Abstract
Deciphering the genetic code is a fundamental process in all living organisms. In many bacteria, AUA codons are deciphered by tRNAIle2 bearing lysidine (L) at the wobble position. L is a modified cytidine introduced post-transcriptionally by tRNAIle-lysidine synthetase (TilS). Some bacteria, including Mycoplasma mobile, do not carry the tilS gene, indicating that they have established a different system to decode AUA codons. In this study, tRNAIle2 has been isolated from M. mobile and was found to contain a UAU anticodon without any modification. Mycoplasma mobile isoleucyl-tRNA synthetase (IleRS) recognized the UAU anticodon, whereas Escherichia coli IleRS did not efficiently aminoacylate tRNAIle2UAU. In M. mobile IleRS, a single Arg residue at position 865 was critical for specificity for the UAU anticodon and, when the corresponding site (W905) in E. coli IleRS was substituted with Arg, the W905R mutant efficiently aminoacylated tRNA with UAU anticodon. Mycoplasma mobile tRNAIle2 cannot distinguish between AUA and AUG codon on E. coli ribosome. However, on M. mobile ribosome, M. mobile tRNAIle2UAU specifically recognized AUA codon, and not AUG codon, suggesting M. mobile ribosome has a property that prevents misreading of AUG codon. These findings provide an insight into the evolutionary reorganization of the AUA decoding system.
INTRODUCTION
Accurate translation of the genetic code is required to synthesize proteins correctly. Determination of the mechanisms by which tRNAs decode all sense codons on mRNAs is essential to our understanding of this basic principle in all living organisms. Base modifications at the first (wobble) anticodon position of tRNAs play critical roles in deciphering cognate codons (1,2). In sense codons, with the exception of AUR (R = A or G), two-codon sets ending in a purine (NNR) specify identical amino acids because modified uridines (U*) at the wobble position pair not only with A but also with G by classical wobble pairing (3). In AUR codons, however, isoleucine (Ile) and methionine (Met) are encoded by AUA and AUG, respectively. Hence, AUR codons must be deciphered separately by different tRNA species. The decoding system for the AUA codon relies strictly on the wobble modifications of tRNAs (1).
In eukaryotes, tRNAIle bearing pseudouridine (Ψ) or inosine (I) at the wobble position decodes the AUA codon by forming a Ψ-A or I-A pair (4). In almost all bacteria and some organelles, the AUA codon is deciphered by tRNAIle bearing lysidine (L or 2-lysylcytidine) at the wobble position (5–7). In most archaea, the AUA codon is deciphered by tRNAIle bearing 2-agmatinylcytidine (agm2C or agmatidine) at the wobble position (8,9). Because of the chemical similarity between L and agm2C, conjugation of lysine or agmatine to the C2 carbon of the cytosine induces a tautomeric conversion of the base from enamine to imine, enabling the base to pair with adenosine instead of guanosine at the third position of codons (8). In bacteria and archaea, the precursor form of tRNAIle bearing a CAU anticodon behaves as tRNAMet. In fact, this precursor is charged with Met and decodes the AUG codon (8,10,11). Thus, these single modifications confer both Ile-specificity and AUA decoding ability on tRNAsIle in bacteria and archaea. L and agm2C are modified cytidines that are post-transcriptionally introduced by the essential enzymes tRNAIle-lysidine synthetase (TilS) (12,13) and tRNAIle-agmatidine synthetase (TiaS) (8,14), respectively.
As more genomic sequences have been completed, bacteria lacking a tilS homologue and archaea lacking a tiaS homologue have been discovered (13,15). Detailed inspection of tDNAs in these organisms revealed the presence of a unique tDNAIle with a TAT anticodon in place of the CAT anticodon found in most bacteria and archaea that carry tilS or tiaS (13,16–19) (Supplementary Table S1). tDNAs with the TAT anticodon are also found in environmental metagenome sequences. To date, 371 species of unique tDNAIleTAT sequences have been identified in a metagenome database (19). Most of these presumably originated in bacteria, although the possibility of eukaryotic origins cannot be excluded. These observations suggest that a system for deciphering AUA codons by tRNAs without wobble-modified cytidines has been established in some bacteria and archaea. Investigation of the alternative decoding system will provide insights into the evolution and underlying mechanisms of the decoding system, particularly that of the AUA codon.
We used Mycoplasma mobile as a model organism to investigate the molecular details of the AUA decoding system in the absence of wobble modifications. Mycoplasma mobile is a flask-shaped piscine mycoplasma (∼1 × 0.3 μm); the ability of M. mobile to glide rapidly has been extensively studied (20). The single circular genome consists of 777 kb and contains 635 protein coding sequences and 28 tRNA genes (21). It was reported, in this organism, that natural mistranslation occurs because of having aminoacyl-tRNA synthetases lacking editing activity (22). Mycoplasma mobile carries tDNAIle with the TAT anticodon and has no homologue of tilS (15). Two main issues must be considered when investigating the AUA decoding system in this organism. First, the mechanism underlying the recognition of tRNAIle2 bearing the UAU anticodon by isoleucyl-tRNA synthetase (IleRS) must be determined. As Escherichia coli IleRS does not recognize the UAU anticodon efficiently (23), the substrate specificity of M. mobile IleRS must be different to allow recognition of the UAU anticodon and efficient isoleucylation. Second, the mechanism by which M. mobile tRNAIle2 distinguishes the AUA codon from the near-cognate AUG codon on the ribosome must be determined because the UAU anticodon can potentially recognize the AUG codon by forming a U·G pair, according to the classical wobble rule (3).
MATERIALS AND METHODS
Bacterial cell culture
Mycoplasma mobile 163 K was cultured in slightly modified Aluotto medium (pH 7.8), consisting of 2.1% heart infusion broth, 0.56% yeast extract, 10% horse serum and 50 μg/ml ampicillin at 25°C (24). Escherichia coli A19 strain was cultured in LB medium at 37°C.
tRNA isolation
Mycoplasma mobile cells (20 g) were washed once with phosphate-buffered saline, suspended in 50 ml of lysis buffer (50 mM of sodium acetate and 10 mM of magnesium acetate), mixed with 43 ml of water-saturated phenol and shaken for 3 h at room temperature. The solution was centrifuged, and chloroform was added to the aqueous phase to a concentration of 20%. After vigorous shaking, the mixture was centrifuged, and the RNAs were precipitated from the aqueous phase with 2-propanol. Escherichia coli total RNA was extracted using the same method. Mycoplasma mobile and E. coli tRNAIle2 were isolated from the respective total RNA preparations by reciprocal circulating chromatography (RCC) (25). Oligodeoxynucleotide probes (Supplementary Table S5), modified at the 5′-termini by addition of an ethylcarbamate amino linker (Sigma-Aldrich), were covalently immobilized on NHS-activated Sepharose 4 Fast Flow (GE healthcare) and packed in tip columns, which were then attached to an RCC device.
Mass spectrometric analysis of M. mobile tRNAIle2UAU
liquid chromatography–mass spectrometry (LC/MS) analysis of tRNA was performed essentially as described previously (26). The isolated M. mobile tRNAIle2 (1.6 pmol) was digested at 37°C for 30 min in a reaction mixture consisting of 20 mM ammonium acetate (pH 5.3) and 50 U of RNase T1 (Epicentre), or 10 mM ammonium acetate (pH 7.7) and 1 ng of RNase A (Ambion). After digestion, an equal volume of 0.1 M triethylamine acetate (pH 7.0) was added to the reaction solution. The digest (500–800 fmol) was analysed using a linear ion trap-orbitrap hybrid mass spectrometer (LTQ Orbitrap XL, Thermo Fisher Scientific) equipped with a custom-made nanospray ion source, and a splitless nano–high-performance liquid chromatography system (Dina, KYA Technologies) with a C18 capillary column. To identify pseudouridine (Ψ), 1.6 pmol of M. mobile tRNAIle2 was cyanoethylated by mixing with 15 μl of 41% ethanol/1.1 M of triethylamine acetate (pH 8.6) and 2 μl of acrylonitrile, followed by incubation at 70°C for 2 h (27). The mixture was then digested with RNase T1 and analysed by LC/MS as previously described.
Recombinant IleRS
Because the TGA codon encodes Trp, and not the stop signal, in M. mobile (21), the entire M. mobile ileS gene, in which all TGA codons were replaced with TGG codons, was synthesized in vitro using 30 DNA oligomers (92–110mers) with overlapping sequences (Supplementary Table S4). Codon usage was optimized for efficient expression in E. coli. The oligomers were ligated to the full-length recombinant M. mobile ileS gene by polymerase chain reaction in three steps using PrimeSTAR HS DNA Polymerase (TaKaRa). The polymerase chain reaction product was excised from the agarose gel, extracted and cloned into the NcoI and BamHI sites of the pET28a expression vector (Novagen).
The mutations W905R, W905K and W905L were introduced into the E. coli IleRS expression plasmid (kindly provided by Dr Y. Shimizu) (28) using the PrimeSTAR Mutagenesis Basal Kit (TaKaRa) and a set of primers shown in Supplementary Table S5.
The hexahistidine-tagged M. mobile or E. coli IleRS were expressed in E. coli Rosetta (DE3) at 30°C or 37°C, respectively, in the presence of 0.1 mM of IPTG(Isopropyl β-D-1-thiogalactopyranoside). Wild-type and mutant E. coli IleRS and wild-type M. mobile IleRS were purified using HisTrap HP (GE healthcare) or HisTALON Superflow Cartridges (Clontech). The pooled IleRS proteins were dialysed against buffer consisting of 50 mM HEPES–KOH (pH 7.5), 100 mM KCl and 1 mM dithiothreitol (DTT). Glycerol was added to a final concentration of 30%, and the IleRS solutions were flash-frozen in liquid nitrogen and stored at −80°C. Protein concentration was determined using a Bio-Rad protein assay kit (Bio-Rad) with bovine serum albumin as a standard.
Preparation of modified tRNA transcripts
For in vitro transcription, template DNAs for the M. mobile and E. coli tRNAIle2 genes bearing CAU, UAU and GAU anticodons without CCA termini were synthesized using the class III T7 promoter and synthetic DNA oligonucleotides shown in Supplementary Table S5. Each tRNA was transcribed at 37°C in a reaction mixture consisting of 40 mM Tris–HCl (pH 8.0), 24 mM MgCl2, 5 mM DTT, 2 mM spermidine, 0.01% Triton X-100, 2 mM each dNTP, 10 mM GMP, 10 μg/ml template DNA and T7 RNA polymerase (29). The CCA terminus was added to each transcribed tRNA by the E. coli CCA-adding enzyme as described (30). Each transcript was purified by electrophoresis on a 10% polyacrylamide gel containing 7 M urea and eluted in buffer consisting of 300 mM sodium acetate (pH 5.2), 0.1% sodium dodecyl sulphate and 1 mM ethylenediaminetetraacetic acid (pH 8.0). The t6A modification was introduced into each tRNA at 37°C for 1.5 h in a reaction mixture consisting of 100 mM HEPES–KOH (pH 7.5), 20 mM MgCl2, 300 mM KCl, 5 mM DTT, 10 mM adenosine triphosphate (ATP), 25 mM NaHCO3, 1 mM l-threonine, transcribed tRNA and 1.5 μM each YrdC, YgjD, YjeE and YeaZ (31). For tRNAs bearing the CAU anticodon, L modification was carried out at 37°C for 1.5 h in a reaction mixture containing 100 mM HEPES–KOH (8.6), 50 mM KCl, 2 mM DTT, 50 μM l-lysine, transcribed tRNA and 1.5 μM TilS (32). At each step of the reaction, tRNAs were extracted with Tripure Isolation Reagent (Roche), desalted with PD10 (GE Healthcare Life Sciences) and precipitated with ethanol. Finally, each tRNA was desalted by drop dialysis with MF-MEMBRANE FILTERS (Millipore, 0.025 μm VSWP membrane). The frequency of t6A and L introduced in each tRNA was estimated by LC/MS analysis as previously described.
Kinetic analysis of isoleucylation
Isoleucylation of each tRNA was carried out at 37°C in 30 μl of reaction mixture consisting of 100 mM HEPES–KOH (pH 7.5), 5 mM MgCl2, 10 mM KCl, 1 mM DTT, 2 mM ATP, 30 μM L-[U-14C]Ile (8.14–9.62 GBq/mmol) (Moravek Biochemicals), 0.125–8 μM tRNA and 21–100 nM recombinant IleRS. The initial rate of isoleucylation was measured at five or six different tRNA concentrations between 0.125 and 8 μM. At different time points, a 7 μl aliquot was spotted onto a Whatman 3MM filter disc. The discs were washed twice with 5% trichloroacetic acid and dried with ethanol. Radioactivity was then measured by liquid scintillation counting. The kinetic parameters Km and kcat were calculated based on non-linear least square fitting of the initial velocities at the different tRNA concentrations using GraphPad PRISM 5 (GraphPad Software).
Ribosome preparation
Mycoplasma mobile cells were ground in liquid nitrogen for 12 min using a mortar and pestle and then suspended in lysis buffer consisting of 20 mM HEPES–KOH (pH 7.5), 10 mM magnesium acetate, 30 mM ammonium chloride and 6 mM β-mercaptoethanol. The lysate was centrifuged at 7000 r.p.m. for 20 min at 4°C, and the supernatant was ultracentrifuged at 22 000 r.p.m. for 45 min at 4°C. Seventy units of DNase I were then added to the supernatant, followed by ultracentrifugation at 40 000 r.p.m. for 4 h. The ribosomal pellet was suspended in lysis buffer. The 70S ribosomes were obtained by sucrose density gradient centrifugation basically as described (33,34). The crude ribosomal fraction was layered on top of a 10–40% sucrose density gradient and separated by ultracentrifugation in a SW-28 Rotor (Beckman) at 20 000 r.p.m. for 14 h at 4°C. Fractions were collected using a Piston Gradient Fractionator (BIOCOMP). The 70S ribosomal fraction was ultracentrifuged at 45 000 r.p.m. for 41 h, the supernatant was removed and the 70S ribosomal pellet was suspended in lysis buffer.
A-site tRNA binding assay
The A-site tRNA binding assay was performed as described with slight modification (35,36). The mRNA containing the SD sequence and the AUA (or AUG) codon at the A-site was designed as follows; 5′-GGG UUA ACU UAA GUA AGG AGG UAU ACU GAG AUA(or AUG) UAA CUG CAG AAA AAA-3′ (A-site codons underlined). The template DNAs for the mRNAs were constructed using synthetic DNAs listed in Supplementary Table S5 and transcribed in vitro with T7 RNA polymerase. The mRNA containing the CGU codon at the A-site was used as a negative control. The 5′ terminus of the tRNA was dephosphorylated by bacterial alkaline phosphatase (TAKARA BIO) and phosphorylated with [γ-32P] ATP by T4 polynucleotide kinase (Toyobo). First, the ribosomal P-site was occupied with tRNAGlu in a 10 μl of mixture consisting of 3 pmol of 70S ribosome, 16 pmol tRNAGlu, 1 μg mRNA, 50 mM Tris–HCl (pH 7.5), 6.5 mM MgCl2, 60 mM KCl, 1 mM DTT and 0.5 mM spermine and incubated at 37°C for 20 min. Then, 1.6 pmol of the 5′-32P-labelled tRNA in 10 μl of 50 mM Tris–HCl (pH 7.5), 6.5 mM MgCl2, 60 mM KCl, 1 mM DTT and 0.5 mM spermine was added to the mixture and incubated at 37°C for 20 min. At the end of the reaction, a 20 μl of aliquot was spotted onto a nitrocellulose membrane filter (Advantec) and washed with buffer consisting of 50 mM Tris–HCl (pH 7.5), 6.5 mM MgCl2, 60 mM KCl, 1 mM DTT and 0.5 mM spermine. The amount of bound tRNA was calculated by measuring the count of Cerenkov lights with a liquid scintillation counter using the [32P]-labelled tRNA as a tracer.
RESULTS
Isolation and analysis of M. mobile tRNAIle2
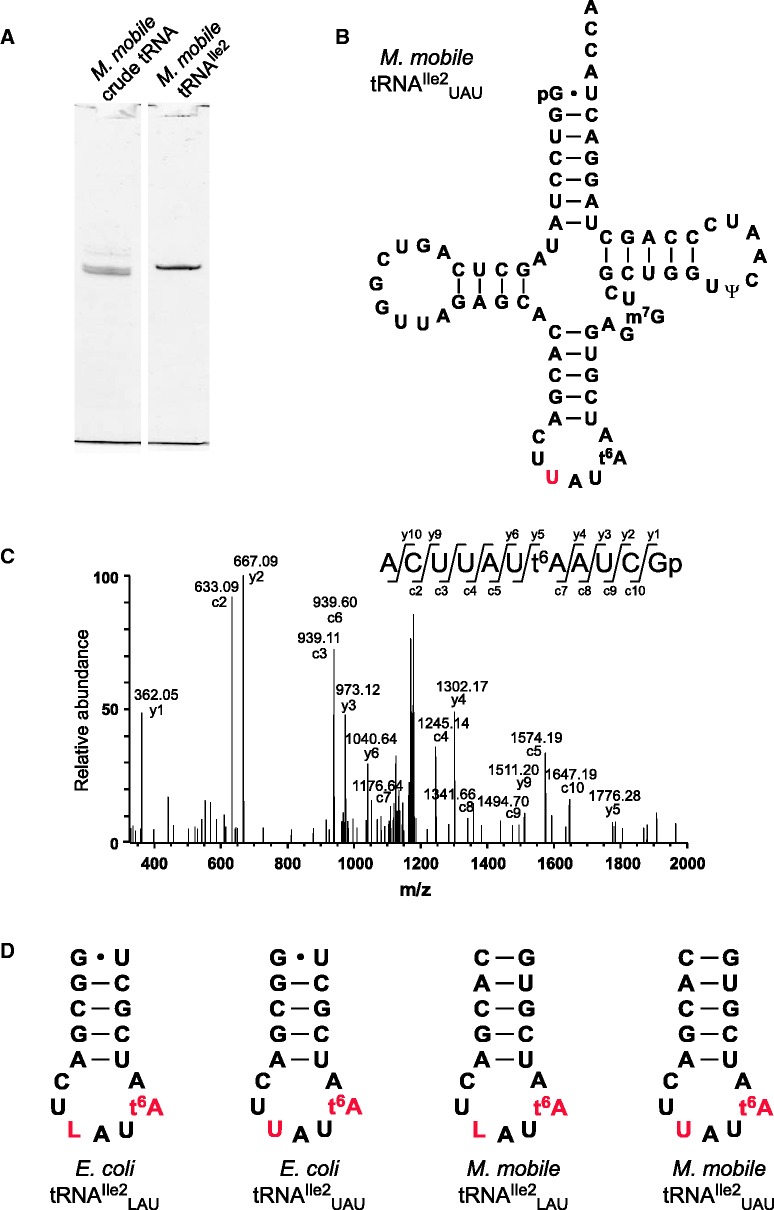
We first isolated tRNAIle2 from M. mobile total RNA (Figure 1A) by RCC (25). The tRNAIle2 was digested with G-specific RNase T1 or pyrimidine-specific RNase A and then analysed by LC/MS (Supplementary Figure S1A, Supplementary Tables S2 and S3). No modification was detected at the wobble position (Figure 1B and C and Supplementary Figure S1A) while N6-threonylcarbamoyladenosine (t6A) was identified at position 37 and 7-methylguanoisne (m7G) was partially introduced at position 47 (Figure 1B, Supplementary Table S2 and S3). To detect pseudouridine (Ψ), which is a mass-silent modification, tRNAIle2 was treated with acrylonitrile to cyanoethylate Ψ before LC/MS analysis. The cyanoethylated RNA fragment (positions 54–64) was identified (Supplementary Figure S1B) and probed by collision-induced dissociation (CID) (Supplementary Figure S1C); cyanoethylated Ψ was identified at position 55 (Figure 1B). Similar to other RNA fragments that lack Ψ, the anticodon-containing fragment (positions 31–41) showed low cyanoethylation efficiency (Supplementary Figure S1B), which is likely the result of non-specific cyanoethylation of uridines (37). We concluded that the wobble uridine of M. mobile tRNAIle2 remained unmodified.
Figure 1.
Isolation and analysis of M. mobile tRNAIle2UAU. (A) Purification of M. mobile tRNAIle2UAU by RCC. The isolated tRNA was resolved by 10% polyacrylamide gel electrophoresis and detected by staining with SYBR Gold. (B) Cloverleaf structure of M. mobile tRNAIle2UAU with the post-transcriptional modifications N6-threonylcarbamoyladenosine (t6A), 7-methylguanoisne (m7G) and pseudouridine (Ψ). U34 is shown in red. (C) CID spectrum of the anticodon-containing fragment of RNase T1-digested M. mobile tRNAIle2UAU. The parent ion for CID is m/z 1218.49. c- and y-series product ions are shown. (D) The anticodon stem–loop sequences of the synthetic tRNAs used for kinetic analyses of isoleucylation: E. coli tRNAIle2LAU, E. coli tRNAIle2UAU, M. mobile tRNAIle2LAU and M. mobile tRNAIle2UAU. L and U at the wobble position and t6A at position 37 are shown in red. The complete sequences are shown in Supplementary Figure S2.
Kinetic characterization of M. mobile IleRS
It is known that E. coli IleRS does not efficiently recognize tRNAIle with the UAU anticodon (23). However, M. mobile IleRS must be able to efficiently recognize tRNAIle with the UAU anticodon to translate the AUA codon. To investigate this further and to determine whether M. mobile IleRS recognizes the LAU anticodon, recombinant M. mobile IleRS was expressed in E. coli. t6A at position 37 plays an important role in efficient isoleucylation of tRNAIle by E. coli IleRS (38). Native M. mobile tRNAIle2UAU and E. coli tRNAIle2LAU were isolated for use as substrates in kinetic analysis. In addition, four species of tRNAsIle containing t6A and/or L were constructed by in vitro transcription and modification (Figure 1D and Supplementary Figure S2): E. coli tRNAIle2LAU, E. coli tRNAIle2UAU, M. mobile tRNAIle2LAU and M. mobile tRNAIle2UAU. t6A was enzymatically introduced by YrdC, YgjD, YjeE and YeaZ (31), and L was introduced by TilS (12,32) because it is impossible to estimate the tRNAIle variants without t6A for isoleucylation. E. coli tRNAIle2UAU and M. mobile tRNAIle2LAU are tRNA variants to examine the specificity of IleRSs. Each tRNA was digested with RNase T1 and analysed by LC/MS, which showed that the frequency of each modification was >90% (Supplementary Figure S3). The kinetic parameters of isoleucylation of each substrate by M. mobile or E. coli IleRS were then assessed.
In the E. coli system, both native and synthetic tRNAsIle2 with the LAU anticodon showed efficient activity, indicating that only two modifications, t6A and L, are sufficient for full isoleucylation (Table 1). Interestingly, the Km of the synthetic tRNA was lower than that of the native molecule (Table 1). When the wobble base was replaced with U, Ile-accepting activity of tRNAIle2UAU decreased significantly, with high Km and low kcat values (Table 1). The relative kcat/Km of tRNAIle2UAU showed only 1.1% of the activity of native tRNAIle2LAU. The relative kcat/Km of the LAU anticodon normalized to that of the UAU anticodon was 360. By contrast, in the M. mobile system, both native and synthetic tRNAsIle2 with the UAU anticodon were efficiently isoleucylated by M. mobile IleRS, as expected (Table 1). In addition, M. mobile IleRS recognized tRNAIle2 with the LAU anticodon as efficiently as native tRNAIle2UAU (Table 1). This observation was surprising because L modification does not occur in M. mobile. The relative kcat/Km value for the LAU anticodon normalized to that of the UAU anticodon was 3.6. In terms of the relative kcat/Km values, M. mobile IleRS showed 100-fold greater preference for the UAU anticodon compared with E. coli IleRS.
Table 1.
Kinetic parameters of isoleucylation by IleRS
| IleRS | Substrate tRNA | Km (μM) | kcat (×10−3 s−1) | kcat/Km (relative) | LAU/UAU of kcat/Km (relative) | |
|---|---|---|---|---|---|---|
| M. mobile WT | M. mobile tRNAIle2UAU (native) | 0.70 ± 0.11 | 270 ± 10 | 1.0 | ||
| M. mobile tRNAIle2LAU (synthetic) | 0.40 ± 0.02 | 310 ± 20 | 2.0 | } | 3.6 | |
| M. mobile tRNAIle2UAU (synthetic) | 1.0 ± 0.2 | 230 ± 20 | 0.57 | |||
| E. coli WT | E. coli tRNAIle2LAU (native) | 1.4 ± 0.4 | 260 ± 30 | 1.0 | ||
| E. coli tRNAIle2LAU (synthetic) | 0.25 ± 0.02 | 180 ± 10 | 3.9 | } | 360 | |
| E. coli tRNAIle2UAU (synthetic) | 8.3 ± 1.5 | 17 ± 4 | 0.011 | |||
| E. coli W905R | E. coli tRNAIle2LAU (synthetic) | 0.37 ± 0.07 | 130 ± 30 | 1.9 | } | 18 |
| E. coli tRNAIle2UAU (synthetic) | 1.9 ± 0.3 | 39 ± 2 | 0.11 |
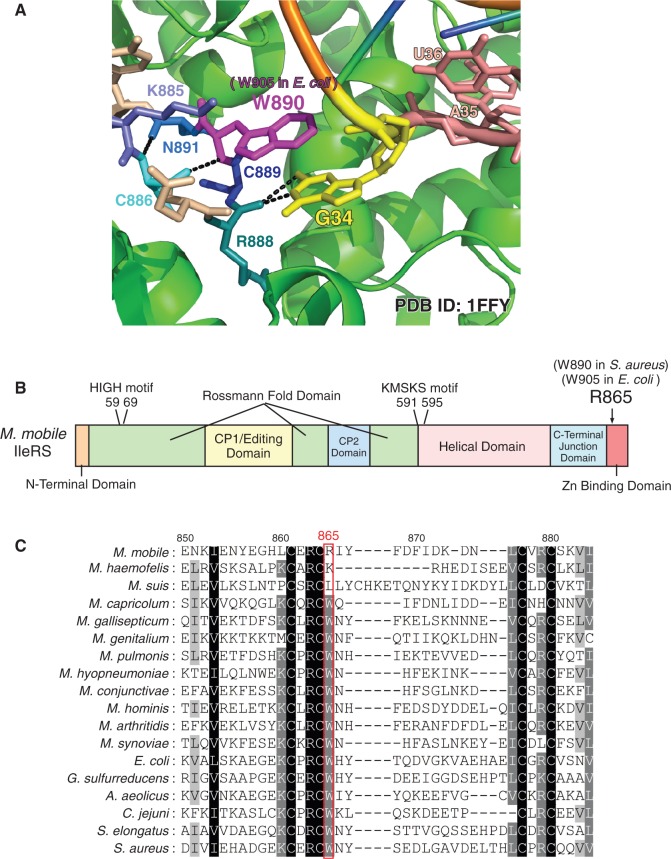
Arg865 in M. mobile IleRS is critical for UAU anticodon specificity
Based on bacterial phylogeny, the AUA decoding system in M. mobile was most likely established after Tenericutes (Mycoplasma species) branched out from the ancestral clade (Supplementary Figure S4). This hypothesis is consistent with the fact that tilS homologues are widely distributed in all clades of bacteria and in other Mycoplasma species. Thus, M. mobile IleRS might have acquired specificity for the UAU anticodon during Tenericutes evolution. To investigate the molecular basis of the UAU anticodon specificity, we attempted to identify the residue critical for recognition of the uracil base at the wobble position. In the crystal structure of Staphylococcus aureus IleRS complexed with E. coli tRNAIle1 (Figure 2A) (39), Arg888, Cys889 and Trp890 in the Zn-binding domain (Figure 2B) reside adjacent to the wobble position. The carbonyl group in the backbone of Arg888 interacts with the wobble base and Trp890 stacks to the wobble base. When we looked at the alignment of Zn-binding domains in IleRSs from various bacteria, including Mycoplasma species (Figure 2C), the Trp residue at position 890 in S. aureus IleRS (Trp905 in E. coli IleRS) is conserved in species bearing a tilS homologue. The corresponding site is replaced with Arg (Arg865) in M. mobile, with Lys in Mycoplasma haemofelis (Lys852) and with Leu in Mycoplasma suis (Leu878); neither species has a tilS homologue. This analysis suggested the possibility that M. mobile IleRS acquired specificity for the UAU anticodon by a single amino acid mutation. To investigate this hypothesis, a mutant E. coli IleRS was constructed in which Trp905 was replaced with Arg (W905R). Kinetic analysis revealed that the W905R mutant recognized tRNAIle2 bearing the UAU anticodon with reduced Km and increased kcat values, resulting in a 10-fold increase in relative kcat/Km value, compared with that for the tRNA bearing the LAU anticodon (Table 1). Specifically, an ∼4-fold decrease in the Km indicates that the W905R mutation enhanced recognition of the tRNA containing the UAU anticodon. The mutation did not affect specificity for tRNAIle2 bearing the LAU anticodon. The relative kcat/Km value for the LAU anticodon normalized to that for the UAU anticodon was 18. A 20-fold enhancement of specificity for the UAU anticodon, compared with wild-type IleRS, was acquired with a single amino acid substitution.
Figure 2.
Structure and sequence alignment of bacterial IleRS. (A) Close-up view of the wobble position in the crystal structure of S. aureus IleRS complexed with E. coli tRNAIle1 (PDB ID: 1FFY) (39). The numbering of residues is based on S. aureus IleRS. (B) Domain structure of M. mobile IleRS. The domain names are based on the reference (39). (C) Sequence alignment of the Zn-binding domains of bacterial IleRSs. The numbering of residues is based on M. mobile IleRS. M. capricolum, Mycoplasma capricolum; M. gallisepticum, Mycoplasma gallisepticum; M. pulmonis, Mycoplasma pulmonis; M. hyponeumoniae, Mycoplasma hyponeumoniae; M. conjunctivae, Mycoplasma conjunctivae; M. hominis, Mycoplasma hominis; M. arthritidis, Mycoplasma arthritidis; M. synoviae, Mycoplasma synoviae; G. sulfurreducens, Geobacter sulfurreducens; A. aeolicus, Aquifex aeolicus; C. jejuni, Campylobacter jejuni; S. elongatus, Synechococcus elongatusi.
In M. haemofelis and M. suis, the site corresponding to Trp890 in S. aureus IleRS is substituted with Lys and Leu, respectively (Figure 2C). Using E. coli tRNAIle2 variants bearing anticodons GAU, LAU, UAU and CAU as substrates, we examined initial rate of isoleucylation by E. coli IleRS WT, W905R, W905K, W905L and M. mobile IleRS (Supplementary Figure S5). Similar to the W905R mutant (Table 1, Supplementary Figure S5B), the W905K mutant recognized tRNAIle2 with UAU anticodon (Supplementary Figure S5C). In contrast, E. coli IleRS W905L mutant did not recognize the UAU anticodon efficiently (Supplementary Figure S5D), as observed in the wild-type of E. coli IleRS (Supplementary Figure S5A). Taken together, these results show that the basic residues in the Zn-binding domains of IleRS in the two Mycoplasma species play a critical role in recognition of the UAU anticodon. The specificity of M. suis IleRS is considered in the discussion.
In this experiment, we also estimate the CAU anticodon specificity of these IleRSs. tRNAIle2CAU is a precursor of tRNAIle2LAU, which is a substrate for MetRS (10). Thus, tRNAIle2CAU is not recognized by E. coli IleRS WT (Supplementary Figure S5A). Both W905R and W905K mutants of E. coli IleRS recognized tRNAIle2CAU as efficiently as tRNAIle2UAU (Supplementary Figure S5B and C), whereas M. mobile IleRS did not aminoacylate tRNAIle2CAU (Supplementary Figure S5E).
Specific AUA decoding at the A-site of the M. mobile ribosome
With the UAU anticodon in tRNAIle2, mistranslation of the AUG as Ile is possible. An A-site tRNA binding experiment was performed to examine AUR codon recognition of M. mobile tRNAIle2UAU on the ribosome (Figure 3). As positive controls, the binding of E. coli tRNAs to the AUR codons at the A-site of the E. coli 70S ribosome was examined. As expected, E. coli tRNAMet specifically bound to the AUG codon, and not to AUA, whereas E. coli tRNAIle2 bound the AUA codon, and not AUG (Figure 3). In contrast to the results with E. coli tRNAIle2, M. mobile tRNAIle2 recognized not only the AUA codon, but also the AUG codon via U·G wobble pairing (Figure 3), indicating that M. mobile tRNAIle2 cannot distinguish between AUA and AUG codon on the E. coli ribosome. However, at the A-site of the M. mobile ribosome, M. mobile tRNAIle2UAU bound to the AUA codon, and not to AUG (Figure 3). These findings suggest that some property of the M. mobile ribosome precludes misreading of the AUG codon by tRNAIle2UAU.
Figure 3.
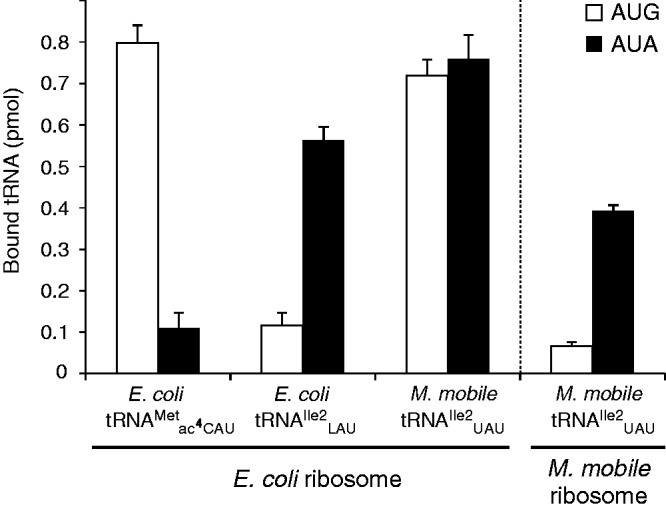
Recognition of AUA and AUG codons at the ribosomal A-site by tRNAs. Non-enzymatic A-site binding of AUG (white bars) and AUA (black bars) codons by E. coli tRNAMet, E. coli tRNAIle2 and M. mobile tRNAIle2 on the E. coli or M. mobile ribosome. Values represent mean ± standard deviation (SD) of three independent analyses.
DISCUSSION
To the best of our knowledge, nine bacterial species dispersed in three clades lack a tilS homologue and carry the tRNAIleTAT gene (Supplementary Table S1). According to the bacterial phylogeny (Supplementary Figure S4) constructed on the basis of 16S rRNA genes, the AUA decoding system without L modification should have emerged independently in each clade. In this study, we showed that M. mobile has IleRS that efficiently recognizes the UAU anticodon of tRNAIle. In M. mobile IleRS, the Arg residue at position 865 (Lys in M. haemofelis) is critical for UAU anticodon specificity. Structural analysis will be necessary to elucidate the molecular basis of recognition of the wobble uridine by the basic residues in mycoplasma IleRSs. Based on the isoleucylation of a series of E. coli tRNAIle2 variants, both W905R and W905K mutants of E. coli IleRS recognized tRNAIle2CAU as efficiently as tRNAIle2UAU (Supplementary Figure S5B and C), suggesting that basic residues at position 905 in the Zn-binding domains allow IleRS to recognize pyrimidines at the wobble position of tRNAIle2. M. mobile IleRS, however, did not aminoacylate tRNAIle2CAU (Supplementary Figure S5E). Thus, M. mobile IleRS should have another measure to discriminate the UAU anticodon from the CAU anticodon. In contrast, the W905L mutant of E. coli IleRS did not show the efficient UAU anticodon recognition. M. suis IleRS might have acquired other mechanism to recognize the UAU anticodon. Several residues near the wobble position might establish the specificity to the UAU anticodon. Otherwise, the UAU anticodon might be modified in M. suis.
On the other hand, Bifidobacterium and Neorickettsia species (Supplementary Table S1) that lack a tilS homologue have unique IleRSs that originate from a eukaryotic IleRS found in eukaryotes and archaea. Based on sequence, IleRSs can be classified into two types in all domains of life (40–43). The majority of bacteria and mitochondria possess the bacterial type IleRS, whereas the eukaryotic type IleRS is commonly found in eukaryotes and archaea. Bacterial and eukaryotic IleRSs can be distinguished by their characteristic C-terminal domains, which are required for anticodon recognition. As noted earlier in the text, some bacteria possess the eukaryotic IleRS, which is assumed to have been transferred horizontally from eukaryotes (40). As no structural information is available on the eukaryotic IleRS, the mechanism underlying UAU anticodon recognition in species that lack L modification remains to be determined. However, in eukaryotes, a minor tRNAIle bearing the ΨAΨ anticodon is a substrate for IleRS, indicating that the eukaryotic IleRS in bacteria might recognize the UAU (U*AU) anticodon.
It is intriguing that M. mobile IleRS efficiently recognized the LAU anticodon in vitro despite the fact that L modification does not occur in this organism. Phylogenetic analysis suggests that M. mobile lost tilS and tRNAIleCAT genes after segregation of Tenericutes from the ancestral clade (Supplementary Figure S4). From this perspective, it makes sense that M. mobile IleRS recognizes the LAU anticodon, which most likely was a substrate in the ancestral species. This theory also suggests that the AUA decoding system might have been reorganized through a transition state using both tRNAIleLAU and tRNAIleUAU. Some bacteria carry the tRNAIleTAT gene despite the presence of a tilS homologue (Supplementary Table S1). This suggests that the AUA codons are deciphered redundantly by both tRNAIleLAU and tRNAIleUAU in these bacteria. Inspection of the IleRSs in these species (Supplementary Figure S6) showed that a Trp residue in the Zn-binding domain interacts with the wobble base in Cyanothece sp. (PCC 7424) and Ferrimonas balearica (DSM 9799), whereas an Arg residue occurs at the corresponding site in Lactobacillus casei (BL23 and BD-II). This observation implies that tRNAIleUAU might be a poor substrate for IleRS in the former two species, whereas IleRS might have evolved to recognize the UAU anticodon efficiently in the latter two. In that case, tilS might no longer be an essential gene in L. casei.
The suppressor strain of a Bacillus subtilis (Firmicutes) mutant with a temperature-sensitive tilS (tilSts) has been isolated (18). This suppressor carries a mutant tRNAIle1GAT gene, in which the wobble G is substituted with T. In the absence of TilS activity, AUA codons are translated into Ile by the suppressor tRNAIle1 containing the UAU anticodon. This clearly demonstrates that reorganization of the AUA decoding system may have been accompanied by a shift from LAU to UAU in anticodon utilization. However, for efficient isoleucylation of the suppressor tRNAIle1, B. subtilis IleRS must recognize the UAU anticodon. It is interesting to note that B. subtilis IleRS has an Arg residue in the Zn-binding domain that interacts with the wobble base (Supplementary Figure S6), suggesting that tRNAIle containing the UAU anticodon could potentially be recognized in B. subtilis. As previously described, L. plantarum and L. casei, which belong to the same clade as B. subtilis (Firmicutes), have IleRSs with an Arg residue that could potentially recognize the UAU anticodon. They also contain a tRNAIleTAT gene despite the presence of a tilS homologue. It would not be surprising if Firmicutes lacking a tilS homologue are discovered in the near future.
During reorganization of the AUA decoding system, the ribosome decoding site may have evolved to prevent misreading of the AUG codon by tRNAIle2UAU. In fact, at the A-site of the M. mobile ribosome, tRNAIle2UAU specifically distinguished between the AUA and the AUG codons in vitro. In the cell, as tRNAMet strongly recognizes the AUG codon, potential misreading of the AUG codon by tRNAIle2UAU can be reduced to a certain extent by competition with tRNAMet. However, ribosome-mediated prevention of AUG misreading might be required to maintain robust and accurate decoding of the AUA codon. The ribosomal A-site in the 30S subunit contains highly conserved bases in 16S rRNA and some ribosomal proteins, including S12. Specific substitutions in 16S rRNA or ribosomal proteins might be responsible for this specificity in the M. mobile 30S subunit.
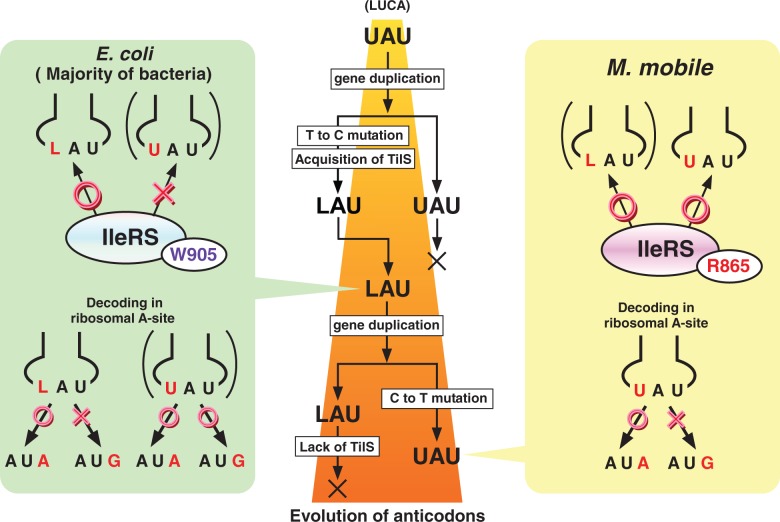
Finally, we will discuss the evolutionary reorganization of the AUA decoding system (Figure 4). Considering that the AUA codon is deciphered by anticodons with different wobble modifications in the three domains of life, the AUA codon might have been deciphered by the UAU anticodon lacking a wobble modification in the last universal common ancestor (LUCA). In such a system, tRNAIleUAU would have been efficiently isoleucylated by IleRS. Because the single amino acid substitution of Arg for Trp conferred specificity for the UAU anticodon on E. coli IleRS, it is reasonable to assume that the primitive IleRS in LUCA originally had an Arg residue at the site that recognizes the wobble base. In LUCA, the AUG codon might have been mistranslated as Ile by tRNAIleUAU. Decoding fidelity could have been maintained to a certain extent by kinetic competition with tRNAMet, and the ribosome might have been involved in preventing AUG misreading, as observed in M. mobile. Duplication of the tRNAIleTAT gene, followed by T-to-C transition at the wobble position in one of the genes, could have generated the tRNAIleCAT gene. Acquisition of tilS in the ancient clade of bacteria would have generated the capacity to modify the tRNA and create the LAU anticodon. As tilS is a family of ATP pyrophosphatases (13), tilS might have originated from this family. Decoding of AUA by the LAU anticodon is an ideal system for prevention of misreading of the AUG codon. At this stage of evolution, two tRNAIles containing the LAU and UAU anticodons would have redundantly decoded the AUA codons. When IleRS acquired the Trp residue (Trp905 in E. coli) at the site recognizing the wobble base, tRNAIleUAU lost the capacity for isoleucylation and the gene disappeared. The majority of bacteria exhibit properties characteristic of this stage of evolution. In some branches of bacterial clades, including Tenericutes, the tRNAIleCAT gene was duplicated and a C-to-T transition at the wobble position of one gene generated the tRNAIleTAT gene. Once IleRS evolved by altering its sequence, including Arg865 in the case of M. mobile, to recognize tRNAIleUAU efficiently, tRNAIleUAU gained the capacity for efficient isoleucylation. At this stage, the AUA codon is redundantly deciphered by two tRNAsIle containing the LAU and UAU anticodons; some bacteria exhibit properties characteristic of this stage. When tilS disappeared, L was lost. Ribosomal prevention of AUG misreading by the UAU anticodon, as observed in this study, might play a critical role in reorganization of the AUA decoding system.
Figure 4.
Model for reorganization of the AUA decoding system during bacterial evolution. The evolution of anticodons in tRNAIle is described in the centre panel. IleRS recognition and codon recognition by tRNAIle2 in E. coli (and the majority of bacteria) and M. mobile are described in left and right panels, respectively.
In conclusion, the system of decoding of the AUA codon by tRNAIle lacking a wobble modification has been established in M. mobile based on two main factors. IleRS acquired specificity for the UAU anticodon by a single Trp-to-Arg substitution, and the ribosome acquired the capacity to prevent misreading of the AUG codon by the UAU anticodon.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5 and Supplementary Figures 1–6.
FUNDING
Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan (to T.S.); New Energy and Industrial Technology Development Organization (NEDO) (to T.S.). Funding for open access charge: Japan Ministry of Education, Science, Sports and Culture.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Y. Sakaguchi for mass spectrometric analysis and Dr Takeo Suzuki and Dr S. Kimura for technical support. They are also grateful to the other members of the Suzuki laboratory for their valuable advice and discussion.
REFERENCES
- 1.Suzuki T. Fine-Tuning of RNA Functions by Modification and Editing. New York: Springer-Verlag; 2005. Biosynthesis and function of tRNA wobble modifications; pp. 24–69. [Google Scholar]
- 2.Curran JF. Modified Nucleosides in Translation. Washington, DC: ASM press; 1998. pp. 493–516. [Google Scholar]
- 3.Crick FH. Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- 4.Senger B, Auxilien S, Englisch U, Cramer F, Fasiolo F. The modified wobble base inosine in yeast tRNAIle is a positive determinant for aminoacylation by isoleucyl-tRNA synthetase. Biochemistry. 1997;36:8269–8275. doi: 10.1021/bi970206l. [DOI] [PubMed] [Google Scholar]
- 5.Muramatsu T, Yokoyama S, Horie N, Matsuda A, Ueda T, Yamaizumi Z, Kuchino Y, Nishimura S, Miyazawa T. A novel lysine-substituted nucleoside in the first position of the anticodon of minor isoleucine tRNA from Escherichia coli. J. Biol. Chem. 1988;263:9261–9267. doi: 10.1351/pac198961030573. [DOI] [PubMed] [Google Scholar]
- 6.Weber F, Dietrich A, Weil JH, Maréchal-Drouard L. A potato mitochondrial isoleucine tRNA is coded for by a mitochondrial gene possessing a methionine anticodon. Nucleic Acids Res. 1990;18:5027–5030. doi: 10.1093/nar/18.17.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harada F, Nishimura S. Purification and characterization of AUA specific isoleucine transfer ribonucleic acid from Escherichia coli B. Biochemistry. 1974;13:300–307. doi: 10.1021/bi00699a011. [DOI] [PubMed] [Google Scholar]
- 8.Ikeuchi Y, Kimura S, Numata T, Nakamura D, Yokogawa T, Ogata T, Wada T, Suzuki T. Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea. Nat. Chem. Biol. 2010;6:277–282. doi: 10.1038/nchembio.323. [DOI] [PubMed] [Google Scholar]
- 9.Mandal D, Köhrer C, Su D, Russell SP, Krivos K, Castleberry CM, Blum P, Limbach PA, Söll D, RajBhandary UL. Agmatidine, a modified cytidine in the anticodon of archaeal tRNA(Ile), base pairs with adenosine but not with guanosine. Proc. Natl Acad. Sci. USA. 2010;107:2872–2877. doi: 10.1073/pnas.0914869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muramatsu T, Nishikawa K, Nemoto F, Kuchino Y, Nishimura S, Miyazawa T, Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988;336:179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- 11.Köhrer C, Srinivasan G, Mandal D, Mallick B, Ghosh Z, Chakrabarti J, Rajbhandary UL. Identification and characterization of a tRNA decoding the rare AUA codon in Haloarcula marismortui. RNA. 2008;14:117–126. doi: 10.1261/rna.795508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soma A, Ikeuchi Y, Kanemasa S, Kobayashi K, Ogasawara N, Ote T, Kato J, Watanabe K, Sekine Y, Suzuki T. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell. 2003;12:689–698. doi: 10.1016/s1097-2765(03)00346-0. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki T, Miyauchi K. Discovery and characterization of tRNAIle lysidine synthetase (TilS) FEBS Lett. 2010;584:272–277. doi: 10.1016/j.febslet.2009.11.085. [DOI] [PubMed] [Google Scholar]
- 14.Terasaka N, Kimura S, Osawa T, Numata T, Suzuki T. Biogenesis of 2-agmatinylcytidine catalyzed by the dual protein and RNA kinase TiaS. Nat. Struct. Mol. Biol. 2011;18:1268–1274. doi: 10.1038/nsmb.2121. [DOI] [PubMed] [Google Scholar]
- 15.Silva FJ, Belda E, Talens SE. Differential annotation of tRNA genes with anticodon CAT in bacterial genomes. Nucleic Acids Res. 2006;34:6015–6022. doi: 10.1093/nar/gkl739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Randau L, Pearson M, Söll D. The complete set of tRNA species in Nanoarchaeum equitans. FEBS Lett. 2005;579:2945–2947. doi: 10.1016/j.febslet.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 17.Elkins JG, Podar M, Graham DE, Makarova KS, Wolf Y, Randau L, Hedlund BP, Brochier-Armanet C, Kunin V, Anderson I, et al. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc. Natl Acad. Sci. USA. 2008;105:8102–8107. doi: 10.1073/pnas.0801980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabret C, Dervyn E, Dalmais B, Guillot A, Marck C, Grosjean H, Noirot P. Life without the essential bacterial tRNA Ile2-lysidine synthetase TilS: a case of tRNA gene recruitment in Bacillus subtilis. Mol. Microbiol. 2011;80:1062–1074. doi: 10.1111/j.1365-2958.2011.07630.x. [DOI] [PubMed] [Google Scholar]
- 19.Abe T, Ikemura T, Sugahara J, Kanai A, Ohara Y, Uehara H, Kinouchi M, Kanaya S, Yamada Y, Muto A, et al. tRNADB-CE 2011: tRNA gene database curated manually by experts. Nucleic Acids Res. 2011;39:D210–D213. doi: 10.1093/nar/gkq1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyata M. Unique centipede mechanism of Mycoplasma gliding. Annu. Rev. Microbiol. 2010;64:519–537. doi: 10.1146/annurev.micro.112408.134116. [DOI] [PubMed] [Google Scholar]
- 21.Jaffe JD, Stange-Thomann N, Smith C, DeCaprio D, Fisher S, Butler J, Calvo S, Elkins T, FitzGerald MG, Hafez N, et al. The complete genome and proteome of Mycoplasma mobile. Genome Res. 2004;14:1447–1461. doi: 10.1101/gr.2674004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Boniecki MT, Jaffe JD, Imai BS, Yau PM, Luthey-Schulten ZA, Martinis SA. Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc. Natl Acad. Sci. USA. 2011;108:9378–9383. doi: 10.1073/pnas.1016460108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nureki O, Niimi T, Muramatsu T, Kanno H, Kohno T, Florentz C, Giegé R, Yokoyama S. Molecular recognition of the identity-determinant set of isoleucine transfer RNA from Escherichia coli. J. Mol. Biol. 1994;236:710–724. doi: 10.1006/jmbi.1994.1184. [DOI] [PubMed] [Google Scholar]
- 24.Aluotto BB, Wittler RG, Williams CO, Faber JE. Standardized bacteriologic techniques for the characterization of Mycoplasma species. Int. J. Syst. Bacteriol. 1970;20:35–58. [Google Scholar]
- 25.Miyauchi K, Ohara T, Suzuki T. Automated parallel isolation of multiple species of non-coding RNAs by the reciprocal circulating chromatography method. Nucleic Acids Res. 2007;35:e24. doi: 10.1093/nar/gkl1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki T, Ikeuchi Y, Noma A, Sakaguchi Y. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 2007;425:211–229. doi: 10.1016/S0076-6879(07)25009-8. [DOI] [PubMed] [Google Scholar]
- 27.Mengel-Jørgensen J, Kirpekar F. Detection of pseudouridine and other modifications in tRNA by cyanoethylation and MALDI mass spectrometry. Nucleic Acids Res. 2002;30:e135. doi: 10.1093/nar/gnf135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 29.Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomari Y, Suzuki T, Watanabe K, Ueda T. The role of tightly bound ATP in Escherichia coli tRNA nucleotidyltransferase. Genes Cells. 2000;5:689–698. doi: 10.1046/j.1365-2443.2000.00360.x. [DOI] [PubMed] [Google Scholar]
- 31.Deutsch C, El Yacoubi B, de Crécy-Lagard V, Iwata-Reuyl D. Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J. Biol. Chem. 2012;287:13666–13673. doi: 10.1074/jbc.M112.344028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikeuchi Y, Soma A, Ote T, Kato J, Sekine Y, Suzuki T. molecular mechanism of lysidine synthesis that determines tRNA identity and codon recognition. Mol. Cell. 2005;19:235–246. doi: 10.1016/j.molcel.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Kitahara K, Suzuki T. The ordered transcription of RNA domains is not essential for ribosome biogenesis in Escherichia coli. Mol. Cell. 2009;34:760–766. doi: 10.1016/j.molcel.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Komoda T, Sato NS, Phelps SS, Namba N, Joseph S, Suzuki T. The A-site finger in 23 S rRNA acts as a functional attenuator for translocation. J. Biol. Chem. 2006;281:32303–32309. doi: 10.1074/jbc.M607058200. [DOI] [PubMed] [Google Scholar]
- 35.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 36.Kirino Y, Yasukawa T, Ohta S, Akira S, Ishihara K, Watanabe K, Suzuki T. Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease. Proc. Natl Acad. Sci. USA. 2004;101:15070–15075. doi: 10.1073/pnas.0405173101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakurai M, Yano T, Kawabata H, Ueda H, Suzuki T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat. Chem. Biol. 2010;6:733–740. doi: 10.1038/nchembio.434. [DOI] [PubMed] [Google Scholar]
- 38.Niimi T, Nureki O, Yokogawa T, Hayashi N, Nishikawa K, Watanabe K, Yokoyama S. Recognition of the Anticodon Loop of tRNAIle 1 by Isoleucyl-tRNA Synthetase from Escherichia coli. Nucleos. Nucleot. 1994;13:1231–1237. [Google Scholar]
- 39.Silvian LF, Wang J, Steitz TA. Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin. Science. 1999;285:1074–1077. [PubMed] [Google Scholar]
- 40.Wolf YI, Aravind L, Grishin NV, Koonin EV. Evolution of aminoacyl-tRNA synthetases–analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- 41.Fournier GP, Andam CP, Alm EJ, Gogarten JP. Molecular evolution of aminoacyl tRNA synthetase proteins in the early history of life. Orig. Life Evol. Biosph. 2011;41:621–632. doi: 10.1007/s11084-011-9261-2. [DOI] [PubMed] [Google Scholar]
- 42.Brown JR, Doolittle WF. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc. Natl Acad. Sci. USA. 1995;92:2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woese CR, Olsen GJ, Ibba M, Soll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.