Abstract
We previously showed that mRNAs synthesized from three genes that naturally lack introns contain a portion of their coding sequence, known as a cytoplasmic accumulation region (CAR), which is essential for stable accumulation of the intronless mRNAs in the cytoplasm. The CAR in each mRNA is unexpectedly large, ranging in size from ∼160 to 285 nt. Here, we identified one or more copies of a 10-nt consensus sequence in each CAR. To determine whether this element (designated CAR-E) functions in cytoplasmic accumulation of intronless mRNA, we multimerized the most conserved CAR-E and inserted it upstream of β-globin cDNA, which is normally retained/degraded in the nucleus. Significantly, the tandem CAR-E, but not its antisense counterpart, rescued cytoplasmic accumulation of β-globin cDNA transcripts. Moreover, dinucleotide mutations in the CAR-E abolished this rescue. We show that the CAR-E, but not the mutant CAR-E, associates with components of the TREX mRNA export machinery, the Prp19 complex and U2AF2. Moreover, knockdown of these factors results in nuclear retention of the intronless mRNAs. Together, these data suggest that the CAR-E promotes export of intronless mRNA by sequence-dependent recruitment of the mRNA export machinery.
INTRODUCTION
Different steps in gene expression such as transcription, RNA processing, mRNA export and mRNA surveillance are carried out by specific machineries, which are extensively coupled to one another both functionally and physically (1–4). For example, the conserved TREX mRNA export complex associates with the spliceosome and is recruited to mRNA during splicing (5,6). Consistent with this splicing-dependent recruitment of TREX, spliced mRNA typically accumulates in the cytoplasm more efficiently than its cDNA counterpart (6,7). β-globin provides a particularly striking example of this coupling, as the cDNA is largely retained/degraded in the nucleus, whereas the spliced mRNA is rapidly and efficiently exported (8–11). The TREX complex, conserved from yeast to human, contains the multi-subunit THO complex and the proteins UAP56, Aly and CIP29 (6,7,12,13). The human TREX complex is recruited to the 5′-end of mRNAs during splicing via an interaction between Aly and the cap-binding complex (14). In yeast, the TREX complex is recruited to mRNA during transcription and 3′-end formation (15–17). Recently, studies in both yeast and human revealed that the Prp19 complex is also involved in a network of coupled interactions involving transcription, splicing and mRNA export (18–20). The Prp19 complex contains a number of proteins conserved from yeast to human, including Prp19, Cdc5l, Xab2 and Crnkl1 (21). The splicing factor U2AF2 was recently found to associate with the Prp19 complex and function in coupling transcription to splicing (19).
In contrast to splicing-dependent mRNA export, little is known about how naturally intronless mRNAs in higher eukaryotes are exported in the absence of splicing. Most of the work has been done with naturally intronless viral mRNAs, revealing that these mRNAs bind to specific cellular or viral proteins that recruit the mRNA export machinery (22–26). In the case of naturally intronless cellular mRNAs, histone H2A contains an element that binds to SR proteins and recruits the mRNA export receptor Nxf1 (27–31). However, H2A mRNA is unique among cellular mRNAs, lacking a polyA tail. In Drosophila, the homolog of human U2AF2 associates with naturally intronless mRNAs and functions in their export (32).
To further understand the mechanism for exporting naturally intronless mRNAs, we previously examined three of these mRNAs, HSPB3, IFNα1 and IFNβ1 (10). We found that the TREX complex associates with the intronless mRNA and their export is blocked by knockdown of TREX components (UAP56 and Thoc2) or Nxf1 (10). We also showed that a portion of the coding region, which we named cytoplasmic accumulation regions (CARs), is essential for stability/export of these mRNAs. Insertion of the CARs upstream of β-globin cDNA promotes the cytoplasmic accumulation of the cDNA transcript, whereas the antisense of the CARs does not (10). These data are consistent with a previous study showing that a portion of the coding region of c-Jun mRNA, which is also naturally intronless, has the same function as the CARs that we identified (33). For the purpose of this study, we refer to the c-Jun element as a CAR. In all of the intronless mRNAs, the size of the CAR is unexpectedly large, ranging from 162 to 402 nt. Further work is required to understand how CARs function in promoting cytoplasmic accumulation of naturally intronless mRNAs.
Here, we report the identification of a 10-nt consensus element C[CA]AG[ATC][TA][CG][CG]TG, designated CAR-E (CAR Element) found in the CARs in HSPB3, c-Jun, IFNα1 and IFNβ1 mRNAs. We show that tandem duplication of the CAR-E, but not the antisense or a mutated counterpart, promotes cytoplasmic accumulation of β-globin cDNA. Our data indicate that components of the TREX complex, the Prp19 complex and U2AF2 associate with the tandem CAR-E and function in mRNA export. Together, our data suggest that naturally intronless mRNAs bypass splicing-dependent export via specific elements that recruit the mRNA export machinery.
MATERIALS AND METHODS
Identification of the CAR-E
To identify sequences that function in export of naturally intronless mRNAs, we used the MEME algorithm (34) (http://meme.nbcr.net) to search for overrepresented motifs in the four CARs. Using several different approaches, we identified similar or overlapping motifs that were specifically enriched in the CARs but not in their antisense or in the sequences surrounding the CARs. For example, in one approach, we looked for 13-nt motifs, searching for any number of elements in the four CARs. In another approach, we looked for 10-nt motifs looking for any number of motifs using a combination of sense and antisense of the four CARs. The motifs generated via MEME were then further analyzed using FIMO (35) to search for the motifs in the antisense of the CARs or in size-matched sequences surrounding each CAR. Using a P-value output threshold set at 1 × 10−4, the motif was not present in these negative control sequences. For further analysis, we used the most conserved motif (CCAGTTCCTG), which had the lowest P-value (2.37 × 10−6) in our MEME search.
Constructs and antibodies
Wild-type (WT) β-globin containing its natural introns and β-globin cDNA plasmids were described (10). The tandem CAR-E and CAR-EAS β-globin cDNA plasmids were constructed by inserting a DNA sequence (5′ CCAGTTCCTG × 16 3′) into the Hind III site upstream of the cDNA. To generate the CAR-E mutant 1–5 constructs, oligonucleotides containing each dinucleotide mutation were synthesized and inserted into the HindIII site upstream of the β-globin cDNA construct. To generate the Slu7 and DDX3x plasmids, the cDNA and HA-tag were first amplified from Slu7 or DDX3 cDNA plasmids (36,37) by PCR with the following primers: Slu7-F-5′-TTTGGTACCATGTACCCATACGACGTCCCAGACTACGCTTCAGCCACAGTTGTAGATGCA; Slu7-R-5′-AAACTCGAGCTACTGTCCAAGGAAAGAGGCCAT; DDX3x-F-5′-TTTGGTACCATGTACCCATACGACGTCCCAGACTACGCTAGTCATGTGGCAGTGGAAAAT; DDX3x-R-5′-ATACTCGAGTCAGTTACCCCACCAGTCAACCCCCTG. The PCR products were then inserted into the pcDNA5/FRT/TO vector (Invitrogen) at the KpnI and XhoI sites to generate the cDNA versions of the constructs. The same PCR products were also inserted into the KpnI and XhoI sites of pcDNA5/FRT/TO vector containing the tandem CAR-E. Constructs were verified by DNA sequencing. Polyclonal antibodies against Xab2 (Proteintech, 1:800), U2AF2 (Santa Cruz, 1:800), AQR (Bethyl, 1:2000) and monoclonal antibodies against the HA tag (Covance, 1:2000), EGFP (Origene, 1:1000) and tubulin (Sigma, 1:10 000) were used for western blots.
Cell culture, transfection and HeLa nuclear microinjection
HeLa cells were cultured in Dulbecco’s modified eagle medium supplemented with 10% fetal bovine serum. Transfection was carried out in MatTek plates using 1 µg of each plasmid and Lipofectamine 2000. For siRNA transfection, 1.25 µl of 40 µM siRNAs (Smartpool from Dharmacon) was added after mixing with 1 µl lipofectamine 2000. To determine the knockdown efficiency for CRNKL1 and ISY1, the following primers were used for RT–PCR: CRNKL1-RT-F-5′-TGAGGACGTCGATGAGAGTG, CRNKL1-RT-R-5′-GCAGAGCTGGGAAATGAACT and ISY1-RT-F-5′-ACTGGTGCGAAGGAAGAAAA, ISY1-RT-R-5′-TCAAAATGGCAGTGCAAGTC. For microinjection of CMV constructs, a 10-µl mixture containing 2 µl 70-kDa Dextran (Molecular Probes) and 50 ng/µl of each plasmid DNA was microinjected into HeLa cell nuclei. For fluorescence in situ hybridization (FISH), samples were rinsed once with 1 × phosphate buffered saline (PBS) and fixed with 4% paraformaldehyde in PBS for 15 min, and then permeabilized in PBS containing 0.1% Triton X-100. After three rinses with 1 × PBS, the samples were rinsed twice with 1 × SSC/50% formamide, fluorescent probe was added and hybridization was performed overnight at 37°C. The FISH probe was a 70-nt DNA oligonucleotide complementary to the vector sequence downstream of the multiple cloning site and pre-labeled at the 5′-end with Alexa Fluor 546 NHS ester and HPLC-purified. Probe sequence: 5′-AAGGCACGGGGGAGGGGCAAACAACAGATGGCTGGCAACTAGAAGGCACAGTCGAGGCTGATCAGCGGGT. Images were taken with a Nikon TE2000E Inverted Fluorescence Microscope.
Purification of RNPs
Templates for in vitro transcription of the tandem CAR-E or mutant tandem CAR-Em2 were amplified from the β-globin CAR-E and CAR-Em2 cDNA plasmids using the following primers: F-5′-TGGAGGTCGCTGAGTAGTGC and R-5′-TTCCATGGTGGCGGCGGTACCAA. For in vitro transcription, 1 µg of the PCR product was transcribed with 50 units of T7 RNA polymerase (New England Biolabs) for 90 min in reaction mixtures containing 32P-UTP and biotin-16-UTP (Roche). For RNP assembly, 2 µg of CAR-E or CAR-Em2 RNA was incubated for 2 h at 30°C in a 2-ml reaction mixture containing 600 µl of HeLa nuclear extract, 600 µl splicing dilution buffer (20 mM HEPES at pH 7.9, 100 mM KCl), 3.2 mM MgCl2, 20 mM creatine phosphate di-Tris salt and 0.5 mM ATP (13). The mixtures were then loaded on Sephacryl S-500 columns and the peak fractions containing the RNP were pooled and purified by binding to streptavidin agarose (Thermo Scientific). Total proteins were eluted from equivalent amounts of 32P-labeled RNA from each mRNP and TCA precipitated. For mass spectrometry, total proteins were subjected to in-solution trypsin digestion overnight. Tryptic peptides were analyzed by nanoscale-microcapillary reversed phase liquid chromatography–tandem mass spectrometry (LC–MS/MS) essentially as described (38). MS/MS spectra were searched using the SEQUEST algorithm (39) against a concatenated forward/reverse mouse IPI (ver 3.60) database as described (40) with dynamic modification of methionine oxidation. All peptide matches were filtered based on mass deviation, charge, XCorr and dCn to a target peptide false discovery rate of 1% using Linear Discriminant Analysis to distinguish between forward and reverse hits as described previously (41). For Table 1 and Supplementary Table S1, the proteins were categorized based on their best-known function. We found similar numbers of peptides for CBP20 and CBP80 in the CAR-E versus CAR-Em2 mRNP and used these proteins for peptide number comparisons with other proteins listed in Table 1. Contaminants such as keratin, tubulin, ribosomal and translation-related proteins were omitted. In addition, proteins >300 amino acids with total peptide numbers ≤3 were omitted.
Table 1.
Proteins present in tandem CAR-E and CAR-Em2 RNPs
| CAR-E |
CAR-Em2 |
Mol weight (kDa) | |||
|---|---|---|---|---|---|
| Unique | Total | Unique | Total | ||
| TREX complex | |||||
| THOC2 | 29 | 37 | 1 | 1 | 183 |
| THOC5 | 13 | 16 | 79 | ||
| THOC6 | 7 | 9 | 38 | ||
| THOC3 | 5 | 5 | 39 | ||
| THOC1 | 5 | 5 | 76 | ||
| UAP56/URH49 | 13 | 15 | 49 | ||
| Prp19 complex and U2AF | |||||
| Xab2 | 9 | 12 | 1 | 1 | 100 |
| AQR | 10 | 13 | 2 | 2 | 171 |
| CDC5L | 4 | 4 | 1 | 1 | 92 |
| Prp19 | 6 | 6 | 55 | ||
| U2AF2 | 7 | 11 | 54 | ||
| Cap-binding complex | |||||
| NCBP1 | 18 | 28 | 7 | 24 | 92 |
| NCBP2 | 3 | 3 | 3 | 6 | 18 |
| Examples of proteins enriched in CAR-Em2 RNP | |||||
| PTBP1 | 10 | 40 | 57 | ||
| ADAR | 9 | 11 | 17 | 59 | 136 |
| DICER1 | 1 | 1 | 12 | 22 | 219 |
| KHSRP | 8 | 14 | 73 | ||
| NOLC1 | 1 | 1 | 8 | 13 | 74 |
A subset of proteins detected by mass spectrometry in the tandem CAR-E and CAR-Em2 RNPs is shown. The number of total and unique peptides and molecular weight of the proteins are indicated.
RESULTS
A consensus element in CARs promotes cytoplasmic accumulation of RNA
To identify sequences that function in cytoplasmic accumulation of naturally intronless mRNAs, we used a variety of criteria to search for overrepresented motifs within the four CARs using the MEME algorithm (34) (see ‘Materials and Methods’ section). Our analysis led to the identification of a 10-nt consensus element, designated the CAR-E (CAR-Element) that is present in one to four copies in each CAR (Figure 1A). The positions of the CAR-E in each CAR are shown in Figure 1B. The sequence of each CAR-E (colored nucleotide) and non-conserved flanking regions are shown in Figure 1C. We previously showed that the antisense of the CARs are non-functional in export (10). Significantly, no CAR-Es are present in the antisense of the CARs at the same P-value output threshold (1 × 10−4) as used to identify the CAR-Es in the sense strand of the CARs. Moreover, no CAR-Es were detected in size-matched regions outside of the CARs in each naturally intronless mRNA at this threshold. Based on these observations, we pursued the CAR-E further.
Figure 1.
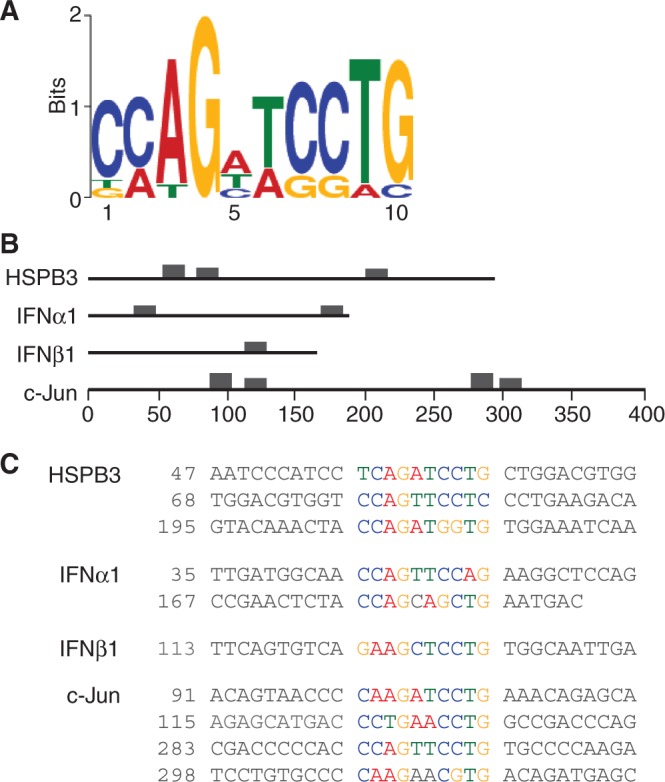
CARs in naturally intronless mRNAs contain a conserved motif. (A) The 10-nt consensus element (CAR-E) identified in the CARs from HSPB3, c-Jun, IFNα1 or IFNβ1 is shown. (B) The locations of the CAR-Es in each CAR are indicated by the boxes. The height of box is proportional to −log(P-value), truncated at the height for a motif with a P-value of 1 × 10−10 (34). The numbering is relative to the first nucleotide of each CAR sequence. (C) The sequences of each of the CAR-Es (colored bases) surrounded by flanking sequence (black bases) are shown.
To examine the role of the CAR-E in naturally intronless mRNA export, we mutated either one or more of the CAR-Es in HSPB3 mRNA. Although we made complete substitutions of all of these elements, the mutations had no apparent effect on cytoplasmic accumulation of HSPB3 mRNA (data not shown). In addition, we made a construct in which all three of the CAR-Es were mutated and five additional sequences that resembled the CAR-Es were mutated, but no effect of these mutations was observed (see ‘Discussion’ section). Thus, as an alternative approach to determine whether the CAR-E plays a role in the cytoplasmic accumulation of intronless mRNAs, we inserted 16 tandem copies of the most conserved CAR-E (CCAGTTCCTG from c-Jun) or its antisense counterpart (CAR-EAS) upstream of β-globin cDNA (Figure 2A). These constructs were transfected into HeLa cells and FISH was carried out to determine the nucleocytoplasmic distribution of β-globin RNA. Constructs encoding β-globin cDNA alone or WT β-globin containing its two natural introns were analyzed as controls. Consistent with previous work (8–11), WT β-globin mRNA, but not the cDNA transcript, efficiently accumulated in the cytoplasm (Figure 2B). Significantly, insertion of the tandem CAR-E resulted in accumulation of β-globin cDNA transcripts in the cytoplasm (Figure 2B). In contrast, β-globin cDNA transcripts containing the CAR-EAS had a nucleocytoplasmic distribution that was similar to β-globin cDNA transcripts alone (Figure 2B). Specifically, as readily seen in the low magnification fields, the CAR-EAS cDNA and cDNA transcripts alone were largely retained/degraded in the nucleus (Figure 2C). We note that the low level of FISH signal observed from the CAR-EAS cDNA and cDNA transcripts is not due to differences in transfection efficiency, as shown by western blot of the transfection control, EGFP (Figure 2D). In addition, our previous work showed that the transcript levels of different CMV constructs are the same at 5 min after microinjection, regardless of the transcribed sequence (9,10).
Figure 2.
Tandem CAR-E promotes cytoplasmic accumulation of β-globin cDNA and β-globin protein expression. (A) Schematic of CMV-β-globin cDNA construct indicating the position where the CAR-E and CAR-EAS were inserted. The start codon (ATG), HA tag and BGH polyA signal (pA) are indicated. The sizes of each of the exons (in nucleotides) are shown. (B) FISH was used to determine the nucleocytoplasmic distribution of the indicated transcripts 24 h after transient transfection of the respective constructs into HeLa cells. DAPI staining was used to identify the nucleus. Scale bar: 10 µm. (C) Low magnification of the data shown in panel B. (D) Western analysis at 24 h (Day 1) or 45 h (Day 2) after transient transfection of the indicated constructs into HeLa cells. HA-β-globin, co-transfection control EGFP and tubulin (loading control) are indicated.
Consistent with our results showing enhanced export of β-globin cDNA containing the CAR-E, western analysis 1 or 2 days after transfection showed that high levels of protein were expressed from both WT and the CAR-E constructs, whereas little protein was detected from the cDNA or CAR-EAS construct (Figure 2D). These results indicate that the consensus element is functional in promoting cytoplasmic accumulation of mRNA and production of protein. We note that insertion of 6 or 10 copies of the CAR-E promoted expression of β-globin cDNA, but the levels were significantly higher with 16 copies. Thus, we used the 16-copy CAR-E construct for further studies. To determine whether the effect of the tandem CAR-E element might be general, we inserted it upstream of two other cDNAs, Slu7 and DDX3x (Supplementary Figure S1). In both cases, the CAR-E resulted in increased mRNA export and greater levels of protein than observed with the corresponding cDNA alone (Supplementary Figure S1), suggesting that the function of the CAR-E may be general.
To further analyze the tandem CAR-E, we introduced dinucleotide mutations across each copy of the CAR-E and inserted this sequence upstream of β-globin cDNA (Figure 3A). These constructs were transfected into HeLa cells and western analysis was used to assay the levels of β-globin protein. After 1 day of transfection, no protein was detected from CAR-E mutants m1-m4 and less protein was detected from CAR-E mutant m5 versus either WT β-globin or the CAR-E cDNA (Figure 3B). Similar results were obtained 2 days after transfection (Figure 3B). Consistent with these data, FISH analysis showed that β-globin RNA containing the CAR-E mutants 1–5 either did not accumulate (mutant 1–4) or partially accumulated (mutant 5) in the cytoplasm, whereas β-globin RNA containing the CAR-E was mostly detected in the cytoplasm (Figure 3C). These data indicate that the CAR-E promotes cytoplasmic accumulation of RNA.
Figure 3.
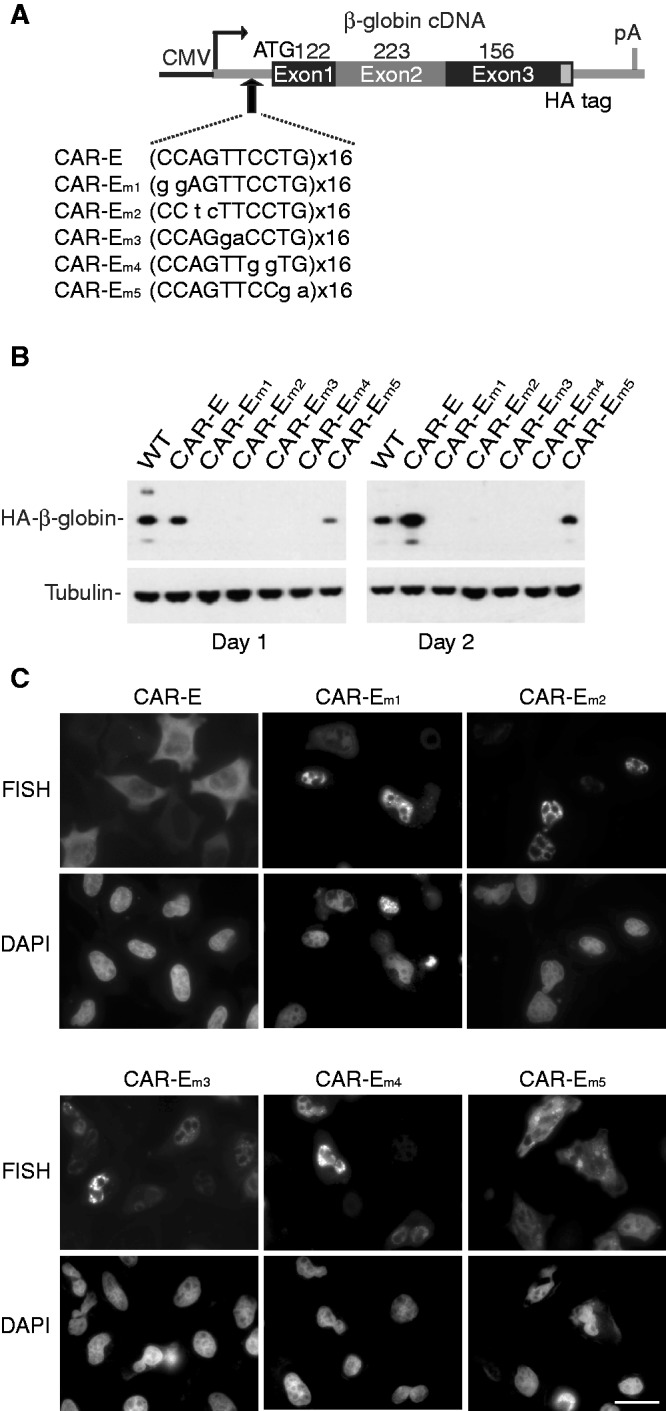
Tandem CAR-E containing dinucleotide substitutions is non-functional in RNA export. (A) Schematic of CMV-β-globin cDNA construct showing sequences of the CAR-E and each of the mutants that were inserted into the cDNA. Mutated nucleotides are shown in lower case. (B) Western analysis at 24 h (Day 1) or 45 h (Day 2) after transient transfection of the indicated constructs into HeLa cells. HA-β-globin and tubulin (loading control) are indicated. (C) Nucleocytoplasmic distribution of CAR-Em1-5 transcripts was determined by FISH. DAPI staining was used to identify the nucleus. Scale bar: 10 µm.
The tandem CAR-E RNA associates with mRNA export factors
We next sought to identify proteins that associate with the tandem CAR-E, using CAR-Em2 as a negative control. To do this, we biotinylated the RNAs and isolated the RNPs by binding to streptavidin agarose (42). Total proteins from equivalent amounts of these RNPs were then analyzed by mass spectrometry. Not unexpectedly, both types of RNPs contained a large number of RNA-related proteins, including those associated with the spliceosome, the polyadenylation machinery as well as hnRNP proteins (Supplementary Table S1). Of particular relevance to this study, we found that TREX complex components, including THOC2, THOC5, THOC6, THOC3, THOC1 and UAP56, were specifically enriched in the CAR-E RNP compared with the CAR-Em2 RNP (Table 1). Interestingly, we also observed a strong enrichment in the CAR-E RNP for components of the Prp19 complex and U2AF2 (Table 1), both of which function in mRNA export (18,32). Both the CAR-E and CAR-Em2 RNPs contained similar numbers of peptides derived from the CAP-binding complex proteins (Table 1), consistent with the presence of the cap on both the CAR-E and CAR-Em2 RNAs. However, in contrast to the CAR-E, the CAR-Em2 RNP was not enriched in mRNA export factors, but was instead enriched in proteins such as PTB, ADAR, KHSRP and other RNA-binding proteins (Table 1 and Supplementary Table S1). Together, these data suggested that the reason that the tandem CAR-E, but not the tandem CAR-Em2, facilitates cytoplasmic accumulation of β-globin cDNA is because the CAR-E recruits mRNA export factors.
Knockdown of Prp19 components or U2AF2 results in nuclear retention of naturally intronless mRNAs
Previously, we showed that TREX components (UAP56 and THOC2) and the mRNA export receptor Nxf1 were required for cytoplasmic accumulation of HSPB3, IFNα1 and IFNβ1 (10). On the basis of our mass spectrometry data of the CAR-E RNP, we next sought to determine whether Prp19 complex components or U2AF2 also functioned in cytoplasmic accumulation of naturally intronless mRNAs. We were especially interested in these factors because we recently found that several of the Prp19 complex components (e.g. XAB2, CRNKL1, CDC5L and PPIE) specifically associate with UAP56 (13). Moreover, recent work in Saccharomyces cerevisiae showed that the Prp19 complex is required for TREX occupancy at intronless genes and that components of this complex interact both genetically and biochemically with the TREX complex (12). Finally, it is known that U2AF2 associates with UAP56 (43) and functions in intronless mRNA export in Drosophila (32). To determine whether Prp19 components or U2AF2 function in cytoplasmic accumulation of naturally intronless mRNAs, we microinjected the IFNβ1 construct driven by the CMV promoter (Figure 4A) into the nuclei of cells knocked down using siRNAs against the Prp19 components Xab2, Aqr, Isy1, Crnkl1 (Figure 4B–I) or U2AF2 (Figure 4J and K). Cells transfected with non-targeting siRNA were used as a negative control. We note that we used microinjection rather than transfection of the CMV constructs for this analysis because we were examining knockdown cells. However, in a great deal of previous work [e.g. (10,11)] and in our present study, we found that expression of CMV constructs is the same for both microinjection and transfection. Significantly, this analysis revealed that IFNβ1 was retained in the nucleus when each of the Prp19 components or U2AF2 was knocked down (Figure 4C, E, G, I and K). These data indicate that Prp19 components and U2AF2 function in export of IFNβ1 mRNA. We obtained the same results when HSPB3 or IFNα1 driven by the CMV promoter were microinjected into the Prp19 component or U2AF2 knockdown cells (Supplementary Figure S2). Knockdown of other Prp19 components, including Prp19 itself, CCDC16, PPIE and PLRG1, resulted in partial or no nuclear retention of intronless mRNA (data not shown). However, the observation that knockdown of several Prp19 complex components and U2AF2, which associates with the Prp19 complex, results in nuclear retention of each of the naturally intronless mRNAs that we tested indicates a role for these factors in intronless mRNA export.
Figure 4.
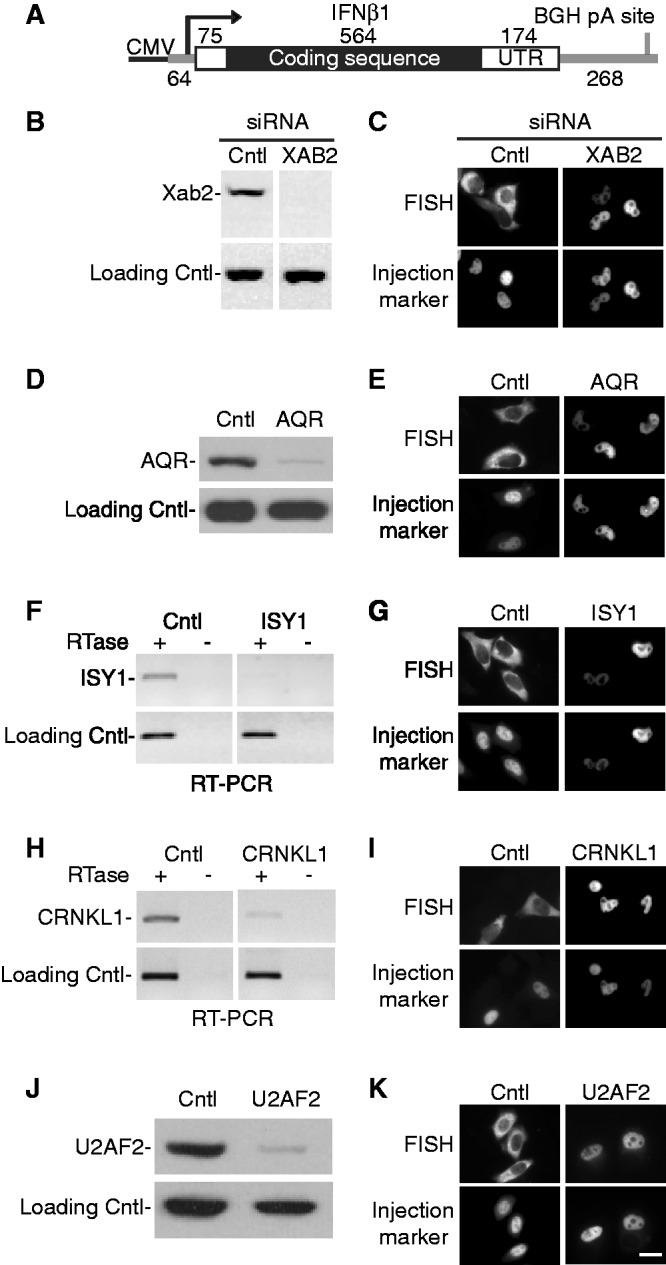
RNAi of Prp19 complex components or U2AF2 blocks export of naturally intronless IFNβ1 mRNAs. (A) Schematic of CMV-IFNβ1 construct used for microinjection. The length of the 5′-UTR, coding region and 3′-UTR are indicated. Vector sequences are shown as gray lines. BGH pA: bovine growth hormone polyA site. (B, D, J) Western blots showing levels of XAB2 (B), AQR (D) or U2AF2 (J) after transfecting HeLa cells with targeting or non-targeting (Cntl) siRNAs as indicated. Tubulin was used as a loading control. (F and H) RT–PCR showing levels of ISY1 or CRNKL1 mRNAs after transfecting HeLa cells with targeting or non-targeting (Cntl) siRNAs as indicated. Endogenous DNAJB1 was amplified by PCR as a loading control. Reverse transcription was carried out in reactions that contained (+) or lacked (−) reverse transcriptase (RTase) followed by PCR. (C, E, G, I, K) CMV-IFNβ1 naturally intronless construct was microinjected into nuclei of the indicated knockdown or control cells. FISH was carried out to determine the nucleocytoplasmic distribution of the RNAs. Dextran 70 kDa was used an injection marker. Scale bar: 10 µm.
DISCUSSION
In this study, we identified a 10-nt consensus sequence, the CAR-E, which is common to the CARs that we previously identified in three naturally intronless mRNAs (10) and is also present in the CAR found in c-Jun mRNA (33). The CAR-E is not present in sequences that do not promote export, such as the antisense of the CARs or regions surrounding the CARs. Evidence that the CAR-E is functional was provided by the observation that insertion of tandem copies of the CAR-E upstream of β-globin cDNA rescued the cytoplasmic accumulation of this cDNA transcript, which is normally retained/degraded in the nucleus. In contrast, the antisense of the tandem CAR-E was not functional in this assay. In addition, several dinucleotide mutations abolished the function of the tandem CAR-E, indicating that the effect we observed is not due to a random sequence but is instead specific to the CAR-E sequence. The tandem CAR-E is similar to CARs in that they both promote cytoplasmic accumulation of RNA. This effect is distinguished from elements that enhance RNA stability, such as the ENE in the Kaposi’s sarcoma-associated herpesvirus, which stabilizes the RNA but does not affect RNA localization or protein production (44,45).
Despite the functionality of the multimerized CAR-E in promoting export of cDNA transcripts, we were unable to observe an export defect when the elements were mutated in their natural context in the CAR. However, our previous work showed that the antisense of the CARs have no activity in mRNA export (10). Thus, the sense strand of the CARs must contain elements or structures that promote cytoplasmic accumulation of the intronless mRNAs. One of these elements may be the CAR-E that we identified here, but additional sequences remain to be identified. Recently, a new, curated resource called the Intronless Gene Database was reported (46). It contains 687 human intronless genes, which represents ∼3% of the genome (46,47). In a global in silico analysis of these genes, we identified six 10-nt motifs that were present in >90% of the intronless genes with one highly similar to CAR-E. It is possible that one or more of these motifs function in intronless mRNA export and/or that a structure(s) is involved. These possibilities require further investigation.
Consistent with the possibility that the CAR-Es function in intronless mRNA export, we found that the tandem CAR-E, but not the mutant CAR-Em2, assembles into an RNP that is enriched in the TREX mRNA export machinery as well as Prp19 complex components and U2AF2. Interestingly, both the Prp19 complex and U2AF2 were previously found to function in the intronless mRNA export pathway in yeast and Drosophila, respectively (18,32). Moreover, knockdown of TREX components, Prp19 complex components (Xab2, Aqr, Crnkl1 or Isy1), or U2AF2 resulted in nuclear retention of the naturally intronless mRNAs [(10), this study]. Knockdown of other Prp19 complex components, such as Prp19, CCDC16, PPIE and PLRG1, had partial or no effect on export of intronless mRNAs (data not shown). These results may be explained by functional redundancy as observed for UAP56 and URH49 (9,13,48); insufficient knockdown to observe a robust phenotype or some of the Prp19 components may form different complexes with distinct functions.
In addition to the enrichment of TREX and Prp19 complex components, we observed that many other proteins, including splicing factors, were enriched in the CAR-E RNP (Supplementary Table S1). As splicing promotes export of mRNAs generated by splicing, an interesting possibility is that splicing factors also play a role in export of naturally intronless mRNAs. This possibility is supported by previous work showing a role for SR proteins in histone mRNA export (49) and the studies showing a role for UAP56 (43,50), U2AF2 (32) and the Prp19 complex in intronless mRNA export (18). Another possible explanation for the association of splicing factors with the CAR-E mRNP is that these factors participate in protein–protein interactions with export factors. We note that the TREX components Aly and CIP29 were not detected in the CAR-E mRNP. Extensive additional studies are needed to determine whether other factors enriched in the CAR-E RNP are involved in export of naturally intronless mRNAs.
Our data, together with previous work, suggest that the mRNA export machinery can be recruited either in a sequence-dependent manner (this study) or in a splicing-dependent manner (7). Thus, the CAR-E may have general utility for enhancing the expression of cDNAs without the need for insertion of an intron.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–2.
FUNDING
Funding for open access charge: The National Institutes of Health [GM043375 to R.R.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Marlene Winkelbauer-Hurt, Eric Folco, Yong Yu and Tomohiro Yamazaki for useful discussions and comments on the manuscript. They also thank the Nikon Imaging Center at Harvard Medical School for assistance with microscopy.
REFERENCES
- 1.Cramer P, Srebrow A, Kadener S, Werbajh S, de la Mata M, Melen G, Nogues G, Kornblihtt AR. Coordination between transcription and pre-mRNA processing. FEBS Lett. 2001;498:179–182. doi: 10.1016/s0014-5793(01)02485-1. [DOI] [PubMed] [Google Scholar]
- 2.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 3.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reed R, Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 2002;108:523–531. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]
- 7.Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell. Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 8.Lu S, Cullen BR. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA. 2003;9:618–630. doi: 10.1261/rna.5260303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dias AP, Dufu K, Lei H, Reed R. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat. Commun. 2010;1:97. doi: 10.1038/ncomms1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei H, Dias AP, Reed R. Export and stability of naturally intronless mRNAs require specific coding region sequences and the TREX mRNA export complex. Proc. Natl Acad. Sci. USA. 2011;108:17985–17990. doi: 10.1073/pnas.1113076108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valencia P, Dias AP, Reed R. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl Acad. Sci. USA. 2008;105:3386–3391. doi: 10.1073/pnas.0800250105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;28:28. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 13.Dufu K, Livingstone MJ, Seebacher J, Gygi SP, Wilson SA, Reed R. ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes Dev. 2010;24:2043–2053. doi: 10.1101/gad.1898610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng H, Dufu K, Lee C-S, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5' end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 15.Abruzzi KC, Lacadie S, Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson SA, Cubberley G, Bentley DL. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3' end processing factor Pcf11. Mol. Cell. 2009;33:215–226. doi: 10.1016/j.molcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zenklusen D, Vinciguerra P, Wyss JC, Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chanarat S, Seizl M, Straber K. The Prp19 complex is a novel transcription elongation factor required for TREX occupancy at transcribed genes. Genes Dev. 2011;25:1147–1158. doi: 10.1101/gad.623411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.David CJ, Boyne AR, Millhouse SR, Manley JL. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev. 2011;25:972–983. doi: 10.1101/gad.2038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuraoka I, Ito S, Wada T, Hayashida M, Lee L, Saijo M, Nakatsu Y, Matsumoto M, Matsunaga T, Handa H, et al. Isolation of XAB2 complex involved in pre-mRNA splicing, transcription, and transcription-coupled repair. J. Biol. Chem. 2008;283:940–950. doi: 10.1074/jbc.M706647200. [DOI] [PubMed] [Google Scholar]
- 21.Chan SP, Kao DI, Tsai WY, Cheng SC. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- 22.Boyne JR, Colgan KJ, Whitehouse A. Herpesvirus saimiri ORF57: a post-transcriptional regulatory protein. Front Biosci. 2008;13:2928–2938. doi: 10.2741/2898. [DOI] [PubMed] [Google Scholar]
- 23.Chen IH, Sciabica KS, Sandri-Goldin RM. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 2002;76:12877–12889. doi: 10.1128/JVI.76.24.12877-12889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guang S, Felthauser AM, Mertz JE. Binding of hnRNP L to the pre-mRNA processing enhancer of the herpes simplex virus thymidine kinase gene enhances both polyadenylation and nucleocytoplasmic export of intronless mRNAs. Mol. Cell. Biol. 2005;25:6303–6313. doi: 10.1128/MCB.25.15.6303-6313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toth Z, Stamminger T. The human cytomegalovirus regulatory protein UL69 and its effect on mRNA export. Front Biosci. 2008;13:2939–2949. doi: 10.2741/2899. [DOI] [PubMed] [Google Scholar]
- 26.Wang W, Cui ZQ, Han H, Zhang ZP, Wei HP, Zhou YF, Chen Z, Zhang XE. Imaging and characterizing influenza A virus mRNA transport in living cells. Nucleic Acids Res. 2008;36:4913–4928. doi: 10.1093/nar/gkn475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y, Carmichael GG. The mouse histone H2a gene contains a small element that facilitates cytoplasmic accumulation of intronless gene transcripts and of unspliced HIV-1-related mRNAs. Proc. Natl Acad. Sci. USA. 1997;94:10104–10109. doi: 10.1073/pnas.94.19.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y, Wimler KM, Carmichael GG. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 1999;18:1642–1652. doi: 10.1093/emboj/18.6.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 30.Gilbert W, Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol. Cell. 2004;13:201–212. doi: 10.1016/s1097-2765(04)00030-9. [DOI] [PubMed] [Google Scholar]
- 31.Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, Lian LY, Wilson SA, Allain FH. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J. 2006;25:5126–5137. doi: 10.1038/sj.emboj.7601385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanchette M, Labourier E, Green RE, Brenner SE, Rio DC. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF50 in the nuclear export of intronless mRNAs. Mol. Cell. 2004;14:775–786. doi: 10.1016/j.molcel.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 33.Guang S, Mertz JE. Pre-mRNA processing enhancer (PPE) elements from intronless genes play additional roles in mRNA biogenesis than do ones from intron-containing genes. Nucleic Acids Res. 2005;33:2215–2226. doi: 10.1093/nar/gki506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chua K, Reed R. Human step II splicing factor hSlu7 functions in restructuring the spliceosome between the catalytic steps of splicing. Genes Dev. 1999;13:841–850. doi: 10.1101/gad.13.7.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee CS, Dias AP, Jedrychowski M, Patel AH, Hsu JL, Reed R. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 2008;36:4708–4718. doi: 10.1093/nar/gkn454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villen J, Beausoleil SA, Gygi SP. Evaluation of the utility of neutral-loss-dependent MS3 strategies in large-scale phosphorylation analysis. Proteomics. 2008;8:4444–4452. doi: 10.1002/pmic.200800283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc.Mass Spectr. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 40.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 41.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 43.Fleckner J, Zhang M, Valcarcel J, Green MR. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 1997;11:1864–1872. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- 44.Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol. Cell. 2006;24:943–953. doi: 10.1016/j.molcel.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 45.Conrad NK, Steitz JA. A Kaposi's sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 2005;24:1831–1841. doi: 10.1038/sj.emboj.7600662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Louhichi A, Fourati A, Rebai A. IGD: a resource for intronless genes in the human genome. Gene. 2011;488:35–40. doi: 10.1016/j.gene.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 47.Grzybowska EA. Human intronless genes: functional groups, associated diseases, evolution, and mRNA processing in absence of splicing. Biochem. Biophys. Res. Commun. 2012;424:1–6. doi: 10.1016/j.bbrc.2012.06.092. [DOI] [PubMed] [Google Scholar]
- 48.Pryor A, Tung L, Yang Z, Kapadia F, Chang TH, Johnson LF. Growth-regulated expression and G0-specific turnover of the mRNA that encodes URH49, a mammalian DExH/D box protein that is highly related to the mRNA export protein UAP56. Nucleic Acids Res. 2004;32:1857–1865. doi: 10.1093/nar/gkh347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 50.Luo MJ, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



