Abstract
The transcriptional co-activator BOB.1/OBF.1 was originally identified in B cells and is constitutively expressed throughout B cell development. BOB.1/OBF.1 associates with the transcription factors Oct1 and Oct2, thereby enhancing octamer-dependent transcription. In contrast, in T cells, BOB.1/OBF.1 expression is inducible by treatment of cells with PMA/Ionomycin or by antigen receptor engagement, indicating a marked difference in the regulation of BOB.1/OBF.1 expression in B versus T cells. The molecular mechanisms underlying the differential expression of BOB.1/OBF.1 in T and B cells remain largely unknown. Therefore, the present study focuses on mechanisms controlling the transcriptional regulation of BOB.1/OBF.1 and Oct2 in T cells. We show that both calcineurin- and NF-κB-inhibitors efficiently attenuate the expression of BOB.1/OBF.1 and Oct2 in T cells. In silico analyses of the BOB.1/OBF.1 promoter revealed the presence of previously unappreciated combined NFAT/NF-κB sites. An array of genetic and biochemical analyses illustrates the involvement of the Ca2+/calmodulin-dependent phosphatase calcineurin as well as NFAT and NF-κB transcription factors in the transcriptional regulation of octamer-dependent transcription in T cells. Conclusively, impaired expression of BOB.1/OBF.1 and Oct2 and therefore a hampered octamer-dependent transcription may participate in T cell-mediated immunodeficiency caused by the deletion of NFAT or NF-κB transcription factors.
INTRODUCTION
Regulated gene expression is a complex process, as different signals need to be integrated in a cell-type-specific manner in accordance with the particular developmental stage and activation state. This complexity is achieved by the architecture of a given promoter and/or enhancer and therefore by the integrated action of different transcription factors in conjunction with recruited co-activators or -repressors. These proteins act together on promoter DNA finally leading to the formation of specific transcriptional complexes based on the DNA sequence they bind as well on the activity of each component itself.
The octamer element ATGCAAAT is one of such DNA sequences and plays an important role in mediating promoter activity of a large array of ubiquitous and lymphocyte-specific genes. Octamer-dependent transcription is achieved in first line by transcription factors that belong to the Oct family. The selectivity of Oct factors to octamer sequences and their transcriptional activity can be enhanced by the recruitment of either ubiquitously expressed or cell type-specific co-activators. For instance, the histone H2B promoter activity depends on Oct1 (Pou2f1) and its interaction with the transcriptional co-activator OCA-S, a protein complex containing GAPDH as a key component, whose expression is highly increased during the S phase of the cell cycle (1). In lymphocytes, the transcriptional co-activator BOB.1/OBF.1 (B cell Oct binding factor 1/Oct binding factor 1; Pou2af1) is responsible for the cell type-specific octamer-dependent transcription. BOB.1/OBF.1 is recruited to DNA by the interaction with Pit-1/Oct1,2/Unc-86 domains of the ubiquitously expressed Oct1 or the lymphocyte specific factor Oct2 (Pou2f2) (2–8), the two Oct family members expressed in lymphocytes (9).
However, not all octamer-regulated promoters depend on the presence of BOB.1/OBF.1 (10,11). The ability of Oct1 or Oct2 to recruit BOB.1/OBF.1 to the DNA might be conferred by different octamer sequences that favor or disfavor the ternary complex formation of these proteins at the octamer motif (12). In addition, we and others demonstrated that the presence of BOB.1/OBF.1 enables Oct factors to bind to unfavorable non-consensus octamer motifs (13,14). Together, the lymphocyte-specific regulation of octamer-dependent transcription depends on an appropriate DNA sequence, on the activity of Oct1 and Oct2 transcription factors and on the presence of the transcriptional co-activator BOB.1/OBF.1. Furthermore, the latter is posttranslationally modified by phosphorylation at Ser184, which is required for its constitutively or inducible transcriptional activity in B or T cells, respectively (15).
The importance of octamer-dependent transcription is underlined by the phenotypes of Oct1-, Oct2- and BOB.1/OBF.1-deficient mice. The deletion of the ubiquitously expressed Oct1 protein leads to embryonic lethality (16), and deletion of the lymphocyte specific Oct2 protein causes death of newborn mice shortly after birth (17). Fetal liver cell transfer into immuno-compromised mice revealed that Oct1 is dispensable for B cell development and function (18). In contrast, Oct2-deficient B cells are unable to differentiate into immunoglobulin-secreting cells (17). This phenotype is similar to that observed for BOB.1/OBF.1-deficient mice. Although viable, these mice are unable to form germinal centers on administration of T cell-dependent antigens. Hence, the production of secondary immunoglobulins is severely compromised (19–21). Besides missing germinal centers, BOB.1/OBF.1−/− mice show multiple defects at several stages of B cell development (22–24). Although the relevance of Oct proteins and BOB.1/OBF.1 for B cell development and function cannot be dismissed, these proteins are also important for T cells. Functional octamer motifs could be detected within the promoter regions of the chemokine receptor CCR5 (25) as well as IL2 (26–30) and IL4 (28,31,32) genes. Also, the IFNγ promoter contains an octamer motif that is bound by Oct proteins together with BOB.1/OBF.1. As a consequence, the secretion of IFNγ by BOB.1/OBF.1-deficient TH1 cells is reduced to a level that disabled these mice to efficiently combat a Leishmania major infection (33). Given the importance of the octamer-dependent transcription for B and T cell-development and function, it is, on the one hand, important to search for octamer-dependent target genes and, on the other, to understand the regulatory mechanisms underlying the octamer-dependent transcription itself.
Regulation of transcription is one major mechanism to determine the capacity of a given protein. The promoters of ubiquitously expressed Oct1 gene or the lymphocyte-specific Oct2 gene have not been described until today. In contrast, the BOB.1/OBF.1 promoter was extensively studied to investigate its cis-acting elements controlling its activity in B cells (34–36) where BOB.1/OBF.1 is constitutively expressed at all stages of B cell development (37), albeit at different levels. The highest expression of BOB.1/OBF.1 was found in germinal center B cells. Accordingly, signals important for germinal center formation, like the stimulation with anti-CD40 antibodies plus IL4, are able to increase the expression of BOB.1/OBF.1 in vitro (38,39). In contrast, in T cells BOB.1OBF.1 expression is inducible by treatment of T cells with Phorbol 12-myristate 13-acetate (PMA)/Ionomycin (P/I) or by antigen receptor engagement (15,40), suggesting that different signals and possibly also transcription factors might be responsible for the expression of BOB.1/OBF.1 in B versus T cells. Interestingly, also the expression of the lymphocyte-specific Oct2 protein is upregulated in activated T cells with a kinetic similar to that obtained for BOB.1/OBF.1 (41,42). As the expression and function of Oct proteins together with BOB.1/OBF.1 was also found to be important for the control of TH1 and TH2 immune responses (33), the present study focuses on the regulation of octamer-dependent transcription in T cells. Our analyses revealed the involvement of the Ca2+/calmodulin-dependent phosphatase calcineurin (CN), as Oct2 and BOB.1/OBF.1 expression are efficiently suppressed in the presence of the immunosuppressant cyclosporin A (CsA) or by the siRNA-mediated suppression of CN-A. On T cell receptor (TCR) stimulation, CN turned out to be a key signaling molecule essential for the induction of both Nuclear Factor of Activated T cells (NFAT) and NF-κB transcription factors (43–45). CN dephosphorylates NFAT transcription factors enabling them to translocate into the nucleus where they control the expression of numerous genes essential for T cell activation and differentiation (46). In addition, CN is involved in the regulation of the TCR-induced NF-κB signaling pathway by regulating the complex assembly of Carma1, Bcl10 and Malt1 by dephosphorylating Bcl10. Carma1, Bcl10 and Malt1 complex formation is an essential prerequisite for the activation of the IκB-kinase complex that in turn phosphorylates the inhibitor of NF-κB (IκB) leading to its proteasomal degradation. Consequently, cytosolic NF-κB becomes released and translocates into the nucleus where it regulates, similar to NFAT, the promoter activity of numerous genes crucial for T cell development, differentiation and function (46). Therefore, both signaling pathways were analysed for their capacity to mediate Oct and/or BOB.1/OBF.1 gene expression and were identified as important regulators of octamer-dependent transcription in T cells.
MATERIALS AND METHODS
Cell lines and culture
The Jurkat-4 x Octamer-Luc cell line was generated by electroporation of Jurkat cells with a luciferase reporter construct bearing four copies of the octamer motif cloned earlier (47) together with a plasmid expressing the Puromycin resistant gene (pSV-Puro) and selected by Puromycin. Jurkat-NEMO−/− was described (48). Jurkat cells and derivates (Jurkat-4 × Octamer-Luc, Jurkat-NEMO−/−) and φNX amphotropic retrovirus producer cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Invitrogen), containing 10% fetal calf serum (FCS) (Biochrome), 2 mM of L-glutamine, 100 U/ml of penicillin and 100 µg/ml of streptomycin (Gibco, Invitrogen) and 50 µM of β-mercaptoethanol at 37°C and 5% CO2. A3.01 T cells and Namalwa B cells as well as primary CD4+ cells were cultured in RPMI-1640 medium containing 10% FCS, 2 mM of L-glutamine, penicillin/streptomycin and 50 µM of β-mercaptoethanol at 37°C and 5% CO2.
CD4+ T cell isolation
Primary CD4+ T cells were isolated from lymph nodes of mice either by positive or negative selection using magnetic microbead technology (Miltenyi).
Mice
C57BL/6 wild-type and TNFRI/p65 double-deficient mice (and all their wild-type/heterozygous combinations) were generated and obtained from our breeding facility. The NFATc1/c2 double knockout mice were generated by crossing the Nfatc2−/− mouse (49) with the Nfatc1flx/flx [described in (50)] × Cd4-Cre mouse. Mice were analysed 3–4 or 10–12 weeks after birth, respectively.
Cell stimulation
Cells were stimulated in the presence of the following agents: 100 ng/ml of CsA and 200 ng/ml of FK506 (CN inhibitors), 1, 2 or 4 µM of Bay-117082 (a NF-κB inhibitor) as indicated, 25 ng/ml of PMA, 500 ng/ml of Ionomycin, 200 nM of OH-tamoxifen (OHT) (all obtained from Sigma-Aldrich), 15 or 30 µM of SB203580 (Upstate) as indicated, 4 µg/ml of αCD3 and 0.5 µg/ml of αCD28 (BD). The NEMO-binding peptide and the NFAT inhibitory peptide 11 R-VIVIT were obtained from Calbiochem.
Promoter sequence analyses
In silico search for binding sites in BOB.1/OBF.1 promoters of different species was performed using the MatInspector tool (Matrix Family Library Version 8.3) of the Genomatrix software tool (Genomatrix Company).
Electrophoretic mobility shift assays
Preparation of whole cell extracts for electrophoretic mobility shift assays (EMSA) and the protocol of the EMSA procedure have been described earlier (33). The used oligonucleotides bearing the appropriate transcription factor binding site were annealed and subsequently labeled using 32P-αdCTP in a fill in reaction. Sequences are presented in the Supplementary Table S1.
Promoter cloning
The 1500 and 500 bp BOB.1/OBF.1 promotor constructs were cloned into the pTKL/2 vector containing the HSV-thymidine kinase promoter (−105 to +52) from the pBLCAT2 in front of the firefly luciferase coding region. The HSV-thymidine kinase promoter was excised by a restriction endonuclease digest using HindIII and BglII and replaced by the 1500 bp or 500 bp BOB.1/OBF.1 promoter constructs cloned via genomic polymerase chain reaction (PCR) using the following primers: mBOB.1/OBF.1prom 1500 bp 5′ (HindIII): GCC AGG AAG CTT AGG GGT TGA G; mBOB.1/OBF.1prom 1500 bp 3′ (BglII): GCC TTT TCT CTT TGA AGC AGA GAT CTT GGC TTC TTT ACT. For amplification of the 500 bp promoter fragment, the same 3′ primer as for the 1500 bp fragment was used in combination with the following primer: mBOB.1/OBF.1prom 500 bp 5′ (HindIII): GAC CAA TGG TAA GCT TAG TCC TGC. Cloning of the BOB.1/OBF.1 promoter mBOB.1/OBF.1prom Δ500 bp construct was achieved using the following combination of primers for genomic PCR: mBOB.1/OBF.1prom 1500 bp 5′ (HindIII) as indicated earlier in the text, together with mBOB.1/OBF.1prom Δ500 bp 3′ (BglII): CCA TTT ACA GGA CAG ATC TTA CCA TTG GTC.
The BOB.1/OBF.1 promoter mBOB.1/OBF.1prom Δ500 bp + TATA construct was generated by cloning of an insert that was amplified by genomic PCR using the following primers: mBOB.1/OBF.1prom 1500 bp 5′ (HindIII): GCC AGG AAG CTT AGG GGT TGA G and mBOB.1/OBF.1prom Δ500 bp 3′ (HindIII): GCA GGA CTA AGC TTA CCA TTG GTC, into the HindIII site of the pTATA vector harboring the TATA box of the HSV-thymidine kinase promoter from pBLCAT2 (−38 to +52) in front of the firefly luciferase coding region.
In vitro mutagenesis
Mutations of the predicted NFAT/NF-κB sites within the BOB.1/OBF.1 promoter were generated using the QuickChange™ Mutagenesis kit (Promega). Primers were ordered from Biomers, and sequences are provided in Supplementary Table S4.
Transfection of cells
Transfections of Jurkat T and Namalwa B cells were performed by electroporation (Bio-Rad) with 450 V and 250 µF in phosphate buffered saline. The expression vector for the constitutive active version of CREB (c2CREB) is a kind gift of G. Tiel (51). Expression vectors for NFATc1 and RelA/p65 have been described (52,53). The pRL-CMV plasmid (Renilla Luciferase control reporter vector; Promega) was cotransfected in all experiments and used for normalization of different transfection efficiencies in the individual experiments. For suppression of CN A isoforms by siRNA, Jurkat T cells were transfected with 75 nM of each SMARTpool siRNA (CnBa: PPP3CA # L008300-0005; CnBb: PPP3CB # L009704-0005) or with the appropriate concentration of control siRNA (OnTARGETplus # D001810-01-05) using the Nucleofection Kit V (Amaxa/Lonza). The cells were subsequently incubated for 72 h before analysis. Used SMARTpool siRNA were obtained from Dharmacon.
Retroviral infection of cells
For virus production, the retroviral vectors expressing NF-ATc1/αA-ER, NF-ATc1/αC-ER (54), IKK2-EE, IKK2-KD (55) or the respective empty vectors were transfected into the amphotropic φNX retrovirus producer cells by use of the calcium phosphate method. Supernatant containing the retrovirus was collected 24 h and 48 h after transfection and used to infect A3.01 T cells in the presence of 8 µg/ml of polybrene. At 2 consecutive days, spin infection was performed at 2700 rpm (1300g) and 37°C for 120 min. Positive cells were selected with Zeocin until all cells were nearly 100% positive for green fluorescent protein.
Western blots
For western blot analysis, 5–10 µg of total protein extracts were separated on 12.5% polyacrylamid gels and transferred onto nitrocellulose membranes (Schleicher&Schuell). Membranes were blocked (TBS, 0.1% Tween, 5% milk); stained with anti-BOB.1/OBF.1 (Sigma), Oct2, ERK2, IKK2 (Santa Cruz Biotechnology) and NFATc1 (Alexis) antibodies followed by incubation with HRP-coupled secondary antibodies (Pierce); and visualized by enhanced chemiluminescence (Pierce).
Reverse transcriptase-PCR
Total RNA was isolated using Trizol reagent (Qiagen) and reverse transcribed using moloney murine leukemia virus reverse transcriptase (Roche). Quantitative PCR were performed using qPCR-SYBR-Green (Roche). Primers ordered from Biomers were designed using ‘Universal Probe Library Assay Design Center’ Software (Roche). Primer sequences are given in Supplementary Table S2. Quantification of gene regulation was performed by the ΔΔCp method using RPL13 as house-keeping gene.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) experiments were performed using the ChIP-IT® Express Chromatin Immunoprecipitation Kit and the Re-ChIP-IT® magnetic chromatin re-immunoprecipitation kit from Active Motif, according to the manufacturer’s protocol with slight modifications: after enzymatic digestion for 10 min, the chromatin was sheared by two cycles of sonication (10 pulses each cycle) in the same buffer. The chromatin was precleared for 2 h with protein G microbeads (Invitrogen) and then incubated with rabbit polyclonal anti-p65 antibody sc-372 (2 µg/ml; Santa Cruz Biotechnology), mouse monoclonal anti-NFATc1 antibody 7A6 (4 µg/ml; Alexis), mouse IgG (Dianova) or normal rabbit serum (Pierce) at the appropriate concentrations for 4 h followed by incubation with protein G microbeads for 1 h. The amount of precipitated DNA was evaluated by quantitative PCR using the Roche Light Cycler LC480. The used primers and primer positions are depicted in Figure 6 and presented in Supplementary Table S3. The relative amount of precipitated DNA was calculated using the following formula: E(crossing point 1/10 total input − crossing point sample) and is depicted as amount of precipitated genomic DNA relative to that precipitated by control antibodies (mouse IgG or normal rabbit serum). E = efficiency of the PCR determined by serial dilutions of total input.
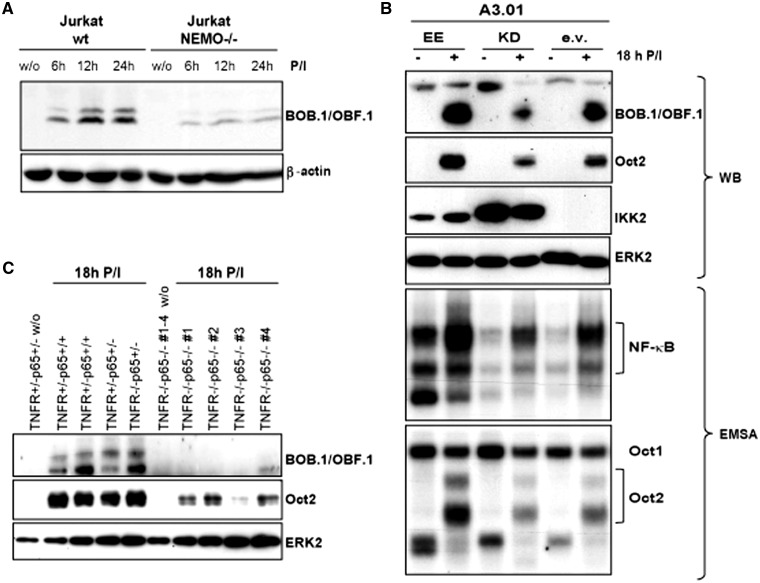
Figure 6.
NF-κB activity influences BOB.1/OBF.1 and Oct2 expression levels in T cells. Wild-type (wt) or NEMO-deficient (NEMO−/−) Jurkat T cells were stimulated for the time indicated with P/I or left untreated. The expression of BOB.1/OBF.1 was analysed by western blotting. (B) Human A3.01 T cells were retrovirally infected using vectors expressing either a constitutive active version of IKK2 (EE), a kinase dead version (KD) or the empty vector (ev). After selection, cells were stimulated with P/I for 18 h and subsequently analysed for the protein expression levels of BOB.1/OBF.1 and Oct2 by western blotting (WB). The detection of IKK or ERK2 expression levels served as internal control or loading control experiments, respectively. Additionally, the same extracts were used in EMSA to monitor the NF-κB and Oct2 binding activity to DNA using labeled consensus sites. (C) Primary CD4+ T cells of mice of the indicated genotype were either left untreated or stimulated with P/I for 18 h. Afterwards, the protein expression levels of BOB.1/OBF.1 and Oct2 were analysed in immunoblots.
RESULTS
NFAT and NF-κB inhibitors restrain the inducible expression of BOB.1/OBF.1 and Oct2 in T cells
To elicit the signaling pathways controlling the octamer-dependent transcription in T cells, human Jurkat T cells were stably transfected with a luciferase reporter construct harboring four consecutive sequences bearing the octamer binding motif ATGCAAAT in front of a luciferase gene. As expected, the octamer activity was strongly upregulated on treatment with P/I but remained unaffected by stimulation with either P or I alone, suggesting the requirement of a combined calcium influx and protein kinase C activation for octamer-dependent transcription in T cells (Figure 1A). Several transcription factors are known to depend on such a combinatorial calcium and protein kinase C signaling, including NF-κB and NFAT. To further specify the signaling pathways controlling octamer-dependent transcription, we used a panel of specific pharmacological inhibitors. Indeed, complete suppression of the inducible octamer activity was seen when the cells were pre-treated with the CN inhibitor CsA. Also the NF-κB inhibitor Bay11-7082 and the p38 inhibitor SB203580 interfered with the inducible octamer activity, although less efficiently. These data suggest that Ca2+/CN, NF-κB and mitogen activated protein kinase-dependent signaling pathways are involved in the regulation of octamer-dependent transcriptional activity (Figure 1A).
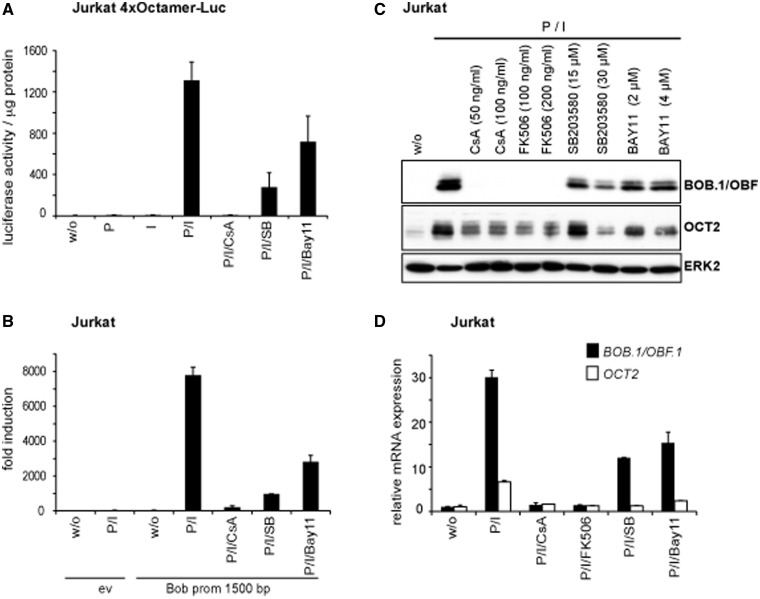
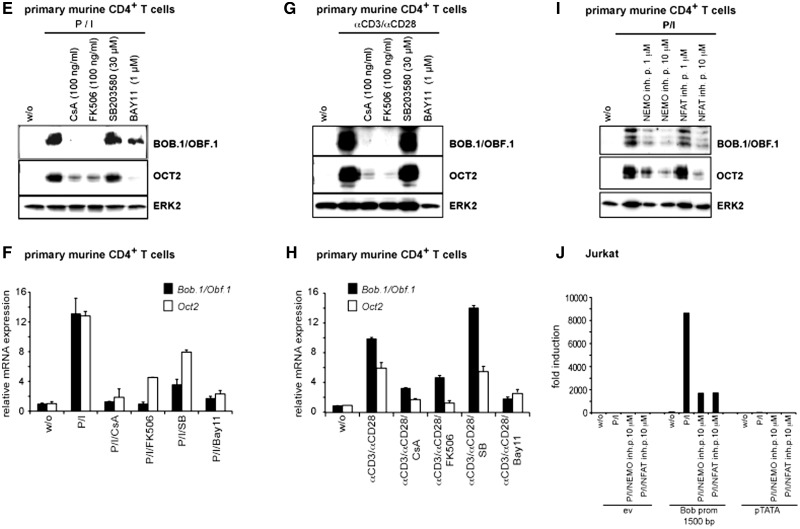
Figure 1.
Inhibition of octamer function by CsA, p38 and NF-κB inhibitors in T cells. (A) A luciferase reporter construct bearing four copies of the consensus octamer element was stabily transfected into Jurkat T cells. Cells were either left untreated or stimulated with P, I alone or together (P/I), or were pretreated with CsA (100 ng/ml), SB203580 (20 µM) or Bay11-7082 (2 µM) before the stimulation with P/I. Mean values ± s.d. of five experiments are shown. (B and J) The 1500 bp BOB.1/OBF.1 promoter construct cloned in front of a luciferase gene was transfected together with the appropriate empty vector (ev) into Jurkat T cells that were subsequently treated as described in (A) or as indicated. The determined relative luciferase activity of the empty vector without stimulation was set one, and the fold increase of the promoter activity was calculated. Mean values ± s.d. of three (in B) and two (in J) independent experiments are shown. The analyses of the pTATA promoter construct served as internal control. (C–I) Jurkat or isolated primary CD4+ T cells were treated as indicated. After 18 h of stimulation, cells were harvested. One part was used for protein extraction and immunoblotting (C, E, G, I), and the other half was used for RNA isolation and quantitative RT-PCR (D, F, H) for the detection of BOB.1/OBF.1 and Oct2 mRNA expression levels that were determined relative to the expression of the housekeeping gene RPL13. Analyses of HPRT mRNA expression also normalized to RPL13 expression served as internal control (data not shown). The analyses of ERK2 expression (C, E, G, I) served as loading control.
We next wondered whether the same pharmacological inhibitors could influence the activity of the known BOB.1/OBF.1 promoter. Therefore, a reporter construct harboring the previously described 1500 bp BOB.1/OBF.1 promoter (34) was transiently transfected into Jurkat T cells, which were either left untreated or pre-treated with inhibitors for CN, NF-κB or p38 for 30 min before the stimulation with P/I. These experiments revealed that the BOB.1/OBF.1 promoter is sensitive to all of the analysed inducers and inhibitors, similar to the octamer-dependent reporter (Figure 1B). To explore whether protein expression of BOB.1/OBF.1 mirrors its promoter activity, Jurkat T cells and primary CD4+ cells isolated from lymph nodes of C57BL/6 wild-type mice were either left untreated or pre-treated for 30 min with the inhibitors used in the previous experiments and subsequently stimulated with P/I. Interestingly, we found a striking coregulation of Oct2 and BOB.1/OBF.1 in Jurkat (Figure 1C and D) and primary CD4+ T cells (Figure 1E and F). Thus, the same inducers and inhibitors control the expression of Oct2 and BOB.1/OBF.1 at protein and mRNA levels. However, although the BOB.1/OBF.1 expression was completely abolished by treatment of Jurkat or CD4+ T cells with CsA even at low concentrations (50–100 ng/ml), low levels of Oct2 expression were still detectable (Figure 1C and E). In contrast, the NF-κB inhibitor Bay11-7082 had only a moderate influence on BOB.1/OBF.1 and Oct2 expression in Jurkat T cells even at high concentrations (2–4 µM), whereas in primary CD4+ T cells, the inhibition of the NF-κB pathway led to an almost complete abrogation of Oct2 expression and to a marked reduction in BOB.1/OBF.1 expression at low concentrations (1 µM) of the inhibitor. The insufficient block of BOB.1/OBF.1 and Oct2 expression by Bay11-7082 might be owing to a residual NF-κB activity, as seen in EMSA experiments (Supplementary Figure S1). However, higher concentrations of the inhibitor Bay11-7082 were toxic for primary T cells. Also, pre-treatment of primary CD4+ T cells with CN inhibitors CsA or FK506 or by the NF-κB inhibitor Bay11-7082 completely inhibited the strong BOB.1/OBF.1 and Oct2 induction seen after the more physiological stimulation with anti-CD3 and anti-CD28 antibodies (Figure 1G and H). The p38 inhibitor SB-203580 had a moderate effect on BOB.1/OBF.1 and Oct2 expression only at higher concentrations (Figure 1C), suggesting a rather minor and/or indirect effect of p38 on the regulation of octamer-dependent transcription in T cells. Interestingly, EMSA studies (Supplementary Figure S1) revealed that the NF-κB inhibitor Bay11-7082 inhibits additionally NFAT binding to DNA and, vice versa, the CN inhibitors CsA and FK506 interfere with NFAT binding but also with the NF-κB signaling pathway, as it was described recently (44,45). Therefore, the observed effect of CsA and FK506 on BOB.1/OBF.1 expression is not only mediated by inhibiting NFAT activity, as these compounds also affect the activity of NF-κB, but rather by the combined activity of NFAT and NF-κB. However, treatment of murine primary CD4+ T cells or transfected Jurkat cells with specific NFAT and NF-κB signaling pathway inhibitors, like the NFAT inhibitory peptide 11 R-VIVIT and the NF-κB essential modulator (NEMO)-binding peptide, respectively, clearly demonstrates that both pathways are involved in the regulation of BOB.1/OBF.1 expression via the regulation of BOB.1/OBF.1 promoter activity (Figure 1I and J).
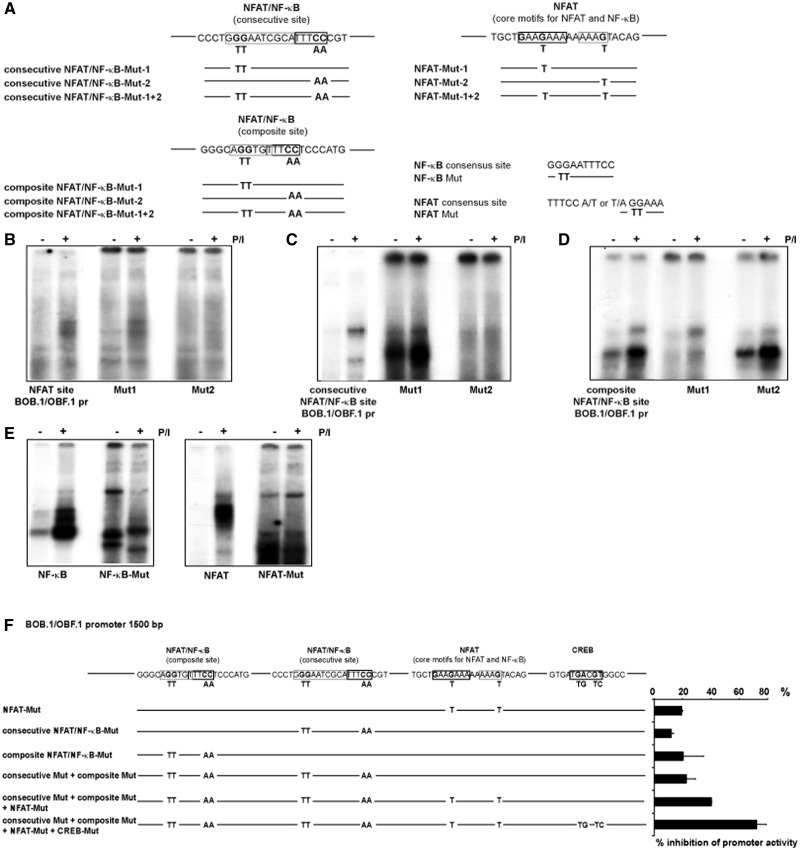
The BOB.1/OBF.1 promoter contains several binding sites for NFAT and NF-κB
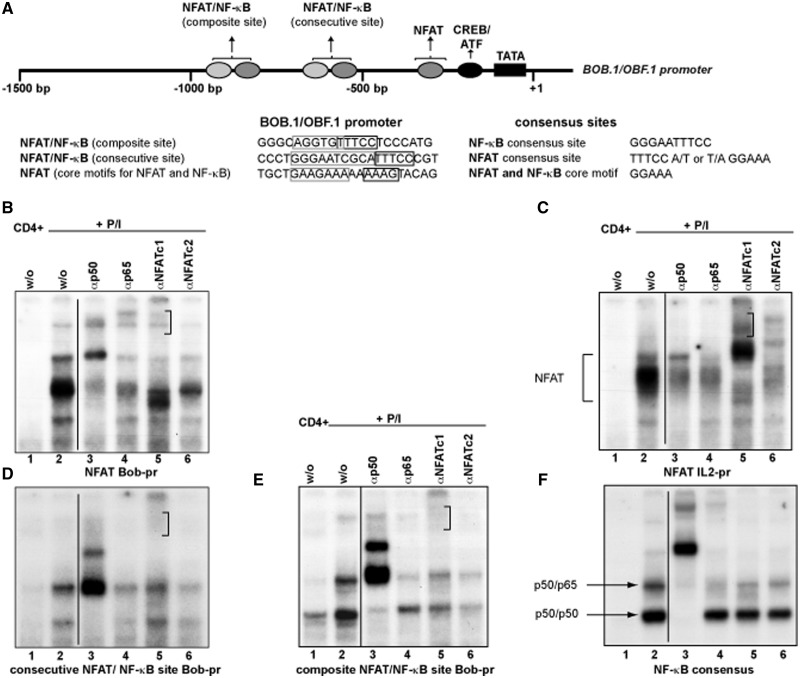
Previously, the BOB.1/OBF.1 promoter was analysed to identify regulatory elements responsible for its activity in B cells (34–36). In those studies, a functional cAMP responsive element-binding protein (CREB)/Activating Transcription Factor (ATF) site could be identified, which is crucial for the B lymphocyte specific activity of the promoter. In addition, a sequence related to a NFAT binding site was described, however without any functional relevance for the B cell-specific BOB.1/OBF.1 promoter activity. In search for possible additional transcription factor binding sites using the Genomatrix Software, we identified two combined NFAT/NF-κB binding sites within the BOB.1/OBF.1 promoter. Both newly identified sites are conserved between rat, mouse and man, suggesting a possible functional relevance of these regulatory elements. In contrast to B cells, where a function of the predicted NFAT binding site has not been revealed (34), an inducible complex formation could be detected after P/I treatment of primary CD4+ T cells. This complex resembles that formed at the NFAT site of the IL2 promoter used in control experiments (Figure 2B, lane 2; and C, lane 2). In addition, an inducible complex formation could also be observed using the newly identified combined composite and consecutive NFAT/NF-κB sites of the BOB.1/OBF.1 promoter as probes (Figure 2D and E, lane 2). In supershift experiments, antibodies against NF-κB p50 were able to compete against the observed inducible complexes formed at the predicted NFAT/NF-κB binding sites within the BOB.1/OBF.1 promoter (Figure 2D and E, lane 3) in a similar way like anti-p50 antibodies prevent p50 binding to a consensus NF-κB site (Figure 2F, lane 3). Interestingly, anti-p50 antibodies are also able to interfere with the inducible complex formed at the predicted NFAT site of the BOB.1/OBF.1 promoter (Figure 2B, lane 3) and with the NFAT binding observed at the IL2 promoter site (Figure 2C, lane 3). Similarly, anti-p65 antibodies interfere with binding to the consecutive and composite NFAT/NF-κB sites, which is comparable with the competition seen at the consensus NF-κB site (Figure 2D, E and F, lane 4). Additionally, anti-p65 antibodies also interfere with the complex formation on the NFAT site of the BOB.1/OBF.1 and IL2 promoters (Figure 2B and C, lane 4). The use of NFATc1 antibodies reduces the binding to the NFAT and NFAT/NF-κB sites of the BOB.1/OBF.1 promoter and generates supershifted complexes (marked by clamp in Figure 2B, D and E, lane 5). Also, NFATc2 antibodies prevent complex formation to these specific sites (Figure 2B, D and E, lane 6). Control experiments using IgG antibodies or normal rabbit serum revealed the specificity of antibody binding (Supplementary Figure S2). Moreover, unlabeled oligonucleotides bearing the consensus NFAT or NF-κB binding site could efficiently compete against labeled NFAT or NFAT/NF-κB sites identified in the BOB.1/OBF.1 promoter (Supplementary Figure S2). Together, our data suggest that NFAT and NF-κB family members are able to bind to all of these newly identified potential cis-acting elements of the BOB.1/OBF.1 promoter in vitro.
Figure 2.
Inducible complex formation on potential NFAT and NF-κB sites within the BOB.1/OBF.1 promoter. (A) Schematic representation of the murine BOB.1/OBF.1 promoter. Indicated are the positions of the TATA box, of the CREB/ATF binding site and the positions/sequences of the potential NFAT and NF-κB sites. (B–F) Primary murine CD4+ T cells were either left untreated or stimulated for 18 h with P/I. Whole protein extracts were prepared and analysed in EMSA. In supershift experiments, the indicated antibodies were used.
Newly identified NFAT/NF-κB binding sites contribute to BOB.1/OBF.1 promoter activity
To demonstrate the importance of the potential NFAT or NFAT/NF-κB binding site for the BOB.1/OBF.1 promoter activity, mutation analyses were performed. In these experiments, specific point mutations were introduced to prevent either NFAT or NF-κB binding to these sites (Figure 3). Point mutation (G→T; Figure 3A) within the previously identified NFAT motif within the BOB.1/OBF.1 promoter did not lead to alterations in complex binding to this site. However, introduction of a point mutation (G→T) within the second core motif for NFAT and NF-κB clearly prevents inducible by P/I complex formation, indicating the importance of this residue within this potential transcription factor binding site (Figure 3B). Within the consecutive NFAT/NF-κB binding site of the BOB.1/OBF.1 promoter, both mutations, preventing NF-κB (GG→TT) and NFAT binding (CC→AA), abolish inducible complex formation on treatment of T cells with P/I (Figure 3C). Therefore, both sites possibly contribute to the inducible BOB.1/OBF.1 promoter activity in T cells. In contrast, although mutations of critical residues within the NF-κB motif (GG→TT) of the composite NFAT/NF-κB site within the BOB.1/OBF.1 promoter lead to a clear reduction of complex binding on P/I treatment of CD4+ T cells, mutations within the potential NFAT binding site (CC→TT) slightly enhance the ability of complex binding to this site (Figure 3D). For comparison, the mutated consensus NFAT and NF-κB sites were also analysed (Figure 3E).
Figure 3.
Mutation analyses of potential NFAT and NF-κB sites within the BOB.1/OBF.1 promoter. (A) Potential NFAT and NF-κB sites within the BOB.1/OBF.1 promoter as well as consensus NFAT and NF-κB site were mutated as indicated and used (B–E) in EMSA experiments together with whole cell extracts of isolated murine CD4+ T cells that were either left untreated or were stimulated for 18 h with P/I as indicated. (F) Potential NFAT and NF-κB sites within the BOB.1/OBF.1 promoter that was cloned in front of a luciferase gene were mutated as indicated. Mutated constructs were transfected into Jurkat T cell that were subsequently stimulated with P/I over night. The percentage inhibition of promoter activity was calculated relative to the activity of the wild-type 1500 bp BOB.1/OBF.1 promoter was set as 100%.
Additionally, the introduction of mutations into the 1500 bp BOB.1/OBF.1 promoter construct that was cloned in front of a luciferase gene and transfected into Jurkat T cells (Figure 3F) revealed that all three newly identified NFAT and NFAT/NF-κB site are important for full BOB.1/OBF.1 promoter activity, as mutation of each reduces BOB.1/OBF.1 promoter activity by ∼15–20%. Mutation of all three NFAT/NF-κB sites led to a reduction of BOB.1/OBF.1 promoter activity by 40%. Interestingly, additional mutation of the previously identified CREB/ATF site, important for BOB.1/OBF.1 promoter activity in B cells, almost completely abrogates BOB.1/OBF.1 promoter activity in T cells.
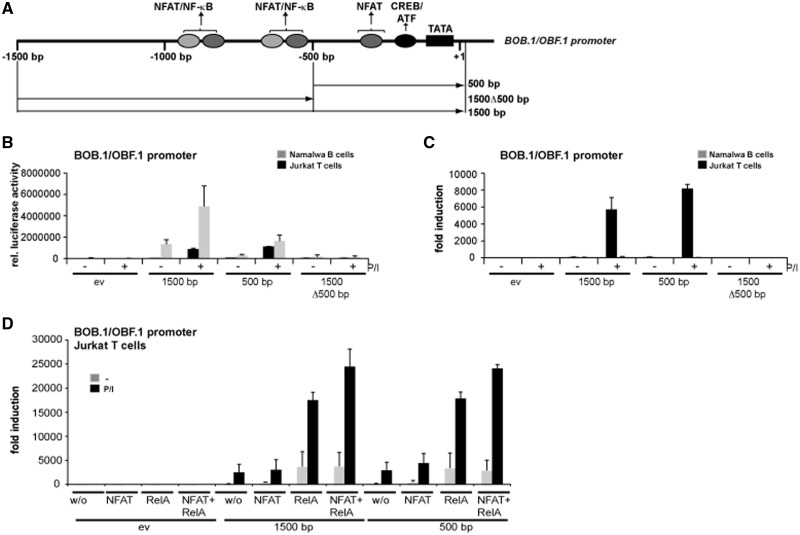
A 500 bp sequence is necessary and sufficient for the highest inducible BOB.1/OBF.1 promoter activity in Jurkat T cells
For the full activity of the BOB.1/OBF.1 promoter in B cells, a sequence spanning 1500 bp was identified (34). To define regions important for the inducible activity in T cells, additionally a shorter promoter construct was generated encompassing ∼500 bp upstream of the start site of transcription of the BOB.1/OBF.1 gene (Figure 4A). To compare the behavior of these constructs with respect to the basal and inducible BOB.1/OBF.1 promoter activity in B and T cells, the constructs were transfected into Namalwa B or Jurkat T cells. In accordance with previous findings (34), the 1500 bp promoter showed the highest basal activity in Namalwa B cells that could be further increased (3-fold) by treatment of cells with P/I (Figure 4B and C). In B cells, the activity of 500 bp construct was ∼3 times lower when compared with the 1500 bp fragment with respect to basal and inducible promoter activity. As expected, in T cells, no basal BOB.1/OBF.1 promoter activity could be observed. However, it was strongly induced when cells were treated with P/I (Figure 4C). Notably, in T cells, the 500 bp promoter construct showed the highest inducibility after P/I treatment (Figure 4B and C). The importance of the 500 bp sequence of the BOB.1/OBF.1 promoter was further underscored by deleting this promoter element, which led to the complete loss of basal and inducible BOB.1/OBF.1 promoter activity in B and T cells (Figure 4B and C). This loss of activity was not caused by the deletion of regulatory elements necessary for the recruitment of the basal transcriptional complex, as the fusion of the same promoter construct to a TATA box element of the thymidine kinase promoter did also not lead to enhanced BOB.1/OBF.1 promoter activity in T cells (Supplementary Figure S3).
Figure 4.
The BOB.1/OBF.1 promoter spanning 500 bp is necessary and sufficient for full inducible activity in T cells. (A) Schematic representation of the BOB.1/OBF.1 promoter. The positions of the analysed cis-elements and of the promoter fragments used in reporter assays are indicated. (B and C) Namalwa B and Jurkat T cells were transiently transfected with the empty vector (ev) or with reporter constructs bearing either the 1500 bp or the 500 bp BOB.1/OBF.1 promoter construct or a deletion mutant of the longer version were the first 500 bp are missing (1500Δ500 bp). The relative luciferase activity (B) or the fold induction (C) of different promoter fragments was determined without or after stimulation of cells with P/I, were in the second case the fold induction was determined relative to the luciferase activity of the empty vector without stimulation that was set to one. (D) Jurkat T cells were transiently transfected with either the empty vector or the 1500 or 500 bp BOB.1/OBF.1 promoter constructs as indicated, either alone or together with expression vectors for NFATc1, RelA or combination of both. Transfected cells were either left untreated or stimulated subsequently with P/I. Next day, the cells were harvested to determine the relative luciferase activity. The relative luciferase activity resulting from the transfection of the empty vector without induction was set to one. The fold induction relative to the empty vector was calculated and is depicted. (B–D) Shown are the mean values ± s.d. of three independently performed experiments.
To investigate the contribution of NFAT and NF-κB to the BOB.1/OBF.1 promoter activity in T cells, Jurkat cells were transfected with the 500 bp or 1500 bp BOB.1/OBF.1 promoter constructs, together with expression vectors for NFATc1 or RelA/p65. Although overexpression of NFATc1 had only a moderate effect, overexpression of RelA/p65 already significantly enhanced the inducible activities of the 1500 and 500 bp promoters. Coexpression of both factors together leads to a further increase of BOB.1/OBF.1 promoter activity, although this effect was modest (Figure 4D). Again, the 500 bp promoter construct was sufficient to achieve full BOB.1/OBF.1 promoter activity induced by NFAT/NF-κB overexpression.
In B cells, the CREB/ATF site was found to be important for BOB.1/OBF.1 promoter activity. To investigate whether this site is also important for the inducible expression of BOB.1/OBF.1 in T cells, the 1500 bp promoter construct was transfected into Jurkat T cells, together with expression vectors coding for NFATc1, RelA/p65 or a constitutive active version of CREB either alone or in different combinations of these vectors. Also in T cells, the CREB/ATF site seems to be of relevance, as overexpression of a constitutive active CREB protein leads to a significant increase of BOB.1/OBF.1 promoter activity that was further enhanced by overexpression of NFAT and NF-κB (Supplementary Figure S4). These data together with our data obtained from EMSA and transfection experiments in that the CREB site was inactivated indicate that this CREB site is also of considerable importance for BOB.1/OBF.1 promoter activity in T cells.
Modulation of NFAT activity interferes with the expression levels of BOB.1/OBF.1 and Oct2
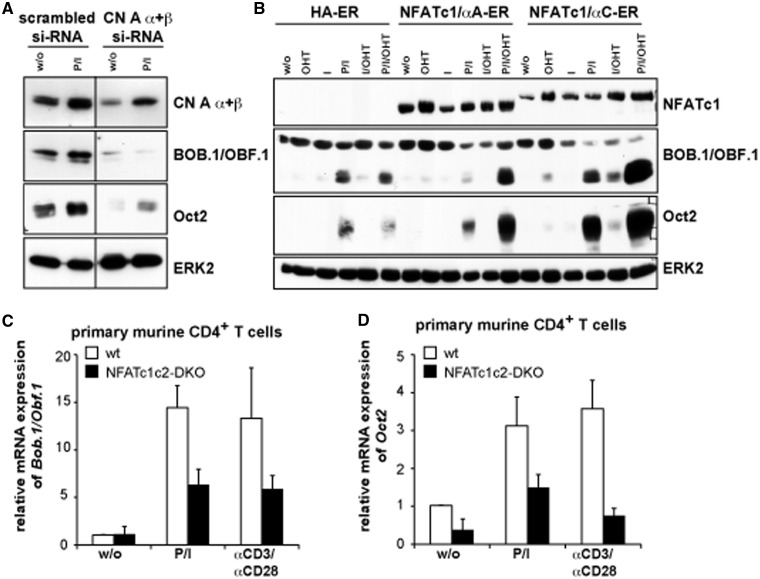
One major regulator of NFAT activity is the Ca2+/calmodulin-dependent protein phosphatase CN that dephosphorylates cytosolic NFAT proteins. NFAT, in turn, translocates into the nucleus where it binds to DNA and regulates gene expression, a process that can be efficiently inhibited by CsA. Down modulation of CN by Amaxa transfection of siRNA directed against the isoforms α and β of the catalytic subunit CN A revealed a clear dependence of BOB.1/OBF.1 and Oct2 expression on CN activity (Figure 5A). Obviously, Amaxa transfection of the indicated siRNA itself pre-activated Jurkat T cells, as BOB.1/OBF.1 and Oct2 expression could be detected even in the unstimulated state. However, the expression of both proteins could be further enhanced by P/I treatment. Although the downregulation of CN expression was incomplete, a clear inhibition of inducible BOB.1/OBF.1 and Oct2 expression could be observed, indicating the importance of CN for BOB.1/OBF.1 and Oct2 expression in T cells.
Figure 5.
CN and NFAT factors control the inducible BOB.1/OBF.1 and Oct2 expression in T cells. (A) Jurkat T cells were nucleoporated either with scrambled siRNA or siRNA pools directed against the α and β isoforms of CN A subunit (CN A α + β). Subsequently, cells were stimulated for 8 h with P/I. Whole cell extracts were prepared and used for protein expression analyses of CN A α + β, BOB.1/OBF.1 or Oct2 by immunoblotting. The detection of ERK2 expression served as loading control. (B) Human A3.01 T cells were infected retrovirally with vectors expressing either the NF1ATc1/αA-ER or NFATc1/αC-ER or just the empty control vector (HA-ER). Afterwards, cells were either left untreated or treated with OHT, I, P/I or with combinations of these inducers as indicated. After 18 h of stimulation, cells were harvested and subjected for immunoblotting, using primary antibodies against NFATc1, BOB.1/OBF.1 and Oct2. The detection of ERK2 expression levels served as loading control. (C and D) Primary CD4+ T cells were isolated from wild-type (wt) or mutant mice in which the expression of NFATc1 and NFATc2 was simultaneously deleted in T cells specifically (NFATc1c2-double knock out). After stimulation of cells with P/I or with αCD3 + αCD28 antibodies for 18 h cells were harvested to analyse BOB.1/OBF.1 and Oct2 expression at mRNA levels by quantitative RT-PCR. The relative mRNA expression levels are expressed relative to that determined in unstimulated wild-type cells that were set as one. The experiments were performed twice in triplicates and using different dilutions of the cDNA reveling the same result. One representative experiment is depicted as mean values ± s.d.
In peripheral T cells, mainly two members of the NFAT family are expressed, NFATc1 and NFATc2. On T cell activation, the NFATc1 isoform NFATc1/αA is predominantly expressed (56). To study the influence of NFAT on BOB.1/OBF.1 or Oct2 expression, human A3.01 T cells were retrovirally infected using vectors expressing either the NF1ATc1/αA or NFATc1/αC isoform that were fused to a part of the modified hormone binding domain of the estrogen receptor α (ERα) (54), which is controlled by OH-Tamoxifen (OHT). Although the OHT treatment of cells infected with the empty vector left BOB.1/OBF.1 and Oct2 expression unaffected, a significant increase in their protein expression level could be observed when NFATc1/αA or NFATc1/αC became functional active in the presence of OHT (Figure 5B).To test whether the absence of endogenous NFAT protein expression would influence BOB.1/OBF.1 and Oct2 expression, CD4+ T cells were isolated from Nfatc2−/−xNfatc1flx/flxxCD4-Cre mice in which all T cells are devoid of the expression of NFATc1 and NFATc2. After stimulation of purified CD4+ T cells with either P/I or αCD3+αCD28 for 18 h, the mRNA expression of BOB.1/OBF.1 and Oct2 was analysed by quantitative PCR. The mRNA levels of BOB.1/OBF.1 and Oct2 were sensitive to the combined ablation of NFATc1 and NFATc2 protein expression (Figure 5C and D), indicating the importance of these factors for octamer-dependent transcription in T cells.
Modulation of NF-κB activity affects the expression levels of BOB.1/OBF.1 and Oct2
Next, we investigated the influence of the NF-κB activity on BOB.1/OBF.1 or Oct2 expression. First, NEMO-deficient or wild-type Jurkat cells were stimulated with P/I. Western blot analysis revealed a clear dependence of BOB.1/OBF.1 expression on the presence of NEMO in T cells (Figure 6A). NEMO is essential for the canonical NF-κB signaling, as its deficiency leads to a specific block of IKK (IκB kinase) activity (48). Therefore, these findings clearly indicate a contribution of NF-κB to the regulation of BOB.1/OBF.1 gene transcription.
In another approach, human A3.01 T cells were retrovirally transduced using vectors expressing either a constitutive active version (EE) or a kinase dead mutant (KD) of IKK2 (55) (Figure 6B). In EMSA, the modulation of NF-κB activity was monitored. Overexpression of a kinase dead version of IKK2 led to a clear reduction in the inducible NF-κB binding activity, whereas overexpressing a constitutive active IKK2 led to an enhanced basal and inducible NF-κB activity. In EMSA, a clear correlation between NF-κB and Oct2 binding activity to the appropriate consensus DNA binding sites could be observed. Again, western blot analyses revealed a clear dependence of the expression level of BOB.1/OBF.1 and Oct2 on the level of NF-κB activity. To analyse the effect of defective NF-κB activity on BOB.1/OBF.1 and Oct2 expression in vivo, we have used RelA/p65-deficient mice that were bred into the TNFR type 1 (TNFRI; Tumor Necrosis Factor Receptor I)-deficient background to overcome the TNF-α-induced liver toxicity and embryonic lethality (57) observed in RelA/p65−/− mice (58). CD4+ T cells were isolated from lymph nodes of mice deficient for TNFRI and RelA/p65 (double knock out) or from mice that were wild-type or heterozygous for one or the other or both genes and stimulated with P/I for 18 h. When the expression of RelA/p65 was abolished, the expression of BOB.1/OBF.1 was almost completely restrained, whereas Oct2 protein is still detectable, although the level of expression was severely reduced (Figure 6C). Together, these data indicate an essential role of NF-κB signaling for the induction of Oct2 and BOB.1/OBF.1 expression in T cells.
To address the question whether the loss or gain of NF-κB or NFAT transcription factors leads to changes in expression and/or binding activity of the other factor, we have used the advantage of CD4+ T cells obtained from mice bearing genetic mutations either for NFATc1/NFATc2 or for TNFRI/p65. Additionally, we overexpressed NFATc1A and RelA/p65 in Jurkat T cells (Supplementary Figure S5). Interestingly, these experiments revealed that the deletion or overexpression of NFAT does not significantly lead to changes neither in NF-κB p65 expression as seen in western blot analyses nor in NF-κB binding activity analysed in EMSA experiments using a consensus NF-κB binding site (Supplementary Figure S5A and C). However, deletion of RelA/p65 in primary CD4+ T cells leads to a clear reduction of NFATc1 expression and consequently to a considerable reduction in NFAT binding activity to the DNA motif of the IL2 promoter. Vice versa, overexpression of RelA/p65 in Jurkat cells clearly enhances NFAT binding activity (Supplementary Figure S5B and C). These data indicate that the observed effect on BOB.1/OBF.1 and Oct2 expression in TNFRI/p65 double deficient murine CD4+ T cells and in human Jurkat or A3.01 T cells expressing various mutants interfering with NF-κB signaling (Figure 6) could be mediated, at least in part, also by changes in NFAT activity.
Analyses of contribution of NFAT and NF-κB to the BOB.1/OBF.1 promoter activity in vivo
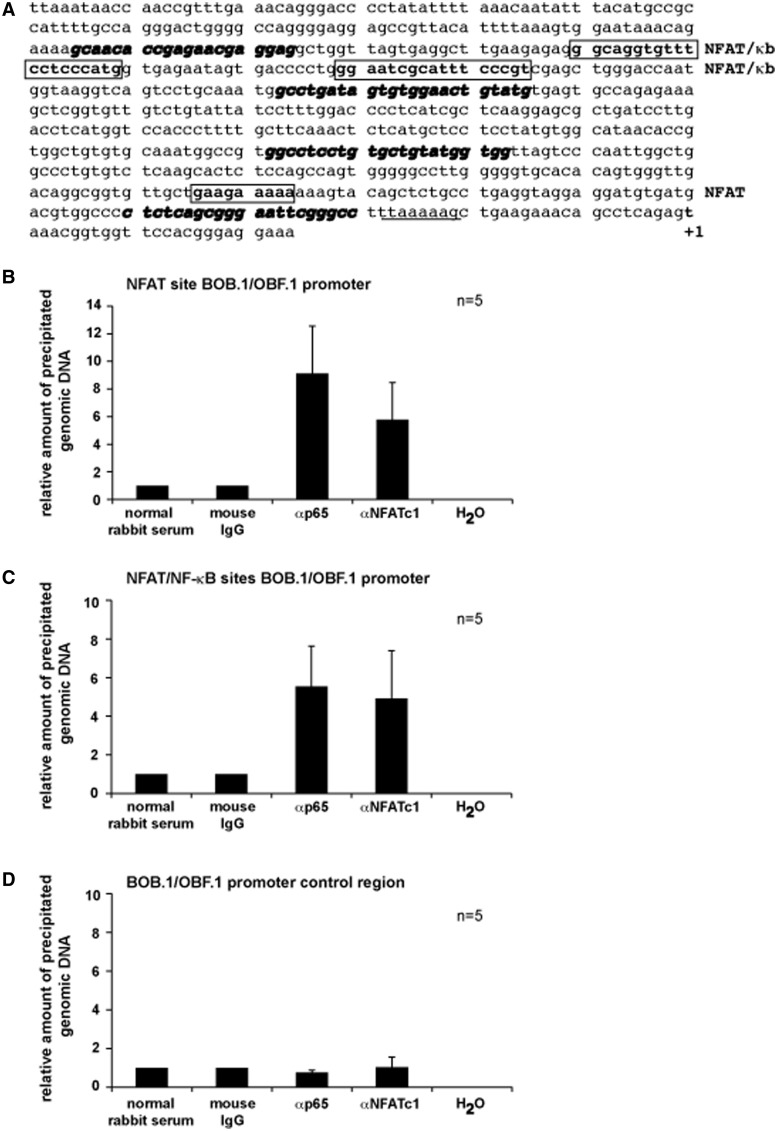
To clarify whether the observed NFAT and NF-κB effects on BOB.1/OBF.1 and Oct2 expression are direct or rather indirect, primary CD4+ T cells were used in ChIP experiments in which a fragment encompassing 189 bp from the first 500 bp of the BOB.1/OBF.1 promoter containing the already described NFAT binding site was analysed. Additionally, a second fragment of 161 bp was analysed containing the combined NFAT/NF-κB sites described here. The locations of analysed binding sites and of primers used for amplification of genomic DNA are depicted in Figure 7A. After induction with P/I, both fragments of the BOB.1/OBF.1 promoter could be efficiently enriched by NFATc1 and RelA/p65 antibodies (Figure 7B and C), indicating that NFAT and NF-κB transcription factors exert a direct effect on the BOB.1/OBF.1 promoter.
Figure 7.
NFAT and NF-κB transcription factors bind to different BOB.1/OBF.1 promoter regions in vivo. (A) The nucleotide sequence of the analysed mouse BOB.1/OBF.1 promoter region is depicted. The NFAT and the predicted combined NFAT/NF-κB sites are shown. The position of used primers for amplification of DNA fragments after chromatin immunoprecipitation are shown with bold, italic letters. Underlined letters indicate the position of the TATA box. The start site of transcription is marked as +1. (B and C) Murine primary CD4+ T cells were treated for 18 h with P/I. The chromatin was cross-linked, sheared and immunoprecipitated using the indicated antibodies. Immunoprecipitations using mouse IgG or normal rabbit serum served as negative controls. Immunoprecipitated DNA was purified and used as template in quantitative PCR reactions using primers as indicated in (A) for the detection of fragments bearing either the NFAT site (B) or both combined NFAT/NF-κB sites (C). (D) As an internal control, precipitated DNA was amplified using primers (Supplementary Table S3) located upstream of analysed potential NFAT and NF-κB sites of the BOB.1/OBF.1 promoter.
DISCUSSION
Stimulation of the TCR triggers the activation of different signaling cascades leading finally to the activation of NFAT, NF-κB and AP-1, which act together with several other transcription factors to orchestrate gene expression important for T cell function, survival, proliferation and differentiation. Oct2 and BOB.1/OBF.1 are among these induced genes in activated T helper cells. Owing to their ability to act in concert at the octamer motif identified within the promoter region of the IFNγ gene (33), BOB.1/OBF.1 and Oct2 are important for the balanced TH1 and TH2 cytokine secretion by T helper cells. While investigating signaling pathways, which regulate octamer-dependent transcription, we found that the expression of BOB.1/OBF.1 and Oct2 critically depends on the expression and function of CN. Down modulation or inhibition of the Ca2+/calmodulin-dependent serine/threonine-specific protein phosphatase using either siRNA or CsA attenuated the expression of both proteins. The fact that CsA affects NFAT and NF-κB activity (43–45) prompted us to investigate the involvement of both signaling cascades in the regulation of BOB.1/OBF.1 and Oct2 expression in T cells. Using NEMO and NFAT inhibitory peptides, blocking the NF-κB and the NFAT pathway in a specific manner, revealed that both transcriptional activities are involved that process.
Indeed, we found several transcription factor binding sites within the BOB.1/OBF.1 promoter region conserved between mouse and man that can be bound on stimulation by both NFAT and NF-κB family members in vitro and in vivo. The most distally located ‘composite’ NFAT/NF-κB site (see Figure 2A) contains the classical NFAT core motif TTTCC that is also found in the distal promoter of the IL2 gene and an adjacent overlapping NF-κB binding motif AGGTGT that was recently described as an active NF-κB site within the human prostatic acid phosphatase gene promoter (59). Composite NFAT/NF-κB binding sites are described for numerous of promoters. For those identified within the HIV LTR and the human IL8 promoter binding of NFATc2 as a homodimer has been detected (60,61). Others have shown that NFATc2 competes for binding with NF-κB to the composite NFAT/NF-κB site within the HIV LTR (62), indicating that either NFAT or NF-κB factors bind to this site. Indeed, there is no experimental evidence for a heterodimer formation between NFAT and NF-κB family members. Although in supershift experiments, antibodies against NF-κB and NFAT are able to prevent inducible complex formation on this composite NFAT/NF-κB site, introduction of specific mutations revealed that rather NF-κB than NFAT transcription factors are bound to this site.
The second combined, ‘consecutive’ NFAT/NF-κB site also contains the classical NFAT core motif TTTCC that is followed at the 5′ site by the sequence 5′-GGGAATCGCA-3′. The latter represents the canonical NF-κB decamer from position 1 to 6 and bears also a cytidine at position 9. At such consecutive sites, NFAT and NF-κB complexes can be formed simultaneously, not competing for binding among each other. However, in Re-ChIP experiments, we were not able to show the mutual binding of NFATc1 together with RelA/p65 (data not shown) to this site. Yet, both the NF-κB and the NFAT motif are important for transcription factor binding, as individual mutation of both motifs prevents inducible by P/I complex formation.
The third important site for the regulation of BOB.1/OBF.1 promoter activity is the already described non-consensus NFAT binding site GAAGAAA that has no functional relevance for the BOB.1/OBF.1 promoter activity in B cells (34). In contrast, in T cells, an inducible complex formation could be detected to this site in vitro. Moreover, a 500 bp BOB.1/OBF.1 promoter construct bearing this site was sufficient for full inducible promoter activity in T cells that could be further enhanced by cotransfection of expression vectors for NFATc1 and RelA/p65. Cotransfection of both expression vectors together could further increase the promoter activity in comparison with that achieved by an individual overexpression of each factor, indicating that both cooperate in the regulation of the 500 bp BOB.1/OBF.1 promoter activity. Additionally, supershift experiments and chromatin immunoprecipitations revealed a binding of both, NFAT and NF-κB, to this part of the promoter. Hence, a not-yet identified NF-κB binding site should be present in close proximity to the mentioned non-canonical NFAT site. Because of a high homology between the DNA binding domains of NF-κB and NFAT family members, they generate a similar conformation and therefore contact identical nucleotides at the core motif GGAAA (63). The non-canonical NFAT site GAAGAAA within the BOB.1/OBF.1 promoter is followed by the sequence AAAAAAG (Figure 2A). Both sites share high similarity with the core sequence GGAAA to which both factors, NF-κB and NFAT, are able to bind. The second motif AAAG seems to be of biologically relevance, as the point mutation G→T prevents inducible complex binding in T cells. Possibly, this part of the BOB.1/OBF.1 promoter is essential for the cooperative action of NFAT and NF-κB necessary for the high inducible activity of the 500 bp promoter.
Additionally, the CREB/ATF binding site, important for the promoter activity in B cells (34), seems also to be relevant in T cells, as overexpression of an active version of CREB further increased the NFAT/NF-κB induced BOB.1/OBF.1 promoter activity in Jurkat T cells, whereas mutation of this site abrogates significantly BOB.1/OBF.1 promoter activity. Therefore, the observed decreasing effect of the p38 inhibitor SB203580 on the BOB.1/OBF.1 promoter and on the activity of octamer-dependent transcription in general could be mediated by the inhibition of CREB and/or ATF-1 that can also bind to the same site, as both factors need p38 kinase function for full transcriptional activity (64). Otherwise, an inhibition of p38 could affect NFAT proteins, as p38-mediated signaling activates NFATc1 (NFAT2) promoter activity and also increases its mRNA stability (65).
Together, here we identified three different functional NF-κB binding sites close to NFAT binding sites important for BOB.1/OBF.1 promoter activity. One of these NF-κB sites (identified in the composite NFAT/NF-κB site) was recently identified as an alternative NF-κB binding site different from the canonical, the second (identified in the consecutive NFAT/NF-κB site) shares high homology to the canonical site; however, there are variations within the second κB half-site, and the third (around the previously identified NFAT site) displays similarities to just the core motif to which NFAT and NF-κB proteins can bind. Indeed, deviations from the consensus NF-κB site were described for several promoters leading finally to the modulation of the affinity of different factors of the family to the appropriate site (66) and also influences the composition of the interacting heterodimers bound [reviewed in (67)]. This leads to the assumption that the expression of BOB.1/OBF.1 is differentially regulated under certain conditions, like primary or secondary T cell activation or in different T helper subtypes, by different homo- or heteromeric complexes that are bound to one or the other NF-κB site within the promoter.
The fact that the NF-κB sites identified here are in close proximity to NFAT sites and thereby build either composite or consecutive combined NFAT/NF-κB binding motifs suggests that either NFAT or NF-κB complexes or both together are able to transactivate the BOB.1/OBF.1 promoter. This conclusion is in line with our overexpression data, where the modulation of NFAT or NF-κB activity alone has already significant influence on BOB.1/OBF.1 and Oct2 expression. Additionally, the use of specific inhibitory peptides revealed that both the NFAT and the NF-κB signaling pathways are important for BOB.1/OBF.1 promoter activity and BOB.1/OBF.1 expression in T cells. Otherwise, we were not able to show simultaneous binding of both NFAT and NF-κB factors in Re-ChIP experiments. The fact that the deletion of RelA/p65 leads to a complete loss of BOB.1/OBF.1 expression in primary CD4+ T cells, whereas the deletion of NFATc1/c2 decreases BOB.1/OBF.1 expression by ∼50%, indicates that NF-κB activation is possibly of higher importance for sufficient BOB.1/OBF.1 expression in T cells than NFAT factors. But the activity of NFAT and NF-κB depends on Ca2+ oscillation (68), which differs in different cell types, including TH1, TH2 and TH17 T helper subpopulations (69,70). This leads to distinct levels of NFAT and NF-κB nuclear localization and finally to differential cytokine expression by different T helper subtypes. Therefore, it seems likely that also the availability and activity of NFAT or NF-κB family members in different T helper subpopulations or under certain differentiation conditions may determine whether the one or the other or both factors together contribute to the BOB.1/OBF.1 promoter activity in vivo.
In fact, the regulation of BOB.1/OBF.1 expression in T cells seems to be complex, as in natural regulatory T cells (nTreg), its expression is down modulated (71), whereas in T cells, which are energized or suppressed by nTreg, its expression was found >10 times higher than in non-suppressed CD4+ T cells (72). Although we have already described the importance of BOB.1/OBF.1 for balanced TH1/TH2 cytokine secretion (33), the precise regulation of BOB.1/OBF.1 and Oct expression in different T helper subpopulations that possibly affects the induction of a different set of genes, remains to be elucidated.
In summary, by an array of in vitro and in vivo approaches, we provide evidences for the importance of NFAT together with NF-κB transcription factors for the regulation of octamer-dependent transcription in T cells. Further work is required to identify the promoter region and regulatory cis-acting elements responsible for the Oct2 gene regulation. Additionally, it will be interesting to address the question whether the same transcription factors/transcription factor binding sites, identified as important elements for the inducible BOB.1/OBF.1 expression in T cells, have also relevance for its basal and inducible expression in B cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4 and Supplementary Figures 1–5.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) [DFG; TR52, TP A3 to F.B.-S.], [DFG; TR52, TP B2; W. Sander and M. Schell foundations to E.S.], [DFG; SFB 497, TP C5 to T.W.], [DFG; BR-2891/4-1 to C.B.]. Funding for open access charge: Deutsche Forschungsgemeinschaft (DFG) [BR-2891/4-1 to C.B.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are very grateful to Alexander Mayer and to Nina Ushmorova for excellent help and technical assistance.
REFERENCES
- 1.Zheng L., Roeder R.G., Luo Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;114:255–266. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]
- 2.Gstaiger M., Georgiev O., van Leeuwen H., van der Vliet P., Schaffner W. The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with the Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J. 1996;15:2781–2790. [PMC free article] [PubMed] [Google Scholar]
- 3.Gstaiger M., Knoepfel L., Georgiev O., Schaffner W., Hovens C.M. A B-cell coactivator of octamer-binding transcription factors. Nature. 1995;373:360–362. doi: 10.1038/373360a0. [DOI] [PubMed] [Google Scholar]
- 4.Pierani A., Heguy A., Fujii H., Roeder R.G. Activation of octamer-containing promoters by either octamer-binding transcription factor 1 (OTF-1) or OTF-2 and requirement of an additional B-cell-specific component for optimal transcription of immunoglobulin promoters. Mol. Cell. Biol. 1990;10:6204–6215. doi: 10.1128/mcb.10.12.6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo Y., Fujii H., Gerster T., Roeder R.G. A novel B cell-derived coactivator potentiates the activation of immunoglobulin promoters by octamer-binding transcription factors. Cell. 1992;71:231–241. doi: 10.1016/0092-8674(92)90352-d. [DOI] [PubMed] [Google Scholar]
- 6.Luo Y., Roeder R.G. Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol. Cell. Biol. 1995;15:4115–4124. doi: 10.1128/mcb.15.8.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfisterer P., Zwilling S., Hess J., Wirth T. Functional characterization of the murine homolog of the B-cell-specific coactivator BOB.1/OBF.1. J. Biol. Chem. 1995;270:29870–29880. doi: 10.1074/jbc.270.50.29870. [DOI] [PubMed] [Google Scholar]
- 8.Strubin M., Newell J.W., Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- 9.Staudt L.M., Singh H., Sen R., Wirth T., Sharp P.A., Baltimore D. A lymphoid-specific protein binding to the octamer motif of immunoglobulin genes. Nature. 1986;323:640–643. doi: 10.1038/323640a0. [DOI] [PubMed] [Google Scholar]
- 10.König H., Pfisterer P., Corcoran L., Wirth T. Identification of CD36 as the first gene dependent on the B cell differentiation factor Oct2. Genes Dev. 1995;9:1598–1607. doi: 10.1101/gad.9.13.1598. [DOI] [PubMed] [Google Scholar]
- 11.Shore P., Dietrich W., Corcoran L.M. Oct-2 regulates CD36 gene expression via a consensus octamer, which excludes the co-activator OBF-1. Nucleic Acids Res. 2002;30:1767–1773. doi: 10.1093/nar/30.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomilin A., Remenyi A., Lins K., Bak H., Leidel S., Vriend G., Wilmanns M., Scholer H.R. Synergism with the coactivator OBF-1 (OCA-B, BOB-1) is mediated by a specific POU dimer configuration. Cell. 2000;103:853–864. doi: 10.1016/s0092-8674(00)00189-6. [DOI] [PubMed] [Google Scholar]
- 13.Lins K., Remenyi A., Tomilin A., Massa S., Wilmanns M., Matthias P., Scholer H.R. OBF1 enhances transcriptional potential of Oct1. EMBO J. 2003;22:2188–2198. doi: 10.1093/emboj/cdg199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunner C., Wirth T. Btk expression is controlled by Oct and BOB.1/OBF.1. Nucleic Acids Res. 2006;34:1807–1815. doi: 10.1093/nar/gkl131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zwilling S., Dieckmann A., Pfisterer P., Angel P., Wirth T. Inducible expression and phosphorylation of coactivator BOB.1/OBF.1 in T cells. Science. 1997;277:221–225. doi: 10.1126/science.277.5323.221. [DOI] [PubMed] [Google Scholar]
- 16.Wang V.E., Schmidt T., Chen J., Sharp P.A., Tantin D. Embryonic lethality, decreased erythropoiesis, and defective octamer-dependent promoter activation in Oct-1-deficient mice. Mol. Cell. Biol. 2004;24:1022–1032. doi: 10.1128/MCB.24.3.1022-1032.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corcoran L.M., Karvelas M., Nossal G.J.V., Ye Z.S., Jacks T., Baltimore D. Oct-2, although not required for early B-cell development, is critical for later B-cell maturation and for postnatal survival. Genes Dev. 1993;7:570–582. doi: 10.1101/gad.7.4.570. [DOI] [PubMed] [Google Scholar]
- 18.Wang V.E., Tantin D., Chen J., Sharp P.A. B cell development and immunoglobulin transcription in Oct-1-deficient mice. Proc. Natl Acad. Sci. USA. 2004;101:2005–2010. doi: 10.1073/pnas.0307304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schubart D.B., Rolink A., Kosco-Vilbois M.H., Botteri F., Matthias P. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 1996;383:538–542. doi: 10.1038/383538a0. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen P.J., Georgiev O., Lorenz B., Schaffner W. B lymphocytes are impaired in mice lacking the transcriptional co-activator Bob1/OCA-B/OBF1. Eur. J. Immunol. 1996;26:3214–3218. doi: 10.1002/eji.1830261255. [DOI] [PubMed] [Google Scholar]
- 21.Kim U., Qin F.F., Gong S., Stevens S., Luo Y., Nussenzweig M., Roeder R.G. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature. 1996;383:542–547. doi: 10.1038/383542a0. [DOI] [PubMed] [Google Scholar]
- 22.Hess J., Nielsen P.J., Fischer K.D., Bujard H., Wirth T. The B lymphocyte-specific coactivator BOB.1/OBF.1 is required at multiple stages of B-cell development. Mol. Cell. Biol. 2001;21:1531–1539. doi: 10.1128/MCB.21.5.1531-1539.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brunner C., Marinkovic D., Klein J., Samardzic T., Nitschke L., Wirth T. B cell-specific transgenic expression of Bcl2 rescues early B lymphopoiesis but not B cell responses in BOB.1/OBF.1-deficient mice. J. Exp. Med. 2003;197:1205–1211. doi: 10.1084/jem.20022014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samardzic T., Marinkovic D., Nielsen P.J., Nitschke L., Wirth T. BOB.1/OBF.1 deficiency affects marginal-zone B-cell compartment. Mol. Cell. Biol. 2002;22:8320–8331. doi: 10.1128/MCB.22.23.8320-8331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moriuchi M., Moriuchi H. Octamer transcription factors up-regulate the expression of CCR5, a coreceptor for HIV-1 entry. J. Biol. Chem. 2001;276:8639–8642. doi: 10.1074/jbc.M008391200. [DOI] [PubMed] [Google Scholar]
- 26.Shibuya H., Taniguchi T. Identification of multiple cis-elements and trans-acting factors involved in the induced expression of human IL-2 gene. Nucleic Acids Res. 1989;17:9173–9184. doi: 10.1093/nar/17.22.9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunvand M.W., Schmidt A., Siebenlist U. Nuclear factors interacting with the mitogen-responsive regulatory region of the interleukin-2 gene. J. Biol. Chem. 1988;263:18904–18910. [PubMed] [Google Scholar]
- 28.Pfeuffer I., Klein-Heßling S., Heinfling A., Chuvpilo S., Escher C., Brabletz T., Hentsch B., Schwarzenbach H., Matthias P., Serfling E. Octamer factors exert a dual effect on the IL-2 and IL-4 promoters. J. Immunol. 1994;153:5572–5585. [PubMed] [Google Scholar]
- 29.de Grazia U., Felli M.P., Vacca A., Farina A.R., Maroder M., Cappabianca L., Meco D., Farina M., Screpanti I., Frati L., et al. Positive and negative regulation of the composite octamer motif of the Interleukin 2 enhancer by AP-1, OCT-2, and retinoic acid receptor. J. Exp. Med. 1994;180:1485–1497. doi: 10.1084/jem.180.4.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hentsch B., Mouzaki A., Pfeuffer I., Rungger D., Serfling E. The weak, fine-tuned binding of ubiquitous transcription factors to the Il-2 enhancer contributes to its T cell-restricted activity. Nucleic Acids Res. 1992;20:2657–2665. doi: 10.1093/nar/20.11.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chuvpilo S., Schomberg C., Gerwig R., Heinfling A., Reeves R., Grummt F., Serfling E. Multiple closely-linked NFAT/octamer and HMG I(Y) binding sites are part of the interleukin-4 promoter. Nucleic Acids Res. 1993;21:5694–5704. doi: 10.1093/nar/21.24.5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li-Weber M., Salgame P., Hu C., Davydov I.V., Laur O., Klevenz S., Krammer P.H. Th2-specific protein/DNA interactions at the proximal Nuclear Factor-AT site contribute to the functional activity of the human IL-4 promoter. J. Immunol. 1998;161:1380–1389. [PubMed] [Google Scholar]
- 33.Brunner C., Sindrilaru A., Girkontaite I., Fischer K.D., Sunderkotter C., Wirth T. BOB.1/OBF.1 controls the balance of TH1 and TH2 immune responses. EMBO J. 2007;26:3191–3202. doi: 10.1038/sj.emboj.7601742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens S., Wang L., Roeder R.G. Functional analysis of the OCA-B promoter. J. Immunol. 2000;164:6372–6379. doi: 10.4049/jimmunol.164.12.6372. [DOI] [PubMed] [Google Scholar]
- 35.Shen Y., Hendershot L.M. Identification of ERdj3 and OBF-1/BOB-1/OCA-B as direct targets of XBP-1 during plasma cell differentiation. J. Immunol. 2007;179:2969–2978. doi: 10.4049/jimmunol.179.5.2969. [DOI] [PubMed] [Google Scholar]
- 36.Massa S., Junker S., Schubart K., Matthias G., Matthias P. The OBF-1 gene locus confers B cell-specific transcription by restricting the ubiquitous activity of its promoter. Eur. J. Immunol. 2003;33:2864–2874. doi: 10.1002/eji.200323882. [DOI] [PubMed] [Google Scholar]
- 37.Schubart D.B., Sauter P., Massa S., Friedl E.M., Schwarzenbach H., Matthias P. Gene structure and characterization of the murine homologue of the B cell-specific transcriptional coactivator OBF-1. Nucleic Acids Res. 1996;24:1913–1920. doi: 10.1093/nar/24.10.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin X.F., Reichlin A., Luo Y., Roeder R.G., Nussenzweig M.C. OCA-B integrates B cell antigen receptor-, CD40L- and IL 4-mediated signals for the germinal center pathway of B cell development. EMBO J. 1998;17:5066–5075. doi: 10.1093/emboj/17.17.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greiner A., Muller K.B., Hess J., Pfeffer K., Muller-Hermelink H.K., Wirth T. Up-regulation of BOB.1/OBF.1 expression in normal germinal center B cells and germinal center-derived lymphomas. Am. J. Pathol. 2000;156:501–507. doi: 10.1016/S0002-9440(10)64754-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sauter P., Matthias P. The B cell-specific coactivator OBF-1 (OCA-B, Bob-1) is inducible in T cells and its expression is dispensable for IL-2 gene induction. Immunobiology. 1997;198:207–216. doi: 10.1016/S0171-2985(97)80041-1. [DOI] [PubMed] [Google Scholar]
- 41.Kang S.M., Tsang W., Doll S., Scherle P., Ko H.S., Tran A.C., Lenardo M.J., Staudt L.M. Induction of the POU domain transcription factor Oct-2 during T-cell activation by cognate antigen. Mol. Cell. Biol. 1992;12:3149–3154. doi: 10.1128/mcb.12.7.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhargava A.K., Li Z., Weissman S.M. Differential expression of four members of the POU family of proteins in activated and phorbol 12-myristate 13-acetate-treated Jurkat T cells. Proc. Natl Acad. Sci. USA. 1993;90:10260–10264. doi: 10.1073/pnas.90.21.10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clipstone N.A., Crabtree G.R. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- 44.Palkowitsch L., Marienfeld U., Brunner C., Eitelhuber A., Krappmann D., Marienfeld R.B. The Ca2+-dependent phosphatase calcineurin controls the formation of the Carma1-Bcl10-Malt1 complex during T cell receptor-induced NF-kappaB activation. J. Biol. Chem. 2011;286:7522–7534. doi: 10.1074/jbc.M110.155895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frischbutter S., Gabriel C., Bendfeldt H., Radbruch A., Baumgrass R. Dephosphorylation of Bcl-10 by calcineurin is essential for canonical NF-kappaB activation in Th cells. Eur. J. Immunol. 2011;41:2349–2357. doi: 10.1002/eji.201041052. [DOI] [PubMed] [Google Scholar]
- 46.Serfling E., Berberich-Siebelt F., Avots A., Chuvpilo S., Klein-Hessling S., Jha M.K., Kondo E., Pagel P., Schulze-Luehrmann J., Palmetshofer A. NFAT and NF-kappaB factors-the distant relatives. Int. J. Biochem. Cell. Biol. 2004;36:1166–1170. doi: 10.1016/j.biocel.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Annweiler A., Zwilling S., Hipskind R.A., Wirth T. Analysis of transcriptional stimulation by recombinant Oct proteins in a cell free system. J. Biol. Chem. 1993;268:2525–2534. [PubMed] [Google Scholar]
- 48.Harhaj E.W., Good L., Xiao G., Uhlik M., Cvijic M.E., Rivera-Walsh I., Sun S.C. Somatic mutagenesis studies of NF-kappa B signaling in human T cells: evidence for an essential role of IKK gamma in NF-kappa B activation by T-cell costimulatory signals and HTLV-I Tax protein. Oncogene. 2000;19:1448–1456. doi: 10.1038/sj.onc.1203445. [DOI] [PubMed] [Google Scholar]
- 49.Schuh K., Kneitz B., Heyer J., Bommhardt U., Jankevics E., Berberich-Siebelt F., Pfeffer K., Muller-Hermelink H.K., Schimpl A., Serfling E. Retarded thymic involution and massive germinal center formation in NF-ATp-deficient mice. Eur. J. Immunol. 1998;28:2456–2466. doi: 10.1002/(SICI)1521-4141(199808)28:08<2456::AID-IMMU2456>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 50.Bhattacharyya S., Deb J., Patra A.K., Thuy Pham D.A., Chen W., Vaeth M., Berberich-Siebelt F., Klein-Hessling S., Lamperti E.D., Reifenberg K., et al. NFATc1 affects mouse splenic B cell function by controlling the calcineurin—NFAT signaling network. J. Exp. Med. 2011;208:823–839. doi: 10.1084/jem.20100945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Al Sarraj J., Vinson C., Han J., Thiel G. Regulation of GTP cyclohydrolase I gene transcription by basic region leucine zipper transcription factors. J. Cell. Biochem. 2005;96:1003–1020. doi: 10.1002/jcb.20580. [DOI] [PubMed] [Google Scholar]
- 52.Kempe S., Kestler H., Lasar A., Wirth T. NF-kappaB controls the global pro-inflammatory response in endothelial cells: evidence for the regulation of a pro-atherogenic program. Nucleic Acids Res. 2005;33:5308–5319. doi: 10.1093/nar/gki836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chuvpilo S., Jankevics E., Tyrsin D., Akimzhanov A., Moroz D., Jha M.K., Schulze-Luehrmann J., Santner-Nanan B., Feoktistova E., Konig T., et al. Autoregulation of NFATc1/A expression facilitates effector T cells to escape from rapid apoptosis. Immunity. 2002;16:881–895. doi: 10.1016/s1074-7613(02)00329-1. [DOI] [PubMed] [Google Scholar]
- 54.Nayak A., Glockner-Pagel J., Vaeth M., Schumann J.E., Buttmann M., Bopp T., Schmitt E., Serfling E., Berberich-Siebelt F. Sumoylation of the transcription factor NFATc1 leads to its subnuclear relocalization and interleukin-2 repression by histone deacetylase. J. Biol. Chem. 2009;284:10935–10946. doi: 10.1074/jbc.M900465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Denk A., Goebeler M., Schmid S., Berberich I., Ritz O., Lindemann D., Ludwig S., Wirth T. Activation of NF-kappa B via the Ikappa B kinase complex is both essential and sufficient for proinflammatory gene expression in primary endothelial cells. J. Biol. Chem. 2001;276:28451–28458. doi: 10.1074/jbc.M102698200. [DOI] [PubMed] [Google Scholar]
- 56.Serfling E., Chuvpilo S., Liu J., Hofer T., Palmetshofer A. NFATc1 autoregulation: a crucial step for cell-fate determination. Trends Immunol. 2006;27:461–469. doi: 10.1016/j.it.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 57.Alcamo E., Mizgerd J.P., Horwitz B.H., Bronson R., Beg A.A., Scott M., Doerschuk C.M., Hynes R.O., Baltimore D. Targeted mutation of TNF receptor I rescues the RelA-deficient mouse and reveals a critical role for NF-kappa B in leukocyte recruitment. J. Immunol. 2001;167:1592–1600. doi: 10.4049/jimmunol.167.3.1592. [DOI] [PubMed] [Google Scholar]
- 58.Beg A.A., Sha W.C., Bronson R.T., Ghosh S., Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 59.Zelivianski S., Glowacki R., Lin M.F. Transcriptional activation of the human prostatic acid phosphatase gene by NF-kappaB via a novel hexanucleotide-binding site. Nucleic Acids Res. 2004;32:3566–3580. doi: 10.1093/nar/gkh677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giffin M.J., Stroud J.C., Bates D.L., von Koenig K.D., Hardin J., Chen L. Structure of NFAT1 bound as a dimer to the HIV-1 LTR kappa B element. Nat. Struct. Biol. 2003;10:800–806. doi: 10.1038/nsb981. [DOI] [PubMed] [Google Scholar]
- 61.Jin L., Sliz P., Chen L., Macian F., Rao A., Hogan P.G., Harrison S.C. An asymmetric NFAT1 dimer on a pseudo-palindromic kappa B-like DNA site. Nat. Struct. Biol. 2003;10:807–811. doi: 10.1038/nsb975. [DOI] [PubMed] [Google Scholar]
- 62.Macian F., Rao A. Reciprocal modulatory interaction between human immunodeficiency virus type 1 Tat and transcription factor NFAT1. Mol. Cell. Biol. 1999;19:3645–3653. doi: 10.1128/mcb.19.5.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolfe S.A., Zhou P., Dotsch V., Chen L., You A., Ho S.N., Crabtree G.R., Wagner G., Verdine G.L. Unusual Rel-like architecture in the DNA-binding domain of the transcription factor NFATc. Nature. 1997;385:172–176. doi: 10.1038/385172a0. [DOI] [PubMed] [Google Scholar]
- 64.Tan Y., Rouse J., Zhang A., Cariati S., Cohen P., Comb M.J. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 1996;15:4629–4642. [PMC free article] [PubMed] [Google Scholar]
- 65.Wu C.C., Hsu S.C., Shih H.M., Lai M.Z. Nuclear factor of activated T cells c is a target of p38 mitogen-activated protein kinase in T cells. Mol. Cell. Biol. 2003;23:6442–6454. doi: 10.1128/MCB.23.18.6442-6454.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zabel U., Schreck R., Baeuerle P.A. DNA binding of purified transcription factor NF-kappa B. Affinity, specificity, Zn2+ dependence, and differential half-site recognition. J. Biol. Chem. 1991;266:252–260. [PubMed] [Google Scholar]
- 67.Natoli G., Saccani S., Bosisio D., Marazzi I. Interactions of NF-kappaB with chromatin: the art of being at the right place at the right time. Nat. Immunol. 2005;6:439–445. doi: 10.1038/ni1196. [DOI] [PubMed] [Google Scholar]
- 68.Dolmetsch R.E., Xu K., Lewis R.S. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392:933–936. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 69.Fanger C.M., Neben A.L., Cahalan M.D. Differential Ca2+ influx, KCa channel activity, and Ca2+ clearance distinguish Th1 and Th2 lymphocytes. J. Immunol. 2000;164:1153–1160. doi: 10.4049/jimmunol.164.3.1153. [DOI] [PubMed] [Google Scholar]
- 70.Weber K.S., Miller M.J., Allen P.M. Th17 cells exhibit a distinct calcium profile from Th1 and Th2 cells and have Th1-like motility and NF-AT nuclear localization. J. Immunol. 2008;180:1442–1450. doi: 10.4049/jimmunol.180.3.1442. [DOI] [PubMed] [Google Scholar]
- 71.Marson A., Kretschmer K., Frampton G.M., Jacobsen E.S., Polansky J.K., MacIsaac K.D., Levine S.S., Fraenkel E., von Boehmer H., Young R.A. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sukiennicki T.L., Fowell D.J. Distinct molecular program imposed on CD4+ T cell targets by CD4+CD25+ regulatory T cells. J. Immunol. 2006;177:6952–6961. doi: 10.4049/jimmunol.177.10.6952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.