flips to syn conformation and interacts with Ade1 to form A1:A7:A13 triad. The K+ ion is represented as purple sphere.
Formation of a pentad and hexad
Analysis of the trajectory also reveals that Thy5 and Thy17 interact with the quartets resulting in diverse quadruplex geometries. These thymine bases bury their methyl groups into the groove, as the O4 and imino hydrogen atoms of Thy5 and Thy17 hydrogen bond with the N2 amino hydrogen and N3 atom of Gua3 and Gua15 (Figure 5a and b). However, a pentad and hexad is only formed when both O4 and N3 atoms of Thy5/Thy17 are hydrogen bonded to a guanine from the middle quartet. These interactions ensure that the thymine bases are in plane with the quartet. In the crystal structure, Thy17 forms π-π interactions with Ade19. Early during the simulation, the π-stack with Thy17 is lost, and Ade19 forms equivalent interactions with Thy18. Thy17 moves in-plane with the central quartet owing to changes in the backbone torsion angles, to form a pentad (Figure 5c). The first indication of Thy5 and Thy17 interactions with the middle quartet occurs at ∼100 ns, strongly suggesting that these features are not observable in short simulations. During the course of the simulation while Thy17 makes a pentad with the middle quartet, Thy5 vacillates to interact with G-quartet stem in various ways. When Thy5 approaches the plane of the middle quartet, and both Thy5 and Thy17 are simultaneously aligned with the middle quartet, a planar T:(GGGG):T hexad is formed (Figure 5d). This motif is maintained by simultaneous hydrogen bonding of Thy5 with Gua3 and Thy17 with Gua15, which is sampled for ∼3% over the equilibrated trajectory.
Figure 5.
Representation of pentad and hexad alignment. The minimum distance plot of (a) Thy5 and Gua3 and (b) Thy17 and Gua15 from the middle quartet. A pentad is formed when either Thy5 or Thy17 is in plane with the quartet and within hydrogen bonding distance. A hexad is formed when both Thy5 and Thy17 simultaneously align with the middle quartet and form hydrogen bonds. Structural representation of (c) Thy17 interaction through its Watson–Crick face via sheared hydrogen bonds with Gua15 to form pentad with middle quartet. (d) Concurrent alignment of Thy5 and Thy17 with middle quartet through Watson–Crick face forms a T:(GGGG):T hexad. The first 300 ns are highlighted as a yellow box, and an arbitrary line at 3.5 Å is drawn to highlight hydrogen bond formation. The K+ ion is represented as purple sphere.
Role of ions and hydration in maintaining the structure of the quadruplex
This study finds that K+ ions remain positioned between the stacked G-quartets throughout the course of the simulation. There was no exchange of cations between the G-quartet stem and bulk solvent as observed in other quadruplex systems (19,41,42). The formation of the Ade1:Ade13 base pair is briefly coupled by the position of a K+ ion (from bulk) between them. Ade1 and Ade 13 interact with the ion by forming water bridges with the hydration shell around the K+ ion. When Ade13 is within bonding distance with Ade1, the K+ ion leaves, and a stable Ade1 and Ade13 base pair is formed (Supplementary Figure S11). Additional K+ ions stack, albeit briefly, between the base pair and the top quartet and also above the A:A base pair (Figure 6a and b). Similar role of stabilization by ion has also been proposed by Maiti and co-workers (40). Analysis of the trajectory also reveals the presence of K+ ion coordination in the loops, which occurs primarily via phosphate backbone atoms and solvent molecules. The dynamic movement of the backbone phosphate atoms results in the generation of a localized electronegative sink. The loop captures a K+ ion to compensate for the electrostatic repulsion (Figure 6c). Similar stabilization of the loop by a cation has been observed in the crystal structure of the cKit-1 quadruplex (51). An extensive spine of hydration is also observed, similar to those in the crystal structures of quadruplexes (Figure 6d) (51,52). The extensive solvent network mediates interactions between bases from the loop and the G-quartet stem. For example, we observe that water bridges mediate the binding of Thy17 to Gua15 until Thy17 comes into direct hydrogen bonding with Gua15 (Supplementary Figure S12).
Figure 6.
Role of bulk water and ions in stabilization of the topology. Additional K+ ions positioned (a) between the terminal quartet and the A:A diad and (b) on top of the A:A diad. This is analogous to cation bonding between the quartets. (c) K+ ion coordination with the backbone atoms and water molecules. Ion capture in the loops occurs when an electronegative sink is generated owing to the close proximity of phosphate backbone atoms. The ion helps in stabilizing the inter-phosphate repulsion. (d) Spine of hydration as observed in a snapshot during MD simulation. The hydration networks are similar to those observed in quadruplex crystal structures. The K+ ion is represented as purple sphere, whereas the solvent (water) is coloured red.
Clustering
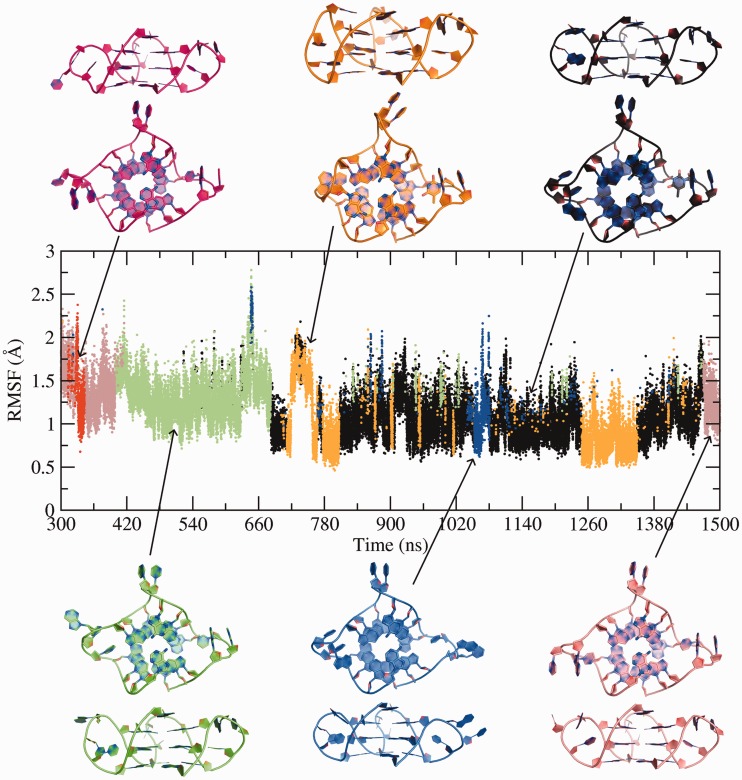
Clustering analysis aims to identify the main substates sampled during the simulation. A 3.0 Å RMSD cut-off identified six clusters (Figure 7). The top three clusters account for ∼86% of the sampled conformational space. The simulation is highly heterogeneous, and diverse structural scaffolds are observed. In the pink cluster, π-stacking interactions dominate in loop1 and loop3. This cluster appears ∼300 ns and then again towards the end of the simulation at ∼1450 ns. The conformation of loop1 and loop3, observed in this pink cluster (at 1481 ns), closely resembles the loops of the X-ray/NMR structures (Figure 8). The red cluster briefly appears within the pink cluster, when the π-stacking in loop1 is lost. Thy6 faces outward into the solvent. However, this is a short-lived conformation, and the loops reform their Thy:Ade:Thy stacking geometry quickly. A stable A:A base pair and a pentad (Thy17) contribute to the conformation adopted in the green cluster. Ade7 in loop1 is positioned towards the solvent, similar to Ade1 in the crystal structure. The jump observed at ∼720 ns, in the orange cluster, is a result of the formation of the A:A:A triad on top of the terminal quartet. The position of Ade7 stretches and constrains loop1. It places Thy6 in plane with the middle quartet. However, the distortions in the phosphate backbone prevent Thy6 to form hydrogen bonds with the middle quartet. Meanwhile, Thy17 interacts with the middle quartet to form a pentad. The orange cluster reappears at ∼1240 ns, highlighting a similar constrained conformation of loop1. The black cluster is defined by the formation of A:A base pair and a pentad (Thy17) along with other π-stacking arrangements within the loops. In the blue cluster, the interactions of loop3 with the quartet are lost, and the bases orient towards the solvent. The structures observed in the orange and blue clusters are transient conformations that lie in the more stable black cluster.
Figure 7.
Ensemble of conformations identified via clustering analysis. RMSD-based clustering, with a cut-off of 3.0 Å, over equilibriated trajectory identified six clusters. Cartoon representations of top and side view of cluster centres are illustrated. The orange and blue clusters are transitional substates observed within the black cluster, whereas the red cluster appears within the pink cluster.
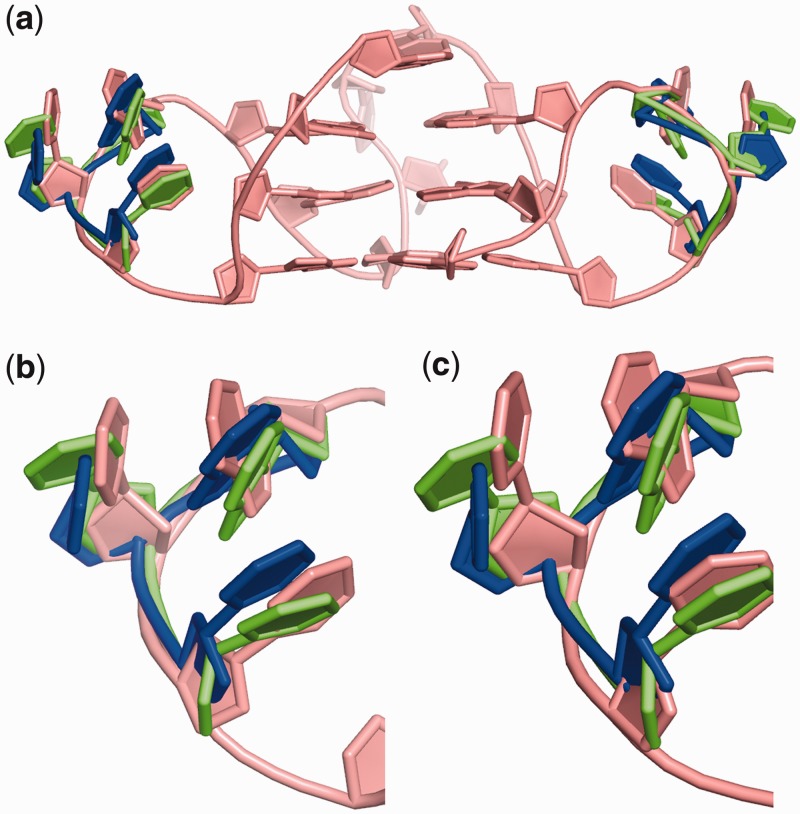
Figure 8.
Comparison of the loops in the (a) simulated structure (pink) with the X-ray (blue) and the NMR structure (green). The superimposition highlights the similarity between the structures from (b) loop1 and (c) Loop3. The snapshot from the simulation was extracted at 1481 ns.
Comparison with experimental data
The present analysis focuses on the maximally sampled base pairing events that modify the topology of the quadruplex DNA. Many of these conformations are unprecedented with regard to the simulation of intramolecular quadruplexes. An important point is that the simulation, despite showing a wide range of loop geometries, is capable of spontaneously sampling loop structures that are similar to those in the currently available atomistic experiments. Clustering analysis helped in identifying a sub-state (at 1481 ns) where the conformation of loop1 and loop3 closely resembles the NMR and crystal structure. A superimposition of the identified conformation with NMR and crystal structures yielded an RMSD of <2.0 Å (Figure 8). Importantly, the individual loops 1 and 3 occasionally also transiently sample such geometry in the other parts of the trajectory, which indicates that the experimental loop geometry, although not being the most stable one, is within the low-energy region of the simulation. Loop2 is not sampling this geometry, as it is constrained by the A1:A13 base pairing. This, however, does not a priori mean a disagreement with experiment. Although A1 in the X-ray structure is involved in crystal contacts, there is a significant non-equivalence between the NMR and simulated sequences, namely, in the NMR experiment, the flanking sequence is 5′-TA- instead of 5′-A-. There is thus no free adenine 5′-OH group in the NMR structure so that the intramolecular hydrogen bond stabilizing the adenine in the syn conformation cannot form. Owing to this, the adenine in the NMR structure is in a high-anti χ region, which is common in DNA.
Our study further reports non-canonical base pairing. The A:A pair formed by Ade1 and Ade13 has a Watson–Crick N1 hydrogen bond, which has been reported in RNA structures such as those in PDB structures id 1LNG and 1HR2 (53,54). Interactions of Ade7 with the backbone of Ade1 are typical of interactions commonly observed in RNA structures (55). Theoretically, an A:A pair with Watson–Crick hydrogen bonding can form an A:A:A triad only through trans sheared Hoogsteen bonds (49). This is the same architecture that is observed in the present simulation. A comparable structure with an A:A:A triad has also been observed in the 30 S ribosomal subunit (PDB id: 1FJG) (56). The T:(GGGG):T hexad, which forms briefly when Thy5 and Thy17 are simultaneously aligned with the middle quartet, has been observed in the structure of a bimolecular quadruplex (PDB id 1XCE) (57). The π-stacking interactions within the loops have been observed in our previous simulation of quadruplex multimers and are consistent with several experimental observations of loop geometries including those seen in quadruplex-ligand interactions (20,58,59). The frequent sampling of stacking interactions between Ade-Thy and Thy-Thy observed in the loops suggests that these interactions contribute to stabilizing quadruplex geometry. These interactions have been evident in several other previous simulations of quadruplex (60). The α/γ torsion angles of the loop bases observed in the present work are also in agreement with the α/γ angles as observed in the crystal structures (22).
DISCUSSION AND CONCLUSIONS
The present study provides an unprecedented dynamic atomistic picture of the propeller-type loops, covering 1.5 µs time window with ps-scale time resolution. It is fair to admit that the dynamics (mainly balance between different substates) may be affected by the approximations of the simulation force field, as single-stranded nucleic acids topologies are specifically challenging for force-field description (23,61,62). This may contribute to the fact that the experimental geometry of the loops, albeit sampled in the simulations transiently, is not the dominant simulation sub-state. Obviously, also the simulation time scale of the present study is still far from full convergence; therefore, we cannot claim that we see ‘correct’ populations of the substates. For both these reasons, the relative balance of different structures seen in the simulation should not be over-interpreted, although we suggest that the simulation likely samples relevant substates. However, a clear result of our simulation is that the conformational space of the loops is rich and more dynamic than we have assumed to date. The simulation results suggest that the loops may exist as a dynamic continuum of interconverting substates, which would be difficult to fully capture by available experimental methods. In addition, we show that long simulations are needed to characterize the quadruplex DNA loop dynamics sufficiently exhaustively and without a visible bias from the starting structure. Considering just the first 300 ns (which is still a much longer trajectory than the majority of those analysed in preceding studies) would be, regarding the loop dynamics, rather misleading.
One of the most interesting structural observations here is end capping of a quadruplex with the terminal adenine base. Such interaction has also been observed in a guanine and adenine-rich quadruplex (63). Stacking of a G-quartet by a Ade(anti):Ade(syn) base pair is also evident in this structure as well as in another bimolecular quadruplex (57,63). Interaction of a pyrimidine base with an Ade(syn):A(anti) base pair to form a triad has also been reported (63). However, to the best of our knowledge, this is the first observation of a stable terminal A:A base pair and an A:A:A triad in a quadruplex simulation. The A:A:A triad with similar bond directionality has been observed in an experimental RNA structure (64). The current basis for quadruplex-targeted drug design is in large part that end stacking on G-quadruplexes is the principal mode of interaction for extended heteroaromatic ligands (65). The A:A stacks in our simulation corroborate with the fact that π-stacking interactions over the terminal quartet stabilize the quadruplex topology. Also, the A:A pairing on the terminal quartet provides a large surface area for ligand binding. This enables molecules whose chromophores have a larger surface area than, for example, a diamido-acridine moiety, to bind to the ends of a quadruplex (22). It is, however, fair to admit that in our simulations to a certain extent, the A1:A13 interaction can be facilitated by the specific 5′-A1 single-nucleotide end capping, which supports the A1 syn conformation via the 5′-end-specific 5′-OH…N3 intramolecular H-bond. This adenine is involved in crystal packing in the X-ray structure, and in the NMR structure, it is not preceeded by any nucleotide so that the intramolecular hydrogen bond cannot be formed.
While exploring the conformational flexibility of the TTA loop, neither the pairing of the guanines nor the stacking geometry of the quartets was altered by the linking topology of the external loops. We observe that although the G-quartet stem of the quadruplex remains rigid, the bases in the loops can form base-pairing alignments that further stabilize the quadruplex. Higher-order pairing alignments such as a pentad, a hexad or a heptad, resulting from interactions of adenines with a G-quartet, have been observed in the 93del aptamer and other quadruplex-forming structures (47,66–68). These alignments are formed when the major groove edge of adenine forms sheared trans Hoogsteen bonds with guanine in a G-quartet (47,63). Hexad alignment with thymine as the interacting base has been reported in the NMR solution structure of the bimolecular quadruplex d(GCGGTTGGAT) (57). This structure involves the Watson–Crick face of a thymine forming sheared hydrogen bonds with a guanine in quartet. This interaction effectively perturbs the quadruplex shape from a circular towards an expanded toroid architecture (57). The sugar conformations in the hexad, in our simulation, remain predominantly in C2′-endo pucker. A notable exception occurs in Thy17 (Supplementary Figure S9). The change in sugar pucker to C3′-endo in Thy17 allows it to become in-plane with the central G-quartet and hydrogen-bond with Gua15. This switch in sugar pucker is to avoid severe steric overlap with the sugar in Thy18. All the guanine nucleotides in the quadruplex are in a BI conformation except for Gua15 (Supplementary Figures S2–S4). The inter-phosphate repulsion is a dominant factor that causes a switch from BI to a BII conformation (46). The flexibility in Gua15 facilitates Thy17 interaction with the middle quartet and maintains stable interaction for much of the simulation time. The 29% sampling of BII conformation by Gua3 suggests that the broadening of the groove is brief, and therefore, the interaction of Thy5 with the middle quartet is not as evident as that of Thy17. It has been suggested that the BI form is intrinsically of lower energy than the BII form owing to the more favourable orientation of the phosphodiester backbone (46,69–71). However, the energy required to populate the BII conformation can be gained from other energy contributions such as base stacking or hydration.
The K+ ions sandwiched between the G-quartet are retained in antiprismatic bipyramidal coordination. The exterior of the quadruplex also coordinates with the ions via an extensive water network (Figure 6). Coordination networks of a nucleic acid with the solvent are observed when non-canonical base pairing play an important role in stabilizing tertiary structures (72). In accordance with this, we observe that formation of the adenine cap is stabilized by a water-K+ bridge (Supplementary Figure S11). Furthermore, K+ ions have also been involved in stabilization of loops. A water bridge is also formed between Thy17 and Gua15, suggesting that solvent plays a role in maintaining intra-molecular contacts within the quadruplex (Supplementary Figure S12).
The present 1.5 µs simulation of a human telomeric quadruplex DNA represents 750 000 frames of structural data. Empirical analysis of this vast data set may result in the loss of important ensembles. Therefore, the trajectory was partitioned into clusters of structures that share similar conformational space. The RMSD-based clustering analysis groups the trajectory into six distinct conformations. These clusters have been correlated with the trajectory after 300 ns to infer the conformational pathway in the part of the simulation, which is already not visibly affected by the starting configuration (Figure 7). The clusters coloured pink, green and black have low median values, highlighting metastable regions. The orange, blue and green clusters can form within the black cluster, suggesting an inter-convertible conformation. The red cluster observed within the pink cluster, which appears both at the start and end of the simulation, suggests the presence of a transition state in the metastable pink cluster. The clustering analysis makes it clear that although the trajectory has equilibrated, it has not yet converged, indicating that the conformational sampling of loops is not complete.
The simulation reflects the flexible and dynamic nature of loops as observed in shorter timescale simulations. The loops during the simulation exhibit conformations that resemble those observed in the solution and crystalline forms. Amongst the loops, loop1 and loop3 transiently show arrangement consistent with the crystal/NMR structure (Figure 8). Ade-Thy stacking in loop2 is lost as a result of the formation of the Ade1:Ade13 base pair over the terminal quartet. The distinct structural scaffolds that are observed in the present simulation have been experimentally reported in other G-quadruplex and RNA structures. However, these conformations have not been reported in a human telomeric quadruplex. The ability of loops to interact with the G-quartet stem in different ways is a significant feature of the current simulation. This provides insight into new conformational topologies and geometries, which may be captured and used by drugs designed for stabilizing quadruplex in human telomeric regions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–14.
FUNDING
‘CEITEC - Central European Institute of Technology’ [CZ.1.05/1.1.00/02.0068] from European Regional Development Fund and by the Grant Agency of the Czech Republic [P208/11/1822] (to J.S.); a Cancer Research UK programme (to S.N.). Funding for open access charge: CCRCB startup fund.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
S.H. would like to thank the COST-MP0802 Program and a CCRCB startup grant.
REFERENCES
- 1.O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nature Rev. Mol. Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saldanha SN, Andrews LG, Tollefsbol TO. Assessment of telomere length and factors that contribute to its stability. Eur. J. Biochem. 2003;270:389–403. doi: 10.1046/j.1432-1033.2003.03410.x. [DOI] [PubMed] [Google Scholar]
- 3.Sahin E, DePinho RA. Axis of ageing: telomeres, p53 and mitochondria. Nature Rev. Mol. Cell Biol. 2012;13:397–404. doi: 10.1038/nrm3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neidle S, Parkinson GN. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003;13:275–283. doi: 10.1016/s0959-440x(03)00072-1. [DOI] [PubMed] [Google Scholar]
- 5.Neidle S. Therapeutic Applications of Quadruplex Nucleic Acids. Boston, MA: Academic Press; 2012. pp. 109–117. [Google Scholar]
- 6.Adrian M, Heddi B, Phan AT. NMR spectroscopy of G-quadruplexes. Methods. 2012;57:11–24. doi: 10.1016/j.ymeth.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Gray RD, Chaires JB. Isothermal folding of G-quadruplexes. Methods. 2012;57:47–55. doi: 10.1016/j.ymeth.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Randazzo A, Spada G, Silva M. Topics in Current Chemistry. Berlin/Heidelberg: Springer; 2012. pp. 1–20. [Google Scholar]
- 9.Yuan G, Zhang Q, Zhou J, Li H. Mass spectrometry of G-quadruplex DNA: formation, recognition, property, conversion, and conformation. Mass Spectrom. Rev. 2011;30:1121–1142. doi: 10.1002/mas.20315. [DOI] [PubMed] [Google Scholar]
- 10.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 11.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heddi B, Phan AT. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011;133:9824–9833. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- 13.Hud NV, Plavec J. In: Quadruplex Nucleic Acids. Neidle S, Balasubramanian S, editors. The Royal Society of Chemistry; 2006. pp. 100–130. [Google Scholar]
- 14.Ellis RJ, Minton AP. Cell biology: join the crowd. Nature. 2003;425:27–28. doi: 10.1038/425027a. [DOI] [PubMed] [Google Scholar]
- 15.Xue Y, Kan ZY, Wang Q, Yao Y, Liu J, Hao YH, Tan Z. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J. Am. Chem. Soc. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- 16.Parkinson GN, Cuenca F, Neidle S. Topology conservation and loop flexibility in quadruplex-drug recognition: crystal structures of inter- and intramolecular telomeric DNA quadruplex-drug complexes. J. Mol. Biol. 2008;381:1145–1156. doi: 10.1016/j.jmb.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 17.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Haider SM, Autiero I, Neidle S. Surface area accessibility and the preferred topology of telomeric DNA quadruplex–ligand complexes. Biochimie. 2011;93:1275–1279. doi: 10.1016/j.biochi.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Reshetnikov RV, Sponer J, Rassokhina OI, Kopylov AM, Tsvetkov PO, Makarov AA, Golovin AV. Cation binding to 15-TBA quadruplex DNA is a multiple-pathway cation-dependent process. Nucleic Acids Res. 2011;39:9789–9802. doi: 10.1093/nar/gkr639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haider S, Parkinson GN, Neidle S. Molecular dynamics and principal components analysis of human telomeric quadruplex multimers. Biophys. J. 2008;95:296–311. doi: 10.1529/biophysj.107.120501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Šponer J, Cang X, Cheatham TE., III Molecular dynamics simulations of G-DNA and perspectives on the simulation of nucleic acid structures. Methods. 2012;57:25–39. doi: 10.1016/j.ymeth.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neidle S, Parkinson GN. Quadruplex DNA crystal structures and drug design. Biochimie. 2008;90:1184–1196. doi: 10.1016/j.biochi.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 23.Fadrná E, Špačková Na, Sarzyñska J, Koča J, Orozco M, Cheatham TE, III, Kulinski T, Šponer J. Single stranded loops of quadruplex DNA as key benchmark for testing nucleic acids force fields. J. Chem. Theory Comput. 2009;5:2514–2530. doi: 10.1021/ct900200k. [DOI] [PubMed] [Google Scholar]
- 24.Perez A, Luque FJ, Orozco M. Dynamics of B-DNA on the microsecond time scale. J. Am. Chem. Soc. 2007;129:14739–14745. doi: 10.1021/ja0753546. [DOI] [PubMed] [Google Scholar]
- 25.Reshetnikov R, Golovin A, Spiridonova V, Kopylov A, Šponer J. Structural dynamics of thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) quadruplex DNA studied by large-scale explicit solvent simulations. J. Chem. Theory Comput. 2010;6:3003–3014. doi: 10.1021/ct100253m. [DOI] [PubMed] [Google Scholar]
- 26.Banáš P, Sklenovský P, Wedekind JE, Šponer J, Otyepka M. Molecular mechanism of preQ1 riboswitch action: a molecular dynamics study. J. Phys. Chem. B. 2012;116:12721–12734. doi: 10.1021/jp309230v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pérez A, Luque FJ, Orozco M. Frontiers in molecular dynamics simulations of DNA. Acc. Chem. Res. 2011;45:196–205. doi: 10.1021/ar2001217. [DOI] [PubMed] [Google Scholar]
- 28.Stebbeds WJD, Lunec J, Larcombe LD. An in silico study of the differential effect of oxidation on two biologically relevant G-quadruplexes: possible implications in oncogene expression. PLoS One. 2012;7:e43735. doi: 10.1371/journal.pone.0043735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez A, Marchan I, Svozil D, Sponer J, Cheatham TE, III, Laughton CA, Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE, III, DeBolt S, Ferguson D, Seibel G, Kollman P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995;91:1–41. [Google Scholar]
- 31.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 32.Hazel P, Parkinson GN, Neidle S. Predictive modelling of topology and loop variations in dimeric DNA quadruplex structures. Nucleic Acids Res. 2006;34:2117–2127. doi: 10.1093/nar/gkl182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darden T, York D, Pedersen L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Phys. Chem. 1993;98:10089–10092. [Google Scholar]
- 34.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 35.Case DA, Cheatham TE, III, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bock HH. Probabilistic models in cluster analysis. Comput. Stat. Data Anal. 1996;23:5–28. [Google Scholar]
- 37.Altona C, Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J. Am. Chem. Soc. 1972;94:8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- 38.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 39.Cang X, Šponer Ji, Cheatham TE., III Insight into G-DNA structural polymorphism and folding from sequence and loop connectivity through free energy analysis. J. Am. Chem. Soc. 2011;133:14270–14279. doi: 10.1021/ja107805r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agrawal S, Ojha RP, Maiti S. Energetics of the human Tel-22 quadruplex−telomestatin interaction: a molecular dynamics study. J. Phys. Chem. B. 2008;112:6828–6836. doi: 10.1021/jp7102676. [DOI] [PubMed] [Google Scholar]
- 41.Akhshi P, Acton G, Wu G. Molecular dynamics simulations to provide new insights into the asymmetrical ammonium ion movement inside of the [d(G3T4G4)]2 g-quadruplex DNA structure. J. Phys. Chem. B. 2012;116:9363–9370. doi: 10.1021/jp304760k. [DOI] [PubMed] [Google Scholar]
- 42.Pagano B, Mattia CA, Cavallo L, Uesugi S, Giancola C, Fraternali F. Stability and cations coordination of DNA and RNA 14-Mer g-quadruplexes: a multiscale computational approach. J. Phys. Chem. B. 2008;112:12115–12123. doi: 10.1021/jp804036j. [DOI] [PubMed] [Google Scholar]
- 43.Arora K, Schlick T. Deoxyadenosine sugar puckering pathway simulated by the stochastic difference equation algorithm. Chem. Phys. Lett. 2003;378:1–8. [Google Scholar]
- 44.Lavery R, Zakrzewska K, Beveridge D, Bishop TC, Case DA, Cheatham TE, III, Dixit S, Jayaram B, Lankas F, Laughton C, et al. A systematic molecular dynamics study of nearest-neighbor effects on base pair and base pair step conformations and fluctuations in B-DNA. Nucleic Acids Res. 2010;38:299–313. doi: 10.1093/nar/gkp834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mura C, McCammon JA. Molecular dynamics of a kappaB DNA element: base flipping via cross-strand intercalative stacking in a microsecond-scale simulation. Nucleic Acids Res. 2008;36:4941–4955. doi: 10.1093/nar/gkn473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madhumalar A, Bansal M. Sequence preference for BI/BII conformations in DNA: MD and crystal structure data analysis. J. Biomol. Struct. Dyn. 2005;23:13–27. doi: 10.1080/07391102.2005.10507043. [DOI] [PubMed] [Google Scholar]
- 47.Majumdar A, Kettani A, Skripkin E, Patel DJ. Pulse sequences for detection of NH2…N hydrogen bonds in sheared G. A mismatches via remote, non-exchangeable protons. J. Biomol. NMR. 2001;19:103–113. doi: 10.1023/a:1008311624772. [DOI] [PubMed] [Google Scholar]
- 48.Cang X, Šponer J, Cheatham TE., III Explaining the varied glycosidic conformational, G-tract length and sequence preferences for anti-parallel G-quadruplexes. Nucleic Acids Res. 2011;39:4499–4512. doi: 10.1093/nar/gkr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zierkiewicz W, Michalska D, Hobza P. Adenine ribbon stabilized by Watson-Crick and Hoogsteen hydrogen bonds: WFT and DFT study. Phys. Chem. Chem. Phys. 2010;12:2888–2894. doi: 10.1039/b920433d. [DOI] [PubMed] [Google Scholar]
- 50.Chou SH, Zhu L, Gao Z, Cheng JW, Reid BR. Hairpin loops consisting of single adenine residues closed by sheared A.A and G.G pairs formed by the DNA triplets AAA and GAG: solution structure of the d(GTACAAAGTAC) hairpin. J. Mol. Biol. 1996;264:981–1001. doi: 10.1006/jmbi.1996.0691. [DOI] [PubMed] [Google Scholar]
- 51.Wei D, Parkinson GN, Reszka AP, Neidle S. Crystal structure of a c-kit promoter quadruplex reveals the structural role of metal ions and water molecules in maintaining loop conformation. Nucleic Acids Res. 2012;40:4691–4700. doi: 10.1093/nar/gks023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haider S, Parkinson GN, Neidle S. Crystal structure of the potassium form of an oxytricha nova G-quadruplex. J. Mol. Biol. 2002;320:189–200. doi: 10.1016/S0022-2836(02)00428-X. [DOI] [PubMed] [Google Scholar]
- 53.Hainzl T, Huang S, Sauer-Eriksson AE. Structure of the SRP19 RNA complex and implications for signal recognition particle assembly. Nature. 2002;417:767–771. doi: 10.1038/nature00768. [DOI] [PubMed] [Google Scholar]
- 54.Juneau K, Podell E, Harrington DJ, Cech TR. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA–solvent interactions. Structure. 2001;9:221–231. doi: 10.1016/s0969-2126(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 55.Zirbel CL, Sponer JE, Sponer J, Stombaugh J, Leontis NB. Classification and energetics of the base-phosphate interactions in RNA. Nucleic Acids Res. 2009;37:4898–4918. doi: 10.1093/nar/gkp468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 57.Webba da Silva M. Experimental demonstration of T:(G:G:G:G):T hexad and T:A:A:T tetrad alignments within a DNA quadruplex stem. Biochemistry. 2005;44:3754–3764. doi: 10.1021/bi0478190. [DOI] [PubMed] [Google Scholar]
- 58.Bazzicalupi C, Ferraroni M, Bilia AR, Scheggi F, Gratteri P. The crystal structure of human telomeric DNA complexed with berberine: an interesting case of stacked ligand to G-tetrad ratio higher than 1:1. Nucleic Acids Res. 2013;41:632–638. doi: 10.1093/nar/gks1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parkinson GN, Cuenca F, Neidle S. Topology conservation and loop flexibility in quadruplex–drug recognition: crystal structures of inter- and intramolecular telomeric DNA quadruplex–drug complexes. J. Mol. Biol. 2008;381:1145–1156. doi: 10.1016/j.jmb.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 60.Sponer J, Spackova N. Molecular dynamics simulations and their application to four-stranded DNA. Methods. 2007;43:278–290. doi: 10.1016/j.ymeth.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fadrna E, Spackova N, Stefl R, Koca J, Cheatham TE, III, Sponer J. Molecular dynamics simulations of Guanine quadruplex loops: advances and force field limitations. Biophys. J. 2004;87:227–242. doi: 10.1529/biophysj.103.034751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Banáš P, Hollas D, Zgarbová M, Jurečka P, Orozco M, Cheatham TE, III, Šponer Ji, Otyepka M. Performance of molecular mechanics force fields for RNA simulations: stability of UUCG and GNRA hairpins. J. Chem. Theory Comput. 2010;6:3836–3849. doi: 10.1021/ct100481h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kettani A, Gorin A, Majumdar A, Hermann T, Skripkin E, Zhao H, Jones R, Patel DJ. A dimeric DNA interface stabilized by stacked A.(G.G.G.G).A hexads and coordinated monovalent cations. J. Mol. Biol. 2000;297:627–644. doi: 10.1006/jmbi.2000.3524. [DOI] [PubMed] [Google Scholar]
- 64.Abu Almakarem AS, Petrov AI, Stombaugh J, Zirbel CL, Leontis NB. Comprehensive survey and geometric classification of base triples in RNA structures. Nucleic Acids Res. 2012;40:1407–1423. doi: 10.1093/nar/gkr810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haider SM, Neidle S, Parkinson GN. A structural analysis of G-quadruplex/ligand interactions. Biochimie. 2011;93:1239–1251. doi: 10.1016/j.biochi.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 66.Phan AT, Kuryavyi V, Ma JB, Faure A, Andreola ML, Patel DJ. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. Proc. Natl Acad. Sci. USA. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang N, Gorin A, Majumdar A, Kettani A, Chernichenko N, Skripkin E, Patel DJ. V-shaped scaffold: a new architectural motif identified in an A x (G x G x G x G) pentad-containing dimeric DNA quadruplex involving stacked G(anti) x G(anti) x G(anti) x G(syn) tetrads. J. Mol. Biol. 2001;311:1063–1079. doi: 10.1006/jmbi.2001.4916. [DOI] [PubMed] [Google Scholar]
- 68.Collie GW, Sparapani S, Parkinson GN, Neidle S. Structural basis of telomeric RNA quadruplex−acridine ligand recognition. J. Am. Chem. Soc. 2011;133:2721–2728. doi: 10.1021/ja109767y. [DOI] [PubMed] [Google Scholar]
- 69.Foloppe N, MacKerell AD. Contribution of the phosphodiester backbone and glycosyl linkage intrinsic torsional energetics to DNA structure and dynamics. J. Phys. Chem. B. 1999;103:10955–10964. [Google Scholar]
- 70.Hartmann B, Piazzola D, Lavery R. BI - BII transitions in B-DNA. Nucleic Acids Res. 1993;21:561–568. doi: 10.1093/nar/21.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hart K, Foloppe N, Baker CM, Denning EJ, Nilsson L, MacKerell AD. Optimization of the CHARMM additive force field for DNA: improved treatment of the BI/BII conformational equilibrium. J. Chem. Theory Comput. 2011;8:348–362. doi: 10.1021/ct200723y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hermann T, Patel DJ. Stitching together RNA tertiary architectures. J. Mol. Biol. 1999;294:829–849. doi: 10.1006/jmbi.1999.3312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.