Abstract
DNA gyrase plays a vital role in resolving DNA topological problems and is the target of antibiotics such as fluoroquinolones. Mycobacterium fluoroquinolone resistance protein A (MfpA) from Mycobacterium smegmatis is a newly identified DNA gyrase inhibitor that is believed to confer intrinsic resistance to fluoroquinolones. However, MfpA does not prevent drug-induced inhibition of DNA gyrase in vitro, implying the involvement of other as yet unknown factors. Here, we have identified a new factor, named Mycobacterium fluoroquinolone resistance protein B (MfpB), which is involved in the protection of DNA gyrase against drugs both in vivo and in vitro. Genetic results suggest that MfpB is necessary for MfpA protection of DNA gyrase against drugs in vivo; an mfpB knockout mutant showed greater susceptibility to ciprofloxacin than the wild-type, whereas a strain overexpressing MfpA and MfpB showed higher loss of susceptibility. Further biochemical characterization indicated that MfpB is a small GTPase and its GTP bound form interacts directly with MfpA and influences its interaction with DNA gyrase. Mutations in MfpB that decrease its GTPase activity disrupt its protective efficacy. Our studies suggest that MfpB, a small GTPase, is required for MfpA-conferred protection of DNA gyrase.
INTRODUCTION
DNA gyrase plays an important role in basic biological functions in bacteria, being involved in DNA replication, transcription and stress responses. It is the only known topoisomerase that introduces negative supercoils in the presence of adenosine triphosphate (ATP) and is involved in resolving DNA topological problems during DNA replication, transcription and recombination (1–3). DNA gyrase is essential in prokaryotes, and its absence in most higher eukaryotes has made it an important drug target in antibacterial chemotherapy (1). Although current studies on DNA gyrase mostly focus on its structure, function and catalytic mechanism, the regulation of this enzyme, especially the protection of its functions, which is critical to biological life, remains to be fully explored.
DNA gyrase is composed of two GyrA and two GyrB subunits, which assemble as an A2B2 heterotetramer (4–6). The crystal structures of the N- and C-terminal domains of the A and B subunits have been solved (7–10). Although structures of gyrase (or topoisomerase IV)-DNA-drug complexes have been published recently (11–14), the structure of the gyrase holoenzyme has not yet been reported. However, the functions of GyrA and GyrB are well understood: GyrA binds DNA, and GyrB is an ATPase. Structural and biochemical studies have shown that the mode of action of DNA gyrase involves a two-gate mechanism (15,16). Gyrase alters the topology of DNA by promoting the passage of one DNA duplex (the transported or ‘T’ segment) through a transient break in a second double-stranded DNA segment (the gate or ‘G’ segment) in an ATP-dependent manner.
Many drugs target DNA gyrase; aminocoumarins, which target GyrB, inhibit the ATPase activity of gyrase, whereas fluoroquinolones stabilize the gyrase-DNA complex by binding at the GyrA-GyrB-DNA interface (1). Quinolones are broad-spectrum antibiotics and the most successful of the drugs that target gyrase; however, their over-use and misuse have led to a loss in their efficacy owing to the development of drug resistance (1,17). Although structural studies have demonstrated that mutations in gyrA and gyrB conferring quinolone resistance play important roles in drug–protein interactions (18,19), the correlation between antibiotic resistance and particular mutation sites in Mycobacterium tuberculosis clinical isolates is not strong (20). This phenomenon suggests that known mutations that confer drug resistance may not be the only factor determining drug resistance; other as yet unknown mechanisms may also be important contributing factors.
The regulation of DNA gyrase is poorly understood; only a few regulatory proteins such as YacG, ParE (the plasmid RK2 toxin protein) and CcdB from Escherichia coli have been identified, all of which, similar to the quinolones, are inhibitors of DNA gyrase activity (21–23). YacG blocks the formation of the gyrase-DNA complex and abolishes its catalytic activity (23), whereas ParE and CcdB target the GyrA subunit of DNA gyrase and stall the gyrase-DNA cleavage complex (21,22). Mycobacterium fluoroquinolone resistance protein A (MfpA), a pentapeptide repeat protein (PRP) family member, has been shown to inhibit DNA gyrase and to be involved in resistance to fluoroquinolones (24). MfpA is chromosomally encoded and was the first PRP that confers endogenous resistance to fluoroquinolones to be crystallized (25). A previous study showed that an mfpA mutation in M. smegmatis caused sensitivity to fluoroquinolones, whereas overexpression of a DNA fragment containing mfpA conferred fluoroquinolone resistance (24). In contrast to this in vivo study, another group has reported that MfpA is unable to protect DNA gyrase from drug-induced damage in a cell-free system (26). In addition, a strain containing the complete coding sequence of mfpA conferred 2-fold less fluoroquinolone resistance than a strain containing mfpA and a 1-kb upstream sequence including the complete open reading frame of its flanking gene Msmeg_1640 (24). As this evidence suggests that Msmeg_1640 cooperates with MfpA to confer resistance to fluoroquinolones, we have named this protein Mycobacterium fluoroquinolone resistance protein B (MfpB) and investigated the mechanism by which MfpB affects MfpA-mediated fluoroquinolone resistance.
Here, we examine the role of MfpB in the regulation of DNA gyrase and its relationship with fluoroquinolone resistance. Using genetic and biochemical studies, we show that MfpB is a GTPase, and that its interaction with MfpA plays an essential role in protecting DNA gyrase from drug interference. Our results shed light on the underlying mechanism of fluoroquinolone resistance and may be useful in the development of new methods for the detection of drug resistance.
EXPERIMENTAL PROCUDEURES
Bacterial strains and culture conditions
In routine culture, liquid cultures of M. smegmatis strains were grown in Middlebrook 7H9 medium (Becton Dickinson) supplemented with 0.2% glycerol (Beijing Modern Eastern Finechemical Co. Ltd., Beijing), 0.05% Tween 80 (Sigma) and 10% albumin, dextrose and saline. As indicated, Sauton minimal medium was used to culture M. smegmatis strains. Single bacterial colonies were cultured on Middlebrook 7H10 medium (Becton Dickinson) containing 10% albumin, dextrose and saline and 0.2% glycerol. Hygromycin (50–75 mg/l for M. smegmatis, 150 mg/l for E. coli; Roche) and kanamycin (25 mg/l for M. smegmatis, 50 mg/l for E. coli; Amresco) were added to the medium as needed. All bacterial strains used in this study are listed in Supplementary Table S1.
Generation of knockout mutant strains, complementation and overexpression strains
Both mfpB and mfpA deletion mutants were created by allelic exchange using a specialized transducing phage delivery system as previously described (27). The 5′-flanking region of mfpB was amplified by polymerase chain reaction (PCR) with the 1640LL/1640LR primer pair, and the 3′-flanking region of mfpB was amplified with the 1640RL/1640RR primer pair. The flanking region of mfpA was generated by amplifying the upstream and downstream regions of mfpA using the 1641LL/1641LR and 1641RL/1641RR primer pairs, respectively (primer sequences are listed in Supplementary Table S2). The resulting cloned arms were ligated with plasmid p0004s (Hsu and Jacobs, unpublished data), digested with Van9I (mfpB) or DraIII (mfpA), and the allelic-exchange plasmids were digested with PacI, and then ligated to PacI-digested phA159 (Hsu and Jacobs, unpublished data). Phage packaging was performed using a MaxPlax packaging extract (Epicenter Biotechnologies, USA), and shuttle plasmids were amplified in the E. coli HB101 strain. The appropriate plasmids were electroporated into M. smegmatis for phage propagation, and target genes were replaced by an allelic exchange marker before selection with hygromycin. Transformants were confirmed by PCR using the primer pairs 1641Inl/1641InR and 1640Inl/1640InR for mfpA and mfpB, respectively. These primers are listed in Supplementary Table S2, and their positions are shown in Figure 1A. Complementation strains were constructed as described previously. Briefly, the full-length sequences of mfpA or mfpB amplified from M. smegmatis genomic DNA were cloned into the integrating vector pMV361 (28) using the primers listed in Supplementary Table S2, and the constructed plasmids were electroporated into knockout strains, yielding C-ΔmfpA and C-ΔmfpB, respectively. To augment MfpB, MfpA and MfpA-MfpB expression, the corresponding fragments were subcloned into pMV261 (28), a non-integrating variant of pMV361 to yield pMV261-mfpB, pMV261-mfpA and pMV261-mfpB/mfpA, respectively.
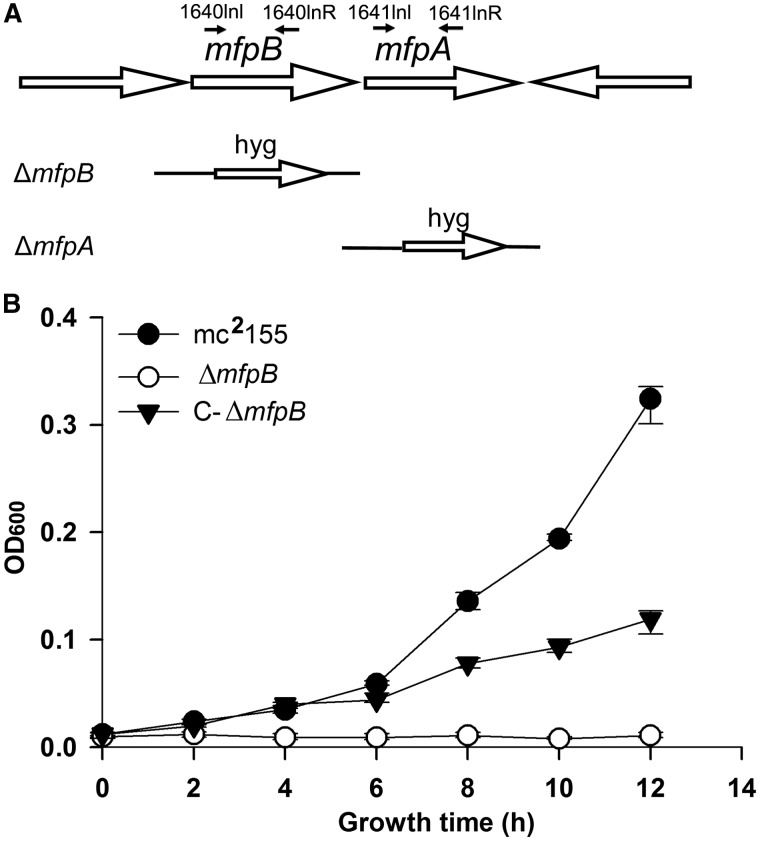
Figure 1.
MfpB is required for fluoroquinolone resistance. (A) Genomic arrangement of the mfpB and mfpA gene loci. Genes are shown as large arrows in their native orientation. Small arrows represent forward and reverse primers used to confirm the knockout mfpB (ΔmfpB) and mfpA (ΔmfpA) strains, respectively. The replacement gene, hygromycin (hyg), is also indicated. (B) Growth rates of mc2155, ΔmfpB and the complementary strain C-ΔmfpB in 7H9 medium with 0.15 mg/l of CIP. Data shown are the means ± standard deviations (SD) from four independent experiments.
Construction of MfpB mutants
Substitutions and deletions of amino acids that are conserved in small GTPases were introduced into the mfpB coding region using a Quick-Change site-directed mutagenesis kit (Stratagene, USA) with suitable vectors (Supplementary Table S2). The pMV261-mfpA/mfpB and pET23-mfpB plasmids were used for overexpression of the mutant proteins in M. smegmatis and protein purification, respectively. pMV261-mfpB/mfpA plasmids for overexpressing mutant proteins were transformed into M. smegmatis, and vectors for expressing mutant proteins to be purified were transformed into E. coli BL21(DE3). The mutation sites are shown later in the text (altered amino acids are highlighted in underlined):
M1: TNVSEG → TNEIEG
M2: PGQRR → PGLRR
M3: RNSAKAA → RNSANAA
M4: GKTTF → GKNNF
M5: GGFGAGKTTFVGA → GGFGAAAAAFVGA
M6: FIVAVNEFDGAPKH → FIVAVAAFAGAPKH
M7: GGFGAGKTTFVGA → GGFF_VGA (deletion of GAGKTT)
Antibiotic susceptibility testing
Normal growth rates of M. smegmatis strains in 7H9 and Sauton media were determined in 20 ml medium in 100 ml flasks by measuring OD600 values at different time points. The susceptibility of the M. smegmatis strains to drugs (ciprofloxacin, CIP; moxifloxacin, MOX; novobiocin, NOV; and trimethoprim, TMP) was determined on microplates. Aliquots (100 µl) of 2-fold serial dilutions of each antibiotic were inoculated with 100 μl of a 105 cells/ml suspension of M. smegmatis on microtitre plates. Plates were incubated at 37°C for 3 days. The OD600 value of cultures was measured using a microplate reader (FLUOstar OPTIMA, BMG LABTECH). All antibiotic susceptibility tests were replicated at least three times. The minimum inhibitory concentration (MIC) was defined as the lowest concentration of drug that inhibited the visible bacterial growth of M. smegmatis after a 2-day incubation (OD600 < 0.05).
Mycobacterial protein fragment complementation and yeast two hybrid assays
The association between MfpB and MfpA in vivo was assessed using mycobacterial protein fragment complementation (M-PFC) (29). Briefly, the coding regions of mfpB and mfpA were amplified separately and cloned into pUAB300 and pUAB400 to construct pUAB300-mfpB and pUAB400-mfpA, respectively. The plasmid pair, pUAB100/pUAB200 was used as a positive control, and the pUAB300/pUAB300 pair was used as a negative control. Plasmid pairs pUAB300/pUAB400-mfpA, pUAB400/pUAB300-mfpB and pUAB300-mfpB/pUAB400-mfpA were respectively co-transformed into M. smegmatis by electroporation. All transformants were selected on TMP to determine the association between MfpB and MfpA. Full-length mfpA and mfpB were PCR amplified using the primers listed in Supplementary Table S2 and cloned into the pGBKT7 and pGADT7 vectors, respectively (Clontech, USA), for yeast two-hybrid (Y2H) assays, which were carried out according to the manufacturer’s instructions.
Cloning, expression and purification of MfpB, MfpA, GyrA and GyrB
The coding regions of mfpB and mfpA were amplified from M. smegmatis genomic DNA and cloned into the expression vector pET23b (+) (Novagen, USA) in-frame fused with a C-terminal His-tag sequence, to give plasmids pET23b-mfpB and pET23b-mfpA, respectively. Similarly, the coding regions of gyrA and gyrB were amplified from M. smegmatis genomic DNA and cloned into the expression vector pACYCDuet1 (Novagen, USA) in-frame fused with an N-terminal His-tag sequence, to give plasmids pACYC-gyrA and pACYC-gyrB, respectively. The final constructs were transformed into BL21 (DE3) for expression and recombinant target proteins were induced by addition of 1-mM isopropyl-β-D-thiogalactopyranoside followed by incubation at 28°C for 8 h. Cells were harvested by centrifugation at 10 000 rpm for 5 min, resuspended in lysis buffer [20 mM of Tris–HCl (pH 8.0), 1 M NaCl, 10% glycerol, 10 mM of imidazole, 0.1% Triton X-100, 1 mM of phenylmethylsulfonyl fluoride (PMSF), 1 mg/ml of lysozyme] and lysed by sonication. Lysates were centrifuged (13 800 rpm, 4°C, 30 min) to remove debris before purification. Supernatants were incubated with Ni-NTA agarose (Qiagen, USA) with rotation (50 rpm) for 3 h at 4°C. Beads were washed three times with washing buffer [20 mM of Tris–HCl (pH 8.0), 0.5 M of NaCl, 10% glycerol, 50 mM of imidazole, 0.1% Triton X-100, 1 mM of PMSF, 25 mM of MgCl2]. Proteins were eluted with elution buffer [50 mM of Tris–HCl (pH 7.6), 0.5 M of NaCl, 25 mM of MgCl2, 10% glycerol, 500 mM of imidazole]. Protein samples for surface plasmon resonance (SPR) analysis were concentrated to ∼10 mg/ml before further purification by gel filtration (Superdex-75 10/30, GE Healthcare). The buffer for gel filtration contained 25 mM of Tris–HCl (pH 8.0) and 150 mM of NaCl. Fractions were collected from the peak. Proteins were stored in a buffer containing 50 mM of Tris–HCl (pH 8.0), 0.5 mM of ethylenediaminetetraacetic acid, 2 mM of dithiothreitol, 25 mM of MgCl2, 100 mM of KCl and 30% glycerol. Protein concentrations were measured using bicinchoninic acid protein assay reagent and a bovine serum albumin standard. Purified proteins were examined using 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis to verify molecular weight and purity.
Isothermal Titration Calorimetry assays
Guanosine diphosphate (GDP), Adenosine diphosphate (ADP), Guanosine 5′-O-thiotriphosphate (GTP-γ-S) and Adenosine 5′-O-thiotriphosphate (ATP-γ-S) (Sigma, USA) were dissolved in isothermal titration calorimetry (ITC) buffer [50 mM of Tris–HCl (pH 7.5), 100 mM of KCl, 1 mM of MgCl2, 1 mM of dithiothreitol (DTT)] to a final concentration of 1 mM. All proteins were dialyzed extensively against ITC buffer at 4°C for 2 days. Protein solutions (∼1.5 ml) were loaded into a Nano ITC-III ITC (Calorimetry Sciences Corporation, USA) cell. The titration syringe (250 µl volume) was filled with GDP, ADP, GTP-γ-S or ATP-γ-S solution. Titrations were carried out using 25 injections of 10 µl each, injected at 3 min intervals with stirring at 150 rpm at 25°C. Data was analysed with NanoAnalyze (Calorimetry Science Corporation).
SPR measurements
SPR was performed on a BIAcore-3000 instrument (Biacore) to assess the interaction between MfpB and MfpA. Purified MfpA-His6 was coated directly on CM5 biosensor chips to achieve ∼1600 response units, then purified GTP-γ-S or GDP-bound MfpB was injected into the chamber in phosphate buffered saline containing 0.05% Tween 20 buffer at 25°C at a flow rate of 30 µl/min. Association and dissociation rates were calculated using a simple 1:1 Langmuir binding model. All experiments were performed in triplicate.
GTPase and ATPase assays
Purified MfpB and its mutants (0–8 μM) were incubated with 15 μM [γ-32P] ATP or 15 μM [γ-32P] GTP (Perkin Elmer, USA) in reaction buffer [50 mM of Tris–HCl (pH 7.5), 100 mM of KCl, 1 mM of MgCl2, 1 mM of DTT, 1 g/l of Azolectin] for 0–360 min at 37°C. At the times indicated, 50 μl samples were removed for further investigation. The concentration of 32Pi released was measured by the charcoal method (30). Briefly, 50 μl of samples were added to 750 μl of 5% (w/v) charcoal (100–400 mesh, Sigma) in 50 mM of NaH2PO4 and vortexed. The charcoal was removed by centrifugation, and the amount of radioactivity present in the supernatant was determined by liquid scintillation counting.
DNA gyrase supercoiling assays
For E. coli gyrase, DNA supercoiling assays were performed using a supercoiling kit (NEB, USA) according to the manufacturer’s instructions. Briefly, 0.4 μg of relaxed plasmid pUC19 and 1 U of E. coli gyrase were mixed in a 30 μl of reaction mixture containing 1 μM GTP unless otherwise indicated. Tests were carried out with various concentrations of purified MfpA with/without MfpB at different concentrations of CIP, GTP, GDP and GTP-γ-S, as indicated. Reactions were incubated at 37°C for 90 min and terminated by addition of sodium dodecyl sulphate to 0.2%. Protease K (0.2%) was then added and incubated for 30 min at 37°C. Reaction mixtures were loaded on a 1% agarose gel for electrophoresis (6 h, 35 V). The gel was stained with ethidium bromide, and the fluorescence of bands was quantified with an Alpha Innotech digital camera and Gelpro software (Media Cybernetics, USA). DNA supercoiling assays with M. smegmatis gyrase were similar to those for E. coli gyrase, except that 0.02 μM M. smegmatis gyrase was used in each reaction, and the incubation time was 60 min. In all, 100 μM of CIP was used to inhibit M. smegmatis gyrase activity as indicated in Figure 7C.
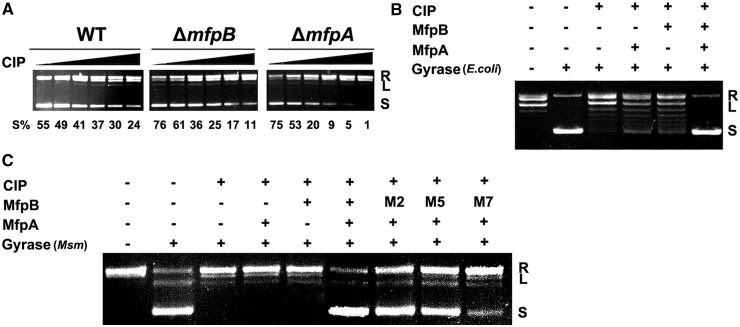
Figure 7.
Cooperative action of MfpB and MfpA protects DNA gyrase against CIP. (A) The effect of CIP on DNA gyrase in mc2155, ΔmfpA and ΔmfpB strains. Whole cell lysates (0.1 mg of total protein) of mc2155 (WT), ΔmfpA and ΔmfpB were incubated with 100 ng of relaxed pUC19 and different concentrations of CIP (0, 2.5, 5, 10, 25 and 50 ng/ml, from left to right) for 6 h. (B) MfpB helps MfpA protect E. coli DNA gyrase against CIP in vitro. Various combinations of 100 ng of relaxed pUC19, 0.5 units of DNA gyrase, 1 μM of MfpA, 2 μM of MfpB and 2 μg/ml of CIP were used in the reaction system. (C) MfpB, but not its mutants M2, M5 and M7, helps MfpA protect M. smegmatis (Msm) DNA gyrase against CIP in vitro. Various combinations of 100 ng of relaxed pUC19, 0.02 μM of Msm DNA gyrase, 0.1 μM of MfpA, 0.2 μM of MfpB or its mutants and 100 μM of CIP were used in the reaction system. Experiments were performed at least three times with consistent results.
RESULTS
MfpB is essential for MfpA-mediated fluoroquinolone-resistance in vivo
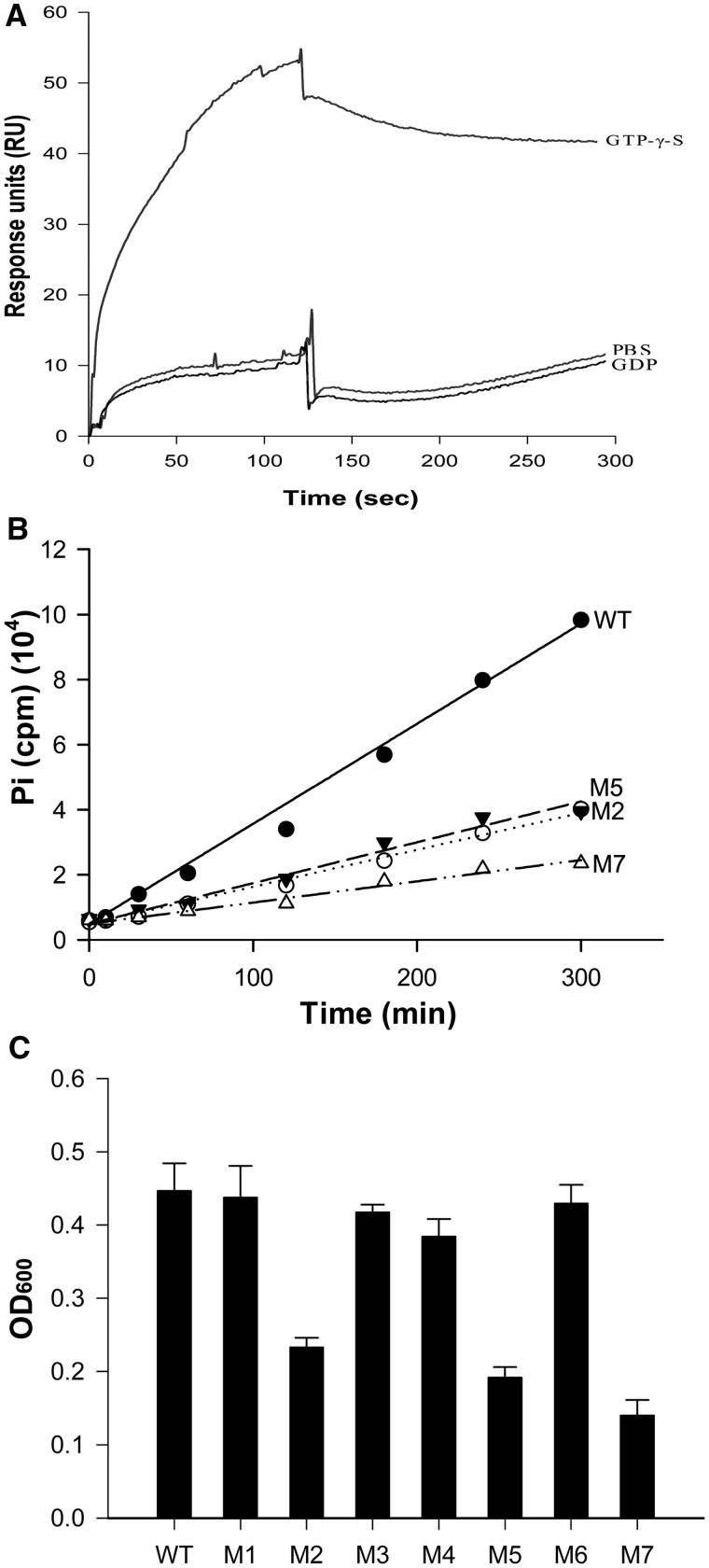
We reasoned that MfpB might be involved in the intrinsic resistance of M. smegmatis to fluoroquinolones. To test this hypothesis, an mfpB-deletion mutant ΔmfpB was constructed in M. smegmatis by specialized transduction (Figure 1A), and a corresponding complementation strain, which contained one copy of M. smegmatis-derived mfpB, was constructed in the mfpB-deficient strain and named C-ΔmfpB. We then examined the growth of the constructed strains in the presence of 0.15 μg/ml of CIP. At this concentration, growth was arrested in the ΔmfpB strain, whereas the wild-type strain grew normally under the same conditions. Moreover, the phenotype of arrested growth in the presence of CIP was partially reversed by complementation using wild-type mfpB from M. smegmatis (Figure 1B). In addition, the growth of ΔmfpB was similar to that of the wild-type in drug-free media (Supplementary Figure S1). Our data show that knockout of the mfpB gene does not influence M. smegmatis growth in either rich or minimal media. These results rule out the possibility that this deficiency in growth was owing to the knockout of MfpB, and indicate that it was owing to treatment with CIP, supporting our hypothesis that MfpB is involved in fluoroquinolone resistance.
An mfpA-deletion mutant strain, ΔmfpA, and its complementary M. smegmatis strain, C-ΔmfpA, were also constructed. As was the case for ΔmfpB, the growth of ΔmfpA was inhibited in the presence of CIP, and the growth defect was reversed in the complemented strain, C-ΔmfpA (Figure 2A). Moreover, as shown in Figure 2B, the MIC of CIP in ΔmfpA was 0.075 μg/ml, the same as that of ΔmfpB, and 4-fold lower than that of the parental mc2155 strain (0.3125 μg/ml), whereas drug susceptibility was recovered in the corresponding complemented strains (Figure 2B). A similar result was also observed when these strains were treated with MOX, but not with NOV, whose target is GyrB (Figure 2B). Together, these results suggest that MfpA and MfpB may be involved in the same drug resistance pathway.
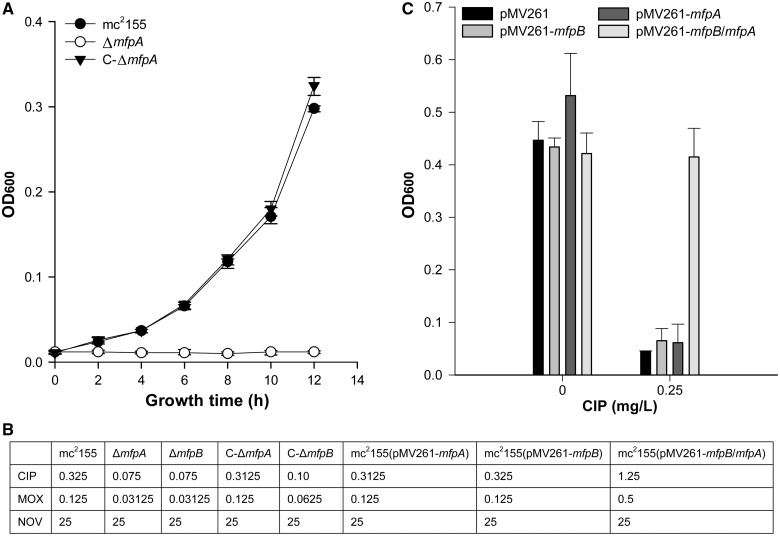
Figure 2.
MfpB is essential for MfpA-mediated fluoroquinolone-resistance in vivo. (A) Growth rates of mc2155, ΔmfpA and the complementary strain C-ΔmfpA in 7H9 medium with 0.15 mg/l of CIP. (B) MICs (mg/l) of CIP, MOX and NOV in different strains. (C) Increased bacterial CIP resistance was only observed when both MfpA and MfpB were overexpressed in the strain, and overexpression of MfpA or MfpB alone did not result in a loss of susceptibility. The resistance levels of mfpB, mfpA and mfpB-mfpA overexpression strains to CIP are indicated as growth in the presence of 0 and 0.25 mg/l of CIP.
To test for possible synergistic effects between MfpA and MfpB, the protein coding sequence of mfpA alone, mfpB alone and mfpA together with mfpB were separately amplified from M. smegmatis and cloned respectively into the pMV261 vector, a multi-copy plasmid with a heat shock protein promoter (Supplementary Table S1). Unexpectedly, in contrast to previous reports, no significant difference was observed in the MIC of CIP against the pMV261-mfpA and pMV261-mfpB strains and that of the wild-type strain containing an empty pMV261 (Figure 2C) (24,25). When mfpA and mfpB were co-expressed, however, there was a 3.8-fold increase in the MIC of CIP compared with its parental strain (Figure 2C). Consistent with this, the M. smegmatis strain containing pMV261-mfpA-MfpB also showed a 4-fold loss in susceptibility to MOX (Figure 2B). Taken together, the aforementioned data suggest that deletion of mfpB enhances susceptibility to fluoroquinolones, and that MfpB may play a role alongside MfpA in innate susceptibility to quinolones.
MfpB interacts with MfpA
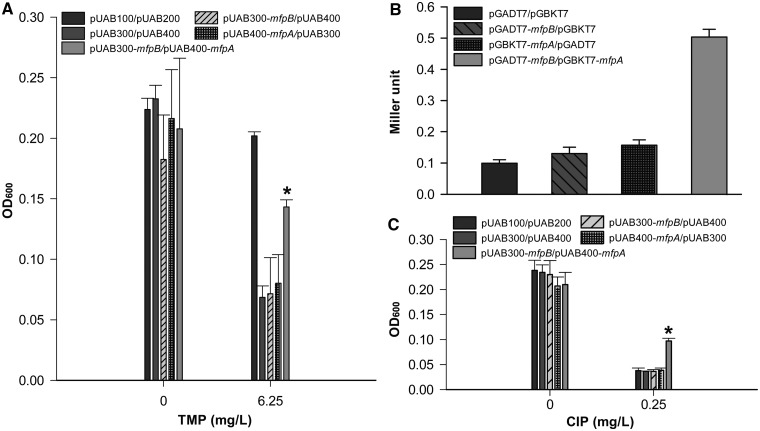
To further define how MfpB affects MfpA-mediated protection of DNA gyrase against fluoroquinolones, a M-PFC (29), which measures protein–protein associations by evaluating TMP susceptibility, was performed to determine whether MfpB interacts physically with MfpA in vivo. The coding regions of mfpA and mfpB were cloned into pUAB400 and pUAB300, respectively, to generate pUAB400-mfpA and pUAB300-mfpB, which were then co-transformed into M. smegmatis. The statistically significant difference in absorption between the resulting strain (mfpB/mfpA) and the negative controls (cells contain plasmid pairs pUAB300 and pUAB400-mfpA, and pUAB300-mfpB and pUAB400) indicates that strain mfpB/mfpA confers resistance to TMP in the presence of 6.25 mg/L TMP (Figure 3A). Furthermore, Y2H confirmed the direct interaction of MfpB and MfpA (Figure 3B). Drug (CIP) susceptibility in mfpA/mfpB, the strain co-expressing pUAB300-mfpB and pUAB400-mfpA, was statistically lower than the negative control strains mfpB/pUAB400 and mfpA/pUAB300 (Figure 3C). These data suggest that fluoroquinolone resistance requires the association of MfpA and MfpB.
Figure 3.
MfpB interacts with MfpA. (A) M-PFC analysis demonstrated that MfpB associates with MfpA. M. smegmatis cells were co-transformed with M-PFC plasmid pairs pUAB100 and pUAB200 as a positive control. Cells harboring plasmid pairs pUAB300 and pUAB400, pUAB300-mfpB and pUAB400, pUAB400-mfpA and pUAB300 were negative controls. The bacterial strain containing pUAB300-mfpB and pUAB400-mfpA was used to detect the interaction between MfpA and MfpB. Bacteria were cultured on 7H9 medium with 6.25 mg/l of TMP. Growth is indicative of protein–protein associations as measured by OD600. (B) Y2H analysis indicates that MfpB interacts with MfpA. Yeast cells were co-transformed with the plasmid pairs pGBKT7 and pGADT7, pGADT7-mfpB and pGBKT7, pGBKT7-mfpA and pGADT7, pGADT7-mfpB and pGBKT7-mfpA, respectively. Quantitative LacZ assays were used to quantitatively test the interaction of MfpB and MfpA. (C) The interaction of MfpB and MfpA confers resistance to CIP. Cultures were treated with 0 or 0.25 mg/l of CIP. Growth is indicative of loss of susceptibility to CIP as measured by OD600. Data shown are means ± standard deviations (SD) from four independent experiments (*P < 0.05).
MfpB is a small GTPase
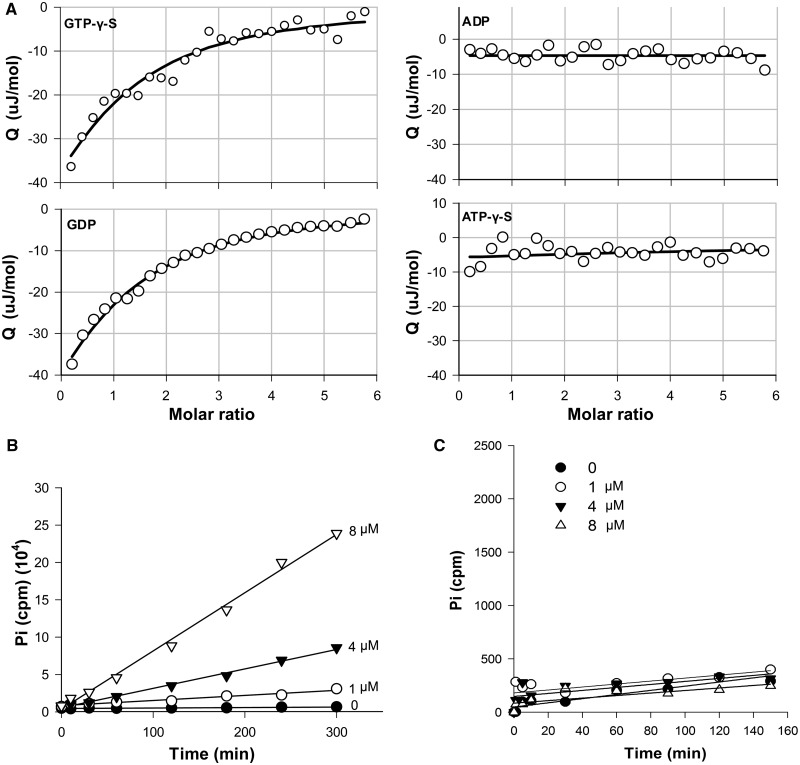
To our knowledge, there are no reports to date on the regulation of PRPs. In Mycobacteria, MfpB has been predicted to be an ATP/GTP-binding protein, but its precise function is unknown. To help define the possible mechanisms underlying MfpB’s involvement in MfpA-mediated fluoroquinolone resistance, we investigated its biochemical characteristics. Using MfpB that had been expressed and purified from E. coli, we first determined the binding capacity of MfpB for GTP-γ-S, ATP-γ-S, GDP and ADP using ITC. MfpB bound both GDP and GTP-γ-S, with Kds and stoichiometries of 0.0105 ± 0.0029 μM/1:1 (N = 0.98 ± 0.07), and 0.0118 ± 0.0032 μM/1:1 (N = 0.94 ± 0.09), respectively, but did not bind ADP or ATP-γ-S (Figure 4A), suggesting that MfpB is a GTP-binding and not an ATP-binding protein. 0, 1, 4 or 8 μM purified MfpB was then incubated with 15 μM [γ-32P] GTP to test its GTPase activity. It catalysed the hydrolysis of GTP, monitored by the release of Pi, in a dose-dependent manner (Figure 4B); the kobs of MfpB with GTP was 1.34 ± 0.26 s−1. Moreover, the ATPase activity of MfpB was also measured, and no hydrolysis of ATP was detected (Figure 4C). As MfpB is homologous to Rab small GTPases (31–33) (Figure S2), the aforementioned results indicate that MfpB is a small GTPase.
Figure 4.
MfpB is a small GTPase. (A) ITC shows that MfpB is a GTP/GDP-binding protein. In all, 10 μM of MfpB was titrated against 0.1 mM of GTP-γ-S, GDP, ADP or ATP-γ-S, stirring at 150 rpm (25°C). Data were analysed by NanoAnalyze software. (B) MfpB is a GTPase. In all, 0, 1, 4 and 8 μM of MfpB were incubated with 15 μM [γ-32P] GTP at 37°C, and 50 μl samples were collected at the times indicated. The concentration of 32Pi released was measured. Data points were fitted to a first-order reaction to obtain kobs. (C) MfpB is not an ATPase. ATPase activities were determined as in (B), except that [γ-32P] GTP was replaced with [γ-32P]ATP.
MfpB’s influence on the interaction of MfpA with DNA gyrase is via its GTPase activity in vivo and in vitro
As we have shown above, MfpB is a GTPase, and thus it is likely that its protection of gyrase from drug-related damage is owing to its GTPase activity. Typical small GTPases usually have three conformation states: inactive GDP-bound, inactive free nucleic acid binding and active GTP-bound (34–36). Here, we determined the binding affinity of GDP-MfpB and GTP-MfpB to MfpA using SPR. Purified MfpA-His6 protein was coated directly onto CM5 biosensor chips, and GTP-γ-S or GDP-bound purified MfpB-His6 was tested. GTP-γ-S bound MfpB, but not GDP-MfpB, interacted with MfpA (Figure 5A).
Figure 5.
The GTPase activity of MfpB is indispensable for protection from CIP interference. (A) GTP-bound MfpB interacts directly with MfpA. SPR was used to determine the binding affinity of GDP-MfpB and GTP-MfpB to MfpA. Purified MfpA-His6 was coated directly on CM5 biosensor chips, then 10 μM purified GTP-γ-S or GDP-bound or nucleotide-free MfpB was injected. (B) MfpB mutants with mutated conserved GTPase domain residues have correspondingly reduced GTPase activities. In all, 4 μM of purified proteins were incubated with 15 μM of [γ-32P]GTP at 37°C, and 50 μl of samples were collected at the times indicated. Hydrolysis of GTP was measured by 32Pi release as described in Figure 4B. (C) GTPase-deficient mutants had decreased growth rates in the presence of 0.25 µg/ml of CIP. Drug resistance was measured as described in Figure 1B.
To further explore the role of GTPase activity in fluoroquinolone resistance, we generated seven MfpB mutants (see Experimental Procedures) in which the conserved GTPase domain residues were mutated, to disrupt GTPase activity. The GTPase activity of three of these mutants, M2, M5 and M7 was reduced, their kobs values decreasing to 0.41 ± 0.11 s−1, 0.46 ± 0.10 s−1 and 0.24 ± 0.05 s−1, respectively, compared with that of the wild-type (1.34 ± 0.26 s−1) (Figure 5B). The GTPase-deficient mutants M2, M5 and M7 also showed decreased growth rates in the presence of CIP (Figure 5C).
MfpB protects the supercoiling activity of DNA gyrase from inhibition by MfpA in vitro
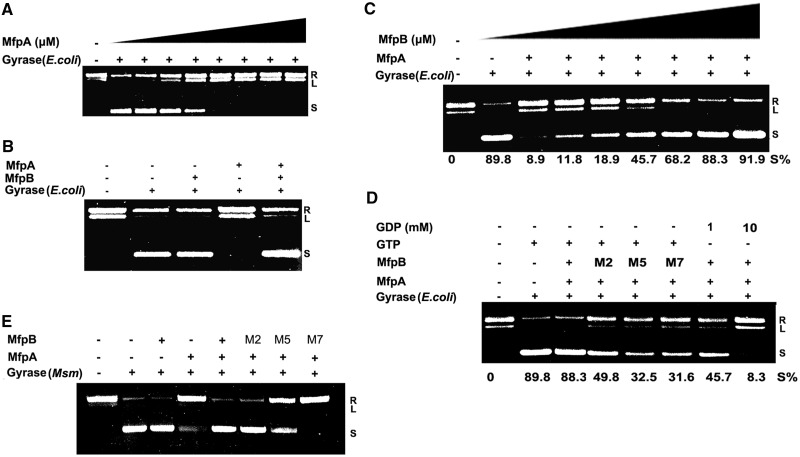
MfpA is required for mycobacterial endogenous fluoroquinolone resistance and has been shown to mimic the structure of DNA and to bind DNA gyrase (25). However, as discussed earlier in the text, MfpA cannot protect gyrase from fluoroquinolone inhibition in vitro. Our data indicate that MfpB interacts directly with MfpA, and that MfpB is sufficient and necessary for CIP resistance in vivo. We hypothesised that MfpB interferes with MfpA’s interaction with DNA gyrase. As MfpA has been shown to affect the function of E. coli gyrase in the same manner as it affects Mycobacterial gyrase (25,26), we used E. coli DNA gyrase for in vitro experiments. We first evaluated the effect of MfpA on the DNA gyrase catalytic reaction. Consistent with previous reports, MfpA inhibited the supercoiling activity of DNA gyrase in a concentration-dependent manner with 2-μM MfpA abrogating 1 unit of gyrase supercoiling activity (Figure 6A). As shown in Figure 6B, MfpB blocked MfpA-mediated inhibition and protected gyrase supercoiling activity, whereas MfpB alone had no effect on the function of DNA gyrase. The effect of MfpB was concentration dependent; the percentage of supercoiled fragments increased from 11.8 to 91.9%, as the concentration of MfpB increased from 0.01 to 5.0 μM (Figure 6C). The GTPase-deficient MfpB mutants M2, M5 and M7 did not efficiently protect E. coli gyrase from inhibition by MfpA; the percentage of supercoiling dropped to 49.8, 32.5 and 31.6%, respectively, compared with the negative control (gyrase only: 89.8%) and the positive control with both MfpB and MfpA in the reaction mixture (88.3%). Moreover, when wild-type MfpB was used, the percentage of supercoiling was 45.7% at a GDP concentration of 1.0 mM and 8.3% at a GDP concentration of 10 mM, indicating that high concentrations of GDP, which can inhibit GTPase activity, perturb MfpB’s action on the MfpA/gyrase complex (Figure 6D). Additionally, intact MfpB, but not mutants M2, M5 and M7, could also protect M. smegmatis gyrase supercoiling activity from MfpA-mediated inhibition (Figure 6E). These results suggest that MfpB can protect gyrase from MfpA-mediated inhibition.
Figure 6.
MfpB protects the supercoiling activity of DNA gyrase from inhibition by MfpA. (A) MfpA inhibits gyrase supercoiling activity in a concentration-dependent manner. One unit of E. coli DNA gyrase and 100 ng of relaxed pUC19 were used in the reactions; MfpA concentrations from 0 to 8 μM were added. (B) MfpB prevents inhibition of E. coli gyrase supercoiling activity by MfpA, but MfpB alone had no effect on the function of DNA gyrase. One unit of E. coli DNA gyrase and 100 ng of relaxed pUC19 were used for the reactions; lane 1, relaxed pUC19; lane 2, DNA gyrase and 0.02 mM of bovine serum albumin; lane 3, DNA gyrase and 2 μM of MfpA; lane 4, DNA gyrase and 2 μM of MfpB; lane 5, DNA gyrase, 2 μM of MfpA and 2 μM of MfpB. (C) The effect of MfpB on protecting E. coli DNA gyrase from MfpA inhibition is concentration-dependent. One unit of DNA gyrase, 100 ng of relaxed pUC19 and 2 μM of MfpA were used in the reaction. Increasing concentrations of MfpB, from 0 to 5.0 μM, were used. (D) MfpB mutants with reduced GTPase activity are unable to protect gyrase from MfpA inhibition. In all, 0.5 units of E. coli DNA gyrase, 1 μM of MfpA, 1 mM of GTP and 2 μM of MfpB or the corresponding mutant protein were used in the assays. A total of 1 (lane 7) or 10 mM (lane 8) GDP were used in the assays to replace GTP in the indicated reactions. (E) MfpB, but not its M2, M5 and M7 mutants, prevents inhibition of M. smegmatis (Msm) gyrase supercoiling activity by MfpA.
Cooperative action of MfpB and MfpA protects DNA gyrase from fluoroquinolones
To confirm the cooperative action of MfpA and MfpB in the protection of DNA gyrase, the effect of fluoroquinolones on DNA gyrase was examined in both the ΔmfpA and ΔmfpB strains. As shown in Figure 7A, in the absence of MfpB, the percentage of supercoiling plasmids decreased to 11% at a CIP concentration of 5 ng/ml, compared with 24% in the wild-type. Strikingly, only 1% of plasmids were supercoiled at the maximum concentration of CIP tested in the absence of MfpA (Figure 7A). Levels of gyrase mRNA in ΔmfpA and ΔmfpB were similar to that in the wild-type strain (Supplementary Figure S3), indicating that mfpA and mfpB do not affect gyrase expression. Moreover, knockout of mfpB did not significantly change mfpA expression (Supplementary Figure S3). Therefore, these results indicate that both MfpB and MfpA are necessary for protecting DNA gyrase against fluoroquinolones in M. smegmatis.
We also investigated how MfpB helps MfpA protect DNA gyrase against fluoroquinolones in vitro. As shown in Figure 7B, the supercoiling activity of E. coli DNA gyrase was disrupted at a CIP concentration of 2.0 μg/ml, and no protective role was detected with either 1.0 μM of MfpA or 2.0 μM of MpfB alone. In contrast, when the same amounts of MpfA and MpfB were mixed together, no inhibitory activity of CIP on DNA gyrase was detected, suggesting that MfpB helps MfpA protect gyrase against fluoroquinolones. Furthermore, wild-type MfpB, but not M2, M5 and M7 mutants, and MfpA together can also protect M. smegmatis gyrase from CIP damage (Figure 7C). Taken together, these results show that MfpB interferes with the interaction of MfpA and gyrase via its GTPase activity both in vitro and in vivo.
DISCUSSION
We have identified a new mycobacterial small GTPase, MfpB, that contributes to resistance against quinolone antibiotics via its involvement with MfpA in the protection of DNA gyrase. This is the first time that a small GTPase has been shown to be involved in the regulation of DNA gyrase and its protection from drug inhibition (37).
MfpA, a recently characterized PRP protein from M. smegmatis that mimics DNA structure and interacts with DNA gyrase, is involved in protecting DNA gyrase from fluoroquinolones, thus conferring resistance (24). However, conflicting evidence concerning the ability of MfpA to protect DNA gyrase against drugs in vivo and in vitro suggests that other factors may be involved in MfpA-related endogenous resistance against fluoroquinolones (24,26). Here, when mfpA and its upstream flanking gene, mfpB, were overexpressed together in M. smegmatis, drug susceptibility decreased (Figure 2), whereas M. smegmatis wild-type strain mc2155 expressing either MfpB (pMV261-mfpB) or MfpA alone (pMV261-mfpA) showed no loss of drug susceptibility. We thus reasoned that MfpB is a factor that participates in MfpA’s protection of DNA gyrase from fluoroquinolone damage.
In contrast to MfpA, Qnrs, which are MfpA paralogs in E. coli (e.g. QnrA1 and QnrB4), have the ability to confer E. coli fluoroquinolone resistance alone and to protect gyrase from drug-related damage (26,38). Alignment of MfpA and E. coli Qnrs reveals major differences in sequence that have previously been shown to be involved in the protection of gyrase against quinolones; deletions in the N- and C-terminal regions of MfpA (Supplementary Figure S4) and the amino acid residue 108 (C in Qnr and R in MfpA) (39). Structure-based alignments show that the intrachain extensions of MfpA (IE1 and IE2) are incomplete compared with the Qnrs of Gram-negative bacteria, possibly affecting its function (39); IE1 and IE2 of the Qnr from Aeromonas hydrophila interact with DNA gyrase and participate in protecting DNA gyrase against inhibition by fluoroquinolones (40). Owing to the absence of these key sequences, the capacity of MfpA to reduce drug toxicity in vivo may be limited. X-ray crystal structures of the MfpA/MpfB complex and MfpA/MfpB-DNA-gyrase would help to provide greater understanding of its mechanism.
To our knowledge, there are no previous reports of the involvement of GTPases in the protection of DNA gyrase against antibiotics. Although MfpB has been predicted to be an ATP/GTP-binding protein based on its sequence (41,42), its precise biological function in Mycobacterial species has not yet been identified. To understand the mechanism by which MfpB is involved in MfpA-mediated fluoroquinolone resistance, we investigated its biochemical characteristics in greater depth. Our data show that MfpB is a GTPase (Figure 4). Small GTPases, like Ras, are well-known as molecular switches that cycle between an inactive GDP- and an active GTP-bound state (34). SPR analysis showed that GTP-MfpB, but not GDP-MfpB, interacts with MfpA, suggesting that GTPase activity is necessary for the interaction of MfpB and MfpA (Figures 5 and 7). Furthermore, when mutations in the GTPase conserved domains of MfpB were introduced to reduce its GTPase activity, the mutated proteins had a decreased capacity to protect gyrase from drug-related damage both in vitro and in vivo (Figures 5 and 7). This suggests that GTPase activity is necessary for MfpB’s function in fluoroquinolone resistance. The nature of the changes that take place in MfpB’s conformation on GTP binding, and how MfpB binds and dissociates from MfpA require further investigation.
Although the physiological role of MfpA/MfpB is still unknown, a recent report has suggested that the PRP protein QnrA3, an MfpA homolog, does confer a selective advantage (43). Fluoroquinolone is reported to give rise to high levels of reactive oxygen species (ROS), and fluoroquinolone-induced bacterial death resulting from DNA disruption is tightly correlated with the production of ROS (44–46). Recently, Ras GTPases have been connected with redox regulation (47,48). It will be interesting to explore the correlation between MfpA/B and ROS and to clarify the biological functions of MfpA/MfpB.
Small GTPases are considered to play a critical role in the physiology and behavior of bacteria (42). For example, the Ras-like small G-protein MglA polarizes Myxococcus xanthus cells by accumulating at the leading cell pole in its active GTP-bound form (49,50), and the GTPase FlhF is necessary for one or more of the steps in flagellar organelle development in polarly flagellated bacteria (51). In addition, some bacterial small GTPases such as elongation factors G and Tu are able to elicit their functions by interacting with RNA and/or ribosomes (52). Considering these reports and its involvement in fluoroquinolone resistance, MfpB may be a critical molecular switch and is regarded as a good candidate for drug screening.
In this study, we have shown that MfpB, a small GTPase, works together with MfpA to protect DNA gyrase from fluoroquinolones. GTP-bound MfpB is active and interacts directly with the active state of MfpA. Intact GTPase activity is necessary to enable the protective role of MfpB. As GTPases are considered to play a critical role in the physiology of Mycobacteria, it will be interesting to see whether MfpB has potential as a candidate for drug screening.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–4 and Supplementary Reference [53].
FUNDING
National Basic Research Program of China [2012CB518700 to K. M.]; National Natural Science Foundation of China [81271891 to J. T., 31270178 and 31070118 to K. M.]; Knowledge Innovation Program of the Chinese Academy of Sciences [KSCX2-EW-J-6 to K. M.]. Funding for open access charge: National Natural Science Foundation of China [81271891 to J. T.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Dr George F. Gao for technical help with protein purification, Dr Hsu and Dr Jacobs for providing p0004s and phA159 and Dr A. J. Steyn for providing the Mycobacterial protein fragment complementation system.
REFERENCES
- 1.Collin F, Karkare S, Maxwell A. Exploiting bacterial DNA gyrase as a drug target: current state and perspectives. Appl. Microbiol. Biotechnol. 2011;92:479–497. doi: 10.1007/s00253-011-3557-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nollmann M, Crisona NJ, Arimondo PB. Thirty years of Escherichia coli DNA gyrase: from in vivo function to single-molecule mechanism. Biochimie. 2007;89:490–499. doi: 10.1016/j.biochi.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 3.Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley DMJ, Cozzarelli NR. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]
- 4.Gellert M, Mizuuchi K, O'Dea MH, Itoh T, Tomizawa J. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl Acad. Sci. USA. 1977;74:4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gellert M, Mizuuchi K, O'Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl Acad. Sci. USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gellert M, O'Dea MH, Itoh T, Tomizawa J. Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc. Natl Acad. Sci. USA. 1976;73:4474–4478. doi: 10.1073/pnas.73.12.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu G, Wu J, Liu W, Zhu D, Hu Y, Deng J, Zhang XE, Bi L, Wang DC. Crystal structure of DNA gyrase B' domain sheds lights on the mechanism for T-segment navigation. Nucleic Acids Res. 2009;37:5908–5916. doi: 10.1093/nar/gkp586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corbett KD, Shultzaberger RK, Berger JM. The C-terminal domain of DNA gyrase A adopts a DNA-bending {beta}-pinwheel fold. Proc. Natl Acad. Sci. USA. 2004;101:7293–7298. doi: 10.1073/pnas.0401595101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morais Cabral JH, Jackson AP, Smith CV, Shikotra N, Maxwell A, Liddington RC. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 10.Wigley DB, Davies GJ, Dodson EJ, Maxwell A, Dodson G. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature. 1991;351:624–629. doi: 10.1038/351624a0. [DOI] [PubMed] [Google Scholar]
- 11.Bax BD, Chan PF, Eggleston DS, Fosberry A, Gentry DR, Gorrec F, Giordano I, Hann MM, Hennessy A, Hibbs M, et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature. 2010;466:935–940. doi: 10.1038/nature09197. [DOI] [PubMed] [Google Scholar]
- 12.Wohlkonig A, Chan PF, Fosberry AP, Homes P, Huang J, Kranz M, Leydon VR, Miles TJ, Pearson ND, Perera RL, et al. Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat. Struct. Mol. Biol. 2010;17:1152–1153. doi: 10.1038/nsmb.1892. [DOI] [PubMed] [Google Scholar]
- 13.Laponogov I, Sohi MK, Veselkov DA, Pan XS, Sawhney R, Thompson AW, McAuley KE, Fisher LM, Sanderson MR. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 2009;16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 14.Laponogov I, Pan XS, Veselkov DA, McAuley KE, Fisher LM, Sanderson MR. Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One. 2010;5:e11338. doi: 10.1371/journal.pone.0011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roca J, Wang JC. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell. 1994;77:609–616. doi: 10.1016/0092-8674(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 16.Wang JC. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 17.Drlica K, Hiasa H, Kerns R, Malik M, Mustaev A, Zhao X. Quinolones: action and resistance updated. Curr. Top. Med. Chem. 2009;9:981–998. doi: 10.2174/156802609789630947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laponogov I, Sohi MK, Veselkov DA, Pan X-S, Sawhney R, Thompson AW, McAuley KE, Fisher LM, Sanderson MR. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 2009;16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 19.Piton J, Petrella S, Delarue M, Andre-Leroux G, Jarlier V, Aubry A, Mayer C. Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase. PLoS One. 2010;5:e12245. doi: 10.1371/journal.pone.0012245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maruri F, Sterling TR, Kaiga AW, Blackman A, van der Heijden YF, Mayer C, Cambau E, Aubry A. A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. J. Antimicrob. Chemother. 2012;67:819–831. doi: 10.1093/jac/dkr566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y, Pogliano J, Helinski DR, Konieczny I. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 2002;44:971–979. doi: 10.1046/j.1365-2958.2002.02921.x. [DOI] [PubMed] [Google Scholar]
- 22.Loris R, Dao-Thi M-H, Bahassi EM, Van Melderen LV, Poortmans F, Liddington RC, Couturier M, Wyns L. Crystal structure of CcdB, a topoisomerase poison from E coli. J. Mol. Biol. 1999;285:1667–1677. doi: 10.1006/jmbi.1998.2395. [DOI] [PubMed] [Google Scholar]
- 23.Sengupta S, Nagaraja V. YacG from Escherichia coli is a specific endogenous inhibitor of DNA gyrase. Nucleic Acids Res. 2008;36:4310–4316. doi: 10.1093/nar/gkn355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montero C, Mateu G, Rodriguez R, Takiff H. Intrinsic resistance of Mycobacterium smegmatis to fluoroquinolones may be influenced by new pentapeptide protein MfpA. Antimicrob. Agents Chemother. 2001;45:3387–3392. doi: 10.1128/AAC.45.12.3387-3392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hegde SS, Vetting MW, Roderick SL, Mitchenall LA, Maxwell A, Takiff HE, Blanchard JS. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science. 2005;308:1480–1483. doi: 10.1126/science.1110699. [DOI] [PubMed] [Google Scholar]
- 26.Merens A, Matrat S, Aubry A, Lascols C, Jarlier V, Soussy CJ, Cavallo JD, Cambau E. The pentapeptide repeat proteins MfpAMt and QnrB4 exhibit opposite effects on DNA gyrase catalytic reactions and on the ternary gyrase-DNA-quinolone complex. J. Bacteriol. 2009;191:1587–1594. doi: 10.1128/JB.01205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bardarov S, Bardarov S, Jr, Pavelka MS, Jr, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs WR., Jr Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148:3007–3017. doi: 10.1099/00221287-148-10-3007. [DOI] [PubMed] [Google Scholar]
- 28.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF, et al. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 29.Singh A, Mai D, Kumar A, Steyn AJ. Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc. Natl Acad. Sci. USA. 2006;103:11346–11351. doi: 10.1073/pnas.0602817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leupold CM, Goody RS, Wittinghofer A. Stereochemistry of the elongation factor Tu X GTP complex. Eur. J. Biochem. 1983;135:237–241. doi: 10.1111/j.1432-1033.1983.tb07643.x. [DOI] [PubMed] [Google Scholar]
- 31.Pereira-Leal JB, Seabra MC. Evolution of the rab family of small GTP-binding proteins. J. Mol. Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- 32.Brown ED. Conserved P-loop GTPases of unknown function in bacteria: an emerging and vital ensemble in bacterial physiology. Biochem. Cell Biol. 2005;83:738–746. doi: 10.1139/o05-162. [DOI] [PubMed] [Google Scholar]
- 33.Caldon CE, March PE. Function of the universally conserved bacterial GTPases. Curr. Opin. Microbiol. 2003;6:135–139. doi: 10.1016/s1369-5274(03)00037-7. [DOI] [PubMed] [Google Scholar]
- 34.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 35.Brinkmann T, Daumke O, Herbrand U, Kuhlmann D, Stege P, Ahmadian MR, Wittinghofer A. Rap-specific GTPase activating protein follows an alternative mechanism. J. Biol. Chem. 2002;277:12525–12531. doi: 10.1074/jbc.M109176200. [DOI] [PubMed] [Google Scholar]
- 36.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 37.Verstraeten N, Fauvart M, Versées W, Michiels J. The Universally Conserved Prokaryotic GTPases. Microbiol. Mol. Biol. Rev. 2011;75:507–542. doi: 10.1128/MMBR.00009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Briales A, Rodriguez-Martinez JM, Velasco C, Diaz de Alba P, Dominguez-Herrera J, Pachon J, Pascual A. In vitro effect of qnrA1, qnrB1, and qnrS1 genes on fluoroquinolone activity against isogenic Escherichia coli isolates with mutations in gyrA and parC. Antimicrob. Agents Chemother. 2011;55:1266–1269. doi: 10.1128/AAC.00927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park KS, Lee JH, Jeong da U, Lee JJ, Wu X, Jeong BC, Kang CM, Lee SH. Determination of pentapeptide repeat units in Qnr proteins by the structure-based alignment approach. Antimicrob. Agents Chemother. 2011;55:4475–4478. doi: 10.1128/AAC.00041-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong X, Bromley EH, Oelschlaeger P, Woolfson DN, Spencer J. Structural insights into quinolone antibiotic resistance mediated by pentapeptide repeat proteins: conserved surface loops direct the activity of a Qnr protein from a gram-negative bacterium. Nucleic Acids Res. 2011;39:3917–3927. doi: 10.1093/nar/gkq1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koonin EV, Aravind L. Dynein light chains of the Roadblock/LC7 group belong to an ancient protein superfamily implicated in NTPase regulation. Curr. Biol. 2000;10:R774–R776. doi: 10.1016/s0960-9822(00)00774-0. [DOI] [PubMed] [Google Scholar]
- 42.Verstraeten N, Fauvart M, Versees W, Michiels J. The universally conserved prokaryotic GTPases. Microbiol. Mol. Biol. Rev. 2011;75:507–542. doi: 10.1128/MMBR.00009-11. second and third pages of table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michon A, Allou N, Chau F, Podglajen I, Fantin B, Cambau E. Plasmidic qnrA3 enhances Escherichia coli fitness in absence of antibiotic exposure. PLoS One. 2011;6:e24552. doi: 10.1371/journal.pone.0024552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goswami M, Mangoli SH, Jawali N. Involvement of reactive oxygen species in the action of ciprofloxacin against Escherichia coli. Antimicrob. Agents Chemother. 2006;50:949–954. doi: 10.1128/AAC.50.3.949-954.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez LJ, Sik RH, Chignell CF. Fluoroquinolone antimicrobials: singlet oxygen, superoxide and phototoxicity. Photochem. Photobiol. 1998;67:399–403. [PubMed] [Google Scholar]
- 46.Wang X, Zhao X, Malik M, Drlica K. Contribution of reactive oxygen species to pathways of quinolone-mediated bacterial cell death. J. Antimicrob. Chemother. 2010;65:520–524. doi: 10.1093/jac/dkp486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferro E, Goitre L, Retta SF, Trabalzini L. The Interplay between ROS and Ras GTPases: physiological and pathological implications. J. Signal Transduct. 2012;2012:365769. doi: 10.1155/2012/365769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heo J, Campbell SL. Mechanism of redox-mediated guanine nucleotide exchange on redox-active Rho GTPases. J. Biol. Chem. 2005;280:31003–31010. doi: 10.1074/jbc.M504768200. [DOI] [PubMed] [Google Scholar]
- 49.Leonardy S, Miertzschke M, Bulyha I, Sperling E, Wittinghofer A, Sogaard-Andersen L. Regulation of dynamic polarity switching in bacteria by a Ras-like G-protein and its cognate GAP. EMBO J. 2010;29:2276–2289. doi: 10.1038/emboj.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Franco M, Ducret A, Mignot T. A bacterial Ras-like small GTP-binding protein and its cognate GAP establish a dynamic spatial polarity axis to control directed motility. PLoS Biol. 2010;8:e1000430. doi: 10.1371/journal.pbio.1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balaban M, Joslin SN, Hendrixson DR. FlhF and its GTPase activity are required for distinct processes in flagellar gene regulation and biosynthesis in Campylobacter jejuni. J. Bacteriol. 2009;191:6602–6611. doi: 10.1128/JB.00884-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caldon CE, Yoong P, March PE. Evolution of a molecular switch: universal bacterial GTPases regulate ribosome function. Mol. Microbiol. 2001;41:289–297. doi: 10.1046/j.1365-2958.2001.02536.x. [DOI] [PubMed] [Google Scholar]
- 53.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotech. 1983;1:784–791. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.